Abstract
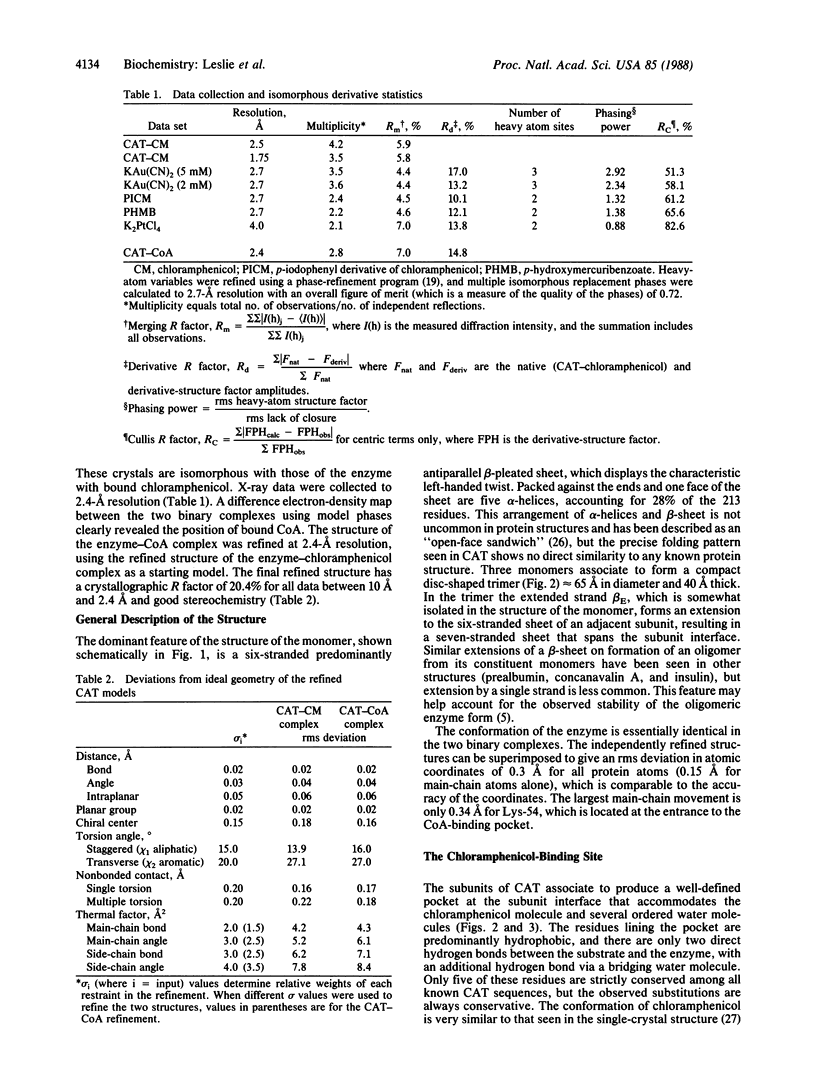
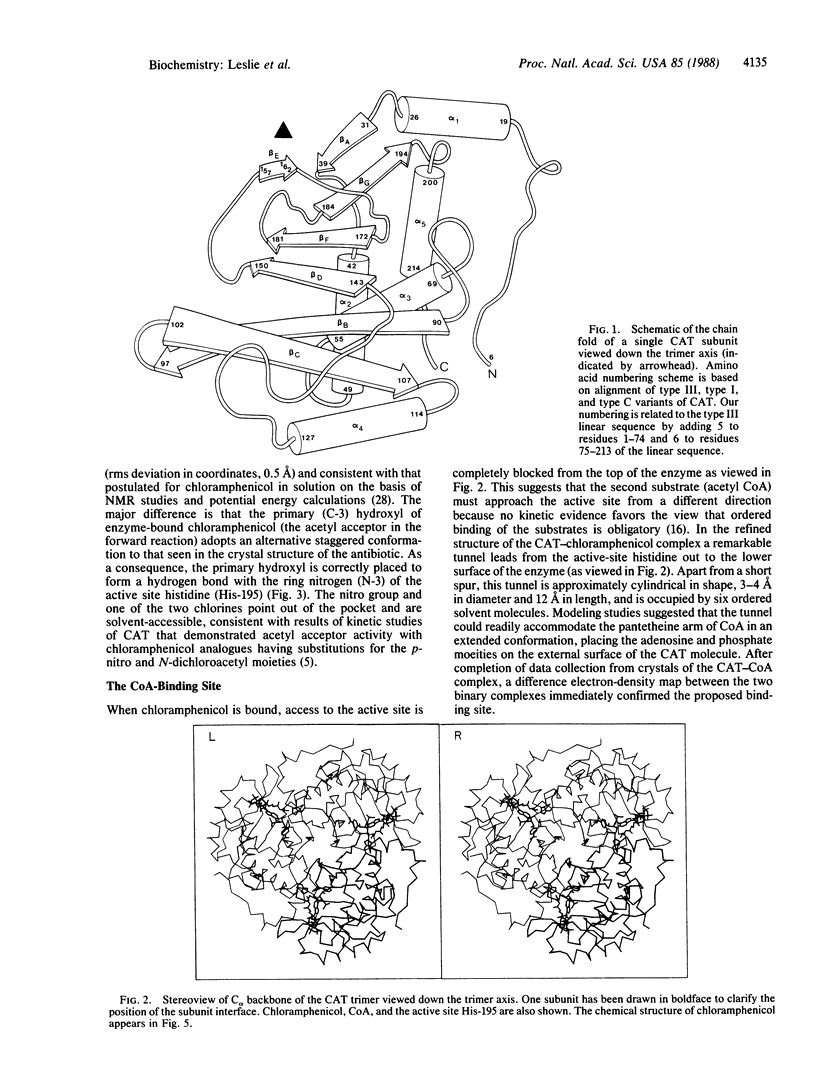
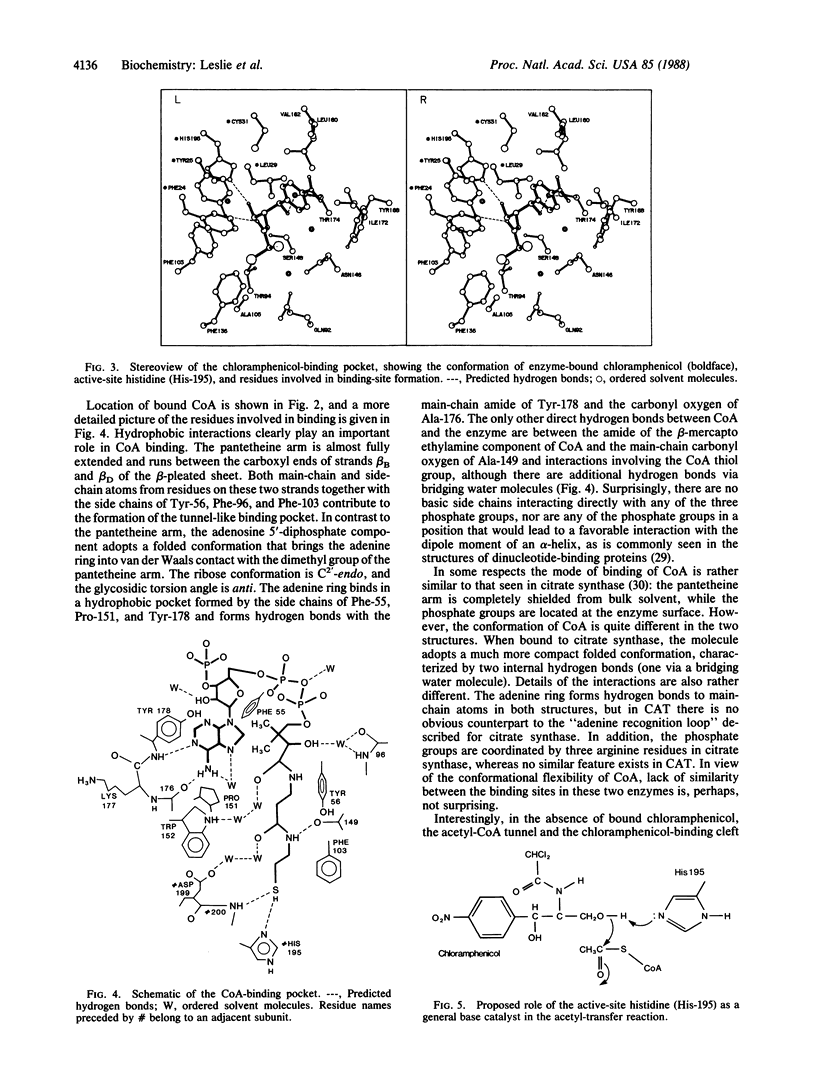
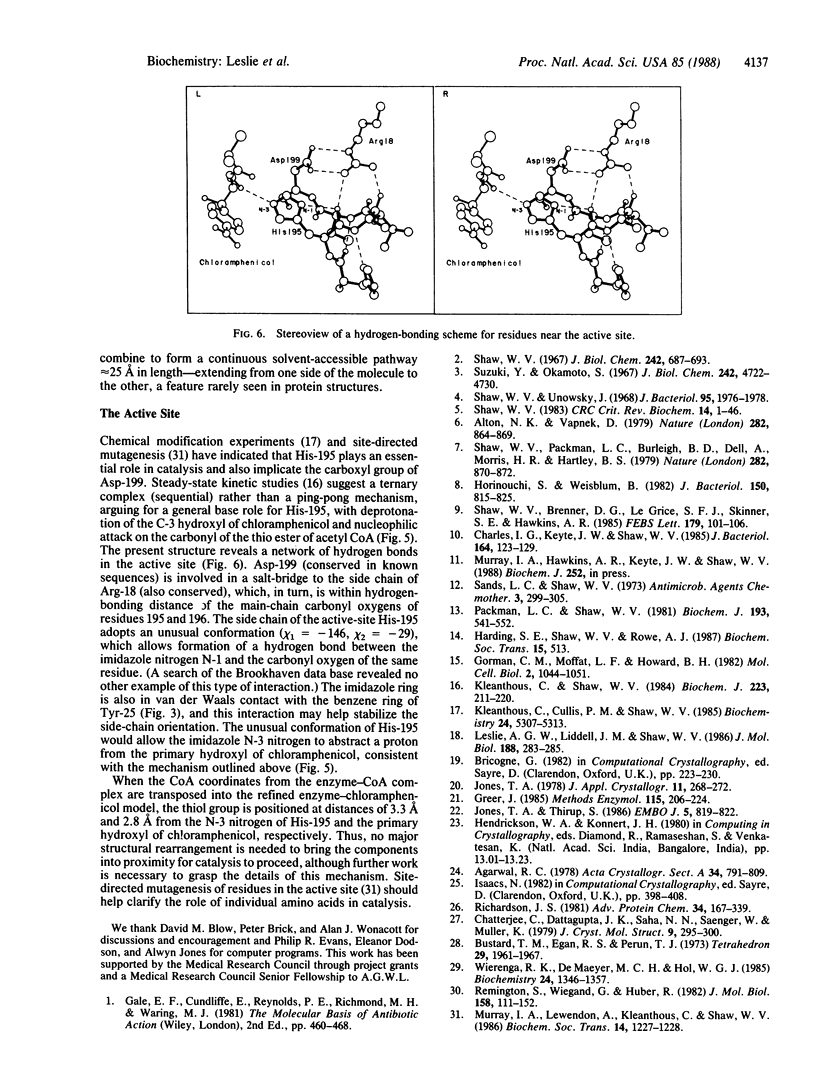
Chloramphenicol acetyltransferase [acetyl-CoA:chloramphenicol O3-acetyltransferase; EC 2.3.1.28] is the enzyme responsible for high-level bacterial resistance to the antibiotic chloramphenicol. It catalyzes the transfer of an acetyl group from acetyl CoA to the primary hydroxyl of chloramphenicol. The x-ray crystallographic structure of the type III variant enzyme from Escherichia coli has been determined and refined at 1.75-A resolution. The enzyme is a trimer of identical subunits with a distinctive protein fold. Structure of the trimer is stabilized by a beta-pleated sheet that extends from one subunit to the next. The active site is located at the subunit interface, and the binding sites for both chloramphenicol and CoA have been characterized. Substrate binding is unusual in that the two substrates approach the active site via clefts on opposite molecular "sides." A histidine residue previously implicated in catalysis is appropriately positioned to act as a general base catalyst in the reaction.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Charles I. G., Keyte J. W., Shaw W. V. Nucleotide sequence analysis of the cat gene of Proteus mirabilis: comparison with the type I (Tn9) cat gene. J Bacteriol. 1985 Oct;164(1):123–129. doi: 10.1128/jb.164.1.123-129.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer J. Computer skeletonization and automatic electron density map analysis. Methods Enzymol. 1985;115:206–224. doi: 10.1016/0076-6879(85)15017-2. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986 Apr;5(4):819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleanthous C., Cullis P. M., Shaw W. V. 3-(Bromoacetyl)chloramphenicol, an active site directed inhibitor for chloramphenicol acetyltransferase. Biochemistry. 1985 Sep 24;24(20):5307–5313. doi: 10.1021/bi00341a006. [DOI] [PubMed] [Google Scholar]
- Kleanthous C., Shaw W. V. Analysis of the mechanism of chloramphenicol acetyltransferase by steady-state kinetics. Evidence for a ternary-complex mechanism. Biochem J. 1984 Oct 1;223(1):211–220. doi: 10.1042/bj2230211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A. G., Liddell J. M., Shaw W. V. Crystallization of a type III chloramphenicol acetyl transferase. J Mol Biol. 1986 Mar 20;188(2):283–285. doi: 10.1016/0022-2836(86)90310-4. [DOI] [PubMed] [Google Scholar]
- Packman L. C., Shaw W. V. The use of naturally occurring hybrid variants of chloramphenicol acetyltransferase to investigate subunit contacts. Biochem J. 1981 Feb 1;193(2):541–552. doi: 10.1042/bj1930541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington S., Wiegand G., Huber R. Crystallographic refinement and atomic models of two different forms of citrate synthase at 2.7 and 1.7 A resolution. J Mol Biol. 1982 Jun 15;158(1):111–152. doi: 10.1016/0022-2836(82)90452-1. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Sands L. C., Shaw W. V. Mechanism of chloramphenicol resistance in staphylococci: characterization and hybridization of variants of chloramphenicol acetyltransferase. Antimicrob Agents Chemother. 1973 Feb;3(2):299–305. doi: 10.1128/aac.3.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V., Brenner D. G., LeGrice S. F., Skinner S. E., Hawkins A. R. Chloramphenicol acetyltransferase gene of staphylococcal plasmid pC221. Nucleotide sequence analysis and expression studies. FEBS Lett. 1985 Jan 1;179(1):101–106. doi: 10.1016/0014-5793(85)80200-3. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase: enzymology and molecular biology. CRC Crit Rev Biochem. 1983;14(1):1–46. doi: 10.3109/10409238309102789. [DOI] [PubMed] [Google Scholar]
- Shaw W. V., Packman L. C., Burleigh B. D., Dell A., Morris H. R., Hartley B. S. Primary structure of a chloramphenicol acetyltransferase specified by R plasmids. Nature. 1979 Dec 20;282(5741):870–872. doi: 10.1038/282870a0. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. The enzymatic acetylation of chloramphenicol by extracts of R factor-resistant Escherichia coli. J Biol Chem. 1967 Feb 25;242(4):687–693. [PubMed] [Google Scholar]
- Shaw W. V., Unowsky J. Mechanism of R factor-mediated chloramphenicol resistance. J Bacteriol. 1968 May;95(5):1976–1978. doi: 10.1128/jb.95.5.1976-1978.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Okamoto S. The enzymatic acetylation of chloramphenicol by the multiple drug-resistant Escherichia coli carrying R factor. J Biol Chem. 1967 Oct 25;242(20):4722–4730. [PubMed] [Google Scholar]


