Abstract
Three cocrystal X-ray structures of the α-ketoheterocycle inhibitors 3–5 bound to a humanized variant of fatty acid amide hydrolase (FAAH) are disclosed and comparatively discussed alongside those of 1 (OL-135) and its isomer 2. These five X-ray structures systematically probe each of the three active site regions key to substrate or inhibitor binding: (1) the conformationally mobile acyl chain-binding pocket and membrane access channel responsible for fatty acid amide substrate and inhibitor acyl chain binding, (2) the atypical active site catalytic residues and surrounding oxyanion hole that covalently binds the core of the α-ketoheterocycle inhibitors captured as deprotonated hemiketals mimicking the tetrahedral intermediate of the enzyme catalyzed reaction, and (3) the cytosolic port and its uniquely important imbedded ordered water molecules and a newly identified anion binding site. The detailed analysis of their key active site interactions and their implications on the interpretation of the available structure–activity relationships are discussed providing important insights for future design.
Introduction
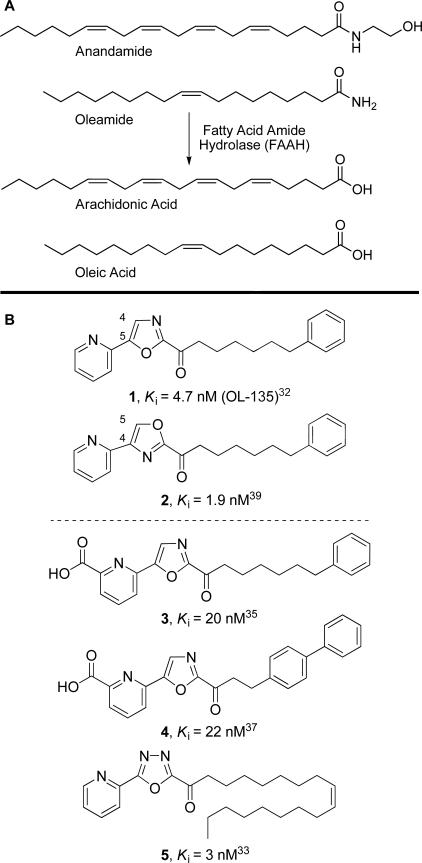
Fatty acid amide hydrolase (FAAHa)1,2 is the enzyme that serves to hydrolyze endogenous lipid amides and ethanolamides3–6 including anandamide7–10 and oleamide11–13 degrading and regulating neuromodulating and signaling fatty acid amides at their sites of action (Figure 1A).4,14 To date, two key classes of inhibitors have been pursued that provide opportunities for the development of FAAH inhibitors with therapeutic potential.15,16 One class is the aryl carbamates and ureas17–29 that irreversibly acylate a FAAH active site serine.28 A second class is the α-ketoheterocycle-based inhibitors30–40 that bind to FAAH through reversible hemiketal formation with an active site serine.
Figure 1.
A) Endogenous substrates of FAAH. B) Inhibitors 1–5 of FAAH.
FAAH belongs to the amidase signature (AS) class of enzymes, serine hydrolases that possesses an unusual Ser–Ser–Lys catalytic triad (Ser241–Ser217–Lys142 in FAAH).41 The catalytic mechanism of FAAH involves the formation of a tetrahedral intermediate, derived from the nucleophilic attack of the catalytic Ser241 residue on the carbonyl group of the substrate. The tetrahedral intermediate collapses to release the amine and the enzyme-bound acyl intermediate. The reaction terminates with a water-mediated deacylation of the enzyme-bound acyl intermediate and release of the free fatty acid with restoration of the active enzyme. FAAH hydrolyzes a wide range of substrates with primary amides being hydrolyzed 2-fold faster than ethanolamides.5 It acts on a wide range of fatty acid chains possessing various levels of unsaturation and lengths, but it preferentially hydrolyzes arachidonoyl or oleoyl substrates (arachidonoyl > oleoyl, 3-fold).5,6
In addition to possessing an atypical catalytic core and central to the discussion herein, FAAH bears a series of channels and cavities that are involved in substrate or inhibitor binding. These include the membrane access channel (MAC) that connects the active site to an opening located at the membrane anchoring face of the enzyme, the cytosolic port that may allow for the exit of hydrophilic products from the active site to the cytosol, and the acyl chain-binding pocket (ABP), which is thought to interact with the substrate's acyl chain during the catalytic reaction.42,43
Following efforts enlisting substrate-inspired inhibitors bearing electrophilic carbonyls,44,45 we described the systematic exploration of a series of potent and selective α-ketoheterocycle-based inhibitors.30–40 In these efforts, initiated at a time when there were still only a handful of such α-ketoheterocycle inhibitors disclosed,46 sufficiently potent, selective, and efficacious FAAH inhibitors were developed to validate FAAH as an important new therapeutic target for the treatment of pain and inflammatory disorders.40
In a recent disclosure, we reported the X-ray crystal structures of two isomeric α-ketoheterocycle inhibitors, 1 (OL-135) and 2 (Figure 1B), bound to FAAH.43 These structures not only established covalent attachment of Ser241 at the inhibitor's electrophilic carbonyl providing stable mimics of the enzymatic tetrahedral intermediate and capturing the atypical active site catalytic residues (Ser241–Ser217–Lys142) in a unique “in action” state, but they further revealed a unique SerOH–π H-bond to the activating heterocycle distinct from active site interactions observed in work with serine proteases.46,47 It also defined a distinguishing acyl chain/membrane access channel flexibility, and revealed an unexpected presence of and prominent role for cytosolic port bound solvent (H2O) in stabilizing inhibitor binding.
Herein, we report the X-ray crystal structures of three additional α-ketoheterocycles, 3–5 (Figure 1B), bound to humanized FAAH that were carefully chosen to further probe the three key regions of the active site contributing to inhibitor and substrate binding: the conformationally mobile acyl chain-binding pocket (ABP) and the membrane access channel (MAC) responsible for fatty acid amide substrate and inhibitor acyl chain binding, the atypical active site catalytic residues and exquisite oxyanion hole that covalently binds to the core of the α-ketoheterocycle, and the cytosolic port and its imbedded H2O molecule. Consequently and complementing the disclosed studies of the isomeric inhibitors 1 and 2,43 the bound inhibitors 3–5 probe the acyl chain-binding pocket with three disparate acyl chains that cover a near maximal difference in length, flexibility, and inhibitor potency, two different core α-ketoheterocycles including a representative member of the more potent oxadiazole-based inhibitors (5) established to provide a near 10–70-fold enhancement over the corresponding oxazole-based inhibitors,33,38 and two related cytosolic port bound aryl substituents that substantially influence inhibitor potency and selectivity, as well as their physical and pharmacokinetic (PK) properties. The detailed analysis of their key active site interactions, the comparison with the prior structures of 1 and 2, and their implications on the interpretation of the available structure–activity relationships (SAR) are discussed herein providing unique insights that may guide future inhibitor design. Because of the comprehensive SAR studies that have been conducted with the α-ketoheterocycle-based inhibitors of FAAH, the corresponding three domains of the inhibitors (acyl chain, activating central heterocycle, and C5 substituent that binds in the cytosolic port) have been shown to exhibit relatively independent contributions to the inhibitor potency or selectivity with parallel results that can be discussed across the series of inhibitors.
In addition to reinforcing the key features of the inhibitor binding observed in the cocrystal structures of 1 and 2 bound to FAAH and revealing new subtle interactions important for future design, these studies additionally reveal that small variations of the central activating heterocycle and its attached C5-substituent can lead to further productive re-orientation of the inhibitor's polar head in the cytosolic port due to interactions with bound water molecules or a putative anion binding site.
Results
The structures of FAAH bound to the α-ketoheterocycle inhibitors 3–5 have been solved at a resolution of The rather high Rmerge for the three structures may be a direct effect of the radiation damage caused by the synchrotron beam intensity and possibly by beam translation along the crystal axes during data collection. However, the overall estimated standard uncertainty (ESU) for Rwork /Rfree in the FAAH–3, FAAH–4, and FAAH–5 structures are only 0.13/0.12, 0.22/0.17, and 0.21/0.17 Å, respectively.
The overall structures of FAAH are nearly identical to the previously published structures of FAAH bound to 1 and 243 (root mean squared deviations based on Cα atoms is about 0.2–0.3 Å) and the small differences are constrained to the subtle active site distinctions discussed below. Unbiased electron density maps defined the orientation of the inhibitors in the active site and confirmed that they are covalently bound to the catalytic Ser241 through reaction with the inhibitor's electrophilic carbonyl. The following description of the bound inhibitors (Figure 2) individually analyzes regions of the enzyme corresponding to the interactions found within the channel/pocket network, the catalytic region composed of the catalytic triad and oxyanion hole, and the cytosolic port.
Figure 2.
Top: Overlay of the portion of the five inhibitors that interact with the cytosolic port (“head”). Bottom: Overlay of the portion of the inhibitors that interact with the acyl chain-binding pocket (“tail”). Inhibitor 3 is shown in cyan, 4 in purple, 5 in green, 143 in dark blue, and 243 in yellow.
Acyl Chain Binding in the Membrane Access Channel/Acyl Chain-Binding Pocket
The phenhexyl chain of bound 3 was found to overlay precisely with the phenhexyl chains of 1 and 2 benefiting from key interactions with the residues lining the hydrophobic channel that pack tightly against the inhibitor forming a cavity complementary in shape to the compounds (Figure 3). Favorable van der Waals interactions are observed with Tyr194, Phe244, Thr377, Leu380, Leu404, Phe432, Thr488, and Val491. The π-system of the bound phenyl group is engaged in an aromatic CH–π type interaction with an aryl ring hydrogen of Phe381, mimicking the stabilizing interactions that support unsaturated fatty acid side chain binding. Phe192, which is oriented to provide a second weak CH–π interaction with the terminal phenyl group of 1,43 rotates in the complex with 3 to accept an aryl CH–π interaction from the pyridyl substituent bound in the cytosolic port. The mobile residues Phe432, Met495, and Met436 adopt the conformation that leads to a broadened and open membrane access channel with truncation of the acyl chain-binding pocket.43 Phe432 makes a key aryl CH–π contact with the inhibitor's phenyl ring while the two methionines orient their sulfur lone pair electrons towards the bound phenyl hydrogens engaging in two aromatic CH–π interactions. These latter three residues and Phe381 appear to provide key anchoring interactions for binding inhibitors related to 1–3, whereas Phe192 appears to swivel to accommodate hydrophobic ligand binding in either the substrate chain binding region or the cytosolic port. Despite the subtle differences discussed above between 1–2 and 3, the comparison of the three complexes reveal that the bound disposition of the phenhexyl chain is identical and independent of the choice of central activating heterocycle or its attached substituents.
Figure 3.
FAAH active site with bound OL-135 (1,43 in dark blue) and 3 (in cyan). The protein backbone is shown in stick representation. Shown here are the phenhexyl chains that overlay precisely and that Phe192 reorients to allow for changes in the oxazole C5 substituent. 2Fo-Fc electron density maps at a contour level of 1.5σ for compound 3 are shown with white meshes.
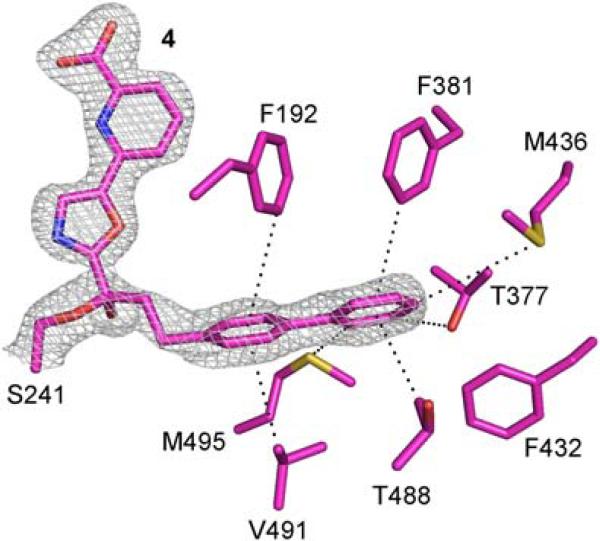
The binding of the biphenylethyl acyl chain of 4 extends into the same cavity up to and terminating at the proximal portion of the channel leading to the membrane (Figure 4). The terminal phenyl group of 4 is bound at precisely the same location and in a nearly identical orientation as the phenyl groups of 1–3. The terminal phenyl group of 4 is rotated ca. 25-30° relative to those of 1–3 in the plane of the ring, it is tilted only slightly (ca. 12°) relative to those of 1–3, and its centroid is displaced by only 0.4 Å (Figure 2 and Supporting Information Figures S1 and S2). These minor changes in the orientation of the bound terminal phenyl group do not alter the nature or the extent of the key interactions with the enzyme (Phe381, Met495, Met436, Thr488) although it does pick up an additional stabilizing interaction with Thr377. In fact, the protein conformation in this region with 4 is practically identical to that found with bound compounds 1–3 including the adoption of the closed acyl chain-binding pocket. The intervening linking phenylethyl chain of 4 and the hexyl chain of 1–3 overlay beautifully with the first two carbons of the two linking chains overlaying nearly identically with one another. The proximal phenyl ring in the linker of 4, which is tilted relative to the distal phenyl ring (ca. 35°), picks up a stabilizing CH–π interaction with an aryl hydrogen of Phe192 and appears to make stabilizing contacts with Val491. These latter two interactions may be mimicking those that bind the Δ5,6-double bond of arachidonoyl substrates and may contribute to the enhanced affinities (typically ca. 3-fold relative to phenhexyl)36,37 of inhibitors bearing this optimized acyl chain.36,37 Notably, the rotated orientation of Phe192 with bound 4 is identical to that observed with 3, where it further benefits from an aryl CH–π interaction with the cytosolic port bound pyridine substituent, and is distinct from the Phe192 orientation observed with 1 and 2.
Figure 4.
FAAH active site with bound 4 (in purple). Hydrophobic residues of FAAH pack tightly around the biphenylethyl chain mimicking that of arachindonoyl substrates. 2Fo-Fc electron density maps at a contour level of 1.0σ for compound 4 are shown with white meshes.
The third inhibitor, 5, possesses an oleyl acyl chain mimicking the nature and size of the prototypical endogenous substrates for FAAH. Although this increase in the length of the acyl chain in such inhibitors decreases their potency (ca. 20-fold), the activating oxadiazole heterocycle in 5 provides a corresponding increase in potency (10π70-fold) relative to an oxazole such that the potency of 5 is roughly equivalent to that of 1 and 2. As such, inhibitor 5 represents only the second such X-ray crystal structure disclosed complementing the initial rat FAAH structure reported that was covalently bound to an arachidonyl phosphonate (PDB code 1MT5).42c This latter structure was conducted with an inhibitor that extended the substrate length by one atom. This subtle distinction, as well as the binding of 5 that is trapped as a deprotonated hemiketal functionally mimicking the tetrahedral intermediate of the enzyme catalyzed reaction (vs uncharged tetrahedral phosphonate), suggests that the structure of the bound complex of 5 with FAAH more closely resembles the enzyme conformation as it acts on endogenous substrates than any preceding structure. Nonetheless, the side chain of 5 and that of the bound arachidonyl phosphonate adopt similar conformations (Figure 5). Despite their differences in atom length from Ser241 (18 atoms vs 21 atoms), both chains terminate at the same location in the acyl chain-binding pocket. The most obvious difference in the binding of the oleyl versus arachidonyl side chains is found at the site of the binding residue Ser241, where the side chains extend into the substrate channel from different angles (ca. 30–35°). No doubt this reflects the distinctions in a bound tetrahedral phosphonate versus the deprotonated hemiketal with 5, as well as the orientation and depth to which they penetrate into the oxyanion hole. Notably, the binding of 5 in this early region of the substrate channel overlays nicely with the side chains of 1–4. However, the binding of the oleyl chain extends into the substrate channel much further than 1–4 and the enzyme adopts a second conformation opening access to the acyl chain-binding pocket (ABP). This bifurcation into two hydrophobic cavities entails a rearrangement of Phe432 and reorientation of Met436 and Met495 that serves to create an extended ABP and reduces the width of the membrane access channel. Thus, the oleyl side chain binding overlays with that observed with 1–4 (Figures 2 and 6), but extends beyond the phenyl binding region into the newly created ABP and adopts a bent, not extended or hairpin conformation with a trajectory of ca. 100° validating early conclusions drawn from the examination of conformationally restricted inhibitors.45 This turn is found at the location of the oleyl Δ9,10-double bond placing its π-unsaturation at nearly the same location as the phenyl groups of 1–4. It benefits from analogous stabilizing interactions involving Phe192, Phe381 and possibly Phe432 through van der Waals and hydrophobic interactions during binding and mimics the binding of the oleamide chain. Furthermore, Phe192 is oriented as it is found in the structures of 1 and 2, differing from the orientation found in 3 and 4, but it is tilted about 20° compared to the 1 or 2 structures, so that the aromatic ring is perpendicular to the plane of the oleyl Δ9,10-double bond π-orbital. Although the distances of the aryl CH's to the π-system are close enough to suggest they may be engaged in aryl CH-π interactions, both their orientation and position are not properly aligned. However, the observations do suggest that Phe192 may be best poised to interact with an arachidonyl Δ5,6-double bond in a manner analogous to its interaction with the internal phenyl group of 4 (see Figure 4). The oleyl Δ9,10-double bond found in 5 and the arachidonyl Δ8,9-double bond of MAP occupy similar sites. To accommodate this, the oleyl chain adopts a gauche versus extended conformation at C6–C9 somewhat analogous to the arachidonyl Δ5,6-double bond. Just as interestingly, the oleyl side chain also appears to adopt gauche versus extended conformations at C11–C14 and C13–C16 mapping onto the arachidonyl Δ11,12 and Δ14,15 double bonds providing an overall bent or curled bound conformation that terminates binding at roughly the same site as MAP. Prior modeling studies enlisting Monte Carlo simulations in conjunction with free energy perturbation calculations (MC/FEP) also projected that the oleyl side chain of 5 would align in the acyl chain-binding pocket and adopt a non-extended conformation similar to that of the FAAH–MAP structure.48
Figure 5.
The overlay of the FAAH–MAP42c (methyl arachidonyl phosphonate) structure in blue and FAAH–5 structure in green shows the distinct trajectory that the two chains take away from the active site region.
Figure 6.
FAAH–MAP42c structure in blue and FAAH–5 structure in green. The distortion in residues 275–278 shown on the right and minor changes to Thr488, Phe432, and Phe381 shown on the left. The opened ABP not present with 1–4 is shown in surface representation on the right panel. 2Fo-Fc electron density maps at a contour level of 1.0σ for compound 5 are shown with white meshes.
These comparison structures highlight several important features that have been shown to impact inhibitor potency. The strategic placement of the terminal phenyl group of 1–4 in proximity to the closed ABP, and the nature of its interactions with FAAH are consistent with both its importance in conveying enhanced potency to the α-ketoheterocycle inhibitors as well as its role in mimicking the π-unsaturation of anandamide (Δ8,9-double bond), oleamide (Δ9,10-double bond), and related fatty acid substrates. Both the presence of π-unsaturation (alkyne > alkene > CH2CH2)30,35,44,45 as well as its stereochemistry (cis > trans)30,35,44,45 within the acyl chain of the inhibitors have been shown to enhance inhibitor affinity. This is consistent with the observed binding of the oleyl chain found in 5.30,35 The terminal phenyl group of the typically more potent biphenylethyl side chain36,37 of 4, and presumably that of a series of related, conformationally restricted side chains containing two phenyl rings,37 lies in the phenyl binding pocket defined by 1–3 supporting the conclusion that it represents a dominant anchoring interaction for such inhibitors.43 Surrounding the phenyl binding region, there is sufficient room and protein flexibility to accommodate the range and character of appended phenyl substituents (m ≥ p > o, FAAHKi) that have been shown to maintain or enhance the affinity of inhibitors closely related to 1–3.36 Inhibitors that contain a shorter linking methylene chain exhibit a progressive and substantially reduced affinity for FAAH failing to fully benefit from the forces that stabilize substrate binding.30,32 Just as significantly, inhibitors that extend beyond this phenyl-binding site also exhibit a progressively diminished binding affinity.30,32 This is observed even with inhibitors that do not contain π-unsaturation or the terminal phenyl group, suggesting that the substantial protein reorganization with opening of the acyl chain-binding pocket to accommodate the longer inhibitors (e.g. oleyl side chain) and/or the inhibitor adoption of non ground state conformations (e.g. gauche vs extended binding of oleyl side chain) offsets potential gains in inhibitor binding derived from their increased size (length). The systematic examination of the terminal phenyl group placement defined that a linking chain length of 5–6 methylenes is optimal for inhibitors such as 1–3, that the biphenylethyl side chain of 4 typically further improves on this, and that terminal phenyl group removal substantially reduces affinity.30,32,36,37,45 Finally and consistent with the hydrophobic nature of the protein in this linking region, introduction of polar atoms into the linker progressively reduces inhibitor affinity (CH2 > S > O > NMe > CH(OH) > SO > SO2 > CONH).36
Oxyanion Hole Interactions
The electron density at the active site unambiguously established that inhibitors 3–5, like 1–2, form covalent complexes with FAAH resulting from Ser241 attack on the electrophilic carbonyl. The resulting tetrahedral hemiketal binds in a deprotonated state with the alkoxide bound tightly in the oxyanion hole defined by the four main-chain amide N-H groups of Ile238, Gly239, Gly240, Ser241, and secondary interactions provided by the side chains of Asp237, Arg243, and Asn498. The oxyanions of 3–5 are located at the center of the oxyanion holes defined by the backbone amides of Ile238–Ser241, and the shortened distances of 2.7–2.9 Å, 2.8–3.0 Å, 3.1–3.3 Å, and 2.6–2.8 Å, respectively, are reflective of oxyanion (–O–) versus protonated hemiketal (–OH) binding (Figure 7). Its axis is perpendicular to the plane of the four amino acids, the γ-oxygen of Ser241 and the bound carbon of the inhibitors are pulled towards the oxyanion hole, and the relevant atoms of the five inhibitors (1–5) are virtually superimposable, Figure 7.
Figure 7.
Superposition of the oxyanion holes from the five structures of FAAH covalently bound to oxa(dia)zole inhibitors 1–5 (1 and 2 taken from ref. 43).
Activating Heterocycle and Cytosolic Port Substituent Binding
This proved to be one of the most interesting regions of the structures to examine. The initial structures of 1 and 2 revealed that the catalytic triad was trapped in an interrupted “in action” state with Lys142 H-bonded to Ser217 that in turn was engaged in an unusual OH–π bond to the activating heterocycle (versus lone pair H-bonded).43 An ordered cytosolic port bound water was found to mediate an indirect and flexible H-bond interaction between Thr236 and the pyridyl nitrogen of the oxazole C5 substituent locking it into one of two possible orientations and providing a key anchoring interaction for such inhibitors. In turn, Thr236 was H-bonded to the protonated Lys142, an integral residue in the Ser241–Ser217–Lys142 catalytic triad. The fact that the pyridyl substituents of both 1 and 2 bound to FAAH superimposably, while their activating oxazole heterocycles were flipped 180° relative to each other, provided a convincing set of observations that supported their defined roles.43
As a result, the bound structures of 3–5 proved especially interesting to examine. The inhibitors 3 and 4 incorporate a 2-pyridyl-6-carboxylic acid as the oxazole C5 substituent. This substituent slightly reduces the inhibitor potency as measured at pH 9,35,37,38 substantially increases FAAH selectivity (vs other serine hydrolases),37,38,49 and significantly increases the inhibitor's intrinsic solubility. The bound disposition of the 5-(2-pyridyl-6-carboxylate)oxazoles in 3 and 4 are identical, the dihedral angle across the two aryl rings is ca. 11–14°, and the pyridyl ring is oriented such that the pyridyl nitrogen is directed toward the oxazole aryl CH rather than oxazole oxygen (anti vs syn) adopting its most stable orientation.48 Like 1 and 2, the pyridine nitrogens of 3 and 4 are in proximity to a cytosolic port ordered water molecule that in turn is H-bonded to Thr236. The distinguishing feature is that the pyridyl-6-carboxylate is displaced relative to the pyridyl rings of 1 and 2. Its nitrogen is now not engaged in a close H-bond to the cytosolic port bound water (3.4–3.5 Å for compound 3 and 4, vs 2.8–2.9 Å and 3.0–3.1 Å for 1 and 2,43 respectively), but the position of the water allows the formation of a new H-bond with the adjacent carboxylate (3.0–3.1 Å distance, Figure 8). Moreover, the carboxylic acid binds to what may be an anion stabilizing site defined by the Gly268–Cys269 backbone amides, and it appears to displace an additional bound active site water molecule. Provocatively, the cytosolic port bound water mediates an indirect H-bond to the active site protonated Lys142 via Thr236 and it is not yet clear whether this distant interaction (protonation) also contributes significantly to the inhibitor affinity. Although the nitrogen atoms experience only a small displacement (0.7 Å vs 1), the plane defined by the bound pyridines is altered with 3 and 4 being drawn toward Phe192, which is now flipped 90° providing a π-interaction with the pyridyl C3 and C4 CH's for 3 and 4. In spite of these minor distinctions, the H-bonding to the ordered cytosolic port water clearly represents a key stabilizing and anchoring interaction. It is known that the putative anion binding site defined by Gly268–Cys269 in the cytosolic port represents a key interaction for a class of FAAH substrates not yet widely appreciated (N-acyl taurines)50 and perhaps even for those yet to be discovered. The endogenous N-acyl taurines, that activate members of the TRP ion channel family and are upregulated 10-fold in FAAH inactivated animals, bear a negatively charged sulphate that has been shown to productively interact with the cytosolic port Gly268 through mutagenesis studies. Thus, mutagenesis of Gly268 to aspartate (G268D) reduced the rate of N-acyl taurine hydrolysis 100- to 1500-fold lower than wild-type FAAH while maintaining wild type levels of N-acyl enthanolamide hydrolysis. It is likely that the inhibitors incorporating the 2-pyridyl-6-carboxylic acid substituent including 3 and 4 are mimicking and stabilized by this endogenous substrate interaction. As such, this is a superb interaction to exploit for altering the physical properties of FAAH inhibitors (e.g. solubility, PK properties), while maintaining or even enhancing inhibitory potency and selectivity. Finally, it is worth highlighting that the inhibitors containing the 2-pyridyl-6-carboxylic acid substituent displayed a consistent and anomalously larger stabilizing effect on the thermal denaturation of the enzyme further suggesting that the carboxylic acid interaction with the anion binding site may be providing a uniquely important stabilization to the bound complexes of such inhibitors.51
Figure 8.
Left: The intricate hydrogen bond network found in the FAAH–3 crystal structure involving the catalytic triad and inhibitors containing a 2-pyridyl-6-carboxylate. 2Fo-Fc electron density maps at a contour level of 1.5σ for compound 3 and water molecules are shown with white meshes. Right: Orientation of Ser217 and the activating heterocycles of compounds 2,43 3, and 5 after superposition of the three structures highlighting the Ser217 π-OH H-bond.
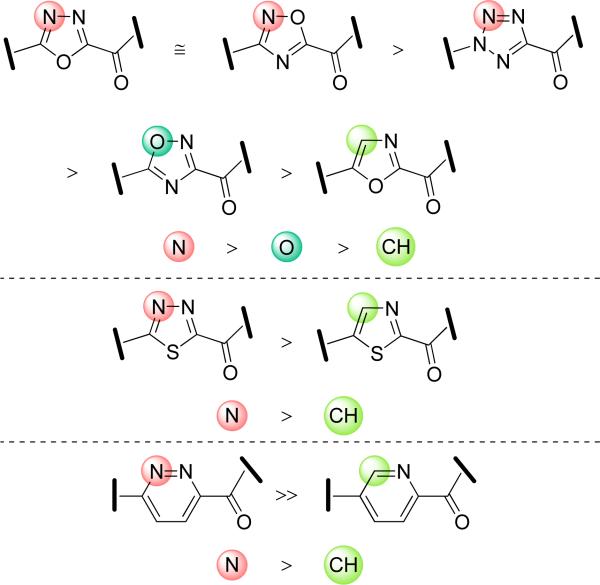
Consistent with the importance of the water mediated H-bond between Thr236 and the more traditional pyridyl substituent, replacing the pyridine nitrogen with a carbon (phenyl vs 2-pyridyl) reduces inhibitor potency 20-fold and even changing its location within the pyridine ring results in a 3–4-fold loss in potency.32,35 A systematic probe of this effect revealed that potency smoothly correlates with the H-bond acceptor properties of the attached C5 heterocycle (e.g. 2-pyridyl = 4-pyrimidyl = 2-oxazole = 2-pyrimidyl > 2-thiazole = 3-pyridazine > 2-furan > 2-thiophene > phenyl), and that it is predictably and subtly influenced by additional substituent effects.30,32,35 Consequently, it is of special note that one of the very few exceptions to this generalization entails removing the nitrogen from the pyridyl group of 3 providing an unexpectedly potent inhibitor (Ki = 5 nM).35 Since altered locations (ortho and para vs meta) of this carboxylic acid on a phenyl substituent were found to be significantly less active, it's unique activity and that of 3 along with the X-ray of 3 depicting the carboxylic acid H-bonding in the anion binding site, indicates that a carboxylic acid placed in this unique location enhances inhibitor binding. What is also clear is that the cytosolic port interactions including that of its ordered water are flexible, potentially accommodating an H-bond acceptor at varied locations, that the interactions are sufficiently strong to account for enhanced inhibitor potencies over well-defined predictions (σp vs –log Ki),34,37,39 and that they serve as a key anchoring interactions capable of substantially influencing inhibitor potency.43
One of the additional most interesting interactions observed at the catalytic core is mediated by Ser217. Rather than lying in the plane of the activating heterocycle and aligned to H-bond to one of its heteroatoms, this residue is located above and oriented toward the center of the heterocycle π-system at a distance of 3.4–3.6 Å (Figure 8). Lys142 is located even further away from the activating heterocycle and is also not spatially aligned for in plane H-bonding. This lack of a stabilizing H-bond with the basic nitrogen of the oxazole is in sharp contrast to the role of the heterocycles first defined in the pioneering efforts with α-ketoheterocycles disclosed by Edwards with serine proteases.47 Like the many cases subsequently explored,46 Edwards observed that the activating heterocycles H-bond through nitrogen to a catalytic residue (typically His) preferentially stabilizing the bound tetrahedral complex. In contrast and given its geometry, the FAAH Ser217 engages in a SerOH–π H-bond with the activating heterocycle. Thus, the role of the activating heterocycle is intrinsically different and this accounts for the remarkable and unanticipated substituent effects observed in our work,34,37,39 where the inhibitor potency actually increases, not decreases, with the addition of electron-withdrawing substituents representing effects that are not observed in the work of Edwards and others.46,47 True to our observations and in contrast to prior works, the heterocycle role is not one of preferential H-bonding stabilization of the tetrahedral adduct via interaction of its proximal basic nitrogen with a core catalytic residue. Rather, its role appears more intimately related to its intrinsic electron-withdrawing character that can be further enhanced by its attached substituents (ρ = 3–3.4 in a Hammett analysis)34,37,39 serving to activate the reactive carbonyl for nucleophilic attack. What is not yet clear is whether the heterocycle simply serves to solvate the catalytic Ser217-OH at the active site or whether this OH–π H-bond provides a preferential stabilizing interaction with the more basic heterocyclic π-system of the tetrahedral adduct. The geometry of the Ser217 H-bond to the π-system of the activating heterocycle is remarkably consistent between the various inhibitors, displaying analogous distances of 3.4–3.6 Å and angles forming on the oxygen between the ring centroid and the serine Cβ atom ranging from 120° and 140°, thus slightly above optimal but still within OH–π H-bond parameters (Figure 8).
Perhaps the most interesting insights emerged from examining this region of the oxadiazole-based inhibitor 5. The pyridine and oxadiazole are also nearly coplanar (ca. 20° dihedral angle) and the sense of the biaxial twist is equal to that observed with 1–4 (Figure 9). The pyridyl ring is oriented in the same direction observed with 1–4 and its nitrogen lies syn to the oxadiazole N4 and anti to the oxadiazole O. Its nitrogen atom lies very close to its position found in 1 and 2, and it is engaged in the same, but an even stronger H-bond with the ordered cytosolic port water (2.6 Å vs 2.8–3.1 Å in 1–2). The second nitrogen of the oxadiazole, that is not found in the oxazole inhibitors (N4 vs C4), is also H-bonded to this same cytosolic port water (3.1–3.2 Å) resulting in the subtle reorientation of the biaryl axis of 5 versus 1 and 2. The net result is that the activating heterocycle and attached pyridine substituent are drawn closer to the catalytic triad including Lys142 as well as Thr236. Although the geometry is not optimally aligned, the OH of Thr236 is now in plane and closer to the oxadiazole N4 nitrogen (3.1–3.3 Å) potentially providing another, albeit less stabilizing, H-bond. This intricate H-bond network involving the cytosolic port water and Thr236 with the pyridyl substituent and activating oxadiazole N4 most likely accounts for the >10-fold increase in inhibitor potency observed with the 1,3,4-oxadiazoles33,38 and its closely related isomers, Figure 10. In several studies, we observed a general and substantial increase in inhibitor potencies if the activating heterocycle incorporated a second weakly basic nitrogen analogous to N4 found in the oxadiazole of inhibitor 5.30,33,38 These trends tracked with the H-bond acceptor properties of the additional heteroatom inserted into the activating central heterocycle (N > O > CH),38 Figure 10. Although several interpretations could be envisioned for such observations, including enhancements in the electron-withdrawing properties of the activating heterocycle, we have also advocated that such heterocycles may participate in an additional stabilizing H-bond interaction at the active site serving as an H-bond acceptor. Even before these X-ray crystallographic studies were available, we suggested this likely involved a mobile H-bond donor at the active site and thought this might involve the protonated Lys142 central to the catalytic triad.30 The structure of 5 bound to FAAH reveals that, in part, this latter interpretation was accurate. The stabilizing H-bonds are derived primarily from the mobile, ordered cytosolic port water mediating the indirect H-bond to Thr236 and its H-bond to the protonated Lys142 as well as a potential direct Thr236 H-bond. This does require a slight reorientation of the oxadiazole plane relative to that observed with oxazole and this appears to partially disrupt the more ideal Ser217 π-bond to the activating heterocycle observed with 1 and 2 (Figure 8). However, even a less optimal geometry for this Ser217 OH–π H–bond interaction is more than compensated for by this dual hydrogen bond interaction of the oxadiazole with the key cytosolic port bound water.
Figure 9.
The cocrystal structures of FAAH–5 (in green) and FAAH–243 (in yellow) display the difference in water H-bonds between an oxadiazole and oxazole core. 2Fo-Fc electron density maps at a contour level of 1.0σ and 1.5σ for compounds 5 and 243 with their water molecules, respectively, are shown with white meshes.
Figure 10.
FAAH inhibitor potency trends from SAR studies on the central activating heterocycle.
Conclusions
Three X-ray cocrystal structures of a carefully chosen set of representative α-ketoheterocycle-based inhibitors of FAAH have been solved and are comparatively examined herein in conjunction with our previously reported cocrystals of 1 and its isomer 2. Each reflects the anticipated reversible covalent addition of the active site Ser241 to the activated carbonyl mimicking the tetrahedral intermediate of the enzyme catalyzed reaction, their comparison allowed features of acyl chain binding in the conformationally mobile membrane access channel and acyl chain-binding pocket to be clarified defining two predominate states (open and closed ABP), and all five bind in a way that establishes a unique role of the activating central heterocycle. This latter role is distinguished from that observed with prior applications of α-ketoheterocycle inhibitors of serine proteases reconciling the large substituent effects found unique to this class of FAAH inhibitors, and each of the five structures display an unusual and now characteristic Ser217 OH–π H-bond. The activating heterocycles are best viewed as electron-withdrawing groups serving to activate the C2 carbonyl on which further substituents can be appended to both increase their intrinsic electron-deficient character and enhance stabilizing cytosolic port interactions. Not only may such substituents be utilized to predictably enhance this electron-deficient character and the active site interactions including that of a putative anion binding site, but even embedded peripheral heteroatoms may serve as H-bond acceptors to engage additional stabilizing cytosolic port interactions mediated by ordered, bound water. Unique to this class of reversible covalent inhibitors and absent in the carbamate and urea-based irreversible inhibitors is the opportunity to define and exploit such dominant cytosolic port interactions for enhancing FAAH affinity and selectivity.
Experimental
Synthesis of Inhibitors 3–5
The inhibitors were prepared in studies disclosed previously.33,35,37,38
FAAH Expression, Purification, and Crystallization
The procedures used were described previously.43 In brief, the N-terminal transmembrane-deleted (ΔTM) form (amino acids 30–579) of the humanized/rat (h/r) FAAH42b gene was expressed in the Escherichia coli BL21 and purified using three chromatography steps including metal affinity, cation exchange, and size exclusion chromatography. The protein sample was concentrated to 25–30 mg/mL in a buffer containing 10 mM Hepes (pH 7.0), 500 mM NaCl, 0.08% n-undecyl-β-d-maltoside (Anatrace), and 2 mM dithiothreitol. Protein concentrations were determined using a reducing-agent compatible BCA protein assay kit (Pierce Biotechnology). The additives xylitol (Sigma) and benzyldimethyl(2-dodecyloxyethyl)-ammonium chloride (Aldrich) were supplemented to the protein sample up to a concentration of 12% and 1%, respectively. After mixing 1:1 the protein solution to the crystallization buffer (30% PEG400, 100 mM Hepes, pH 7.5, and 100 mM NaCl), 6% dimethyl formamide (DMF) and 0.5 mM inhibitor (in DMF) were added to obtain the final crystallization mother liquor. The excess inhibitor precipitated out of solution was spinned down at 16,000g for 3 minutes and discarded. Crystals were grown by sitting drop vapor diffusion at 14 °C in 96-well plates (Innovaplate SD-2; Innovadyne Technologies) and frozen by plunging into liquid nitrogen directly after harvesting. The data for the cocrystal structures of FAAH with 3 and 4 were collected at a temperature of 100 K from a single crystal at the GM/CA-CAT beamline of the Advanced Photon Source (APS, Argonne, IL) using a 10-μm beam collimator. The data for the cocrystal structure of FAAH with 5 was collected at the Stanford Synchrotron Radiation Laboratory (SSRL, Menlo Park, CA) on beamline 11-1. For data reduction we used XDS (FAAH–3, FAAH–5) and HKL2000 (FAAH–4) programs. Structures were solved by molecular replacement using the program Phaser (CCP4 package) and the coordinates of the FAAH–2 structure (PDB code: 2WJ1) as a search model. Structure refinement was performed using the software suite Phenix, Refmac5, and Coot. Chemical parameters for the inhibitors were calculated by the Dundee PRODRG Web server. For the last step of refinement, TLS (Translation/Libration/Screw) parameterization has been applied by dividing each monomer in 8 partitions. Results from data processing and structure refinement are provided in Table 1. The crystal lattices were found in the P3221 space group, containing a FAAH dimer in the asymmetric unit. The structures were determined at a resolution of 1.95 Å (3), 2.25 Å (4), and 2.25 Å (5).
Table 1.
Data Collection and Refinement Statistics of FAAH Crystals.
| |
FAAH–3 |
FAAH–4 |
FAAH–5 |
|---|---|---|---|
| X-ray source | APS-GM/CA-CAT | APS-GM/CA-CAT | SSRL-BL11.1 |
| Crystal data | |||
| Space group | P3221 | P3221 | P3221 |
| Cell dimensions | |||
| a=b, c (Å) | 103.9, 255.4 | 103.4, 254.4 | 104.1, 255.3 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Data collection | |||
| Wavelength (Å) | 1.03312 | 1.03312 | 0.97945 |
| Resolution (Å) | 30.0-1.95(2.02-1.95) | 30.0-2.25(2.33-2.25) | 30.0-2.25(2.32-2.25) |
| Rmerge (%) | 14.0(68.0) | 14.4(45.2) | 11.0(68.9) |
| I / σI | 9.2(2.0) | 8.4(2.8) | 12.2(2.1) |
| Completeness (%) | 98.5(97.7) | 93.6(92.8) | 99.8(99.7) |
| No. of unique reflections | 115337 | 70790 | 76742 |
| Redundancy | 5.5(5.1) | 4.4(3.5) | 4.6(4.5) |
| Refinement | |||
| Resolution (Å) | 1.95(1.97-1.95) | 2.25(2.28-2.25) | 2.25(2.28-2.25) |
| Rwork / Rfree (%) | 15.5(22.7)/18.8(26.5) | 14.4(17.7)/19.8(25.7) | 16.6(25.1)/21.1(33.4) |
| No. atoms | 9589 | 9325 | 9174 |
| Protein | 8431 | 8457 | 8429 |
| Ligand/ion | 58 | 74 | 61 |
| Water | 1100 | 794 | 684 |
| Average B overall (Å2) | 23.6 | 26.2 | 30.6 |
| R.m.s.d. bond length (Å) | 0.011 | 0.012 | 0.010 |
| R.m.s.d. bond angle (°) | 1.254 | 1.306 | 1.213 |
Supplementary Material
Acknowledgments
We gratefully acknowledge the financial support of the National Institutes of Health (DA015648, DLB; DA017259, RCS and BFC) and the Skaggs Institute for Chemical Biology. JG is a Skaggs and ARCS Fellow. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. Use of the Advanced Photon Source at Argonne National Laboratory was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Abbreviations: FAAH, fatty acid amide hydrolase; MAC, membrane access channel; ABP, acyl chain-binding pocket; MAP, methyl arachidonyl phosphonate.
PDB deposition codes : FAAH–3 (3K7F), FAAH–4 (3K83), FAAH–5 (3K84)
Supporting Information Available. Two figures (Figure S1 and S2) providing pair wise structural overlays of bound 1–5.
References
- 1.Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular Characterization of an Enzyme that Degrades Neuromodulatory Fatty Acid Amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 2.Giang DK, Cravatt BF. Molecular Characterization of Human and Mouse Fatty Acid Amide Hydrolases. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2238–2242. doi: 10.1073/pnas.94.6.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patricelli MP, Cravatt BF. Proteins Regulating the Biosynthesis and Inactivation of Neuromodulatory Fatty Acid Amides. Vit. Hormones. 2001;62:95–131. doi: 10.1016/s0083-6729(01)62002-8. [DOI] [PubMed] [Google Scholar]
- 4.Egertova M, Cravatt BF, Elphick MR. Comparative Analysis of Fatty Acid Amide Hydrolase and CB1 Cannabinoid Receptor Expression in the Mouse Brain: Evidence of a Widespread Role for Fatty Acid Amide Hydrolase in Regulation of Endocannabinoid Signaling. Neuroscience. 2003;119:481–496. doi: 10.1016/s0306-4522(03)00145-3. [DOI] [PubMed] [Google Scholar]
- 5.Boger DL, Fecik RA, Patterson JE, Miyauchi H, Patricelli MP, Cravatt BF. Fatty Acid Amide Hydrolase Substrate Specificity. Bioorg. Med. Chem. Lett. 2000;10:2613–2616. doi: 10.1016/s0960-894x(00)00528-x. [DOI] [PubMed] [Google Scholar]
- 6.a Patricelli MP, Cravatt BF. Characterization and Manipulation of the Acyl Chain Selectivity of Fatty Acid Amide Hydrolase. Biochemistry. 2001;40:6107–6115. doi: 10.1021/bi002578r. [DOI] [PubMed] [Google Scholar]; b Lang W, Qin C, Lin S, Khanolkar AD, Goutopoulos A, Fan P, Abouzid K, Meng Z, Biegel D, Makriyannis A. Substrate Specificity and Stereoselectivity of Rat Brain Microsomal Anandamide Amidohydrolase. J. Med. Chem. 1999;42:896–902. doi: 10.1021/jm980461j. [DOI] [PubMed] [Google Scholar]
- 7.Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Isolation and Structure of a Brain Constituent that Binds to the Cannabinoid Receptor. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 8.Martin BR, Mechoulam R, Razdan RK. Discovery and Characterization of Endogenous Cannabinoids. Life Sci. 1999;65:573–595. doi: 10.1016/s0024-3205(99)00281-7. [DOI] [PubMed] [Google Scholar]
- 9.Di Marzo V, Bisogno T, De Petrocellis L, Melck D, Martin BR. Cannabimimetic Fatty Acid Derivatives: The Anandamide Family and Other “Endocannabinoids.”. Curr. Med. Chem. 1999;6:721–744. [PubMed] [Google Scholar]
- 10.Schmid HHO, Schmid PC, Natarajan V. N-Acylated Glycerophospholipids and Their Derivatives. Prog. Lipid Res. 1990;29:1–43. doi: 10.1016/0163-7827(90)90004-5. [DOI] [PubMed] [Google Scholar]
- 11.Boger DL, Henriksen SJ, Cravatt BF. Oleamide: An Endogenous Sleep-Inducing Lipid and Prototypical Member of a New Class of Lipid Signaling Molecules. Curr. Pharm. Des. 1998;4:303–314. [PubMed] [Google Scholar]
- 12.Cravatt BF, Lerner RA, Boger DL. Structure Determination of an Endogenous Sleep-Inducing Lipid, cis-9-Octadecenamide (Oleamide): A Synthetic Approach to the Chemical Analysis of Trace Quantities of a Natural Product. J. Am. Chem. Soc. 1996;118:580–590. [Google Scholar]
- 13.a Cravatt BF, Prospero–Garcia O, Suizdak G, Gilula NB, Henriksen SJ, Boger DL, Lerner RA. Chemical Characterization of a Family of Brain Lipids that Induce Sleep. Science. 1995;268:1506–1509. doi: 10.1126/science.7770779. [DOI] [PubMed] [Google Scholar]; b Lerner RA, Siuzdak G, Prospero–Garcia O, Henriksen SJ, Boger DL, Cravatt BF. Cerebrodiene: A Brain Lipid Isolated From Sleep-Deprived Cats. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9505–9508. doi: 10.1073/pnas.91.20.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a Cravatt BF, Lichtman AH. Fatty Acid Amide Hydrolase: An Emerging Therapeutic Target in the Endocannabinoid System. Curr. Opin. Chem. Biol. 2003;7:469–475. doi: 10.1016/s1367-5931(03)00079-6. [DOI] [PubMed] [Google Scholar]; b Lambert DM, Fowler CJ. The Endocannabinoid System: Drug Targets, Lead Compounds, and Potential Therapeutic Applications. J. Med. Chem. 2005;48:5059–5087. doi: 10.1021/jm058183t. [DOI] [PubMed] [Google Scholar]
- 15.a Ahn K, McKinney MK, Cravatt BF. Enzymatic Pathways that Regulate Endocannabinoid Signaling in the Nervous System. Chem. Rev. 2008;108:1687–1707. doi: 10.1021/cr0782067. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ahn K, Johnson DS, Cravatt BF. Fatty Acid Amide Hydrolase As A Potential Therapeutic Target For The Treatment of Pain and CNS disorders. Exp. Opin. Drug Discov. 2009;4:763–784. doi: 10.1517/17460440903018857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seierstad M, Breitenbucher JG. Discovery and Development of Fatty Acid Amide Hydrolase (FAAH) Inhibitors. J. Med. Chem. 2008;51:7327–7343. doi: 10.1021/jm800311k. [DOI] [PubMed] [Google Scholar]
- 17.a Kathuria S, Gaetani S, Fegley D, Valino F, Duranti A, Tontini A, Mor M, Tarzia G, La Rana G, Calignano A, Giustino A, Tattoli M, Palmery M, Cuomo V, Piomelli D. Modulation of Anxiety Through Blockade of Anandamide Hydrolysis. Nat. Med. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]; b Gobbi G, Bambico FR, Mangieri R, Bortolato M, Campolongo P, Solinas M, Cassano T, Morgese MG, Debonnel G, Duranti A, Tontini A, Tarzia G, Mor M, Trezza V, Goldberg SR, Cuomo V, Piomelli D. Antidepressant-Like Activity and Modulation of Brain Monaminergic Transmission by Blockage of Anandamide Hydrolase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18620–18625. doi: 10.1073/pnas.0509591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jayamanne A, Greenwood R, Mitchell VA, Aslan S, Piomelli D, Vaughan CW. Actions of the FAAH Inhibitor URB597 in Neuropathic and Inflammatory Chronic Pain Models. Br. J. Pharmacol. 2006;147:281–288. doi: 10.1038/sj.bjp.0706510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mor M, Rivara S, Lodola A, Plazzi PV, Tarzia G, Duranti A, Tontini A, Piersanti G, Kathuria S, Piomelli D. Cyclohexylcarbamic Acid 3′- or 4′-Substituted Biphenyl-3-yl Esters as Fatty Acid Amide Hydrolase Inhibitors: Synthesis, Quantitative Structure–Activity Relationships, and Molecular Modeling Studies. J. Med. Chem. 2004;47:4998–5008. doi: 10.1021/jm031140x. [DOI] [PubMed] [Google Scholar]
- 20.a Tarzia G, Duranti A, Tontini A, Piersanti G, Mor M, Rivara S, Plazzi PV, Park C, Kathuria S, Piomelli D. Design, Synthesis, and Structure–Activity Relationships of Alkylcarbamic Acid Aryl Esters, a New Class of Fatty Acid Amide Hydrolase Inhibitors. J. Med. Chem. 2003;46:2352–2360. doi: 10.1021/jm021119g. [DOI] [PubMed] [Google Scholar]; b Tarzia G, Duranti A, Gatti G, Piersanti G, Tontini A, Rivara S, Lodola A, Plazzi PV, Mor M, Kathuria S, Piomelli D. Synthesis and Structure–Activity Relationships of FAAH Inhibitors: Cyclohexylcarbamic Acid Biphenyl Esters with Chemical Modulation at the Proximal Phenyl Ring. ChemMedChem. 2006;1:130–139. doi: 10.1002/cmdc.200500017. [DOI] [PubMed] [Google Scholar]; c Mor M, Lodola A, Rivara S, Vaconido F, Duranti A, Tontini A, Sanchini S, Oiersanti G, Clapper JR, King AR, Tarzia G, Piomelli D. Synthesis and Quantitative Structure–Activity Relationships of Fatty Acid Amide Hydrolase Inhibitors: Modulation at the N-portion of Biphenyl-3-yl Alkylcarbamates. J. Med. Chem. 2008;51:3487–3498. doi: 10.1021/jm701631z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.a Ahn K, Johnson DS, Fitzgerald LR, Liimatta M, Arendse A, Stevenson T, Lund ET, Nugent RA, Normanbhoy T, Alexander JP, Cravatt BF. A Novel Mechanistic Class of Fatty Acid Amide Hydrolase Inhibitors with Remarkable Selectivity. Biochemistry. 2007;46:13019–13030. doi: 10.1021/bi701378g. [DOI] [PubMed] [Google Scholar]; b Johnson DS, Ahn K, Kesten S, Lazerwith SE, Song Y, Morris M, Fay L, Gregory T, Stiff C, Dunbar JB, Jr., Liimatta M, Beidler D, Smith S, Nomanbhoy TK, Cravatt BF. Benzothiophene Piperazine and Piperidine Urea Inhibitors of Fatty Acid Amide Hydrolase (FAAH). Bioorg. Med. Chem. Lett. 2009;19:2865–2869. doi: 10.1016/j.bmcl.2009.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a Abouab–Dellah A, Burnier P, Hoornaert C, Jeunesse J, Puech F. WO 2004/099176 Derivatives of Piperidinyl- and Piperazinyl-alkyl Carbamates, Preparation Methods and Application in Therapeutics (Sanofi) ; b Abouab–Dellah A, Almario GA, Froissant J, Hoornaert C. WO 2005/077898 Aryloxyalkylcarbamate Derivatives, Including Piperidine Carbamates, Their Preparation and Use as Fatty Acid Amide Hydrolase (FAAH) Inhibitors for Treating FAAH-Related Pathologies (Sanofi) ; c Abouab–Dellah A, Almario GA, Hoornaert C, Li AT. WO 2005/070910 1-Piperazine and 1-Homopiperazine Derivatives, Their Preparation and Use as Fatty Acid Amide Hydrolase (FAAH) Inhibitors for Treating FAAH-Related Pathologies (Sanofi) ; d Abouab–Dellah A, Almario GA, Hoornaert C, Li AT. WO 2007/027141 Alkyl(homo)piperazine-carboxylate Derivatives, Their Preparation and Use as Fatty Acid Amide Hydrolase (FAAH) Inhibitors for Treating FAAH-Related Pathologies (Sanofi)
- 23.a Sit SY, Xie K, Deng H. WO2003/06589 Preparation of (Hetero)aryl Carbamates and Oximes as Fatty Acid Amide Hydrolase Inhibitors (Bristol–Myers Squibb) ; b Sit SY, Xie K. WO 2002/087569 Preparation of Bis Arylimidazolyl Fatty Acid Amide Hydrolase Inhibitors for Treatment of Pain (Bristol–Myers Squibb) ; c Sit SY, Conway C, Bertekap R, Xie K, Bourin C, Burris K, Deng H. Novel Inhibitors of Fatty Acid Amide Hydrolase. Bioorg. Med. Chem. Lett. 2007;17:3287–3291. doi: 10.1016/j.bmcl.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 24.a Apodaca R, Breitenbucher JG, Pattabiraman K, Seierstad M, Xiao W. US 2006/0173184 Piperazinyl and Piperidinyl Ureas as Modulators of Fatty Acid Amide Hydrolase (J&J) ; b Apodaca R, Breitenbucher JG, Pattabiraman K, Seierstad M, Xiao W. US 2007/004741 Preparation of Thiadiazolylpiperazine Carboxamides as Modulators of Fatty Acid Amide Hydrolase (FAAH) (J&J) ; c Keith JM, Apodaca R, Xiao W, Seierstad M, Pattabiraman K, Wu J, Webb M, Karbarz MJ, Brown S, Wilson S, Scott B, Tham C-S, Luo L, Palmer J, Wennerholm M, Chaplan S, Breitenbucher JG. Thiadiazolopiperazinyl Ureas as Inhibitors of Fatty Acid Amide Hydrolase. Bioorg. Med. Chem. Lett. 2008;18:4838–4843. doi: 10.1016/j.bmcl.2008.07.081. [DOI] [PubMed] [Google Scholar]; d Karbarz MJ, Luo L, Chang L, Tham C-S, Palmer JA, Wilson SJ, Wennerholm ML, Brown SM, Scott BP, Apodaca RL, Keith JM, Wu J, Breitenbucher JG, Chaplan SR, Webb M. Biochemical and Biological Properties of 4-(3-Phenyl[1,2,4]thiadiazol-5-yl)piperazine-1-carboxylic Acid Phenylamide. A Mechanism-based Inhibitor of Fatty Acid Amide Hydrolase. Anesth. Analg. 2009;108:316–329. doi: 10.1213/ane.0b013e31818c7cbd. [DOI] [PubMed] [Google Scholar]
- 25.a Matsumoto T, Kori M, Miyazaki J, Kiyota Y. WO 2006054652 Preparation of Piperidinecarboxamides and Piperazinecarboxamides as Fatty Acid Amide Hydrolase (FAAH) Inhibitors (Takeda) ; b Matsumoto T, Kori M, Kouno M. WO 2007020888 Preparation of Piperazine-1-carboxamide Derivatives as Brain/Neuronal Cell-Protecting Agents, and Therapeutic Agents for Sleep Disorder (Takeda)
- 26.Ishii T, Sugane T, Maeda J, Narazaki F, Kakefuda A, Sato K, Takahashi T, Kanayama T, Saitoh C, Suzuki J, Kanai C. WO 2006/088075 Preparation of Pyridyl Non-Aromatic Nitrogenated Heterocyclic-1-carboxylate Ester Derivatives as FAAH Inhibitors (Astellas)
- 27.a Moore SA, Nomikos GG, Dickason–Chesterfield AK, Sohober DA, Schaus JM, Ying BP, Xu YC, Phebus L, Simmons RM, Li D, Iyengar S, Felder CC. Identification of a High-Affinity Binding Site Involved in the Transport of Endocannabinoids (LY2183240). Proc. Natl. Acad. Sci. U.S.A. 2005;102:17852–17857. doi: 10.1073/pnas.0507470102. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Alexander JP, Cravatt BF. The Putative Endocannabinoid Transport Blocker LY2183240 is a Potent Inhibitor of FAAH and Several Other Brain Serine Hydrolases. J. Am. Chem. Soc. 2006;128:9699–9704. doi: 10.1021/ja062999h. [DOI] [PubMed] [Google Scholar]
- 28.Alexander JP, Cravatt BF. Mechanism of Carbamate Inactivation of FAAH: Implications for the Design of Covalent Inhibitors and In Vivo Functional Probes for Enzymes. Chem. Biol. 2005;12:1179–1187. doi: 10.1016/j.chembiol.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.b Wang X, Sarris K, Kage K, Zhang D, Brown SP, Kolasa T, Surowy C, El Kouhen OF, Muchmore SW, Briono JD, Stewart AO. Synthesis and Evaluation of Benzothiazole-based Analogues as Novel, Potent, and Selective Fatty Acid Amide Hydrolase Inhibitors. J. Med. Chem. 2009;52:170–180. doi: 10.1021/jm801042a. [DOI] [PubMed] [Google Scholar]; c Myllymaki MJ, Saario SM, Kataja AO, Castillo–Melendez JA, Nevalainen T, Juvonen RO, Jarvinen T, Koskinen AMP. Design, Synthesis, and in vitro Evaluation of Carbamate Derivatives of 2-Benzoxazolyl- and 2-Benzothiazolyl-(3-hydroxylphenyl)methanones as Novel Fatty Acid Amide Hydrolase. J. Med. Chem. 2007;50:4236–4242. doi: 10.1021/jm070501w. [DOI] [PubMed] [Google Scholar]; d Muccioli GG, Fazio N, Scriba GKE, Poppitz W, Cannata F, Poupaert JH, Wouters J, Lambert DM. Substituted 2-Thioxoimidazolidin-4-ones and Imidazolidine-2,4-diones as Fatty Acid Amide Hydrolase Inhibitor Templates. J. Med. Chem. 2006;49:417–425. doi: 10.1021/jm050977k. [DOI] [PubMed] [Google Scholar]; e Saario SM, Poso A, Juvonen RO, Jarvinen T, Salo–Ahen OMH. Fatty Acid Amide Hydrolase Inhibitors from Virtual Screening of the Endocannabinoid System. J. Med. Chem. 2006;49:4650–4656. doi: 10.1021/jm060394q. [DOI] [PubMed] [Google Scholar]; f Minkkila A, Saario SM, Kasnanen H, Leppanen J, Poso A, Nevalainen T. Discovery of Boronic Acids as Novel and Potent Inhibitors of Fatty Acid Amide Hydrolase. J. Med. Chem. 2008;51:7057–7060. doi: 10.1021/jm801051t. [DOI] [PubMed] [Google Scholar]; g Du W, Hardouin C, Cheng H, Hwang I, Boger DL. Heterocyclic Sulfoxide and Sulfone Inhibitors of Fatyy Acid Amide Hydrolase. Bioorg. Med. Chem. Lett. 2005;15:103–106. doi: 10.1016/j.bmcl.2004.10.025. [DOI] [PubMed] [Google Scholar]; h Patricelli MP, Patterson JE, Boger DL, Cravatt BF. An Endogenous Sleep-Inducing Compound Is a Novel Competitive Inhibitor of Fatty Acid Amide Hydrolase. Bioorg. Med. Chem. Lett. 1998;8:613–616. doi: 10.1016/s0960-894x(98)00073-0. [DOI] [PubMed] [Google Scholar]; i Urbach A, Muccioli GG, Stern E, Lambert DM, Marchand–Brynaert J. 3-Alkenyl-2-azetidinones as Fatty Acid Amide Hydrolase Inhibitors. Bioorg. Med. Chem. Lett. 2008;18:4163–4167. doi: 10.1016/j.bmcl.2008.05.081. [DOI] [PubMed] [Google Scholar]; For additional FAAH inhibitors, see: Hart T, Macias AT, Benwell K, Brooks T, D'Alessandro J, Dokurno P, Francis G, Gibbons B, Haymes T, Kennett G, Lightowler S, Mansell H, Matassova N, Misra A, Padfield A, Parsons R, Pratt R, Robertson A, Walls S, Wong M, Roughley S. Fatty Acid Amide Hydrolase Inhibitors. Surprising Selectivity of Chiral Azetidine Ureas. Bioorg. Med. Chem. Lett. 2009;19:4241–4244. doi: 10.1016/j.bmcl.2009.05.097.
- 30.Boger DL, Sato H, Lerner AE, Hedrick MP, Fecik RA, Miyauchi H, Wilkie GD, Austin BJ, Patricelli MP, Cravatt BF. Exceptionally Potent Inhibitors of Fatty Acid Amide Hydrolase: The Enzyme Responsible for Degradation of Endogenous Oleamide and Anandamide. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5044–5049. doi: 10.1073/pnas.97.10.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boger DL, Miyauchi H, Hedrick MP. α-Keto Heterocycle Inhibitors of Fatty Acid Amide Hydrolase: Carbonyl Group Modification and α-Substitution. Bioorg. Med. Chem. Lett. 2001;11:1517–1520. doi: 10.1016/s0960-894x(01)00211-6. [DOI] [PubMed] [Google Scholar]
- 32.Boger DL, Miyauchi H, Du W, Hardouin C, Fecik RA, Cheng H, Hwang I, Hedrick MP, Leung D, Acevedo O, Guimaráes CRW, Jorgensen WL, Cravatt BF. Discovery of a Potent, Selective, and Efficacious Class of Reversible α-Ketoheterocycle Inhibitors of Fatty Acid Amide Hydrolase Effective as Analgesics. J. Med. Chem. 2005;48:1849–1856. doi: 10.1021/jm049614v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung D, Du W, Hardouin C, Cheng H, Hwang I, Cravatt BF, Boger DL. Discovery of an Exceptionally Potent and Selective Class of Fatty Acid Amide Hydrolase Inhibitors Enlisting Proteome-Wide Selectivity Screening: Concurrent Optimization of Enzyme Inhibitor Potency and Selectivity. Bioorg. Med. Chem. Lett. 2005;15:1423–1428. doi: 10.1016/j.bmcl.2004.12.085. [DOI] [PubMed] [Google Scholar]
- 34.Romero FA, Hwang I, Boger DL. Delineation of a Fundamental α-Ketoheterocycle Substituent Effect for Use in the Design of Enzyme Inhibitors. J. Am. Chem. Soc. 2006;128:14004–14005. doi: 10.1021/ja064522b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romero FA, Du W, Hwang I, Rayl TJ, Kimball FS, Leung D, Hoover HS, Apodaca RL, Breitenbucher BJ, Cravatt BF, Boger DL. Potent and Selective α-Ketoheterocycle-Based Inhibitors of the Anandamide and Oleamide Catabolizing Enzyme, Fatty Acid Amide Hydrolase. J. Med. Chem. 2007;50:1058–1068. doi: 10.1021/jm0611509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardouin C, Kelso MJ, Romero FA, Rayl TJ, Leung D, Hwang I, Cravatt BF, Boger DL. Structure–Activity Relationships of α-Ketooxazole Inhibitors of Fatty Acid Amide Hydrolase. J. Med. Chem. 2007;50:3359–3368. doi: 10.1021/jm061414r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimball FS, Romero FA, Ezzili C, Garfunkle J, Rayl TJ, Hochstatter DG, Hwang I, Boger DL. Optimization of α-Ketooxazole Inhibitors of Fatty Acid Amide Hydrolase. J. Med. Chem. 2008;51:937–947. doi: 10.1021/jm701210y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garfunkle J, Ezzili C, Rayl TJ, Hochstatter DG, Hwang I, Boger DL. Optimization of the Central Heterocycle of α-Ketoheterocycle Inhibitors of Fatty Acid Amide Hydrolase. J. Med. Chem. 2008;51:4392–4403. doi: 10.1021/jm800136b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeMartino JK, Garfunkle J, Hochstatter DG, Cravatt BF, Boger DL. Exploration of a Fundamental Substituent Effect of α-Ketoheterocycle Enzyme Inhibitors: Potent and Selective Inhibitors of Fatty Acid Amide Hydrolase. Bioorg. Med. Chem. Lett. 2008;18:5842–5846. doi: 10.1016/j.bmcl.2008.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.a Lichtman AH, Leung D, Shelton CC, Saghatelian A, Hardouin C, Boger DL, Cravatt BF. Reversible Inhibitors of Fatty Acid Amide Hydrolase that Promote Analgesia: Evidence for an Unprecedented Combination of Potency and Selectivity. J. Pharmacol. Exp. Ther. 2004;311:441–448. doi: 10.1124/jpet.104.069401. [DOI] [PubMed] [Google Scholar]; b Chang L, Luo L, Palmer JA, Sutton S, Wilson SJ, Barbier AJ, Breitenbucher JG, Chaplan SR, Webb M. Inhibition of Fatty Acid Amide Hydrolase Produces Analgesia by Multiple Mechanisms. Br. J. Pharmacol. 2006;148:102–113. doi: 10.1038/sj.bjp.0706699. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Palmer JA, Higuera ES, Chang L, Chaplan SR. Fatty Acid Amide Hydrolase Inhibition Enhances the Anti-Allodynic Actions of Endocannabinoids in a Model of Acute Pain Adapted for the Mouse. Neuroscience. 2008;154:1554–1561. doi: 10.1016/j.neuroscience.2008.04.047. [DOI] [PubMed] [Google Scholar]; d Timmons A, Seirestad M, Apodaca R, Epperson M, Pippel D, Brown S, Chang L, Scoot B, Webb M, Chaplan SR, Breitenbucher JG. Novel Ketooxazole Based Inhibitors of Fatty Acid Amide Hydrolase (FAAH). Bioorg. Med. Chem. Lett. 2008;18:2109–2113. doi: 10.1016/j.bmcl.2008.01.091. [DOI] [PubMed] [Google Scholar]; e Schlosburg JE, Boger DL, Cravatt BF, Lichtman AH. Endocannabinoid Modulation of Scratching Response in an Acute Allergenic Model: A New Prospective Neural Therapeutic Target for Pruritus. J. Pharmacol. Exp. Ther. 2009;329:314–323. doi: 10.1124/jpet.108.150136. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Kinsey SG, Long JZ, O'Neal ST, Abdulla RA, Poklis JL, Boger DL, Cravatt BF, Lichtman AH. Blockade of Endocannabinoid-Degrading Enzymes Attenuates Neuropathic Pain. J. Pharmacol. Exp. Ther. 2009;330:902–910. doi: 10.1124/jpet.109.155465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.a McKinney MK, Cravatt BF. Structure and Function of Fatty Acid Amide Hydrolase. Ann. Rev. Biochem. 2005;74:411–432. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]; b McKinney MK, Cravatt BF. Evidence For Distinct Roles in Catalysis For Residues of the Serine–Serine–Lysine Catalytic Triad of Fatty Acid Amide Hydrolase. J. Biol. Chem. 2003;278:37393–37399. doi: 10.1074/jbc.M303922200. [DOI] [PubMed] [Google Scholar]; c Patricelli MP, Cravatt BF. Clarifying the Catalytic Roles of Conserved Residues in the Amidase Signature Family. J. Biol. Chem. 2000;275:19177–19184. doi: 10.1074/jbc.M001607200. [DOI] [PubMed] [Google Scholar]; d Patricelli MP, Lovato MA, Cravatt BF. Chemical and Mutagenic Investigations of Fatty Acid Amide Hydrolase: Evidence for a Family of Serine Hydrolases with Distinct Catalytic Properties. Biochemistry. 1999;38:9804–9812. doi: 10.1021/bi990637z. [DOI] [PubMed] [Google Scholar]; e Patricelli MP, Cravatt BF. Fatty Acid Amide Hydrolase Competitively Degrades Bioactive Amides and Esters through a Nonconventional Catalytic Mechanism. Biochemistry. 1999;38:14125–14130. doi: 10.1021/bi991876p. [DOI] [PubMed] [Google Scholar]
- 42.Ahn K, Johnson DS, Mileni M, Beidler D, Long JZ, McKinney MK, Weerapana E, Sadagopan N, Liimatta M, Smith SE, Lazerwith S, Stiff C, Kamtekar S, Bhattacharya K, Zhang Y, Swaney S, Van Becelaere K, Stevens RC, Cravatt BF. Discovery and Characterization of a Highly Selective FAAH Inhibitor that Reduces Inflammatory Pain. Chem. Biol. 2009;16:411–420. doi: 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mileni M, Johnson DS, Wang Z, Everdeen DS, Liimatta M, Pabst B, Bhattacharya K, Nugent RA, Kamtekar S, Cravatt BF, Ahn K, Stevens RC. Structure-Guided Inhibitor Design for Human FAAH by Interspecies Active Site Conversion. Proc. Natl. Acad. Sci. U.S.A. 2008;105:12820–12824. doi: 10.1073/pnas.0806121105. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Adaptations in a Membrane Enzyme that Terminates Endocannabinoid Signaling. Science. 2002;298:1793–1796. doi: 10.1126/science.1076535. [DOI] [PubMed] [Google Scholar]
- 43.Mileni M, Garfunkle J, DeMartino JK, Cravatt BF, Boger DL, Stevens RC. Binding and Inactivation Mechanism of a Humanized Fatty Acid Amide Hydrolase by α-Ketoheterocycle Inhibitors Revealed from Cocrystal Structures. J. Am. Chem. Soc. 2009;131:10497–10506. doi: 10.1021/ja902694n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patterson JE, Ollmann IR, Cravatt BF, Boger DL, Wong C-H, Lerner AE. Inhibition of Oleamide Hydrolase Catalyzed Hydrolysis of the Endogenous Sleep-Inducing Lipid cis-9-Octadecenamide. J. Am. Chem. Soc. 1996;118:5938–5945. See also: Koutek B, Prestwich GD, Howlett AC, Chin SA, Salehani D, Akhavan N, Deutsch DG. Inhibitors of Arachidonylethanolamide Hydrolysis. J. Biol. Chem. 1994;269:22937–22940.
- 45.Boger DL, Sato H, Lerner AE, Austin BJ, Patterson JE, Patricelli MP, Cravatt BF. Trifluoromethyl Ketone Inhibitors of Fatty Acid Amide Hydrolase: A Probe of Structural and Conformational Features Contributing to Inhibition. Bioorg. Med. Chem. Lett. 1999;9:265–270. doi: 10.1016/s0960-894x(98)00734-3. [DOI] [PubMed] [Google Scholar]
- 46.Maryanoff BE, Costanzo MJ. Inhibitors of Proteases and Amide Hydrolases that Employ an α-Ketoheterocycle as a Key Enabling Functionality. Bioorg. Med. Chem. 2008;16:1562–1595. doi: 10.1016/j.bmc.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 47.a Edwards PD, Meyer EFJ, Vijayalakshmi J, Tuthill PA, Andisik DA, Gomes B, Strimpler A. Design, Synthesis, and Kinetic Evaluation of a Unique Class of Elastase Inhibitors, the Peptidyl α-Ketobenzoxazoles, and the X-ray Crystal Structure of the Covalent Complex Between Porcine Pancreatic Elastase and Ac-Ala-Pro-Val-2-benzoxazole. J. Am. Chem. Soc. 1992;114:1854–1863. [Google Scholar]; b Edwards PD, Bernstein PR. Synthetic Inhibitors of Elastase. Med. Res. Rev. 1994;14:127–194. doi: 10.1002/med.2610140202. [DOI] [PubMed] [Google Scholar]; c Edwards PD, Zottola MA, Davis M, Williams J, Tuthill PA. Peptidyl α-Ketoheterocyclic Inhibitors of Human Neutrophil Elastase. 3. In Vitro and in Vivo Potency of a Series of Peptidyl α-Ketobenzoxazoles. J. Med. Chem. 1995;38:3972–3982. doi: 10.1021/jm00020a011. [DOI] [PubMed] [Google Scholar]; d Edwards PD, Andisik DW, Bryant CA, Ewing B, Gomes B, Lewis JJ, Rakiewicz D, Steelman G, Strimpler A, Trainor DA, Tuthill PA, Mauger RC, Veale CA, Wildonger RA, Williams JC, Wolanin DJ, Zottola M. Discovery and Biological Activity of Orally Active Peptidyl Trifluoromethyl Ketone Inhibitors of Human Neutrophil Elastase. J. Med. Chem. 1997;40:1876–1885. doi: 10.1021/jm960819g. [DOI] [PubMed] [Google Scholar]
- 48.Guimaráes CRW, Boger DL, Jorgensen WL. Elucidation of Fatty Acid Amide Hydrolase Inhibition by Potent α-Ketoheterocycle Derivatives from Monte Carlo Simulations. J. Am. Chem. Soc. 2005;127:17377–17384. doi: 10.1021/ja055438j. See also: Tubert–Brohman I, Acevedo O, Jorgensen WL. Elucidation of Hydrolysis Mechanisms for Fatty Acid Amide Hydrolase and Its Lys142Ala Variant via QM/MM Simulations. J. Am. Chem. Soc. 2006;128:16904–16913. doi: 10.1021/ja065863s.
- 49.Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering Potent and Selective Reversible Inhibitors of Enzymes in Complex Proteomes. Nat. Biotechnol. 2003;21:687–691. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 50.a McKinney MK, Cravatt BF. Structure-Based Design of a FAAH Variant That Discriminates between the N-Acyl Ethanolamine and Taurine Families of Signaling Lipids. Biochemistry. 2006;45:9016–9022. doi: 10.1021/bi0608010. [DOI] [PubMed] [Google Scholar]; b Saghatelian A, McKinney MK, Bandell M, Patapoutian A, Cravatt BF. A FAAH-Regulated Class of N-Acyl Taurines that Activates TRP Ion Channels. Biochemistry. 2006;45:9007–9015. doi: 10.1021/bi0608008. [DOI] [PubMed] [Google Scholar]; c Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Suizdak G, Cravatt BF. Assignments of Endogenous Substrates to Enzymes by Global Metabolite Profiling. Biochemistry. 2004;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 51.Slaymaker IM, Bracey M, Mileni M, Garfunkle J, Cravatt BF, Boger DL, Stevens RC. Correlation of Inhibitor Effects on Enzyme Activity and Thermal Stability for the Integral Membrane Protein Fatty Acid Amide Hydrolase. Bioorg. Med. Chem. Lett. 2008;18:5847–5850. doi: 10.1016/j.bmcl.2008.06.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.