Abstract
To address the role of SGS1 in controlling genome stability we previously identified several “slx” mutants that require SGS1 for viability. Here we report the isolation and characterization of temperature-sensitive (ts) SGS1 alleles in cells lacking SLX4. At the non-permissive temperature (37°C) sgs1-ts slx4 cells progress through S-phase and arrest growth as large-budded cells with at least a 2C DNA content. Analysis of the integrity of the replicated DNA by pulsed-field gel electrophoresis revealed that ChrXII was uniquely altered as it was unable to enter the gel. This defect was specific to the tandem rDNA repeats on ChrXII and occurred as cells progressed through S-phase at 37°C. Reciprocal-shift experiments revealed that viability and ChrXII migration can be restored by allowing Sgs1 to act between G2/M and the subsequent G1 phase. These results suggest that Sgs1 and Slx4 are not required for bulk DNA synthesis but that they play redundant roles in maintaining rDNA structure during DNA replication.
Keywords: SGS1, DNA helicase, DNA replication, rDNA, synthetic-lethal
Introduction
The SGS1 gene of Saccharomyces cerevisiae encodes a member of the RecQ family of DNA helicases. This family includes RECQL, BLM, WRN, RECQL4 and RECQ5 in humans, along with Rqh1 in S. pombe and Dmblm and RecQ5 in D. melanogaster (Puranam and Blackshear 1994; Ellis et al. 1995; Yu et al. 1996; Murray et al. 1997; Stewart et al. 1997; Kitao et al. 1998; Kusano et al. 1999; Sekelsky et al. 1999). These proteins are characterized by a central DNA helicase domain with homology to E. coli RecQ and a large N-terminal domain that is functionally important in yeast (Duno et al. 2000; Mullen et al. 2000). The N-termini of Sgs1 and BLM interact with DNA topoisomerase III (Bennett et al. 2000; Wu et al. 2000; Fricke et al. 2001) while the N-terminus of WRN is unique in that it contains an exonuclease domain (Mushegian et al. 1997; Huang et al. 1998). Like RecQ, the BLM, WRN, and Sgs1 helicases display 3′ to 5′ DNA unwinding activity in vitro (Umezu et al. 1990; Lu et al. 1996; Gray et al. 1997; Karow et al. 1997; Bennett et al. 1998).
Mutations in the RecQ family genes are often associated with genomic instability and sensitivity to DNA damaging agents (German 1963; Hoehn et al. 1975; Gebhart et al. 1988; Gangloff et al. 1994; Watt et al. 1996; Murray et al. 1997; Stewart et al. 1997). In humans three distinct genetic diseases, Bloom’s syndrome, Werner’s syndrome and a subset of Rothmund-Thomson syndrome, are caused by mutations in BLM (Ellis et al. 1995), WRN (Yu et al. 1996), and RECQL4 (Kitao et al. 1998), respectively. In yeast, mutations in SGS1 result in an increase in chromosome missegregation, increased rates of recombination at the rDNA and elsewhere, impaired sporulation and an increased sensitivity to both DNA damaging agents and the DNA synthesis inhibitor hydroxyurea (Gangloff et al. 1994; Watt et al. 1995; Yamagata et al. 1998; Mullen et al. 2000). SGS1 mutants also age prematurely possibly due to an increase in extrachromosomal rDNA circles (ERC’s) (Sinclair and Guarente 1997).
The prototypical member of this family, the E. coli RecQ protein, is required for homologous recombination via the RecF pathway and to inhibit illegitimate recombination (Nakayama et al. 1985; Hanada et al. 1997). Some members of the RecF pathway are required to resume replication following UV-induced DNA damage and RecQ is required to process nascent strands at stalled replication forks (Courcelle et al. 1997; Courcelle and Hanawalt 1999). These data suggest that at least some eukaryotic RecQ proteins might be involved in processing stalled replication forks.
In order to identify the primary target of Sgs1 in yeast we previously isolated a set of mutants that require SGS1 for viability. Growth of these six “slx” mutants requires both the N-terminal and DNA helicase domains of Sgs1, as well as Top3 (Mullen et al. 2001). Most of the SLX genes encode novel proteins that are conserved in other species, and in yeast they associate into three distinct complexes: Slx1-Slx4, Slx5-Slx8, and Mms4-Mus81. Mms4-Mus81 is a heterodimeric structure-specific endonuclease that cleaves 3′ branches from duplex DNA in-vitro (Kaliraman et al. 2001). The Slx1 and Slx4 proteins may also function as a complex since their mutant phenotypes are identical, they co-immunoprecipitate from cell extracts and they interact in a two-hybrid screen (Uetz et al. 2000; Mullen et al. 2001). However, unlike other SLX genes, mutations in SLX1 and SLX4 have no obvious effect on growth rate, DNA damage sensitivity or sporulation efficiency. In this report we describe the isolation and characterization of temperature-sensitive (ts) alleles of SGS1 in the slx4Δ background. We show that SGS1 and SLX4 have overlapping roles in the control of rDNA structure during DNA replication. Eliminating both activities simultaneously does not affect bulk DNA synthesis at the rDNA or elsewhere. However, upon entering S-phase the rDNA loses its ability to migrate in pulsed-field gel electrophoresis (PFGE). This defect has previously been shown to correlate with incompletely replicated chromosomes (Hennessy et al. 1991; Desany et al. 1998). Normal chromosome behavior can be restored if Sgs1 is provided prior to mitosis. Based on the role of RecQ in bacteria, we propose that structural defects in the rDNA of sgs1-ts slx4Δ cells arise from the failure to restart stalled replication forks.
Materials and Methods
Strains, media and plasmid construction
The yeast strains used in this study are listed in Table 1. Strain construction, growth, and transformation followed standard methods (Rose et al. 1990). The sgs1-11::loxP deletion removes all but 15 codons at each end of the SGS1 open reading frame (Mullen et al. 2000). All plasmids were made in pRS vectors (Sikorski and Hieter 1989). Plasmids pNTS1 (sgs1-34), pQTS1 (sgs1-35), & pQTS2 (sgs1-t36) were isolated after PCR mutagenesis of SGS1 in pRS415 and screening for temperature-sensitive growth, as described below. The sgs1-ts alleles were subcloned as XhoI/SacI fragments from pNTSI, pQTS1 and pQTS2 into the corresponding sites of the integrating vectors pRS405 (to create pVK16, 17, and 18, respectively) and pRS404 (to create pVK20, 21, and 22, respectively).
Table 1.
List of Strains
| Name | Genotype | Reference or Source |
|---|---|---|
| CHY125 | MATa ade2-1 ade3::hisG ura3-1 his3-12,15 trp1-1 leu2-3,112 can1-100 | Mullen et al. (2000) |
| K1875 |
MATa ade2-1 ura3-52 his4-260, trp1-H3 leu2-3,112 lys2-ΔBX-CAN1-LYS2 can1 rDNA::ADE2 rDNA::URA3 |
Keil and McWilliams (1993) |
| W303-1a | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 | Thomas and Rothstein (1989) |
| NJY560 |
MATa ade2-1 ade3::hisG ura3-1 his3-12,15 trp1-1 leu2-3,112 can1-100 sgs1-11::loxP slx4-11::loxP [pJM500::SGS1 URA3 ADE3] |
This study |
| NJY1338 | NJY560 but containing [pJM6401::SLX4 URA3 ADE3] instead of pJM500 | This study |
| VKY1227 |
MATa ade2-1 ade3::hisG ura3-1 his3-12,15 trp1-1 leu2-3,112 can1-100 sgs1-11::loxP slx4-11::loxP [pJM6401::SLX4 URA3 ADE3] |
This study |
| NJY1228 | NJY560 LEU2::SGS1 FOAR | This study |
| NJY1229 | NJY560 LEU2::sgs1-34 FOAR | This study |
| NJY1230 | NJY560 LEU2::sgs1-35 FOAR | This study |
| NJY1231 | NJY560 LEU2::sgs1-36 FOAR | This study |
| NJY540 | K1875 sgs1-11::loxP | Mullen et al. (2000) |
| NJY1256 | NJY540 LEU2::SGS1 | This study |
| NJY1257 | NJY540 LEU2::sgs1-34 | This study |
| NJY1248 | NJY540 LEU2::sgs1-35 | This study |
| NJY1249 | NJY540 LEU2::sgs1-36 | This study |
| NJY1250 | NJY540 LEU2::pRS405 | This study |
| NJY531 | CHY125 sgs1::loxP | Mullen et al. (2000) |
| NJY1251 | NJY531 TRP1::pRS404 | This study |
| NJY1252 | NJY531 TRP1::SGS1 | This study |
| NJY1253 | NJY531 TRP1::sgs1-34 | This study |
| NJY1254 | NJY531 TRP1::sgs1-35 | This study |
| NJY1255 | NJY531 TRP1::sgs1-36 | This study |
| RLY1 | NJY560 TRP1::SGS1 FOAR | This study |
| RLY2 | NJY560 TRP1::sgs1-34 FOAR | This study |
| RLY3 | NJY560 TRP1::sgs1-35 FOAR | This study |
| RLY4 | NJY560 TRP1::sgs1-36 FOAR | This study |
Isolation of sgs1 temperature-sensitive (ts) alleles
sgs1-ts alleles of SGS1 were isolated following polymerase chain reaction (PCR) mutagenesis and screening for conditional growth in strain NJY1338 which requires plasmid-borne SGS1 or SLX4 for viability. Two domains of Sgs1 were individually targeted for mutagenesis by amplifying the appropriate regions of SGS1 under mutagenic conditions. The PCR product was then used to repair a plasmid containing the corresponding deletion of SGS1 by in vivo recombination (Muhlrad et al. 1992). Plasmid pSM100 (Mullen et al. 2000) contains a modified SGS1 gene in the LEU2/CEN vector pRS415 (Sikorski and Hieter 1989). A unique NdeI site was engineered at the natural translation start site of SGS1 such that the gene consists of a 153 bp XhoI-NdeI promoter fragment, a 1810 bp NdeI-HindIII fragment encoding the N-terminus and a 2616 bp HindIII- SacI fragment encoding the C-terminus of Sgs1. Using pSM100 as template, the N-terminal domain of Sgs1 (aa 1-652) was targeted for mutagenesis using primer 552 (5′-GGTAACGCCAGGGTTTTCCCA-3′) within pRS415 and anti-sense primer 553 (TTACCTTGCAAAGTTGCATTT) at position 2061 of SGS1 (ATG = position 1). The NTP-binding domain (aa 653-996) of SGS1 was targeted using sense primer 554 (TTGGAAGATGATCTGGAAAAG) at position 1560, and anti-sense primer 555 (TTACCTTGCAAAGTTGCATTT) at position 3214. Gapped versions of pSM100 were prepared by digestion with either NdeI and HindIII (Δ1 - 1810 bp for mutagenesis of the N-terminal domain) or HindIII and EagI (Δ1810 - 2963 bp for mutagenesis of the NTP-domain). Mutagenic PCR (35 cycles) was performed with 0.2 u/μl Taq DNA polymerase (Promega), 100 μM primers, 0.25 mM dNTPs, and 0.25mM to 0.5mM Mn++ in place of Mg++. Gapped plasmids and the PCR products were purified by agarose gel electrophoresis, mixed and used to transform strain NJY1338 (Table 1). Leu+ transformants were selected at 25°C, replica-plated to media containing 5-fluoroorotic acid (5-FOA) at 25°C and then replica-plated to yeast extract/peptone/dextrose (YPD) plates (Rose et al. 1990) to screen for viability at 17°C, 25°C and 37°C. For each domain, 4000 transformants were screened. Three mutants were isolated that were inviable at 37°C. One mutant (sgs1-34) resulted from mutagenesis of the N-terminus and two mutants (sgs1-35, sgs1-36) resulted from mutagenesis of the NTP-domain. The rescued plasmids had a wild-type restriction map, indicating that the gaps were repaired correctly.
DNA damage, recombination and aging assays
To prepare media, methylmethanesulfonate (MMS, final concentration 0.012%) was added to YPD agar just before pouring and the plates were used within 3 days. Cells were scraped from freshly growing plates, resuspended in water and the OD600 was determined. Cells were transferred to a well of a 96-well microtiter plate at OD = 3.0 and serially diluted 1:10 to adjacent wells. A replica-plater was then used to transfer approximately 5 μl from each well to YPD plates containing MMS or no drug. For recombination assays NJY540 cells carrying the integrated versions of the sgs1-ts alleles were maintained on minimal (SD) plates (Rose et al. 1990) lacking adenine, uracil and leucine. Cells were resuspended in water and about 200 cells were spread onto YPD plates containing 20 μg/ml adenine and incubated at 25°C or 37°C. After 3 days, 7 colonies from each plate were individually picked, resuspended in 150 μl water, diluted and a volume containing approximately 400 cells was spread onto a YPD plate and an SD plate containing 5-FOA. After 3 days at 25°C, colonies were counted and the percentage of viable cells that were resistant to 5-FOA (uracil−) was determined. The experiment was repeated three times. Old yeast cells were isolated as described (Smeal et al. 1996).
Microfluorimetry, CHEF gel and density transfer analyses
Yeast strains were grown in liquid YPD broth at 25°C to OD600 = 0.15 and treated with α-factor at 5 μg/ml for 2 hours to arrest the cells in the G1 phase of the cell cycle. Following arrest, the cells were removed by filtration, resuspended in water, filtered, and washed a second time with YPD broth. The cells were then sonicated, resuspended in YPD broth, split in half, and incubated with shaking at either 25°C or 37°C. Aliquots of cells were removed at the indicated times, washed with phosphate-buffered saline (PBS) (Harlow and Lane 1988) and fixed in 70% ethanol for 1 hr. The cells were again washed in PBS, resuspended in PBS containing 0.1 mg/ml RNaseA and incubated at 37°C for 2 hrs. Proteinase K was added to 0.04 mg/ml and the cells were incubated for 1 hr at 55°C. The cells were then washed with PBS, sonicated and incubated with propidium iodide at 100 μg/ml for 1 hr. The cells were diluted 1:4 and sonicated prior to microfluormetric analysis on a Coulter-Epics fluorescence-activated cell sorter (FACS). At least 150,000 events were recorded per sample.
CHEF gel analysis was carried out essentially as described by the manufacturer (Bio-Rad, Hercules, CA). Briefly, the strains were grown in liquid YPD medium at 25°C to OD600 = 0.2 and synchronized with α-factor as above. The culture was released at 37°C and cells collected after 3 or 6 hours. The cells were washed and resuspended in cold 50 mM EDTA. Cells (6 × 108) were then treated as described and loaded into a 1.0% agarose gel (Pulse Field Certified Agarose, BioRad) in 0.5 X TBE and subjected to electrophoresis at 6 V/cm at an included angle of 120° with constant recirculation of the buffer at 12°C. Electrophoresis of chromosomal DNA was performed for 24 hours with an initial switch time of 1 min and final switch time of 2 min. Following electrophoresis the gels were stained with ethidium bromide (0.5 μg/ml), photographed and processed for Southern blotting as described (Sambrook et al. 1989).
The density-transfer assay was performed essentially as described (McCarroll and Fangman 1988). NJY1229 (sgs1-34 slx4Δ) cells were grown at 25°C for seven generations in heavy minimal medium containing 0.1% 13C glucose and 0.01% 15N ammonium sulfate as carbon and nitrogen sources, respectively. The cells were then arrested in G1 with α-factor (5 μg/ml for 3 hours). The cells were then washed, split and released into light rich medium (YPD) under one of the three following conditions: at 25°C with no drug; at 37°C with α-factor (15 μg/ml); or at 37°C with nocodazole (20 μg/ml). Samples (32 × 106 cells) were taken for analysis after the G1 arrest and 3 hrs after release treatment. The DNA was digested with BglII and separated on CsCl gradients (McCarroll and Fangman 1988). The gradients were fractionated, the refractive index of every other fraction was determined and every fraction was dot-blotted and probed with a 32P-labeled rDNA fragment consisting of the 2.7 Kb HindIII fragment from the 5′ end of the 35S transcript fused to the 0.6 kb EcoRI fragment at the 3′ end of the 35S transcript.. A Molecular Dynamics PhosphorImager was used to develop the image which was quantitated using IP LabGel software. The positions of unreplicated DNA, corresponding to a refractive index of 1.4050 (heavy-heavy DNA), and fully replicated DNA, corresponding to a refractive index of 1.4040 (heavy-light DNA), were determined.
Results
Generation of temperature-sensitive alleles of SGS1
To isolate point mutants of SGS1 that result in conditional loss-of-function, we took advantage of the fact that a yeast strain lacking the non-essential gene SLX4 (YLR135W) is dependent on SGS1 for viability (Mullen et al. 2001). We assumed that mutations in either the N-terminal domain or the DNA helicase domain of Sgs1 could be lethal since Sgs1 proteins lacking small portions of the N-terminus or DNA helicase activity were lethal in the slx4Δ background (Mullen et al. 2000). The strain NJY1338 (slx4Δ sgs1Δ pJM6401/SLX4/URA3/ADE3), is unable to grow on media containing 5-FOA since selection against the URA3 plasmid is lethal in this background. Mutagenized SGS1 genes were introduced into NJY1338 on LEU2-based plasmids, swapped for the SLX4 plasmid by growth on 5-FOA at 25°C, and the resulting colonies screened for loss of viability on YPD plates at 37°C. Three independent plasmids were isolated that conferred ts growth. All three sgs1-ts alleles were recessive, as an slx4Δ SGS1 strain containing any of the sgs1-ts plasmids was able to grow at 37°C (data not shown). The sgs1-ts alleles were sequenced and each found to contain a single missense mutation. The sgs1-34 mutation (Q31P) mapped to the N-terminus while sgs1-35 and sgs1-36 (T980I and E987K, respectively) mapped to the more highly-conserved NTP-binding domain. Thus, missense mutations mapping to either the N-terminus or the DNA helicase domain of Sgs1 are capable of generating ts growth in slx4Δ cells.
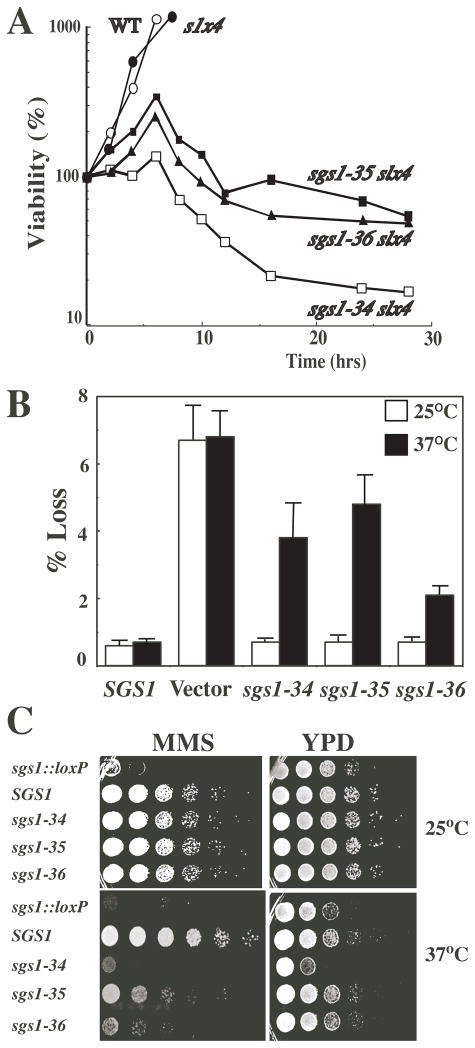
In order to identify allelic differences, each sgs1-ts allele was integrated as the only copy of SGS1 into otherwise wild type (wt) and slx4Δ backgrounds (Table 1). A liquid culture of each sgs1-ts slx4Δ strain was shifted from 25°C to 37°C and at various times cells were removed and plated to measure colony formation at 25°C (Fig. 1A). Double mutant strains displayed an initial increase in viable cell number after which they rapidly lost viability. The sgs1-34 slx4Δ strain exhibited the greatest sensitivity with less than a 2-fold increase in cell number and reduction to 10% viability in 16 hrs. To test whether the thermosensitivity of the sgs1-ts alleles was independent of slx4Δ, we assayed sgs1 phenotypes in the SLX4 background. The recombination phenotype was determined in strain NJY540, which contains the URA3 marker within the rDNA cluster (Keil and McWilliams 1993). The rate of URA3 marker loss was determined following growth on non-selective media at 25°C and 37°C. Strain NJY540 (sgs1Δ) carrying vector alone lost the URA3 marker at 12 times the frequency of wt regardless of temperature (Fig. 1B). This elevation in recombination frequency is similar to that observed previously for the sgs1Δ null strain (Gangloff et al. 1994; Watt et al. 1996; Mullen et al. 2000). In contrast, the sgs1-ts alleles conferred near wt levels of recombination on NJY540 at the permissive temperature and 4- to 10-fold higher levels at 37°C. Thus, with respect to recombination, the sgs1-ts alleles are thermosensitive.
Fig. 1.
Conditional SGS1 activity. A Strains of the indicated genotype were grown to exponential phase in YPD broth at 25°C, transferred to 37°C for the indicated times, and the percentage of viable cells determined. B Recombination rate was determined by stably transforming the indicated SGS1 allele, or vector alone, into strain NJY540 (sgs1::loxP) which contains the URA3 gene integrated at RDN1. The number of uracil auxotrophs was determined following 3 days growth in the absence of selection at the indicated temperature. The percentage of cells undergoing a marker loss event was determined and is presented as the mean ± SD from seven independent colonies. C SGS1-dependent hypersensitivity to DNA damaging agents was determined by resuspending strains of the indicated genotype and spotting them in 10-fold serial dilutions on YPD plates containing MMS or no drug. The plates were photographed following three days at the indicated temperature.
Otherwise wt sgs1-ts strains were next tested for sensitivity to methylmethanesulfonate (MMS). Ten-fold serial dilutions of cells were spotted onto broth plates in the absence or presence of MMS, and the level of growth was observed after 3 days at permissive and restrictive temperatures. As previously observed, the sgs1 deletion strain carrying vector alone grew very poorly in the presence of MMS, while the strain carrying wild type SGS1 grew well at both permissive and restrictive temperatures (Fig. 1C). At 25°C, sgs1-ts strains grew at nearly the wild type rate in the absence or presence of mutagen. In contrast, at 37°C, the growth of all three sgs1-ts mutants was specifically inhibited in the presence of MMS (Fig. 1C), and the level of growth approximated that of the sgs1Δ null strain. As in Fig. 1A, the sgs1-34 allele showed the most severe effect at 37°C. In fact, sgs1-34 grew worse than the null strain on YPD at 37°C (Fig. 1C, bottom right panel). We have previously shown that a strain lacking the first 158 amino acids of Sgs1 (sgs1-ΔN158) displays “hypermorphic” phenotypes relative to the null. For example, the sgs1-ΔN158 stain is more sensitive to MMS than the null strain and grows more slowly than the null strain (Mullen et al. 2000). Since the Q31P mutation falls within the first 158 aa, we presume that sgs1-34 mimics the hypermorphic phenotype of the sgs1ΔN158 allele, but only at the restrictive temperature. Finally, immunoblotting of epitope-tagged Sgs1 proteins revealed nearly equal levels of protein in all three mutant and wt strains grown at 25 or 37°C (data not shown). Based on these assays we conclude that the activity of the three sgs1-ts alleles is greatly reduced at the restrictive temperature.
sgs1-ts alleles affect separate SGS1 functional domains
Sgs1 is composed of two functional domains: the N-terminus (amino acids (aa) 1 - 652) and the DNA helicase domain (aa 645 - 1447). These two domains function in trans to complement sgs1Δ MMS sensitivity and sgs1Δ slx4Δ synthetic lethality (Mullen et al. 2000). We used intragenic complementation to test whether one or both domains were defective in the sgs1-ts alleles. Strain RLY2 (sgs1-34 slx4Δ) was transformed with a series of SGS1 truncation alleles at 25°C and tested for viability by streaking onto YPD plates and observing growth at 37°C. As expected, cells transformed with SGS1 were viable at 37°C while those transformed with vector were not (Table 2). The full-length helicase-defective allele, sgs1-hd, as well as an allele that expresses only the N-terminus (aa 1 - 652), also complemented the lethality of the sgs1-34 slx4Δ strain at 37°C. In contrast, SGS1 alleles encoding N-terminal deletions failed to complement this strain. We conclude that sgs1-34 is defective only in the N-terminal function and retains DNA helicase activity at 37°C. The sgs1-35 and sgs1-36 alleles could not be complemented at the non-permissive temperature by sgs1-hd or by C-terminal truncations. Thus, complementation of these alleles requires an intact helicase domain. Consistent with this result, SGS1 alleles encoding N-terminal deletions of up to 158 aa showed full complemention of sgs1-35 and sgs1-36 (Table 2). Taken together, we conclude that the NTP-domain mutations have impaired DNA helicase activity but retain N-terminal function at 37°C.
Table 2.
Intragenic complementation of sgs1-ts slx4Δ1
| SGS1 allele | Protein | sgs1-34 | sgs1-35 | sgs1-36 | |||
|---|---|---|---|---|---|---|---|
| product 2 | 25°C | 37°C | 25°C | 37°C | 25°C | 37°C | |
| SGS1 | 1 – 1447 | + | + | + | + | + | + |
| sgs1 - hd | 1 – 1447 | + | + | + | − | ND | ND |
| sgs1 - ΔC795 | 1 – 652 | + | + | + | − | + | − |
| sgs1 - ΔN158 | 159 – 1447 | + | − | + | + | + | + |
| vector (pRS415) | none | + | − | + | − | + | − |
Strains RLY2, RLY3, and RLY4 (sgs1Δ slx4Δ TRP1::sgs1-ts) were transformed with the indicated SGS1 truncation alleles in pRS415 at 25°C and tested for viability by streaking on YPD plates and observing growth after three days at 37°C. +, growth; -, no growth. ND: Not Done.
Expressed amino acids of Sgs1. The sgs1-hd allele expresses Sgs1 with the K706A mutation (Lu et al. 1996).
We tested whether the sgs1-ts alleles displayed growth defects in other slxΔ backgrounds at 37°C. As expected from the physical interaction between Slx1 and Slx4, all three sgs1-ts alleles that were isolated in the slx4Δ mutant background were lethal in the slx1Δ background at the non-permissive temperature (Table 3). The growth of sgs1-ts mms4Δ and sgs1-ts mus81Δ double mutants was impaired, although not eliminated at 37°C. Finally, the sgs1-34 slx5Δ double mutant was inviable at 37°C, while sgs1-35 slx5Δ and sgs1-36 slx5Δ strains displayed a growth defect at 37°C. As above, we conclude that the thermosensitivity of the sgs1-ts alleles is independent of the slxΔ background.
Table 3.
Phenotype of sgs1-ts alleles in various SLX mutant backgrounds1
| SGS1 allele | slx1Δ | sgs1Δ | mms4Δ | sgs1Δ | mus81Δ | sgs1Δ | slx4Δ | sgs1Δ | slx5Δ | sgs1Δ |
|---|---|---|---|---|---|---|---|---|---|---|
| 25°C | 37°C | 25°C | 37°C | 25°C | 37°C | 25°C | 37°C | 25°C | 37°C | |
| SGS1 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| sgs1-34 | ++ | − | ++ | + | ++ | + | ++ | − | + | − |
| sgs1-35 | ++ | − | ++ | + | ++ | + | ++ | − | ++ | + |
| sgs1-36 | ++ | − | ++ | + | ++ | + | ++ | − | ++ | + |
The indicated double mutant strains containing plasmid pJM500 (SGS1/URA3) were transformed with the indicated sgs1-ts allele in pRS415 at 25°C and tested for viability by streaking on plates containing 5-FOA and observing growth after three days at 25°C or 37°C.
++, wt growth; + slow growth; -, no growth.
SGS1 and SLX4 are not required for bulk DNA synthesis
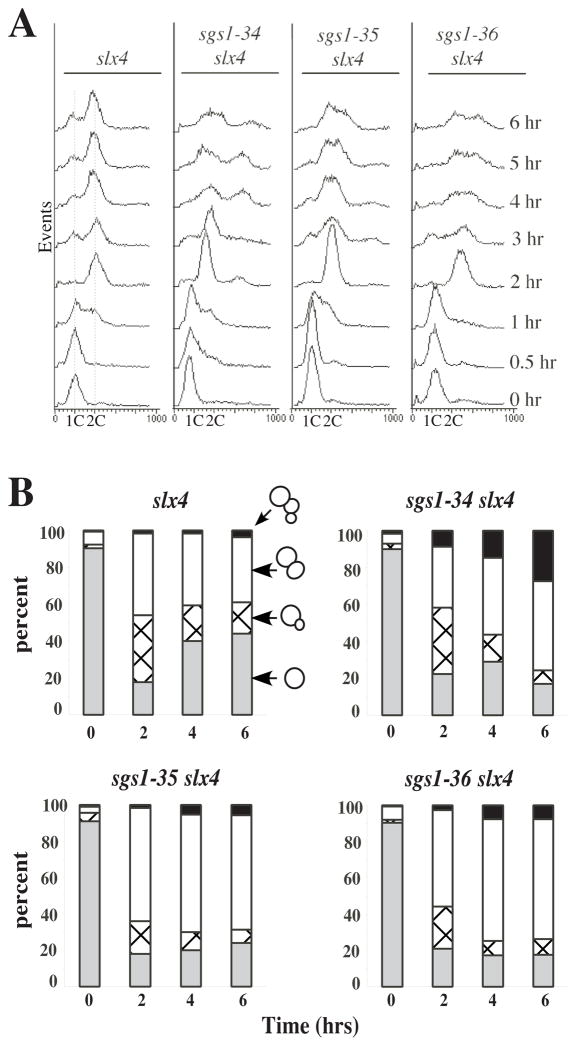
To address the roles of Sgs1 and Slx4 in cell growth, we assayed cell-cycle progression in sgs1-ts slx4Δ cells by flow cytometry. Exponentially growing strains were first arrested with α-factor at the permissive temperature and then released from G1 arrest at 37°C. The slx4Δ strain began DNA synthesis between 0.5 and 1 hour after release, and achieved a 2C DNA content by 2 hours (Fig. 2A). At 3 hours, cells containing 1C DNA content appeared while at later time points there is a loss of synchrony in the culture. Nearly identical profiles were obtained with wt cells at 37°C or single and double mutant combinations at 25°C (data not shown). When the slx4Δ sgs1-34 mutant was released at 37°C, DNA synthesis also began between 0.5 and 1 hour, and 2C DNA content was observed after 3 hours (Fig. 2A). Thus, bulk DNA synthesis was not significantly affected in the double mutant at 37°C. However, unlike single mutants this strain did not produce a 1C peak indicative of G1 cells in the next cell cycle. Instead, the G2 peak broadened and a peak corresponding to 3C to 4C appeared 4 to 6 hours after release. The sgs1-35 and -36 strains behaved similarly: DNA synthesis was completed by 2 hours, they failed to produce G1 cells, and they accumulated greater than 2C DNA content at late times. We note that the increase in DNA content observed 4–6 hours at 37°C appears to coincide with the loss of viability in exponential cultures (Fig. 1A).
Fig. 2.
Cell cycle arrest of sgs1-ts slx4Δ cells. The indicated strains were grown to exponential phase at 25°C and synchronized by arresting growth with α-factor. After release into YPD broth at 37°C, aliquots were removed at the indicated times and fixed in ethanol. A One portion of each aliquot was stained with propidium iodide and analyzed by flow cytometery. B Another portion was microscopically examined for budding morphology: gray, single cells; crosshatch, small budded cells; white, large budded cells; black, aberrant or multiply-budded cells. Data is presented as a percentage of at least 100 cells.
To confirm the cell cycle results, we examined the morphology of these strains growing at 37°C. At time zero all strains were predominantly unbudded, as expected for cells in the G1 phase of the cell cycle (Fig. 2B). After 4 hours at 37°C, slx4Δ cells had lost synchrony and were growing with nearly equal proportions of unbudded, small-budded and large-budded cells. In contrast, double mutant cultures were mostly comprised of large- or multiply-budded cells after 4 - 6 hours at 37°C. DAPI staining of these cells revealed that most large-budded cells contained a single undivided nucleus at the neck of the mother cell (data not shown). This phenotype is similar to that observed when SGS1 expression was repressed in the mms4Δ background (Mullen et al. 2001) and suggests that most cells arrest growth in the late S or G2 phase of the cell cycle. As in the assays described above, the N-terminal domain mutant, sgs1-34, displayed a more severe aberrant budding phenotype than the helicase domain mutants. After 6 hours at 37°C, approximately 30% of these cells were multiply budded compared to 10% or fewer of the helicase domain mutants (Fig. 2B). Taken together, we conclude that DNA synthesis is not significantly inhibited in the absence of SGS1 and SLX4. However, the majority of these cells arrest growth at late S/G2 and undergo additional DNA synthesis after 4–6 hours. The broad 3C - 4C peak may result from incomplete DNA synthesis in cells that are losing viability.
Altered stability of Chromosome XII in sgs1-ts slx4Δ mutants
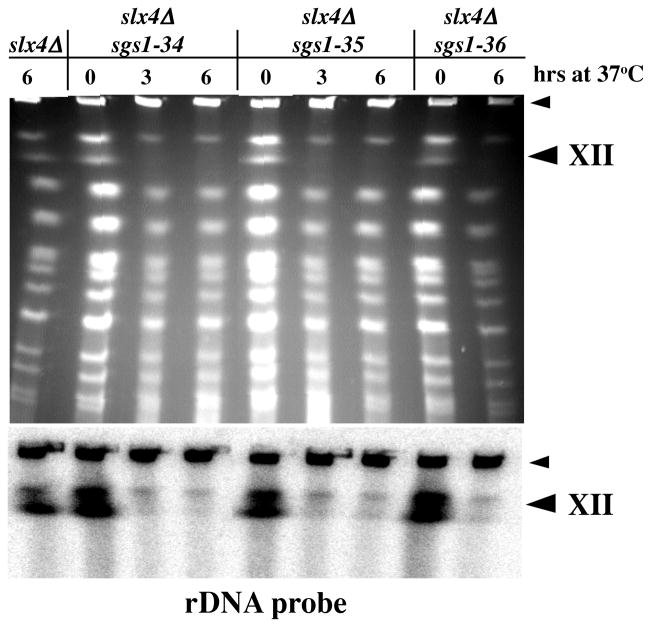
The terminal phenotype of the double mutants suggested a defect late in DNA replication. To identify defects in chromosomal replication, we used PFGE to search for temperature-dependent changes in chromosome structure. Clamped Homogenous Electric Field (CHEF) gels have previously been used to characterize chromosomal abnormalities associated with replicating yeast DNA (Hennessy et al. 1991; Christman et al. 1993; Desany et al. 1998; Shimada et al. 1999). When cells are arrested before or after DNA replication chromosomes migrate normally in this system, however when cells are arrested during DNA replication, for example by treatment with hydroxyurea (HU), chromosomes are unable to enter the gel (Hennessy et al. 1991; Desany et al. 1998). These incompletely replicated chromosomes fail to enter the gel due to DNA replication forks or bubbles that block its electrophoretic migration (Hennessy et al. 1991; Desany et al. 1998; Shimada et al. 1999).
CHEF gels resolve the 16 individual yeast chromosomes into a ladder of 13 bands following ethidium bromide staining. Under the conditions used here Chromosome XII (ChrXII), which contains the 1 Mb rDNA cluster, migrates as the second largest chromosome. This was confirmed by Southern blotting with an rDNA probe which detected ChrXII as a doublet in all strains (Fig. 3, lower panel). Cells were synchronized with α-factor at 25°C, released at 37°C and harvested after 3 and 6 hours for CHEF gel analysis. Under permissive conditions, the pattern of chromosomal bands obtained from wt and mutant strains was identical (Figs. 3, 6A and data not shown). In contrast, when double mutant cells were shifted to 37°C for 3 or 6 hours, there was a slight decrease in the intensity of most bands and a striking loss of ChrXII from the profile (Fig. 3). Southern blotting with an rDNA probe confirmed the identity of the missing band as ChrXII and indicated that it was not co-migrating with other chromosomal bands (Fig. 3, lower panel). This phenotype is specific to the double mutant at restrictive temperature as ChrXII migration was normal in sgs1Δ, slx4Δ and sgs1-ts single mutants at both temperatures (Figs. 3 and 6A). In addition, the phenotype was independent of synchrony since it was also observed in exponentially growing cells upon shift to 37°C (data not shown). We conclude that ChrXII isolated from sgs1-ts slx4Δ strains at 37°C is retained in the well of the gel similar to chromosomes from HU-treated cells.
Fig. 3.
Altered electrophoretic mobility of ChrXII in sgs1-ts slx4Δ strains. The indicated strains were grown to exponential phase at 25°C, arrested with α-factor and released at 37°C for the indicated times. Cells were collected and prepared for CHEF gel electrophoresis in agarose plugs. The gel was stained with ethidium bromide (top) and Southern blotted using an rDNA fragment as probe (bottom). The bottom half of the blot had no signal and is not presented. The large arrowhead indicates the position of ChrXII and the small arrowhead indicates the position of the wells.
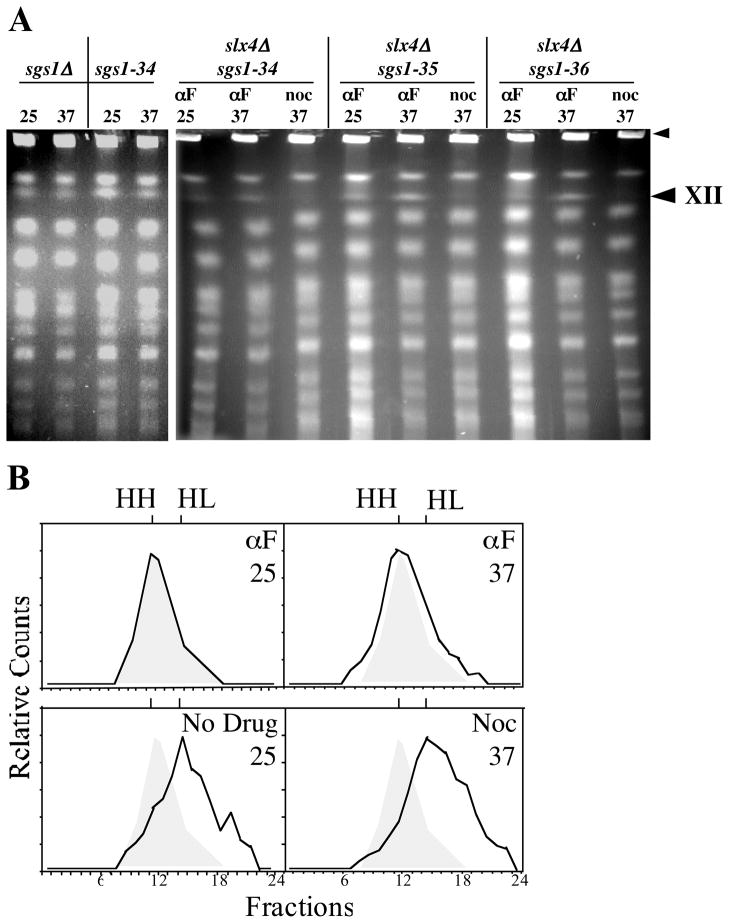
Fig 6.
Replication of the rDNA is required for the altered mobility of ChrXII in sgs1-ts slx4Δ cells. A Cells were synchronized and analyzed as in Fig 3. Left panel: the sgs1Δ and sgs1-34 single mutants were released from αF block and incubated for 6 hours in YPD broth at the indicated temperature. Right panel: the indicated double mutants were released into medium containing α-factor or nocodazole and incubated for 3 hrs at the indicated temperature. Microscopic examination confirmed that 98% of the cells held in α-factor were unbudded and 97% of the cells blocked in nocodazole were large-budded. Cells were harvested and analyzed by CHEF gel electrophoresis. The large arrowhead indicates the position of ChrXII and the small arrowhead indicates the position of the wells. B Density shift analysis of rDNA replication. sgs1-34 slx4Δ cells were grown in heavy minimal medium and arrested with α-factor at 25°C (αF/25). Cells were then washed, and aliquots released at 25°C into light medium (No Drug/25) or at 37°C into light medium containing α-factor (αF/37) or nocodazole (Noc/37). After the indicated treatments the cells were harvested and the DNA was isolated, fractionated by equilibrium gradient density sedimentation, and analyzed by Southern blotting. The positions of unreplicated heavy-heavy (HH) and fully replicated heavy-light DNA (HL) are indicated. The initial HH peak is shown for comparison as a shaded peak in the released samples.
Aberrant migration of chromosome XII requires multiple tandem repeats of rDNA
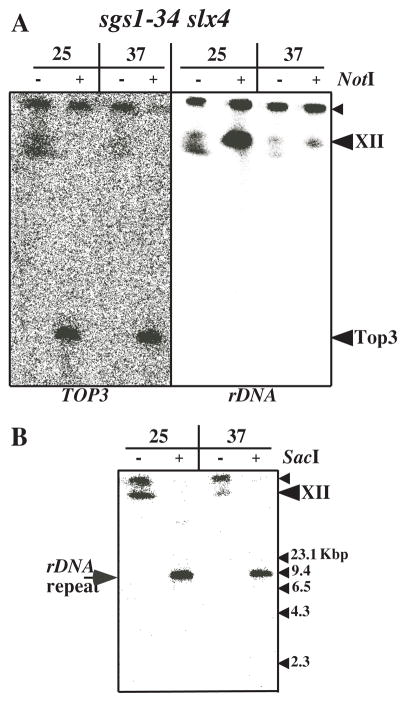
A possible cause of the altered mobility of ChrXII is that the entire chromosome has a unique structure, in which case all portions of the chromosome would be retained in the well. Alternatively, it might be due to the 100–200 tandem repeats of rDNA present on ChrXII, or to a feature unique to each rDNA repeat unit. To test these possibilities, we digested chromosome preparations from sgs1-34 slx4Δ mutants with restriction enzymes prior to CHEF analysis and Southern blotting. NotI digests ChrXII into six large fragments but does not cut within the rDNA array. Southern blotting with probes specific for either rDNA or the flanking DNA would determine whether the migration defect was due to the rDNA cluster or the structure of ChrXII itself. We used the TOP3 gene as a probe for flanking DNA and observed a band migrating at the expected size of 0.25 Mb, regardless of the growth temperature (Fig. 4A, left panel). In contrast, the NotI fragment containing the rDNA failed to migrate into the gel when isolated from cells grown at 37°C (Fig. 4A, right panel). When chromosomes were isolated at 25°C, NotI digestion resulted in a homogeneous rDNA band (approximately 1 Mb) which migrated near the full-length ChrXII bands. Thus, the aberrant migration of ChrXII is specific to the rDNA at the non-permissive temperature.
Fig. 4.
Multiple rDNA repeat units are required for the altered electrophoretic mobility of ChrXII. The sgs1-34 slx4 strain was grown to exponential phase at 25°C, arrested with α-factor and released at 37° for 6 hours. Cells were collected and prepared for CHEF gel electrophoresis in agarose plugs. The plugs were digested with NotI or SacI as indicated. A Following electrophoresis the gel was blotted using the complete TOP3 coding region (left) or an rDNA fragment (right) as probe. B The electrophoresis of chromosomal DNA was performed as above except that the electrophoresis was stopped after 7 hours. The gel was then Southern blotted using an rDNA fragment as probe.
To test whether this aberrant migration was specific to each rDNA repeat or the cluster of repeats, chromosome preparations from sgs1-34 slx4Δ cells were digested with SacI, which cuts once in each of the rDNA repeats. Digestion products were then analyzed as above except the duration of electrophoresis was reduced to prevent the unit length rDNA fragments (9.1 kb) from migrating off the gel. We observed that SacI digestion produced single rDNA fragments that entered the CHEF gel regardless of the growth temperature (Fig. 4B). Thus, the failure of ChrXII to enter the CHEF gel is a characteristic of the rDNA array, not of the repeat unit itself.
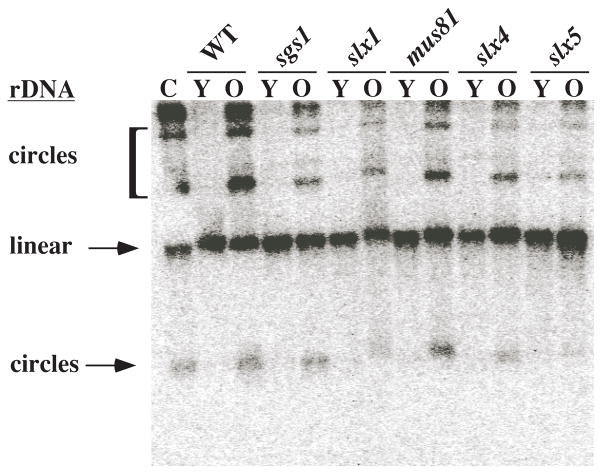
We suspected that the anomalous behavior of rDNA from sgs1-34 slx4Δ strains at the non-permissive temperature might be related to the excision of extra chromosomal rDNA circles (ERCs) in aging cells (Sinclair and Guarente 1997). To examine the effect of slx mutations on the premature excision of ERCs, we measured ERC levels by Southern blotting DNA isolated from young and old (7 - 8 generations) cells with wild-type, sgs1Δ and slxΔ mutant backgrounds (Mullen et al. 2001). As previously reported, there was no evidence of ERCs in any of the young cells; the rDNA was limited to a linear DNA fragment of 23 Kb that arises from the shearing of genomic DNA (Fig. 5). After 7 generations all of the strains produced ERCs, but their formation in sgs1Δ or slxΔ mutants was not quantitatively different from wild type. We conclude that ERC formation does not occur prematurely in slxΔ or sgs1Δ mutants and is therefore unlikely to be involved in the altered structure of the rDNA in the sgs1-ts slx4Δ double mutants.
Figure 5.
Extra-chromosomal rDNA circle formation in sgs1Δ and slxΔ strains. Total DNA was isolated from sorted seven-generation old cells and unsorted young cells. This DNA was subjected to one-dimensional gel electrophoresis and Southern blotted using an rDNA fragment as probe. The average bud scar counts of sorted cells was determined to be between 7 and 8. A control strain (C; top1 top2–4) excises rDNA circles at 25°C and is used as a marker for ERC migration. Y, young cells; O, old cells.
Altered mobility of Chromosome XII requires cell-cycle progression
The altered structure of ChrXII might be due to defects in transcription, recombination, or DNA replication. If it were due to defects in transcription or recombination, it would be expected to be independent of cell-cycle position while a defect in DNA replication should require passage through S-phase. To distinguish between these possibilities, we synchronized the growth of sgs1-ts slx4 cells by arresting them in G1 with α-factor at 25°C. Cells were then released at 37°C in the presence of additional α-factor to hold them in G1, or in the presence of nocodazole to allow them to progress to G2/M. After three hours the cells were harvested for CHEF analysis. As shown in Figure 6A, ChrXII entered the gel when the cells were held in G1 at 37°C, but failed to enter the gel when the cells were allowed to progress through the cell cycle. The simplest explanation for these results is that the rDNA undergoes a structural change during the S-phase of the cell cycle.
Since the FACS analysis presented in Fig. 2A may have failed to detect defects in rDNA replication, we assayed this directly using a density-shift protocol. The sgs1-34 slx4Δ strain was grown in heavy isotopes at 25°C and arrested in G1 with α-factor. The cells were then released from the α-factor block into light medium at 25°C, or at 37°C in the presence of either additional α-factor or nocodozole. After three hours DNA was isolated, fractionated by equilibrium density sedimentation and the fractions were blotted with an rDNA specific probe. As shown in Fig. 6B, DNA isolated prior to release sedimented with a peak at fraction 11 while DNA from cells released at 25°C peaked at fraction 15. These fractions correspond to HH and HL densities, respectively, as judged by refractive index. When the cells were released at 37°C in the presence of additional α-factor its DNA remained HH, while that from cells released at 37°C in the presence of nocodozole peaked at fraction 15 corresponding to HL DNA. CHEF gel analysis of these samples confirmed that ChrXII failed to enter the gel upon release into nocodozole at 37°C (data not shown). We conclude that rDNA synthesis in this strain is not significantly inhibited at 37°C.
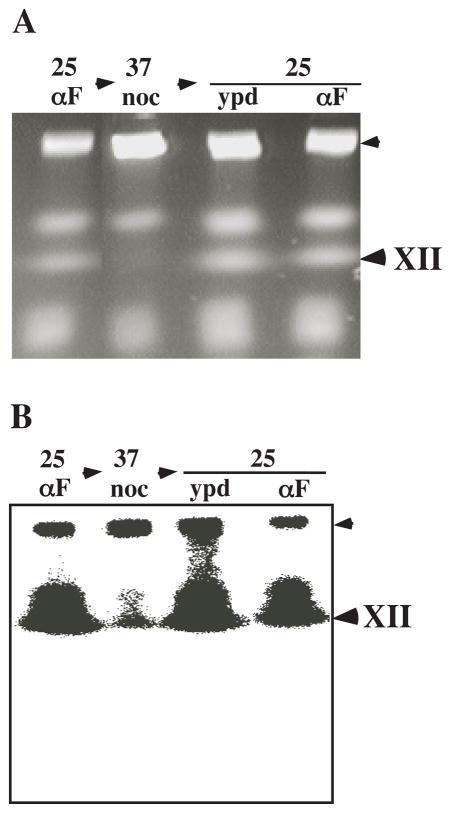
Given that rDNA replication was essentially complete in the absence of these activities, we asked whether Sgs1 activity could be provided following S-phase to correct the defect in ChrXII structure. As above, double mutant cells were arrested in G1 with α-factor and released at 37°C in the presence of nocodozole. Three hours later cells were released from the nocodozole block at 25°C in the presence or absence of α-factor. Samples were taken at each condition for CHEF analysis. As before, ChrXII migration was severely inhibited when cells were allowed to progress through S-phase to the nocodozole block at 37°C (Fig. 7). However, when cells were released from the nocodozole block into YPD at the permissive temperature, ChrXII migration returned to normal (Fig. 7) and cell proliferation continued (data not shown). Moreover, a subsequent S phase is not required to correct ChrXII structure as it migrated normally when cells were returned to the permissive condition in the presence of α-factor. We conclude that Sgs1 is not required for DNA replication and that its function can be provided following bulk DNA synthesis to correct defects in rDNA structure.
Fig. 7.
Reciprocal shift analysis of ChrXII mobility. sgs1-34 slx4Δ cells were arrested with α-factor at 25°C (25/αF) and released at 37°C in the presence of nocodazole (37/noc) for three hours. The cells were then washed and released at 25°C in the absence (25/ypd) or presence (25/αF) of α-factor for three hours. After the indicated treatments cells were harvested and DNA was analyzed by CHEF gel electrophoresis. A Ethidium bromide stain. B Southern blot with rDNA probe.
Discussion
In order to gain further insight into the function of Sgs1 in yeast, we used SLX4 mutants to isolate conditional alleles of SGS1 and search for defects that occur upon Sgs1 inactivation. SLX4 is one of six non-essential genes previously shown to be required for viability in the absence of SGS1 (Mullen et al. 2001). Two other genes isolated in this screen, MMS4 and MUS81, encode a heterodimeric structure-specific endonuclease (Kaliraman et al. 2001). Interestingly, Slx4 functions in a complex with Slx1 (Mullen et al. 2001) which was recently shown to be homologous to a conserved class of UvrC-Intron-type (URI) endonucleases (Aravind and Koonin 2001). Thus, SLX4 and SLX1 appear to encode a heterodimeric endonuclease that functions in a conserved DNA repair or recombination pathway, like Mms4-Mus81.
We isolated SGS1 mutations that map to either the DNA helicase domain (sgs1-35 and sgs1-36) or the N-terminus (sgs1-34). Although these two classes of mutations exhibited phenotypes consistent with previous structure-function analysis (Mullen et al. 2000), they were equally defective in controlling rDNA structure in the slx4Δ background. This supports the idea that Sgs1 requires the Top3-interacting domain, not just DNA helicase activity, for wt function. When sgs1-ts slx4Δ cells were shifted to the non-permissive temperature they replicated their DNA and arrested in late S/G2. These results strongly support the idea that SLX4 and SGS1 are not required for bulk DNA synthesis. The fact that all three SGS1 alleles behaved similarly suggests that these alleles do not exhibit a delay or “leakiness” in their inactivation at 37°C. Moreover, the viability of these cells was dramatically reduced if they were held at 37°C for longer than 5 hours and FACS analysis revealed that additional of DNA synthesis takes place at this time. We suggest that the first round of replication in slx4Δ sgs1-ts cells at 37°C is defective and that a checkpoint transiently arrests the cell cycle in late S/G2. In the absence of repair, adaptation ensues and the cells enter an aberrant, and lethal, cell cycle. The increase in multiply-budded cells and 4C levels of DNA content indicate that cells begin this cell cycle without undergoing a normal mitosis.
Lee and coworkers have reported the isolation of a conditional-lethal allele of SGS1 in the presence of the srs2Δ mutation (Lee et al. 1999). SRS2 encodes a 3′ - 5′ DNA helicase (Rong and Klein 1993) and, in contrast to our results, these authors conclude that at least one of these two DNA helicases is required for DNA replication fork movement in yeast. We used slx4 to screen for sgs1-ts alleles because, like others, we observe only a synthetic-sickness, not synthetic-lethality in the sgs1Δ srs2Δ double mutant (Gangloff et al. 2000; Klein 2001; Mullen et al. 2001). Moreover, the sickness of the sgs1Δ srs2Δ double mutant is dependent on homologous recombination and not a failure to replicate DNA (Gangloff et al. 2000; Klein 2001; McVey et al. 2001). Taken together with the present study, we find no evidence for Sgs1 as a primary replicative DNA helicase. Instead, the function of Sgs1 in DNA replication appears to be required late in DNA replication and may be locus specific.
One molecular defect in slx4Δ sgs1-ts cells at the non-permissive temperature is the loss of structural integrity of the rDNA as assayed by its inability to enter a CHEF gel. Previous work with hydroxyurea and the cdc46 replication mutant suggested that the inability of chromosomal DNA to enter a CHEF gel is due to abnormal structures that result from incomplete replication (Hennessy et al. 1991; Desany et al. 1998). Indeed, we find that this unusual behavior is dependent on slx4Δ sgs1-ts cells progressing through S phase. In contrast to HU-treatment and cdc46 inactivation, in which no chromosomal DNA enters the gel, most of the DNA from slx4Δ sgs1-ts cells at 37°C behaves normally with the exception of the rDNA. Given the specific defect in these cells, we conclude that Slx4 and Sgs1 play an important role late in rDNA replication. Such a result is consistent with the presence of Sgs1 in the nucleolus and its role in maintaining nucleolar structure (Sinclair et al. 1997; Frei and Gasser 2000). Interestingly, defects in DNA topoisomerase I, which is enriched in the nucleolus, also lead to an inability of rDNA to enter a CHEF gel (Muller et al. 1985; Christman et al. 1993). In this case however, when top1Δ cells are grown to saturation, rDNA regains the ability to enter the gel (Christman et al. 1993). Presumably another protein that can relieve torsional stress (e.g., Top2) replaces Top1, albeit with reduced efficiency, thereby completing rDNA replication. It is known that the rDNA has unique topological problems since simultaneously reducing both Top1 and Top2 result in excision of rDNA circles and inhibition of rDNA transcription (Brill et al. 1987; Kim and Wang 1989). Just as Top1 and Top2 have overlapping functions in relieving topological stress at the rDNA, we propose that Sgs1-Top3 and Slx1-4 share responsibility for dealing with another unique aspect of rDNA replication.
In contrast to idea that Sgs1 is directly involved in promoting DNA replication, reciprocal-shift experiments demonstrate that Sgs1 can fulfill its role after most, if not all, DNA has been replicated. It is formally possible that the ability of Sgs1 to act following G2/M is unique to this system; it could be dependent on the absence of Slx4 or the rDNA substrate. We think this is unlikely and suggest that in wild type cells Sgs1 performs its function during and/or after DNA replication. This execution point is consistent with models that propose a role for RecQ proteins in the resolution of stalled replication forks. This idea was originally suggested by the fact that RecQ DNA helicase functions at stalled replication forks in E. coli and that Rqh1 is required for restarting replication in HU-arrested S. pombe (Stewart et al. 1997; Courcelle and Hanawalt 1999). We suggest that Sgs1-Top3 and Slx1-4 function throughout the genome and that the apparent specificity for rDNA reflects the fact that rDNA replication involves numerous stalled replication forks. The replication fork barrier, which prevents forks from progressing into the path of the 35S transcription unit, stalls one fork at every rDNA origin or approximately every 5th rDNA repeat (Walmsley et al. 1984; Brewer and Fangman 1988). In the absence of Sgs1 and Slx4, most rDNA repeats should be replicated completely by the fork moving in the direction of rDNA transcription, however one in five repeats could be defective if the replication block is not properly processed. This may explain why individual rDNA repeats appear to migrate normally in CHEF gels, but the entire rDNA cluster does not. We suggest that each replicon of approximately 5 rDNA repeats contains one partially replicated repeat that inhibits its movement in CHEF gels. While we cannot rule out the possibility that Slx1-4 functions exclusively at the rDNA, we favor the idea that it functions globally, like Cdc46 and Top1, and that its mutant phenotypes are exacerbated at the rDNA.
Early models of replication repair proposed that stalled forks might regress to form Holliday junctions (Higgins et al. 1976). Support for this idea is provided by the fact that the Holliday junction binding proteins RuvAB are required for processing arrested replication forks in E. coli (Seigneur et al. 1998). Interestingly, molecular studies have shown that Holliday junctions form in the rDNA of yeast specifically during S-phase (Zou and Rothstein 1997). The fact that another RecQ family member, the BLM helicase, is capable of branch migrating a Holliday junction suggested that BLM might reverse the formation of Holliday junctions at stalled replication forks (Karow et al. 2000). Sgs1 might play a similar role at the rDNA of yeast where stalled forks are expected to form at every origin. Given this model, the Slx1-4 complex might be required in the absence Sgs1 to inhibit the formation of Holliday junctions or to resolve them with its putative nuclease activity. An alternative model is that the Slx1-4 complex processes subsequent recombination intermediates that occur in the absence of Sgs1. These models predict that the Slx1-4 complex should be enriched in the nucleolus and interact with DNA in a structure-specific manner. These predictions are currently being tested.
Acknowledgments
The authors thank Christina DeCoste for assistance with FACS analysis, Marc Gartenberg for strains and Abram Gabriel for valuable discussions. We also thank Jan Mullen, Bill Fricke, Abram Gabriel, and Marty Nemeroff for helpful comments on the manuscript. This work was supported by NIH grants GM55583 and AG16637.
References
- Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double- strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Noirot-Gros MF, Wang JC. Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J Biol Chem. 2000;275:26898–26905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Sharp JA, Wang JC. Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae. J Biol Chem. 1998;273:9644–9650. doi: 10.1074/jbc.273.16.9644. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- Christman MF, Dietrich FS, Levin NA, Sadoff BU, Fink GR. The rRNA-encoding DNA array has an altered structure in topoisomerase I mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:7637–7641. doi: 10.1073/pnas.90.16.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Carswell-Crumpton C, Hanawalt PC. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duno M, Thomsen B, Westergaard O, Krejci L, Bendixen C. Genetic analysis of the Saccharomyces cerevisiae Sgs1 helicase defines an essential function for the Sgs1-Top3 complex in the absence of SRS2 or TOP1. Mol Gen Genet. 2000;264:89–97. doi: 10.1007/s004380000286. [DOI] [PubMed] [Google Scholar]
- Ellis NA, et al. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Kaliraman V, Brill SJ. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J Biol Chem. 2001;276:8848–8855. doi: 10.1074/jbc.M009719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- Gebhart E, Bauer R, Raub U, Schinzel M, Ruprecht KW, Jonas JB. Spontaneous and induced chromosomal instability in Werner syndrome. Hum Genet. 1988;80:135–139. doi: 10.1007/BF00702855. [DOI] [PubMed] [Google Scholar]
- German J. Bloom Syndrome: A mendelian prototype of somatic mutational disease. Medicine. 1963;72:393–406. [PubMed] [Google Scholar]
- Gray MD, et al. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. RecQ DNA helicase is a suppressor of illegitimate recombination in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1988. [Google Scholar]
- Hennessy KM, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- Higgins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. J Mol Biol. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- Hoehn H, Bryant EM, Au K, Norwood TH, Boman H, Martin GM. Variegated translocation mosaicism in human skin fibroblast cultures. Cytogenetics & Cell Genetics. 1975;15:282–298. doi: 10.1159/000130526. [DOI] [PubMed] [Google Scholar]
- Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′-->5′ exonuclease. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Chakraverty RK, Hickson ID. The Bloom’s syndrome gene product is a 3′-5′ DNA helicase. J Biol Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil RL, McWilliams AD. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics. 1993;135:711–718. doi: 10.1093/genetics/135.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RA, Wang JC. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell. 1989;57:975–985. doi: 10.1016/0092-8674(89)90336-x. [DOI] [PubMed] [Google Scholar]
- Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- Klein H. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics. 2001;157:557–565. doi: 10.1093/genetics/157.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano K, Berres ME, Engels WR. Evolution of the RECQ family of helicases: A drosophila homolog, Dmblm, is similar to the human bloom syndrome gene. Genetics. 1999;151:1027–1039. doi: 10.1093/genetics/151.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Johnson RE, Yu SL, Prakash L, Prakash S. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- Lu J, Mullen JR, Brill SJ, Kleff S, Romeo A, Sternglanz R. Human homologs of yeast DNA helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- McCarroll RM, Fangman WL. Time of replication of yeast centromeres and telomeres. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- McVey M, Kaeberlein M, Tissenbaum HA, Guarente L. The short life Sspan of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics. 2001;157:1531–1542. doi: 10.1093/genetics/157.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Brill SJ. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2000;154:1101–1114. doi: 10.1093/genetics/154.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller MT, Pfund WP, Mehta VB, Trask DK. Eukaryotic type I topoisomerase is enriched in the nucleolus and catalytically active on ribosomal DNA. EMBO J. 1985;4:1237–1243. doi: 10.1002/j.1460-2075.1985.tb03766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JM, Lindsay HD, Munday CA, Carr AM. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol Cell Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian AR, Bassett DE, Jr, Boguski MS, Bork P, Koonin EV. Positionally cloned human disease genes: patterns of evolutionary conservation and functional motifs. Proc Natl Acad Sci USA. 1997;94:5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Irino N, Nakayama H. The recQ gene of Escherichia coli K12: molecular cloning and isolation of insertion mutants. Mol Gen Genet. 1985;200:266–271. doi: 10.1007/BF00425434. [DOI] [PubMed] [Google Scholar]
- Puranam KL, Blackshear PJ. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- Rong L, Klein HL. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268:1252–1259. [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual 1989 [Google Scholar]
- Seigneur M, Bidnenko V, Ehrlich SD, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- Sekelsky JJ, Brodsky MH, Rubin GM, Hawley RS. Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res. 1999;27:3762–3769. doi: 10.1093/nar/27.18.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, et al. Replication factor C3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, plays a role in both replication and damage checkpoints. Mol Biol Cell. 1999;10:3991–4003. doi: 10.1091/mbc.10.12.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;12:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- Smeal T, Claus J, Kennedy B, Cole F, Guarente L. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell. 1996;84:633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Umezu K, Nakayama K, Nakayama H. Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley RM, Johnston LH, Williamson DH, Oliver SG. Replicon size of yeast ribosomal DNA. Mol Gen Genet. 1984;195:260–266. doi: 10.1007/BF00332757. [DOI] [PubMed] [Google Scholar]
- Watt PM, Hickson ID, Borts RH, Louis EJ. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- Wu L, et al. The Bloom’s syndrome gene product interacts with topoisomerase III. J Biol Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Bloom’s and Werner’s syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc Natl Acad Sci USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CE, et al. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]