Abstract
MicroRNAs, central players of numerous cellular processes, regulate mRNA stability or translational efficiency. Although these molecular events are established, the mechanisms regulating microRNA function and expression remain largely unknown. The microRNA let-7i regulates Toll-like receptor 4 expression. Here, we identify a novel transcriptional mechanism induced by the protozoan parasite Cryptosporidium parvum and Gram(−) bacteria-derived lipopolysaccharide (LPS) mediating let-7i promoter silencing in human biliary epithelial cells (cholangiocytes). Using cultured cholangiocytes, we show that microbial stimulus decreased let-7i expression, and promoter activity. Analysis of the mechanism revealed that microbial infection promotes the formation of a NFκB p50-C/EBPβ silencer complex in the regulatory sequence. Chromatin immunoprecipitation assays (ChIP) demonstrated that the repressor complex binds to the let-7i promoter following microbial stimulus and promotes histone-H3 deacetylation. Our results provide a novel mechanism of transcriptional regulation of cholangiocyte let-7i expression following microbial insult, a process with potential implications for epithelial innate immune responses in general.
Keywords: Chromatin/Immunoprecipitation/ChIP, Immunology/Innate Immunity, Immunology/LPS, RNA/MicroRNA, Transcription/C/EBP, Transcription/NFκB, Transcription/Repressor
Introduction
Intrahepatic bile ducts constitute a complex 3-dimensional tubular network, the biliary tract, through which bile is transported to the duodenum. Human bile is sterile under normal physiological conditions (1); however, the biliary tract is periodically exposed to pathogens, including Escherichia coli and the protozoan parasite Cryptosporidium parvum, or pathogen-derived molecules, including Gram-negative bacteria-derived lipopolysaccharide (LPS).2 Upon pathogen recognition, a phenotypic transition occurs through which biliary epithelial cells (cholangiocytes) promote the innate and adaptive immune responses (1–4). Indeed, cholangiocytes express a variety of pathogen recognition receptors and actively participate in the innate immune response through the secretion of cytokines/chemokines (5, 6), expression of adhesion molecules (7–9), and antimicrobial peptides (1, 10). Expression of these immune-associated genes is a highly regulated process to assure that the epithelium recognizes and responds to invading pathogens, but does not induce injury through an inappropriate immune response. Recent reports suggest the microRNA machinery contributes to the regulation of the immune-associated gene expression. MicroRNAs are small (21–23 nt) RNA molecules that target and regulate the stability or translational efficiency of mRNAs (11). These regulatory RNAs are transcribed as mono- or polycistronic primary microRNAs (pri-microRNAs), which are sequentially processed to precursor and the functionally active mature microRNA. The molecular mechanisms regulating the expression of most microRNAs remain largely unknown.
Using a human cholangiocyte cell culture model of biliary cryptosporidiosis, we previously reported that let-7i, a member of the let-7 family of microRNAs, targets Toll-like receptor 4 mRNA and limits the expression of this pathogen molecular pattern receptor resulting in decreased primary and mature let-7i expression (3). These results suggested that the expression of let-7i is responsive to pathogen recognition and regulated through primary-microRNA transcription. In the present report, we describe a novel transcriptional mechanism mediating the regulation of the primary let-7i transcript expressed in cholangiocytes. We demonstrate that the NFκB p50 subunit and the transcription factor CCAAT/enhancer-binding protein β (C/EBPβ), which belongs to the larger family of basic leucine zipper (b-Zip) transcription factors (12), form an inducible silencing complex in let-7i promoter following microbial stimulus. Overexpression or siRNA inhibition of these transcription factors had reciprocal effects on our let-7i promoter-driven luciferase reporter gene, whereas deletion of the putative NFκB binding sites abrogates the observed microbe or NFκB p50-induced decrease in reporter gene expression. Furthermore, under basal conditions, the promoter is associated with acetylated histones and is permissive for transcription; however, following NFκB p50 and C/EBPβ overexpression, this locus is associated with deacetylated histone H3, which promotes chromatin condensation and hence locus silencing. These results suggest that transcription of let-7i is regulated by both p50 and C/EBPβ, which may exist in an inducible inhibitory complex that promotes microRNA silencing, a process with potential implications for epithelial innate immune responses in general.
EXPERIMENTAL PROCEDURES
Cell Culture and Parasite Infection/LPS Treatment
H69 cells (a gift from Dr. D. Jefferson, Tufts University, Boston, MA) are SV40-transformed normal human cholangiocytes originally derived from normal liver harvested for transplant. These cholangiocytes continue to express biliary epithelial cell markers, including cytokeratin 19, γ-glutamyl transpeptidase, and ion transporters consistent with biliary function and have been extensively characterized (13). C. parvum oocysts of the Iowa strain were purchased from a commercial source (Bunch Grass Farm, Deary, ID). Infection with C. parvum was done in culture medium containing DMEM-F12, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA). Before infecting cells, oocysts were treated with a 10% bleach solution, washed, and excysted to release infective sporozoites (14). Cells were infected using a 1:1 sporozoite/cell ratio. LPS was purchased from a commercial source (Invivogen, San Diego, CA), and cells were treated by adding LPS to the medium at a concentration of 200 ng/ml.
Northern Blot
Total RNA from cultured cells was isolated using the conventional method of acid phenol:chloroform extraction using TRI Reagent (Sigma-Aldrich) and subsequent alcohol precipitation. For Northern blot detection, total RNA was separated on a 15% polyacrylamide gel in 1× TBE. Following electrophoresis, the RNA was transferred to a nylon membrane using a semi-dry transfer and then UV cross-linked to the membrane using 120 mJ for 30 s. An oligonucleotide corresponding to the complementary sequence of the pre-let-7i molecule (accession: MI0000434; Sanger microRNA Registry) and containing a T7 primer-binding site was chemically synthesized (Mayo Clinic Molecular Core Facility). This DNA template was used to generate the antisense let-7i probe for the detection of the primary let-7i transcript using an in vitro transcription approach with [α-32P]UTP according to the manufacturer's directions (Ambion, Austin, TX). The labeled normalizing probe, 18 S RNA, was synthesized with an identical approach. The membranes were incubated with 2.5 × 106 cpm per blot overnight at 42 °C. Subsequently, stringent washes were performed in saline-sodium citrate (SSC) buffer, and the membranes were exposed to autoradiography film for 24–48 h.
p50 and C/EBPβ Nuclear Translocation Assay
Nuclear extracts were obtained from control, LPS-treated, and C. parvum-infected H69 cells at 0, 1, and 6 h post-treatment using a Nuclear Extract kit (Active Motif, Carlsbad, CA) in the presence of phosphatase inhibitors. Briefly, the cells were scraped into hypotonic buffer and allowed to swell on ice. The cells were treated with the non-ionic detergent nonidet-P40 and centrifuged. The resultant pellet was resuspended in nuclear lysis buffer, gently rocked for 30 min, and centrifuged. The Bradford Assay for protein concentration was performed on the resultant supernatant (nuclear extract). The C/EBPβ nuclear binding assay was performed using a transcription factor assay kit (Active Motif) according to the manufacturer's protocol. Briefly, the wells of a 96-well plate were pretreated with the C/EBPβ consensus sequence oligonucleotides. 40 μl of binding buffer was added to the wells, and 2 μg of the nuclear extracts were brought to a total of 10 μl with lysis buffer and added to the wells. 1 μg of the provided rat liver nuclear extract was used as a positive control. Following a 1-h incubation the wells were washed three times with TBS-Tween. The primary C/EBPβ antibody was diluted 1:1500 and added to the wells, incubated for 1 h, and washed three times. The secondary anti-rabbit horseradish peroxidase-conjugated antibody was diluted 1:1000 and added to the wells, incubated for 1 h, and washed four times. The provided developing solution was added and incubated for 5 min before the reaction was stopped in the provided stop solution. The absorbance was read on a spectrophotometer at 450 nm with a reference wavelength of 655 nm. The p50 nuclear translocation assay was performed using a similar ELISA-based method (Panomics, Freemont, CA).
RT-PCR
For RT-PCR analysis of PPM1H, MON2, and C12orf61 mRNA expression, total RNAs were isolated as described above. Total RNA (1 μg) was reverse transcribed to cDNA by using a Moloney Murine Leukemia Virus (M-MLV) Reverse Transcriptase kit (Invitrogen). The specific primers used are listed in supplemental Table S1. The β-actin primers were purchased from Ambion. Amplification was performed at 94 °C for 1 min, 53 °C for 1 min, and 72 °C for 1 min for 35 cycles. PCR products were separated by electrophoresis and confirmed by sequencing (Mayo Clinic Molecular Core Facility).
RACE-PCR
H69 cells were transfected with an siRNA targeting a 3′-untranslated region sequence of Drosha mRNA using Fugene HD Transfection Reagent (Roche, Basel, Switzerland). Total RNA from H69 cells was isolated using TRIzol Reagent (Invitrogen), with subsequent mRNA isolation using oligonucleotide dC10T30 resin beads provided in the Oligotex mRNA Kit (Qiagen, Valencia, CA). The isolated mRNA was reverse transcribed following the manufacturer's instruction provided in the GeneRacerTM Kit (Invitrogen). The gene-specific primers listed in supplemental Table S1 were used in conjunction with the supplied 5′ and 3′ gene racer primers.
let-7i Promoter Constructs
PCR was utilized to amplify the putative upstream promoter of let-7i. Genomic DNA was extracted from H69 cells using a standard phenol:chloroform and ethanol precipitation. Initially, primers were designed to amplify ∼2500 base pairs of the putative promoter. The promoter was amplified on a block thermocycler using the high fidelity Cloned Pfu DNA polymerase (Stratagene, La Jolla, CA). Primers were designed with a Xho1 (forward primer) or BglII (reverse primer) restriction site (supplemental Table S1) for cloning into the pGL4.22 luciferase vector (Promega, Madison, WI). Multiple plasmid constructs were isolated from transfected DH5-α chemically competent Escherichia coli cells. Truncations of the promoter fragment were obtained by PCR using the original promoter construct as template. All plasmids were sequenced at the Mayo Clinic DNA Sequencing Core Facility. PCR mutagenesis was also performed to eliminate each of the putative NFκB binding sites. Primers were designed to anneal to sequences flanking the putative NFκB sites, but to exclude 15 base pairs corresponding to the NFκB consensus sequence (supplemental Table S1). Each mutant promoter (Forward and Reverse) was utilized to amplify either the distal or proximal portion of the predicted promoter using Cloned Pfu DNA polymerase. The resultant amplicons were then used as promoter and template to amplify the mutant promoters, which were subsequently cloned into the pGL4.22 luciferase vector.
Luciferase Assay
Human cholangiocytes (H69 or a spontaneously immortalized human cholangiocyte cell line (15)) were grown in culture to 80% confluence. The Dual-Luciferase reporter assay system (Promega) was used. Briefly, the cells were transfected with the let-7i promoter-pGL4.22 constructs, and the TK-Renilla luciferase plasmid was used as a transfection control. As a baseline control, the pGL4.22 empty vector, which contains minimal DNA regulatory elements or transcription factor binding sites, was utilized. Transfections were performed with Fugene HD. For microbial stimulus experiments, the transfected cells were incubated for 6 h and subsequently infected with C. parvum or treated with LPS as described above. For those experiments in which p50 or C/EBPβ was overexpressed, the plasmids pCDNA3.1-p50 or pBABE-Puro-C/EBPβ (Addgene, Cambridge, MA) were co-transfected with the luciferase vectors. Decreased expression of p50 or C/EBPβ was achieved by transfecting the cells with prevalidated siRNAs (Ambion). The cells were transfected with the siRNA using siPORT (Ambion) 24 h prior to transfection with the reporter constructs. Luciferase detection was performed 24 h after reporter construct transfection. Expression was calculated as the relative let-7i driven Firefly luciferase to transfection control Renilla luciferase and presented as fold-change compared with empty vector pGL4.22 Firefly luciferase to Renilla luciferase, which was arbitrarily set to one.
Chromatin Immunoprecipitation
ChIP was performed following the protocol from Upstate. Briefly, H69 cells were cultured in the presence or absence of C. parvum sporozoites or LPS. Chromatin was cross-linked with 1% formaldehyde, cells were lysed, and cell extract sonicated to shear DNA. Sonicated cell extract was either aliquoted as genomic input DNA or immunoprecipitated using p50 or p65 antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Immunoprecipitates containing protein-DNA complexes were washed and heated at 65 °C for 4 h to reverse cross-link and free DNA. DNA was purified using spin columns. Semi-quantitative and quantitative PCR were performed on both genomic input and ChIP DNA. PCR primers were designed to flank the two NFκB consensus sequences in the let-7i promoter (supplemental Table S1). Quantitative PCR of the ChIP products and genomic input DNA was performed by real-time PCR using SYBR green (Roche). The amount of ChIP DNA present in each sample was reported as percentage of genomic input DNA.
Electrophoretic Mobility Shift Assay
EMSA was performed following the manufacturer's instruction (Pierce). Briefly, nuclear proteins were extracted using NE-PER (Pierce) from uninfected, C. parvum-infected, or LPS-treated H69 cells. Complementary oligonucleotides, corresponding to the NFκB consensus sequences in the let-7i promoter, were synthesized (Mayo Clinic Molecular Core Facility), annealed, and biotinylated according to the manufacturer's instruction (Pierce). The nuclear extract was incubated for 20 min at room temperature with the biotinylated oligonucleotides. Unlabeled NFκB consensus oligonucleotides were used as competimers. These competimers were preincubated with the nuclear extract for 5 min prior to the addition of biotinylated oligonucleotides. EMSA supershift was performed using the p50 antibody sc114x (Santa Cruz Biotechnology, Inc.). The antibodies were preincubated with the nuclear extract for 20 min at room temperature prior to adding the biotinylated oligonucleotides. The DNA-protein complex was resolved on a 5% PAGE gel in 0.5× TBE, transferred to a nylon membrane, and cross-linked at 120 mJ/cm2 for 60 s. The biotinylated-labeled oligonucleotides were then detected by chemiluminescence according to the manufacturer's directions.
Immunoprecipitation of p50 and C/EBPβ
H69 cells were grown in 10-cm dishes to 85% confluence and exposed to C. parvum sporozoites or LPS overnight. The isolated nuclei were suspended in PBS with 1% Nonident 0.5%, sodium deoxycholate, and 0.1% SDS supplemented with 1 mm phenylmethylsulfonyl fluoride, leupeptin, and pepstatin at 20 μg/ml. Immunoprecipitations were performed using 200 μg of nuclear extract from uninfected, C. parvum-infected, or LPS-treated H69 cells. Nuclear extracts were precleared with a 50% Sepharose A slurry (Sigma-Aldrich) and then incubated overnight at 4 °C with 4 μg of either the p50 polyclonal antibody sc114X (Santa Cruz Biotechnology) or C/EBPβ (Abcam, Cambridge, MA) antibodies. Antibody-protein complexes were collected with the 50% protein A slurry, washed, and then boiled in sample buffer to remove the antibody-protein complex from the protein A slurry. Samples were then subjected to SDS-PAGE and immunoblotted.
Statistical Analysis
All values are given as mean ± S.E. Means of groups were compared with Student's t test (unpaired) or the ANOVA test when appropriate. p values <0.05 were considered statistically significant.
RESULTS
Characterization of the let-7i Transcript and Genomic Locus
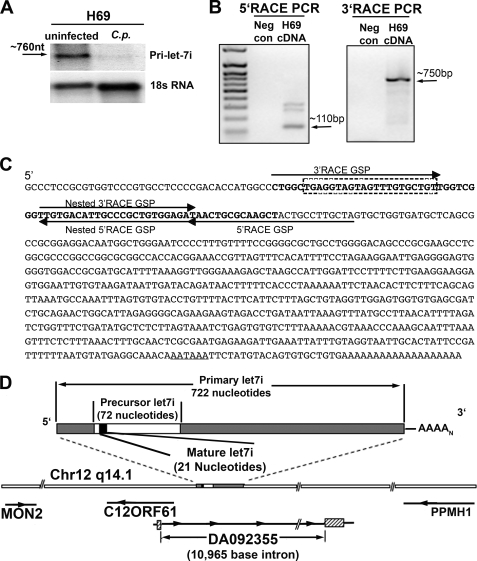
Northern blot detection of primary let-7i from the cholangiocyte cell line H69, revealed an RNA species at ∼750 nucleotides, which is repressed following infection with the protozoon parasite, C. parvum (Fig. 1A). RNA ligase-mediated RACE-PCR was used to amplify the full-length primary microRNA transcript from a polyadenylated-enriched RNA population. Primers for both 5′- and 3′-amplification were designed within the let-7i precursor sequence obtained from the Sanger microRNA registry. 5′-Amplification, and subsequent 5′-nested PCR resulted in several detectable bands on an ethidium bromide-stained DNA electrophoresis gel (Fig. 1B). Each band was gel extracted, cloned into pCR2.1-TOPO sequencing vector, and sequenced. Sequencing confirmed that one band (∼110 bp on agarose gel) corresponded to the precursor sequence and extended this sequence by 39 nucleotides. Nested PCR of our 3′-RACE products resulted in a single 750-bp amplicon. Sequencing confirmed the identity and demonstrated the 3′-end of this transcript terminates in a polyadenylated tail 21 nucleotides after the consensus polyadenylation site (AATAAA), suggesting that this transcript is driven by RNA polymerase II (Fig. 1C).
FIGURE 1.
Identification of the primary let-7i transcript. A, Northern blot for the primary let-7i transcript identified an approximate 760-nucleotide RNA in uninfected H69 cells, whereas C. parvum infection diminishes the expression of this RNA. 18 S rRNA was used as a loading control. B, RACE-PCR, using polyadenylated enriched RNA, was used to identify the primary let-7i transcript. A 100-bp let-7i specific amplicon was identified using 5′-RACE and sequence verified. A single intense band was observed at ∼750 bp in our 3′-nested RACE PCR. This let-7i specific band was sequence verified. C, primary let-7i sequence was identified in cholangiocytes by RACE-PCR with subsequent cloning and sequencing. The 5′ and 3′ gene-specific primers (GSPs) we generated based on the let-7i precursor sequence (bold). The dashed box indicates the mature let-7i sequence, and the predicted polyadenylation signal is underlined. D, let-7i primary transcript is located on chromosome 12 and exists within an intron of an EST (DA092355) identified in human cerebellum. Let-7i is flanked upstream by an open reading frame, which lies in the opposite orientation. MON2, which encodes a large guanine nucleotide exchange factor, terminates ∼6,000 base pairs upstream whereas the protein phosphatase PPM1H lies in the opposite orientation and terminates ∼40,000 base pairs downstream.
let-7i Upstream DNA Elements Are Responsive to Microbial Stimulation
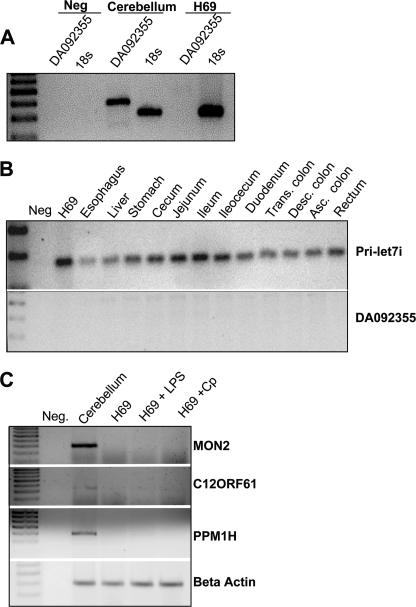
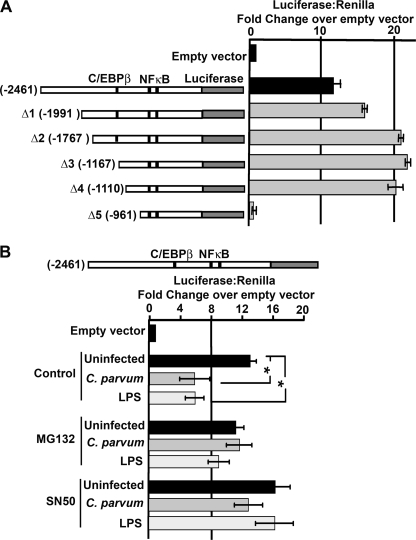
The BLAST-like alignment tool (BLAT, UCSC Genome Bioinformatics) showed that the let-7i gene lies within an expressed sequenced tagged (EST) transcript (DA092355) identified in human cerebellum and lies upstream of the predicted coding sequence, C12ORF61, in the opposite orientation (Fig. 1D). DA092355 was amplified from total cerebellum cDNA; however we were unable to amplify this sequence from cDNA derived from H69 cells (Fig. 2A), or from a panel of GI-associated tissues, whereas the let-7i primary transcript was identified in all the analyzed GI-related tissues (Fig. 2B). This suggests that let-7i is expressed independent of DA092355 in gastrointestinal tissue. The let-7i locus on chromosome 12 is flanked by a predicted open reading frame, C12ORF61, as well as MON2, which encodes a large guanine-nucleotide exchange factors for ADP-ribosylation factor (ARF-GEF), and protein phosphatase 1H (PPM1H), a type 2C protein phosphatase. We next asked whether these flanking genes were expressed in H69 cells and whether their expression was sensitive to microbial stimulus. Attempts to PCR amplify C12ORF61, as well as the annotated sequences, MON2 homolog and PPM1H, was achieved using cDNA derived from the cerebellum, but not from H69 cells under any of the conditions tested (Fig. 2C), suggesting tissue-specific expression of these genes. The let-7i transcript and upstream flanking sequence lies within a predicted CpG island of ∼1500 base pairs. Methylation-sensitive PCR was used to assess methylation of this region in the presence or absence of an infective stimulus. Methylation was not detected in the presence or absence of LPS treatment or C. parvum infection (data not shown). We next tested whether the upstream flanking sequence of let-7i could drive transcription of a luciferase reporter gene. Approximately 2500 base pairs upstream of the identified 5′ terminus of the let-7i primary transcript, as well as truncations of this putative promoter, were cloned into the luciferase reporter plasmid, PGL4.22. Transfection of this plasmid into H69 cells or a spontaneously immortalized human cholangiocyte cell line (15) (data not shown) and a subsequent Dual-Luciferase reporter assay resulted in an over 20-fold increase in luciferase expression compared with the empty pGL4-.22 vector (Fig. 3A). We previously demonstrated that infection of cultured human cholangiocytes with C. parvum or treatment with LPS, results in decreased primary and mature let-7i expression (3). Using our reporter assay, it was demonstrated that C. parvum infection or treatment with LPS represses reporter activity ∼50%, a process that is abrogated when infections were performed in the presence of the NFκB inhibitors MG132 and SN50 (Fig. 3B). Thus, together these findings further support the role of this regulatory sequence as the promoter element for let-7i and suggest a potential regulatory mechanism for the expression of this microRNA.
FIGURE 2.
Expression analyses of genes located on chromosome 12q14. A, rtPCR was used to detect the EST, DA092355, from cerebellum. This transcript was not expressed in H69 cells. 18 S rRNA was amplified as control. B, primary let-7i transcript was detected by rtPCR in all gastrointestinal tissues assessed, whereas the EST DA092355 was not detected in any of these tissues. C, genes flanking let-7i on chromosome 12 including MON2, C12ORF61, and PPM1H, were identified in cerebellum by rtPCR, yet were not detected in basal or microbe-stimulated H69 cells.
FIGURE 3.
Functional analyses of the let-7i Dual-Luciferase reporter assay. A, 2,461 base pairs upstream of the identified let-7i transcription start site were cloned into the luciferase expression plasmid pGL4.22. This putative let-7i promoter drives transcription over 10-fold compared with empty vector control. Serial truncations (Δ1-Δ5) identify a region between −1110 and −961 that is necessary for basal transcription. Two putative NFκB binding sites (−619 and −657) and a C/EBPβ binding site (−1166) were identified within the cloned promoter fragment. B, let-7i promoter-driven luciferase construct is responsive to microbial stimulus. C. parvum infection or LPS treatment reduces luciferase expression over 50% compared with uninfected cells. The observed microbial-induced suppression is diminished following pretreatment of the cells with the NFκB inhibitors MG132 and SN50. Expression was calculated as the relative let-7i-driven Firefly luciferase to transfection control Renilla luciferase and presented as fold-change compared with empty vector pGL4.22 Firefly luciferase to Renilla luciferase, which was arbitrarily set to one. Data are represented as mean ± S.E. from three separate experiments with at least n = 4 for each experiment. *, p < 0.05 compared with control luciferase by ANOVA.
NFκB p50 and C/EBPβ Mediates let-7i Silencing following C. parvum Infection or LPS Treatment
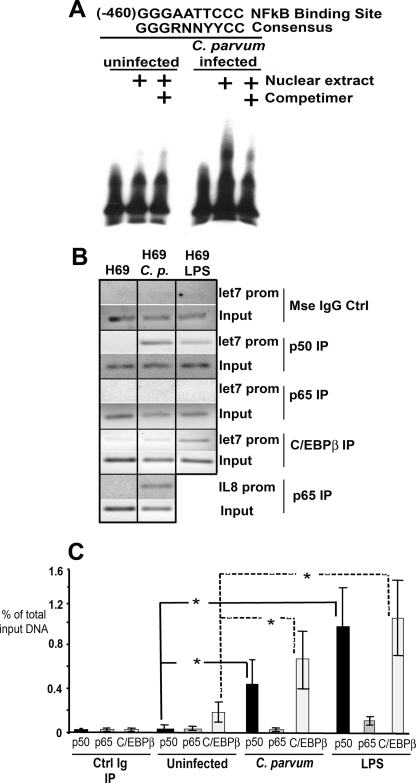
The cloned upstream DNA elements were also analyzed for putative transcription factor binding sites using the web-based transcription factor search site, TFSearch. We identified NFκB and the inflammation-associated transcription factor C/EBPβ as candidate mediators for the silencing effect of C. parvum infection or LPS treatment on this promoter. To validate this initial screening, we performed EMSAs using oligonucleotides containing the predicted NFκB binding sites, The proximal predicted NFκB site demonstrated less mobility through the gel when nuclear extract from C. parvum-infected cells was utilized in this assay, a phenomenon that could be competed away when the experiment was performed in the presence of unlabeled oligonucleotide. These results suggest that the oligonucleotide containing the proximal predicted NFκB site interacted with nuclear proteins following infection (Fig. 4A). Based on this observation, ChIP assays were performed with both an antibody against the p65 and p50 subunits of NFκB. Primers flanking the region of the predicted NFκB binding sites were used to demonstrate that under basal conditions very little p65 or p50 is bound to the promoter. However, following C. parvum infection or LPS treatment, p50 preferentially binds to the promoter elements in the region of the NFκB consensus sequence, raising the possibility that a p50 homodimer interacts with this sequence (Fig. 4B). As a p65-positive control, primers were designed that flanked the NFκB site of the interleukin-8 promoter, a gene known to be responsive to C. parvum infection in an NFκB-dependent manner (5). The NFκB p65 subunit bound to the IL-8 promoter following C. parvum infection (Fig. 4B). Interestingly, ChIP analysis of the putative let-7i promotor using the same primers that flanked the NFκB sites, also demonstrated that the acute phase responsive transcription factor, C/EBPβ, binds to the let-7i promoter following C. parvum infection or LPS treatment (Fig. 4B). We next clarified and quantified the interactions of these transcription factors with the let-7i promoter using quantitative PCR on the ChIP products, and demonstrate increased binding of the NFκB p50 subunit, as well as C/EBPβ, to the let-7i promoter (Fig. 4C). An ELISA-based method was used to demonstrate that following C. parvum infection or LPS treatment, the NFκB p50 subunit translocated to the nucleus (supplemental Fig. S1A), whereas an ELISA-based C/EBPβ binding assay (supplemental Fig. S1B) and immunofluorescence for this protein (supplemental Fig. 1C) demonstrated that this transcription factor is expressed in H69 cells, localizes to the nucleus in uninfected and infected cells, and can interact with the C/EBPβ consensus sequence in vitro in uninfected, C. parvum-infected, or LPS-treated cells. Taken together, these data suggest that C/EBPβ interaction with the let-7i promoter, as demonstrated through ChIP, is a regulated process not dependent on nuclear translocation of C/EBPβ.
FIGURE 4.
Immune-associated transcription factors interact with the putative let-7i promoter following microbial stimulus. A, electrophoretic mobility shift assays were used to demonstrate interactions between nuclear factors and the let-7i promoter following infection. An oligonucleotide containing the proximal NFκB binding site did not shift when the nuclear extract from basal, uninfected cells was used. However, the same oligonucleotide demonstrated less mobility through the gel when the nuclear extract from C. parvum-infected cells was used. This interaction could be competed away using unlabeled oligonucleotide competimer. B, ChIP analysis demonstrated that both NFκB p50 (but not p65) and C/EBPβ interact with the let-7i promoter following microbial stimulus. ChIP analyses for the interleukin-8 promoter were performed as a positive control for p65 with both uninfected and C. parvum-infected H69 cells. C, quantitative PCR ChIP was also performed to verify the increased interaction between the let-7i promoter and the transcription factors NFκB p50 and C/EBPβ. A standard curve was generated using amplicons of a known copy number. The data are presented as the relative amount of the immunoprecipitated fraction compared with total input DNA. Data are represented as mean ± S.E. from four separate experiments. *, p < 0.05 compared with respective uninfected controls by ANOVA.
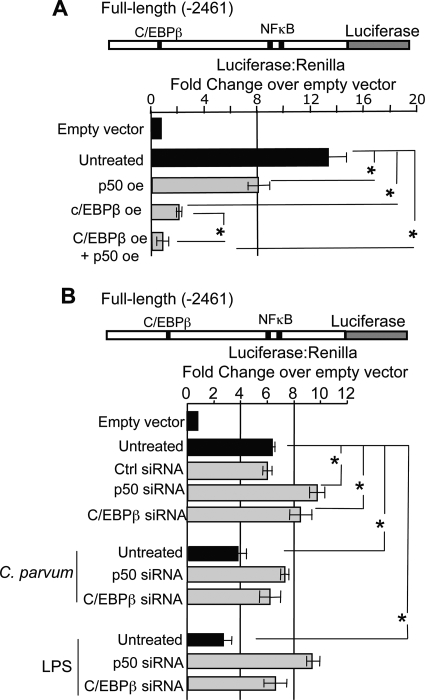
We next addressed whether manipulation of p50 and/or C/EBPβ could affect expression of the luciferase reporter gene. Transfections with pCDNA3.1-p50 plasmid, in which p50 is force expressed from the cytomegalovirus promoter, caused luciferase expression to diminish over 40%, whereas forced expression of C/EBPβ (LAP2) using pBABE-Puro-C/EBPβ reduced luciferase over 85% (Fig. 5A). Concurrent forced expression of these transcription factors further reduced luciferase expression (Fig. 5A). Conversely, diminished p50 or C/EBPβ expression with siRNAs results in a significant increase in reporter gene expression (Fig. 5B). Furthermore, siRNA-induced repression of p50 or C/EBPβ inhibited microbial-induced reduction of luciferase driven by the let-7i promoter (Fig. 5B). Taken together, these results suggest that p50 and C/EBPβ interact with let-7i promoter elements and functionally suppress reporter gene expression.
FIGURE 5.
Manipulation of NFκB p50 and C/EBPβ expression reciprocally affects reporter gene expression. A, the 2,461-bp let-7i promoter increased transcription over 12-fold compared with the pGL4.22 empty vector. Overexpression (oe) of p50 significantly decreased luciferase expression over 25%, whereas C/EBPβ overexpression diminished luciferase expression over 75%. Concurrent overexpression of these transcription factors synergistically represses luciferase expression from this promoter. B, same plasmid construct was used to demonstrate that RNA-mediated silencing of these transcription factors reciprocally affected luciferase expression driven from this promoter. Furthermore, siRNA-induced repression of p50 or C/EBPβ inhibited microbial-induced reduction of luciferase driven by the let-7i promoter. Data are represented as mean ± S.E. from three separate experiments with at least n = 4 for each experiment. *, p < 0.05 between comparisons indicated, using ANOVA.
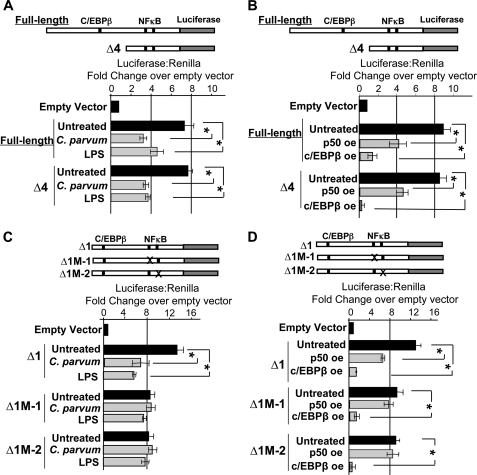
In an effort to demonstrate the functional significance of the predicted C/EBPβ binding sites on the let-7i promoter, a truncation of the full-length promoter, in which the C/EBPβ site is eliminated (Δ4, Fig. 3A), was used in our luciferase reporter system. Elimination of this site had no effect on microbial-induced luciferase suppression (Fig. 6A). Furthermore, this truncation had no effect on the observed luciferase suppression following p50 or C/EBPβ force expression, suggesting that this region of the promoter is not required for microbe-, NFκB p50-, or C/EBPβ-induced repression of let-7i transcription (Fig. 6B). We next asked if the consensus NFκB binding sites within the let-7i promoter affect C. parvum or LPS-induced reduction of reporter gene expression. PCR-based deletions eliminated the predicted NFκB sites, and luciferase assays were performed in the presence or absence of microbial stimulus. In contrast to the promoter with intact NFκB binding sites (Δ1), luciferase expression was not affected in the presence of C. parvum or LPS when either of the two predicted NFκB sites were eliminated (Δ1M-1 and Δ1M-2, Fig. 6C). These results suggest that the NFκB binding sites contribute to pathogen-induced reduction of gene expression. Using the same mutant promoters, we further demonstrate that forced expression of either p50 or C/EBPβ (LAP2) reduced luciferase expression in the wild-type promoter (Δ1) (Fig. 6D). In contrast, luciferase expression is not affected under conditions of p50-forced expression when the NFκB sites are eliminated (Fig. 6D), suggesting that the NFκB sites are involved in p50 suppression of reporter gene expression. Interestingly, C/EBPβ (LAP2) overexpression suppressed reporter gene transcription in each of the reporter constructs tested (Fig. 6D), suggesting that C/EBPβ-dependent silencing of the reporter gene is independent of the putative NFκB binding sites.
FIGURE 6.
The consensus κB binding sites within the let-7i promoter confer responsiveness to microbial stimulus. A, the 2,461 bp promoter and a truncation of this promoter (Δ4), which lacks the predicted C/EBPβ binding site, was used to assess the functional significance of this portion of the cloned promoter. Both the full-length promoter (2,461 bp) and the truncated promoter were responsive to microbial stimulus. C. parvum infection or LPS treatment reduced luciferase expression ∼50% when either promoter construct was used. B, similarly, both the full-length promoter and the truncated promoter were responsive to p50 or C/EBPβ overexpression. Overexpression of p50 reduced luciferase expression ∼50% when either promoter construct was used, whereas C/EBPβ overexpression resulted in a greater than 5-fold reduction in luciferase expression when either promoter construct was used. C, Δ1 construct, which contains both the C/EBPβ and NFκB sites, was used to assess the functional significance of the putative κB binding sites. The Δ1 construct was responsive to microbial stimulus whereas the promoter constructs harboring a deletion of either predicted κB binding site (Δ1M-1 or Δ1M-2) abrogated the microbe-induced suppression of luciferase expression. D, Δ1 construct was also utilized to assess the functional significance of the κB binding sites with respect to NFκB p50 and C/EBPβ overexpression. C/EBPβ overexpression resulted in diminished luciferase expression in each of the promoter constructs. The Δ1 construct was responsive to p50 overexpression. Conversely, p50 overexpression does not significantly affect luciferase expression when the distal or proximal NFκB binding sites are eliminated, suggesting that these sites confer responsiveness to p50 overexpression. Expression was calculated as the relative let-7i-driven firefly luciferase to transfection control Renilla luciferase and presented as fold-change compared with empty vector pGL4.22 firefly luciferase to Renilla luciferase, which was arbitrarily set to one. Data are represented as mean ± S.E. from four separate experiments with at least n = 4 for each experiment. *, p < 0.05 compared with respective untreated controls by ANOVA.
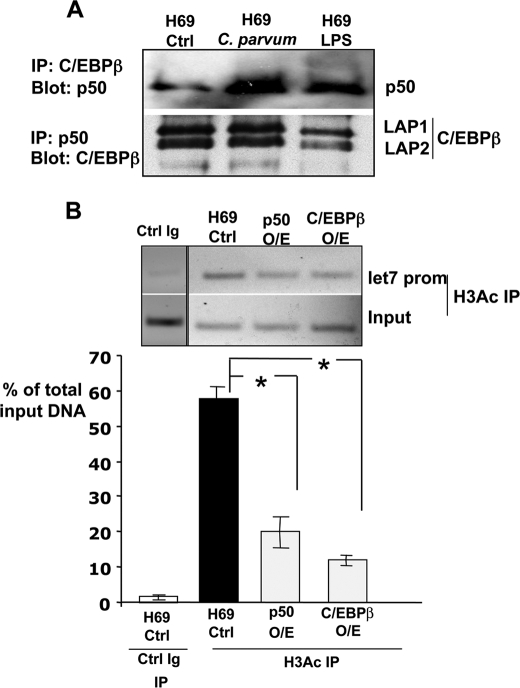
Taken together, our results raise the possibility that C/EBPβ and NFκB p50 subunit may act independently to suppress transcription. However, co-immunoprecipitations studies showed the interaction between C/EBPβ and NFκB p50 following C. parvum infection or LPS treatment (Fig. 7A). These results raise the possibility that whereas each transcription factor can repress transcription from the let-7i promoter independently; they may exist in a repressive complex of as yet unidentified proteins. We next asked whether the let-7i chromatin locus was structurally modified following microbial stimulus. Under basal conditions, we were able to detect acetylated histone H3 in the let-7i locus (Fig. 7B). Interestingly, p50 or C/EBPβ overexpression reduces the acetylated status of histone H3 within the let-7i promoter. Together, these results suggest changes in histone acetylation as the potential transcriptional mechanism used by this p50 and C/EBPβ repression complex to silence the expression of let-7i.
FIGURE 7.
NFκB p50 and C/EBPβ interact and overexpression of either induces let-7i promoter deacetylation. A, immunoprecipitations demonstrate the interactions between NFκB p50 and C/EBPβ. Both the immunoprecipitation of C/EBPβ with subsequent blot for p50 and the converse immunoprecipitation of p50 and blot for C/EBPβ demonstrate that these proteins physically interact in cholangiocytes. Both the Lap1 and Lap2 isoforms were detected in cholangiocytes. B, ChIP analyses for acetylated Histone H3 were performed in both control, uninfected, and in cholangiocytes overexpressing either NFκB p50 or C/EBPβ. Overexpression of either transcription factor significantly (p < 0.01) decreased the detection of acetylated Histone H3 at the let-7i promoter. The data are presented as the relative amount of the immunoprecipitated fraction compared with total input DNA. Data are represented as mean ± S.E. from three separate experiments. *, p < 0.01 compared with H69 control by ANOVA.
DISCUSSION
The results of our study provide the first direct evidence that microbial stimuli regulate the expression of a microRNA via the inflammation-associated transcription factors NFκB p50 and C/EBPβ. Using a human cholangiocyte model, we have shown that: (i) let-7i is expressed as an independent transcriptional unit in these cells; (ii) C/EBPβ and the p50 subunit, but not the p65 subunit, of NFκB repress transcription through interactions with the let-7i promoter following microbial stimulus; (iii) the consensus κB binding sites confer responsiveness of the reporter gene following microbial stimulus; and (iv) forced expression of either NFκB p50 or C/EBPβ results in chromatin deacetylation and let-7i transcription suppression. Our findings provide novel data regarding transcriptional regulation of microRNA expression in cholangiocytes in response to pathogen recognition, a process that may have implications in immune-related functions and inflammatory responses of epithelia in general and in cholangiocytes in particular.
Cholangiocytes express a variety of pathogen recognition receptors and actively participate in the innate immune response through the secretion of cytokines/chemokines (5, 6) that regulate liver cell function, expression of adhesion molecules (7–9) and antimicrobial peptides (1, 10). These processes require the regulated activation of transcription factors (4, 6, 10). Here we have identified and characterized the full-length primary let-7i transcript, a microRNA we previously demonstrated targets and regulates TLR4 expression in cholangiocytes (3). We further demonstrated that this microRNA is transcriptionally regulated following microbial stimulus, a process we propose conditions the cholangiocyte intracellular microenvironment for enhanced responses to microbial challenges and promotion of the innate immune response.
The extent of microRNA involvement in the initiation or attenuation of inflammatory cascades is not well characterized. However, using a monocytic leukemia cell line, it was demonstrated that several microRNAs are up-regulated upon stimulation with LPS. At least one of the up-regulated microRNAs targets IL-1 receptor-associated kinase (IRAK1), and TRAF6, both with established roles in TLR and proinflammatory cytokine signaling cascades (16). Conversely, using a human cholangiocyte cell culture model of biliary cryptosporidiosis, we reported that infection of cultured human cholangiocytes with C. parvum results in decreased let-7i expression. Decreased let-7i expression resulted in up-regulation of TLR4 in infected cells, and increased NFκB signaling (3). The work presented here provides a generalizable mechanism of transcription factor-dependent reduction of let-7i expression. Hence, it is plausible, if not likely that the transcriptional regulation of microRNA synthesis and microRNA-regulated post-transcriptional pathways contribute to host-cell responses to microbial infection, either through increasing inflammatory signaling in response to pathogens (3) or attenuation of the inflammatory response (16).
Our results demonstrate a functional role for the NFκB p50 subunit and C/EBPβ in modulating let-7i transcription. We demonstrate that these proteins are complexed and interacting with the let-7i promoter following microbial stimulus. Therefore, the spatial distribution of these proteins within the nucleus of microbially stimulated cells and their demonstrated physical interaction support the feasibility of a repressive complex of proteins. Our data support a model in which upon microbial stimulus, a complex of these two proteins represses transcription through chromatin deacetylation, a well-established mechanism for gene silencing. The C/EBP family of transcription factors, together with AP-1 and ATF/CREB, belong to the b-ZIP class of DNA-binding proteins (17). The functional interaction between C/EBPβ and NFκB has been described previously (18–20), including, most recently, the functional interaction between C/EBPβ and the p50 subunit of NFκB in regulating the antiapoptotic protein, Nur77, in testicular Leydig cells (21). This interaction can either synergistically promote the expression of immune or acute-phase response elements but can also repress transcription (20, 22). The ability of C/EBPβ to function as a transcriptional activator or repressor has been attributed to the expression of different C/EBPβ isoforms generated by leaky ribosomal scanning of the primary mRNA (23). Three isoforms have been characterized: liver-enriched transcriptional activator protein 1 (LAP1), liver-enriched transcriptional activator protein 2 (LAP2), and liver-enriched inhibitory protein (LIP). LAP1 and -2 contain dimerization and DNA binding domains, a transcriptional regulatory domain, and function as dimeric transcriptional regulators. LIP lacks the regulatory domain and is thought to function as a dominant negative inhibitor of LAPs (23, 24). The force-expressed C/EBPβ used in our reporter assays (LAP2) contains a conservative ATG to ATC mutation (Met to Ile) that eliminates the LIP start codon and hence is incapable of forming inhibitory LIP (25). Additionally, we were unable to detect the 27-kDa LIP protein in the p50/C/EBPβ immunoprecipitations. We therefore propose, and our data support, that upon microbial insult, C/EBPβ LAP and the p50 subunit of NFκB functionally repress transcription of the microRNA, let-7i, through the induction of chromatin remodeling.
The let-7 family of microRNAs consists of 12 genes that encode nine distinct mature let-7 microRNAs designated let-7a-let-7i. The mature forms of these microRNAs, and the highly similar mir-98, differ from the consensus let-7a by one to four nucleotides. Currently, the transcriptional mechanisms regulating the expression of primary let-7 are not well defined. However, post-transcriptional mechanisms have been implicated in the regulation of mature microRNA expression, including the let-7 family. For instance, post-transcriptional regulation has been demonstrated at the level of Drosha (26, 27) and/or Dicer (28), which enables the expression of either the primary or precursor microRNA, whereas expression of the mature, functional microRNA is inhibited (26). In addition, the regulated processing of microRNAs has been observed in embryonic tissues and human cancers (26). Specifically, Lin-28, a small developmentally regulated RNA-binding protein, was shown to selectively block the processing of precursor let-7 family members in embryonic cells (27). Additionally, methylation-dependent regulation of several microRNAs was observed in human cholangiocarcinoma cell lines force expressing IL-6 (29). We were unable to detect methylation at the let-7i promoter region under basal or microbe-stimulated conditions. Hence, in addition to post-transcriptional regulation of let-7 microRNAs, our current data provide a mechanism of transcription factor-induced repression following microbial insult involving both NFκB p50 and C/EBPβ.
In summary, we have identified and characterized the primary let-7i transcript expressed in human cholangiocytes. Using a cell culture model of cholangiocyte response to pathogens, we have defined a novel transcription mechanism mediating repression of let-7i primary transcript expression, a process that may contribute to the innate immune-associated phenotype, which actively participates in the defense response against pathogens and immune/inflammatory responses in general. It will be of interest to extend these studies to confirm the existence of an inhibitory complex of proteins, identify the full complement of proteins within this repressive complex, determine the extent of microRNAs regulated via this mechanism, and address the functional consequences of this microRNA repression under normal and pathological conditions.
Supplementary Material
Acknowledgment
We thank Deb Hintz for secretarial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK57993 (to N. F. L.), DK76922 (to S. P. O.), and A2071321 (to X. C.). This work was also supported by a grant from the Mayo Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
- LPS
- lipopolysaccharide
- RACE
- rapid amplification of cDNA ends
- ChIP
- chromatin immunoprecipitation assay
- ANOVA
- analysis of variance
- ELISA
- enzyme-linked immunosorbent assay
- EMSA
- electrophoretic mobility shift assay
- IL
- interleukin
- EST
- expressed sequence tag
- C/EBPβ
- CCAAT/enhancer-binding protein β.
REFERENCES
- 1.Harada K., Ohba K., Ozaki S., Isse K., Hirayama T., Wada A., Nakanuma Y. (2004) Hepatology 40, 925–932 [DOI] [PubMed] [Google Scholar]
- 2.Chen X. M., O'Hara S. P., LaRusso N. F. (2008) Immunol. Cell Biol. 86, 497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X. M., Splinter P. L., O'Hara S. P., LaRusso N. F. (2007) J. Biol. Chem. 282, 28929–28938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokoyama T., Komori A., Nakamura M., Takii Y., Kamihira T., Shimoda S., Mori T., Fujiwara S., Koyabu M., Taniguchi K., Fujioka H., Migita K., Yatsuhashi H., Ishibashi H. (2006) Liver Int. 26, 467–476 [DOI] [PubMed] [Google Scholar]
- 5.Chen X. M., Levine S. A., Splinter P. L., Tietz P. S., Ganong A. L., Jobin C., Gores G. J., Paya C. V., LaRusso N. F. (2001) Gastroenterology 120, 1774–1783 [DOI] [PubMed] [Google Scholar]
- 6.Park J., Gores G. J., Patel T. (1999) Hepatology 29, 1037–1043 [DOI] [PubMed] [Google Scholar]
- 7.Cruickshank S. M., Southgate J., Selby P. J., Trejdosiewicz L. K. (1998) J. Hepatol. 29, 550–558 [DOI] [PubMed] [Google Scholar]
- 8.Morita M., Watanabe Y., Akaike T. (1994) Hepatology 19, 426–431 [PubMed] [Google Scholar]
- 9.Scholz M., Cinatl J., Blaheta R. A., Kornhuber B., Markus B. H., Doerr H. W. (1997) Tissue Antigens 49, 640–643 [DOI] [PubMed] [Google Scholar]
- 10.Chen X. M., O'Hara S. P., Nelson J. B., Splinter P. L., Small A. J., Tietz P. S., Limper A. H., LaRusso N. F. (2005) J. Immunol. 175, 7447–7456 [DOI] [PubMed] [Google Scholar]
- 11.Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 12.Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., Nakajima T., Hirano T., Kishimoto T. (1990) EMBO J. 9, 1897–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grubman S. A., Perrone R. D., Lee D. W., Murray S. L., Rogers L. C., Wolkoff L. I., Mulberg A. E., Cherington V., Jefferson D. M. (1994) Am. J. Physiol. 266, G1060–G1070 [DOI] [PubMed] [Google Scholar]
- 14.Chen X. M., LaRusso N. F. (2000) Gastroenterology 118, 368–379 [DOI] [PubMed] [Google Scholar]
- 15.Joplin R., Strain A. J., Neuberger J. M. (1989) In Vitro Cell Dev. Biol. 25, 1189–1192 [DOI] [PubMed] [Google Scholar]
- 16.Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinson C. R., Sigler P. B., McKnight S. L. (1989) Science 246, 911–916 [DOI] [PubMed] [Google Scholar]
- 18.Agrawal A., Cha-Molstad H., Samols D., Kushner I. (2003) Immunology 108, 539–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cha-Molstad H., Young D. P., Kushner I., Samols D. (2007) Mol. Immunol. 44, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 20.Stein B., Cogswell P. C., Baldwin A. S., Jr. (1993) Mol. Cell. Biol. 13, 3964–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Asmar B., Giner X. C., Tremblay J. J. (2009) J. Mol. Endocrinol. 42, 131–138 [DOI] [PubMed] [Google Scholar]
- 22.Ralph W. M., Jr., Liu K., Auborn K. J. (2006) J. Gen. Virol. 87, 51–59 [DOI] [PubMed] [Google Scholar]
- 23.Descombes P., Schibler U. (1991) Cell 67, 569–579 [DOI] [PubMed] [Google Scholar]
- 24.Grimm S. L., Rosen J. M. (2003) J. Mammary Gland Biol. Neoplasia 8, 191–204 [DOI] [PubMed] [Google Scholar]
- 25.Gomis R. R., Alarcón C., Nadal C., Van Poznak C., Massagué J. (2006) Cancer Cell 10, 203–214 [DOI] [PubMed] [Google Scholar]
- 26.Thomson J. M., Newman M., Parker J. S., Morin-Kensicki E. M., Wright T., Hammond S. M. (2006) Genes Dev. 20, 2202–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viswanathan S. R., Daley G. Q., Gregory R. I. (2008) Science 320, 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obernosterer G., Leuschner P. J., Alenius M., Martinez J. (2006) Rna 12, 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng F., Wehbe-Janek H., Henson R., Smith H., Patel T. (2008) Oncogene 27, 378–386 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.