Abstract
Ptf1a, a basic helix-loop-helix transcription factor, plays an indispensable role for cell fate specification of subsets of neurons in the developing central nervous system. However, downstream molecules induced by Ptf1a during neural development have not been well characterized. In the present study, we identified immunoglobulin superfamily molecules, Nephrin and Neph3, as direct downstream targets of Ptf1a. First, the expression domains of Nephrin and Neph3 closely resembled those of Ptf1a in the developing retina, hypothalamus, cerebellum, hindbrain, and spinal cord. Second, Ptf1a bound directly to a PTF-binding motif in the 5′-flanking region of Nephrin and Neph3 genes. Third, Ptf1a activated transcription driven by the 5′-flanking region of these genes. Finally, the expression of Nephrin and Neph3 was lost in Ptf1a-null mice, whereas ectopic expression of Nephrin and Neph3 was induced by forced expression of Ptf1a. We provided further evidence that Nephrin and Neph3 could interact homophilically and heterophilically, suggesting that Nephrin and Neph3 might regulate certain developmental aspects of Ptf1a-positive neurons as homo- or heterooligomers.
Introduction
Ptf1a,3 which encodes a basic helix-loop-helix transcription factor, was first identified as a cell fate determinant of the pancreas (1, 2). Mutations in the human and mouse Ptf1a result not only in the malformation of the pancreas but also in cerebellar agenesis, indicating its involvement in neuronal development (3, 4). Recent studies demonstrated that Ptf1a is expressed in neuronal progenitors in the retina, cerebellum, hindbrain, and spinal cord, most of which are fated to be subsets of GABA (γ-aminobutyric acid)-ergic and glycinergic inhibitory neurons (4–10). In the Ptf1a-null cerebellum, Ptf1a-positive progenitors, which are normally fated to inhibitory neurons, trans-fate to excitatory granule cells (8). Cell fate changes in Ptf1a-null mice were also reported in the retina, hindbrain, and spinal cord (5–7, 9). Conversely, ectopic expression of Ptf1a in neural progenitors of the dorsal telencephalon, which are normally fated to glutamatergic excitatory neurons, confers inhibitory GABA characteristics to these neurons (4). These loss- and gain-of-function experiments suggest that Ptf1a acts as a cell fate switch in the central nervous system. However, how Ptf1a operates to fulfill such a critical role is not known because of a lack of understanding of its downstream targets.
In the present study, we searched for direct downstream targets of Ptf1a, which is likely to be responsible for cell fate specification of Ptf1a neurons. We identified Nephrin and Neph3 as candidates of Ptf1a target molecules. Nephrin and Neph3 are transmembrane proteins of the immunoglobulin superfamily. Nephrin was originally isolated as a gene responsible for the congenital nephrotic syndrome of the Finnish type (11). Neph3 belongs to Neph family, which is composed of Neph1, Neph2, and Neph3 (also called as Kirrel, Kirrel3/mKirre, and Kirrel2/Filtrin, respectively), and Neph family members are structurally related to Nephrin (12). Nephrin and Neph family members can interact homophilically, and in some cases heterophilically, which in turn triggers cell adhesion or signal transduction (13–19). Recent reports showed expression of Nephrin and Neph family molecules in the developing central nervous system (17, 18, 20–22). Although their function in vertebrate neural development is still unclarified, their invertebrate homologs appear to play indispensable roles in synapse formation or axon guidance during development (23, 24).
Here, we provide evidence that Ptf1a directly controls the expression of Nephrin and Neph3. Moreover, Nephrin and Neph3 interacted homophilically and heterophilically. These findings suggest that Nephrin and Neph3, via their interaction, play important roles in Ptf1a-positive neuronal progenitors.
EXPERIMENTAL PROCEDURES
Vectors
pCAG, pCAG-EGFP, pCAG-mCherry, pCAG-IRES-EGFP, and pEF-RL vectors were described previously (25–29). Full-length NephrinB cDNA was cloned from mouse E13.5 cerebellum, and its sequence was deposited into DDBJ with accession number AB513652. For construction of expression vectors of NephrinA (87–3857 of NM_019459), NephrinB (1–3729 of AB513652), and Neph3 (130–2232 of BC052773), cDNAs were subcloned into pCAG vector. A HA or FLAG sequence was inserted between 200 and 201 of NephrinA (NM_019459), between 72 and 73 of NephrinB (AB513652), and between 189 and 190 of Neph3 (BC052773). For construction of expression vectors of Ptf1a (199–1173 of NM_018809), E47 (87–2033 of AF352579), RBPJ (217–1716 of NM_009035), N-Twist (66–572 of NM_033522), Math1 (196–1251 of NM_007500), and Neurogenin1 (232–966 of NM_010896), cDNAs were subcloned into pCAG or pCAG-IRES-EGFP. A HA, FLAG, or Myc tag was added in their N termini by PCR. pGL2-basic vector was obtained commercially (Promega). The 5′-flanking region of Nephrin and Neph3 genes was amplified by PCR from genomic DNA of ICR mice and subcloned into pGL2-basic. The following primers were used for PCR amplification: primer 1 (5′-ATG CAA GCT TCA GAG TGC CAG GAA AGG G-3′), primer 2 (5′-GAC CAA GCT TTG TGT GTA CCC CAA GAT C-3′), and primer 3 (5′-GAC CAA GCT TGT TTG TTG CTG TC-3′). For amplification of the 1.6-kb sequence upstream of the Neph3 start codon (see Fig. 4A), primers 1 and 2 were used as forward and reverse primers, respectively. For the 1.6-kb sequence upstream of the NephrinB start codon or 3.3 kb upstream of the NephrinA start codon (see Fig. 4B), primers 2 and 1, or primers 2 and 3 were used as forward and reverse primer, respectively. The E-box and the TC-box mutations were introduced by PCR-based site-directed mutagenesis.
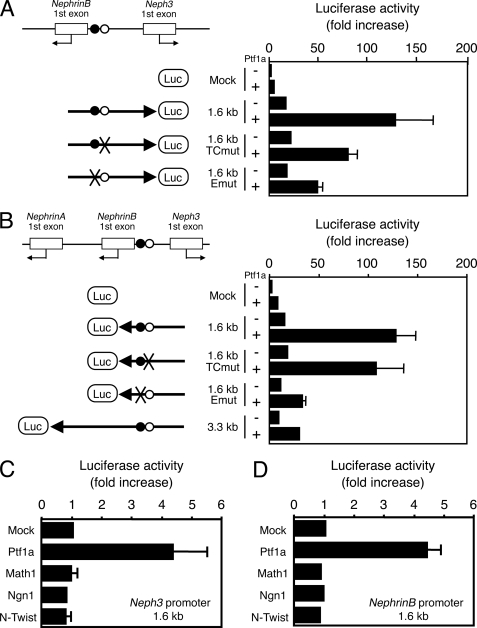
FIGURE 4.
Luciferase reporter assay of the 5′-flanking region of Nephrin and Neph3 genes. A and B, promoter activity of 5′-flanking region of Neph3 (A) or Nephrin (B) driven by Ptf1a was analyzed in COS7 cells. The luciferase activities were normalized against those of co-transfected Renilla luciferase. The results are presented as fold increase relative to mock-transfected sample. Black and white circles on the left indicate the E-box and the TC-box of the PTF1 motif, respectively. The E-box and the TC-box mutations were introduced the same as in Fig. 3. C and D, luciferase reporter constructs together with Ptf1a, Math1, Neurogenin1, or N-Twist expression vector were transfected into COS7 cells, and luciferase activities in the lysates were analyzed. Values are the means ± S.E. (error bars) of three to five independent transfections.
Antibodies
Anti-Ptf1a antibody was described previously (9, 30). Anti-FLAG (M2, Sigma), anti-Myc (9E10, Santa Cruz Biotechnology), anti-HA (3F10, Roche Applied Science), alkaline phosphatase (AP)-conjugated anti-mouse IgG (Bio-Rad), AP-conjugated anti-rat IgG (Jackson Laboratories), AP-conjugated anti-digoxigenin (DIG, Roche Applied Science), peroxidase-conjugated anti-DIG (Roche Applied Science), anti-GFP (Molecular Probes), and biotin-conjugated anti-rabbit IgG (Jackson Laboratories) antibodies were obtained commercially.
In Situ Hybridization and Immunohistochemistry
The Ptf1acre mouse line was described previously (1). In situ hybridization was performed using DIG-labeled RNA probes of Nephrin (2738–3764 of NM_019459) and Neph3 (1037–2129 of BC052773). For fluorescent detection, samples were incubated with a peroxidase-conjugated anti-DIG antibody, biotinylated-tyramide (PerkinElmer Life Sciences), and Alexa Fluor 594-streptavidin (Molecular Probes). To couple with immunostaining, sections were first hybridized with DIG-RNA probes and immunostained with an anti-Ptf1a (30) or an anti-GFP antibody.
In Utero Electroporation
In utero electroporation into the cerebral cortex of E14.5 mouse embryos was described previously (26). Expression vectors were introduced into E14.5 ICR mice, and pregnant mice were killed 1 or 2 days later for further analyses.
Chromatin Immunoprecipitation Assay
Cerebellum from E11.5 ICR mice was dissected out, and chromatin immunoprecipitation assay was performed as described previously (31). Endogenous Ptf1a was immunoprecipitated with an anti-Ptf1a antibody (9). The amount of co-precipitated DNA fragments was analyzed by quantitative PCR using a LightCycler (Roche Applied Science). Primers used in quantitative PCR were as follows: PTF1 motif forward (5′-GCC AGG AGT TCA GAT TTA GGT G-3′) and reverse (5′-AGG ATG GGA AAA CCA ACA GAC-3′); 1.5 kb 5′ of the PTF1 forward (5′-TCC CTC TCA CCC ACT CAC AG-3′) and reverse (5′-AGT TCA CAC TGG GTC CAA GC-3′); 1.5 kb 3′ of the PTF1 forward (5′-TAC ACA CAC TCG GGA TGC TG-3′) and reverse (5′-CCA AAC AGG GCT GTA ATG GAC-3′).
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed following the manufacturer's protocol in the LightShift Chemiluminescent EMSA kit (Pierce). Nuclear extracts of HA-Ptf1a-, FLAG-E47-, or Myc-RBPJ-transfected COS7 cells were prepared as described previously (32). Forward and 3′-biotinylated reverse oligonucleotides were purchased commercially (GeneDesign). The sequence of oligonucleotides is shown in Fig. 3B. Nuclear extracts were incubated with 200 fmol of annealed biotinylated oligonucleotides. For the supershift assay, the nuclear extracts were preincubated with anti-HA, anti-FLAG, or anti-Myc antibody. Biotinylated oligonucleotides were detected by streptavidin-AP (Jackson Laboratories) and an AP substrate, CDP star (Roche Applied Science), and chemiluminescence was analyzed by RAS3000 mini (Fuji film).
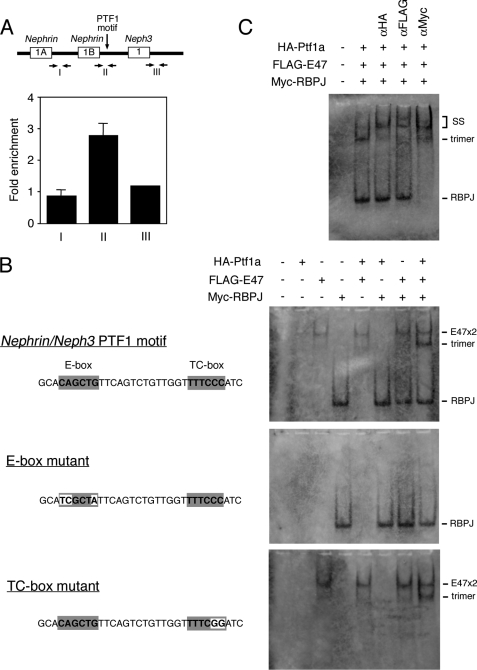
FIGURE 3.
Interaction of Ptf1a with the 5′-flanking region of Nephrin and Neph3 genes. A, Ptf1a chromatin immunoprecipitation assay was performed on chromatin isolated from E11.5 cerebellum. The amount of precipitated DNA fragments around the 5′-flanking region of Nephrin and Neph3 genes was analyzed by quantitative PCR. PCR primers encompassing the PTF1 site (II) or at a distance of 1.5 kb from this site (I and III) were used. The amount of PCR products in immunoprecipitates with anti-Ptf1a relative to those without antibodies is shown. Values are the means ± S.E. (error bars) of three samples. B, EMSA using oligonucleotides of wild type PTF1 motif (top panel), E-box mutant (middle panel), and TC-box mutant (bottom panel), in the presence of HA-Ptf1a, FLAG-E47, and/or Myc-RBPJ. Sequences of the oligonucleotides are shown on the left. C, EMSA supershift analysis. EMSA was performed using oligonucleotides with wild type PTF1 motif, HA-Ptf1a, FLAG-E47, and Myc-RBPJ, in the presence of an anti-HA, an anti-FLAG, or an anti-Myc antibody.
Luciferase Reporter Assay
Luciferase reporter assay was performed following the manufacturer's protocol in the Dual Luciferase Reporter Assay system (Promega). pGL2-basic vector constructs and pEF-RL with or without pCAG-Ptf1a were transfected into COS7 cells by FuGENE 6 (Roche). 48 h later, the activity of the firefly and Renilla luciferases was analyzed by Luminometer (Promega).
Affinity Probe in Situ Assay
An affinity probe in situ assay using Neph3-AP was performed as described previously (18). Binding of Neph3-AP was detected by AP activity using nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate as substrates. To measure the binding affinity of Neph3-AP to COS7 cells transfected with HA-Neph3 or HA-NephrinA expression vector, cells were incubated with an AP substrate, p-nitrophenyl phosphate, for 2 h. Increase of A405 in each sample relative to that in mock-transfected samples was analyzed.
Immunoprecipitation and Western Blotting
COS7 cells were transfected with expression vectors by FuGENE 6 or a standard calcium phosphate method. Immunoprecipitation and Western blotting were performed as described previously (13).
RESULTS
Nephrin and Neph3 Genes Have a PTF1-binding Motif in Their 5′-Flanking Region
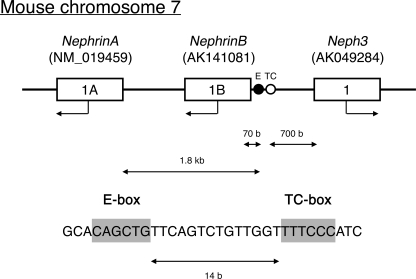
Ptf1a is a component of a trimeric transcription factor, PTF1, which interacts with the PTF1-binding motif, comprising an E-box (CAG/CCTG) and a TC-box (TTTCCC) separated by one or two helical turns (33–35). In an attempt to identify novel downstream targets of Ptf1a, we took advantage of a data base, DBTSS, to screen for genes containing PTF1 motif in their 5′-flanking regions. As a result, we found a PTF1 motif located in the shared 5′-flanking region of Nephrin and Neph3 genes on mouse chromosome 7 (Fig. 1). The Nephrin gene has two isoforms, NephrinA and NephrinB, resulting from utilizing alternative first exons, 1A and 1B. Thus, the PTF1 motif is located 700 bases upstream of the first exon of Neph3, and 70 bases and 1.8 kb upstream of exon 1B and exon 1A of Nephrin, respectively.
FIGURE 1.
PTF motif found in the 5′-flanking region of Nephrin and Neph3 gene. Black and white circles on the upper line indicate E-box and TC-box of the PTF1 motif, respectively. The sequence of the PTF motif is shown in the bottom.
Nephrin and Neph3 Are Expressed in Ptf1a-positive Domains in the Developing Central Nervous System
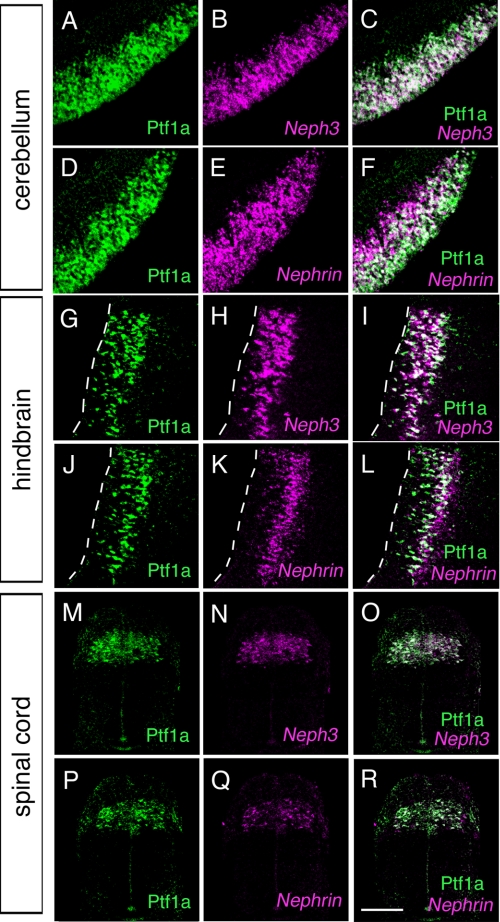
If Nephrin and Neph3 are downstream targets of Ptf1a, they would be expressed in Ptf1a-positive domains. To examine this possibility, we first analyzed the expression of Nephrin and Neph3 in the developing central nervous system. Nephrin and Neph3 were expressed in overlapping regions where Ptf1a is expressed, in the cerebellum and hindbrain at E12.5, in the spinal cord at E10.5, and in the retina and hypothalamus at E13.5 (Fig. 2, supplemental Fig. 1, and data not shown). Many Ptf1a-positve cells appeared to co-express Nephrin and Neph3 (Fig. 2). Moreover, Nephrin and Neph3 were co-expressed in the same cells, which was confirmed by double labeling of Nephrin and Neph3 (data not shown). We further examined the expression of two isoforms of Nephrin in the developing cerebellum by quantitative PCR. Consistent with the previous report (36), comparable amounts of endogenous NephrinA and B were expressed in the developing cerebellum, whereas adult kidney predominantly expressed NephrinA (supplemental Fig. 2). These results raise the possibility that Ptf1a may control the expression of Nephrin and Neph3.
FIGURE 2.
Expression of Ptf1a, Neph3, and Nephrin in the developing cerebellum, hindbrain, and spinal cord. Immunostaining by anti-Ptf1a (A, C, D, F, G, I, J, L, M, O, P, and R), together with in situ hybridization of Neph3 (B, C, H, I, N, and O) or Nephrin (E, F, K, L, Q, and R) on coronal sections of E12.5 cerebellum (A–F), E12.5 dorsal hindbrain (G–L), and E10.5 spinal cord (M–R) are shown. Dorsal is upward. In the cerebellar sections (A–F), midline is toward the right, and lateral is toward the left. In the sections of the hindbrain (G–L), medial is toward the left. Scale bar, 100 μm.
Ptf1a Binds to the Promoter Region of Nephrin and Neph3 Genes That Contains PTF1 Motif
Because Nephrin and Neph3 genes contain the PTF1 motif in their 5′-flanking region, it is highly possible that Ptf1a controls the transcription of these genes by direct binding to the PTF1 motif. We examined this possibility by chromatin immunoprecipitation assay to test whether Ptf1a in the developing cerebellum associates with the 5′-flanking region of Nephrin and Neph3 genes in vivo. Anti-Ptf1a antibody immunoprecipitated DNA fragments centered on the PTF1 motif, but not 1.5 kb upstream or downstream of the motif (Fig. 3A). This interaction could also be detected in Ptf1a-overexpressed cerebral cortex (supplemental Fig. 3). We further examined whether the interaction could be reconstituted in vitro. Beres et al. (37) reported that PTF1 complex in the pancreas comprised Ptf1a, a class A basic helix-loop-helix protein, such as E47 or HEB, and RBPL. In the developing central nervous system, RBPJ instead of RBPL is expressed and interacts with a PTF1-binding motif in vitro (37, 38). Thus, we performed EMSA using oligonucleotides containing the PTF1 motif and a combination of Ptf1a, E47, and RBPJ (Fig. 3B). RBPJ or an E47 dimer could bind the PTF motif alone. However, addition of all three proteins resulted in a new shifted band associated with a heterotrimeric complex composed of Ptf1a/E47/RBPJ. The presence of Ptf1a/E47/RBPJ trimer in the shifted band was confirmed by the fact that incubation with an antibody for each component all led to a supershift of the band (Fig. 3C). Mutations in E-box abolished Ptf1a binding, whereas the mutated TC-box had no effect in its binding, suggesting that E-box is necessary for the binding of Ptf1a to the PTF1 motif (Fig. 3B). Taken together, these results demonstrated in vivo and in vitro that Ptf1a binds to the promoter region of Nephrin and Neph3 genes and this binding is dependent on the PTF1 motif.
Ptf1a Activates Transcription from the Nephrin and Neph3 Promoter
We next tested whether Ptf1a binding to the promoter region of Nephrin and Neph3 leads to trans-activation of downstream genes by using a luciferase reporter system. Expression of Ptf1a in COS7 cells strongly activated a luciferase reporter driven by the 5′-flanking region of Nephrin and Neph3 containing the PTF1 motif in both directions (Fig. 4, A and B). Note that transcriptional activation by Ptf1a could take place from both promoter fragments upstream of exon 1A and exon 1B of the Nephrin gene, albeit weaker in the former. Mutations in the E-box or TC-box, particularly the E-box, notably reduced the level of Ptf1a-dependent transactivation, suggesting that activation of the promoter depends on the successful binding of Ptf1a to the PTF1 motif. Some other members of basic helix-loop-helix, such as Math1, Neurogenin1, or N-Twist, did not enhance Ptf1a transactivity, and Ptf1a did not activate c-fos promoter (Fig. 4, C and D, and data not shown). Furthermore, co-expression of Ptf1a and E47 caused synergistic enhancement of Neph3 promoter activation (supplemental Fig. 4). To confirm the dependence of Neph3 promoter activation on Ptf1a in vivo, we performed a luciferase assay in the developing cerebellum and cerebral cortex, which are Ptf1a-positive and Ptf1a-negative areas, respectively. As shown in supplemental Fig. 4, Neph3 promoter was highly activated in the developing cerebellum compared with the cerebral cortex, indicating a correlation of the presence of Ptf1a and high activation of the Neph3 promoter. These results suggest that Ptf1a activates transcription from Nephrin and Neph3 promoter.
Ptf1a Is Essential for Expression of Nephrin and Neph3 in Vivo
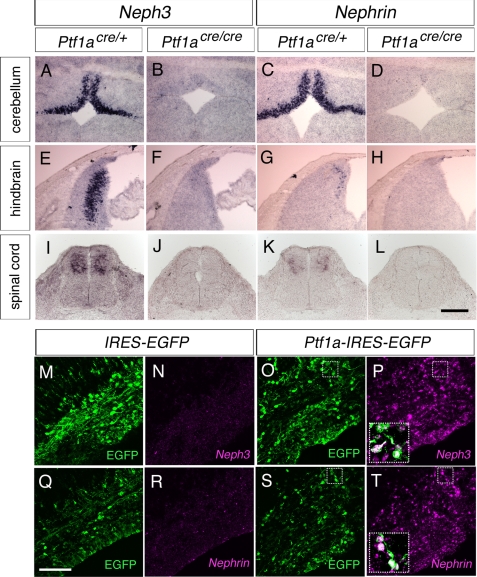
Ptf1a interacted with the PTF1 motif, thereby inducing transactivation of the Nephrin/Neph3 promoter, raising the possibility that Ptf1a controls the expression of Nephrin and Neph3 in vivo. To assess this possibility, we first examined the expression of Nephrin and Neph3 in Ptf1a-null mice (Fig. 5, A–L, and supplemental Fig. 1). The expression of Nephrin and Neph3 was lost in the developing cerebellum, hindbrain, spinal cord, retina, and hypothalamus of the homozygotes. We next performed gain-of-function experiments by ectopically expressing Ptf1a in the developing cerebral cortex, where Ptf1a, Nephrin, and Neph3 are not endogenously expressed (data not shown). We found that ectopic Ptf1a triggered a notable induction of Nephrin and Neph3 expression (Fig. 5, M–T), but not the other Neph family members Neph1 and Neph2 (data not shown). Because Ptf1a activated transcription from exon 1A and 1B of the Nephrin gene (Fig. 4B), we further examined whether both Nephrin isoforms are induced by ectopic expression of Ptf1a in the cerebral cortex. As expected, we found induction of NephrinA and B by Ptf1a (supplemental Fig. 2). These results together suggest that Ptf1a controls the expression of two isoforms of Nephrin and Neph3 in the developing central nervous system.
FIGURE 5.
Nephrin and Neph3 expression in Ptf1a-null mice and Ptf1a-misexpressed cerebral cortex. A–L, expression of Nephrin and Neph3 in Ptf1a-null mice. Expression of Neph3 (A, B, E, F, I, and J) and Nephrin (C, D, G, H, K, and L) was analyzed by in situ hybridization on coronal sections of E13.5 cerebellum (A–D), E13.5 dorsal hindbrain (E–H), and E10.5 spinal cord (I–L). Ptf1acre/+ mice (A, C, E, G, I, and K) and Ptf1acre/cre mice (B, D, F, H, J, and L) were used. Dorsal is upward. Scale bar, 100 μm. M–T, expression of Nephrin and Neph3 in Ptf1a-overexpressed cerebral cortex at E16.5. Expression of Neph3 (N and P) and Nephrin (R and T) were analyzed by in situ hybridization on coronal sections of IRES-EGFP-transfected (M, N, Q, and R) or Ptf1a-IRES-EGFP-transfected (O, P, S, and T) cerebral cortex. Expression of EGFP on the same sections was analyzed by immunostaining by anti-GFP (M, O, Q, and S). Insets in P and T are high magnification views of merged images outlined in O, P, S, and T. Dorsal is upward. Scale bar, 100 μm.
Interaction between Nephrin and Neph3
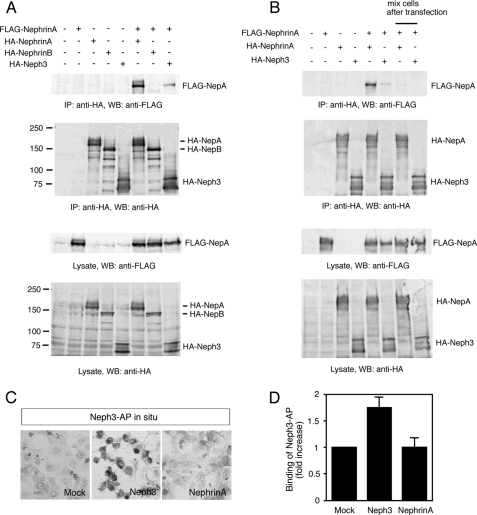
So far we have shown that Ptf1a simultaneously activates the transcription of Nephrin and Neph3 in subsets of neural precursors. This raises the possibility that Nephrin and Neph3 may interact and in turn control certain aspects of the fate of these cells. Indeed, heterophilic interactions between Nephrin, and Neph1 or Neph2 have been previously demonstrated (13–16). However, a Nephrin-Neph3 interaction has not been reported. We therefore investigated any potential interaction between Nephrin and Neph3 by co-immunoprecipitation experiments in COS7 cells. As shown in Fig. 6A, FLAG-NephrinA was co-precipitated with co-expressed HA-Neph3 or HA-NephrinA, but not with HA-NephrinB. To examine whether NephrinA and Neph3 interact in cis on the same cell, or in trans on different cells, we performed a similar experiment using a mixture of cells transfected with each construct separately (Fig. 6B). However, NephrinA was co-precipitated with neither Neph3 nor itself in this condition. These results imply that NephrinA interacts with Neph3 as well as itself in cis. To investigate whether NephrinA-Neph3 may interact in trans, we performed an affinity probe in situ assay using Neph3-AP fusion protein (Fig. 6, C and D). Consistent with a previous report (18), Neph3-AP bound to Neph3-expressing cells, but not to NephrinA-expressing cells.
FIGURE 6.
Homophilic and heterophilic interaction of Nephrin and Neph3. A, interaction of FLAG-NephrinA with HA-NephrinA, HA-NephrinB, or HA-Neph3 by co-immunoprecipitation experiments in COS7 cells. B, interaction between NephrinA and Neph3. Left six lanes, 2.5 μg of the indicated vectors was co-transfected into COS7 cells by FuGENE 6. 24 h later, cells were detached, spread onto the same plates again, and cultured for another 24 h for co-immunoprecipitation assay. Right two lanes, 5 μg of FLAG-NephrinA, HA-NephrinA, or HA-Neph3 was transfected into COS7 cells. 24 h later, the cells were collected, and half of FLAG-NephrinA-cells were mixed with half of HA-NephrinA-cells or HA-Neph3-cells and incubated for another 24 h for co-immunoprecipitation assay. Note that expression of each protein in the lysate in the left six lanes is comparable with that in the right two lanes. C, affinity probe in situ assay of Neph3-AP. Binding of Neph3-AP to mock-, HA-Neph3-, or HA-NephrinA-transfected COS7 cells is shown. D, binding affinities of Neph3-AP to COS7 cells transfected with mock, HA-Neph3, or HA-NephrinA vector. All values are the means of five to six independent transfections ± S.E. (error bars).
It has been shown in invertebrate that trans-interactions between Nephrin and Neph family mediate cell-cell adhesion (23, 24). We thus examined whether NephrinA and Neph3 could mediate cell adhesion. As expected, only Neph3-expressing cells formed cell aggregates, but no aggregation was detected between Neph3- and NephrinA-expressing cells and between NephrinA-expressing cells (supplemental Fig. 5). These data, taken together, showed that NephrinA-Neph3 interactions take place in cis but not in trans, whereas Neph3 could interact homophilically in trans, thereby mediating cell adhesion.
DISCUSSION
In the present study, we identified Nephrin and Neph3 as direct downstream targets of Ptf1a in the developing central nervous system. Our finding provides an important clue to unravel as yet unclarified molecular mechanisms underlying cell fate specification of Ptf1a-neurons. Henke et al. (39) recently reported Neurog2, itself a basic helix-loop-helix transcription factor, as a first example of downstream target of Ptf1a in the developing spinal cord. However, in contrast to Ptf1a-null mice, neurogenesis of Ptf1a-neurons is not affected in Neurog2 knock-out mice, raising the possibility that Ptf1a downstream targets other than Neurog2 are also involved in the development of Ptf1a-neurons (7, 40). Compared with Neurog2, Nephrin and Neph3 as transmembrane proteins more likely mediate the effect of Ptf1a by directly influencing the cellular behavior of Ptf1a-neurons. Moreover, Nephrin and Neph3 were more specifically expressed in the Ptf1a-positive domains, implicating their specific function in Ptf1a-neurons (Fig. 2 and supplemental Fig. 1). Ptf1a might also control Nephrin/Neph3 expression outside the nervous system, such as islet cells in the pancreas (20, 41–43).
Ptf1a required co-factors for binding to the promoter region of Nephrin and Neph3 and to induce full promoter activation (Fig. 3 and supplemental Fig. 4). Our in vitro data suggest that E47 facilitates binding of Ptf1a to the promoter via the E-box, and in turn activates Nephrin/Neph3 transcription. The wide expression of E47 in the ventricular zone of the developing central nervous system supports the possibility that it acts as a co-factor for Ptf1a in vivo (data not shown). RBPJ is another factor that is necessary for the binding of Ptf1a to the promoter of Nephrin and Neph3 in vitro. However, differing from E47, RBPJ interacted with the TC-box of the PTF1 motif, and this interaction was not required for Ptf1a binding to the PTF1 motif (Fig. 3). It is possible that a direct interaction between RBPJ and Ptf1a enhances association of Ptf1a to the E-box. Indeed, this possibility is implied by an in vivo study showing that the ability of Ptf1a to interact with RBPJ is crucial for its role in the specification of GABA-ergic neurons in the dorsal spinal cord (38). Ptf1a/E47/RBPJ may not be the only proteins that trans-activate Nephrin and Neph3 genes. For example, NephrinA expression in the adult kidney is controlled by a zinc finger protein, Wt1, which interacts with the 5′-flanking region of exon 1A of Nephrin gene (44). Transcription factors NF-κB and Sp1 are key regulators of Neph3 expression in the kidney podocytes (45).
In the present study, we characterized homophilic and heterophilic interaction of Nephrin and Neph3, which provides insights into their mechanistic cellular function. To our surprise, we failed to detect an interaction between NephrinA-expressing cells, which has been demonstrated in a previous study (19). A possible reason for this discrepancy may be ascribable to differences in the type of cells being used. Conformation of NephrinA necessary for trans-homophilic interaction might be regulated by its interaction with cell-type specific cytoplasmic proteins, such as CD2AP, which is expressed in the kidney podocytes but not in the brain (46).
The function of Nephrin and Neph3 in the development of Ptf1a-neurons has yet to be determined. So far, no anatomical or morphological abnormalities have been demonstrated in the brains of NephrinA-null mice, and knock-out mice of NephrinB and Neph3 have not been reported (20). Neph3 is transiently expressed in early postmitotic neurons of the developing spinal cord, and intracellularly Neph3 is localized at the adherens junction between apical processes (17). Miyata et al. (47) reported that early postmitotic neurons maintain their apical processes before departing the ventricle to undergo radial migration. Therefore, one may speculate that Neph3 mediates the adhesion between the Ptf1a-progenitors, which in turn is necessary for their later development, such as neuronal migration, maturation, or differentiation. Alternatively, Neph3 trans-homophilic interaction may induce intracellular signals required for further development of Ptf1a-neurons. The ability of Neph family members in triggering intracellular signaling pathway has been best demonstrated in the case of Neph1. Neph1 overexpression activates the AP-1 promoter, and this activation is further stimulated by phosphorylation of Neph1 by tyrosine kinases (12, 48). Moreover, clustering of Neph1 results in its own tyrosine phosphorylation and its interaction with cytoplasmic proteins (49). Interestingly, the same study showed that Neph1 clustering elicits tyrosine phosphorylation of NephrinA and the recruitment of cytoplasmic proteins to the NephrinA complex (49). If Neph3 acts similarly to Neph1, we may speculate that in Ptf1a-neurons, Neph3 clustering triggered by its trans-homophilic binding may induce intracellular signals both via its own cytoplasmic domain and via its cis-interaction with NephrinA in the same cells. We showed that the other isoform of Nephrin, NephrinB, is equally expressed in the cerebellum. However, its function seems less clear because NephrinB does not possess a signal peptide cleavage site in its amino-terminal region and does not localize in the plasma membrane when overexpressed in COS7 cells (36) (supplemental Fig. 6). One may hypothesize that it competes with NephrinA for cytoplasmic targets, sequestering them from the membrane to the cytosol, acting as a negative modulator for the effect mediated by NephrinA. Deeper understanding of molecular mechanism underlying Ptf1a-controlled neuronal cell fate specification must await clarification of signaling cascades downstream of Nephrin and Neph3 and functional characterization of two isoforms of Nephrin.
Supplementary Material
Acknowledgments
We thank Drs. H. Edland, T. Hirano, H. Itoh, J. Miyazaki, M. Murakami, H. Oyama, H. Tanaka, R. Tsien, C. V. E. Wright, M. Yamada, and Y. Zhu for providing materials; Drs. H. Kanzaki, D. Kawauchi, Y. Kita, H. Kobayashi, H. Sakano, K. Tago, H. Takeuchi, Y. Tanabe, K. Terada, M. Torigoe, M. Yanagida, Y. Yasuda, Y. Yoneda, and Y. Zhu for technical advice; and Drs. H. Kobayashi, Y. Tanabe, and Y. Zhu for critical reading of the manuscript.
This work was supported by Solution Oriented Research for Science and Technology from Japan Science Technology Corporation and grant-in-aids from the Japan Society for the Promotion of Science and Ministry of Education, Culture, Sports, Science, and Technology, Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) AB513652.
- Ptf1a
- pancreas transcription factor
- AP
- alkaline phosphatase
- DIG
- digoxigenin
- E followed by number
- embryonic day
- EMSA
- electrophoretic mobility shift assay
- GFP
- green fluorescent protein
- EGFP
- enhanced GFP
- HA
- hemagglutinin
- GABA
- γ-aminobutyric acid.
REFERENCES
- 1.Kawaguchi Y., Cooper B., Gannon M., Ray M., MacDonald R. J., Wright C. V. (2002) Nat. Genet. 32, 128–134 [DOI] [PubMed] [Google Scholar]
- 2.Krapp A., Knöfler M., Ledermann B., Bürki K., Berney C., Zoerkler N., Hagenbüchle O., Wellauer P. K. (1998) Genes Dev. 12, 3752–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sellick G. S., Barker K. T., Stolte-Dijkstra I., Fleischmann C., Coleman R. J., Garrett C., Gloyn A. L., Edghill E. L., Hattersley A. T., Wellauer P. K., Goodwin G., Houlston R. S. (2004) Nat. Genet. 36, 1301–1305 [DOI] [PubMed] [Google Scholar]
- 4.Hoshino M., Nakamura S., Mori K., Kawauchi T., Terao M., Nishimura Y. V., Fukuda A., Fuse T., Matsuo N., Sone M., Watanabe M., Bito H., Terashima T., Wright C. V., Kawaguchi Y., Nakao K., Nabeshima Y. (2005) Neuron 47, 201–213 [DOI] [PubMed] [Google Scholar]
- 5.Fujitani Y., Fujitani S., Luo H., Qiu F., Burlison J., Long Q., Kawaguchi Y., Edlund H., MacDonald R. J., Furukawa T., Fujikado T., Magnuson M. A., Xiang M., Wright C. V. (2006) Development 133, 4439–4450 [DOI] [PubMed] [Google Scholar]
- 6.Nakhai H., Sel S., Favor J., Mendoza-Torres L., Paulsen F., Duncker G. I., Schmid R. M. (2007) Development 134, 1151–1160 [DOI] [PubMed] [Google Scholar]
- 7.Glasgow S. M., Henke R. M., MacDonald R. J., Wright C. V., Johnson J. E. (2005) Development 132, 5461–5469 [DOI] [PubMed] [Google Scholar]
- 8.Pascual M., Abasolo I., Mingorance-Le Meur A., Martinez A., Del Rio J. A., Wright C. V., Real F. X., Soriano E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5193–5198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada M., Terao M., Terashima T., Fujiyama T., Kawaguchi Y., Nabeshima Y., Hoshino M. (2007) J. Neurosci. 27, 10924–10934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiyama T., Yamada M., Terao M., Terashima T., Hioki H., Inoue Y. U., Inoue T., Masuyama N., Obata K., Yanagawa Y., Kawaguchi Y., Nabeshima Y., Hoshino M. (2009) Development 136, 2049–2058 [DOI] [PubMed] [Google Scholar]
- 11.Kestilä M., Lenkkeri U., Männikkö M., Lamerdin J., McCready P., Putaala H., Ruotsalainen V., Morita T., Nissinen M., Herva R., Kashtan C. E., Peltonen L., Holmberg C., Olsen A., Tryggvason K. (1998) Mol. Cell 1, 575–582 [DOI] [PubMed] [Google Scholar]
- 12.Sellin L., Huber T. B., Gerke P., Quack I., Pavenstädt H., Walz G. (2003) FASEB J. 17, 115–117 [DOI] [PubMed] [Google Scholar]
- 13.Barletta G. M., Kovari I. A., Verma R. K., Kerjaschki D., Holzman L. B. (2003) J. Biol. Chem. 278, 19266–19271 [DOI] [PubMed] [Google Scholar]
- 14.Gerke P., Huber T. B., Sellin L., Benzing T., Walz G. (2003) J. Am. Soc. Nephrol. 14, 918–926 [DOI] [PubMed] [Google Scholar]
- 15.Liu G., Kaw B., Kurfis J., Rahmanuddin S., Kanwar Y. S., Chugh S. S. (2003) J. Clin. Invest. 112, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerke P., Sellin L., Kretz O., Petraschka D., Zentgraf H., Benzing T., Walz G. (2005) J. Am. Soc. Nephrol. 16, 1693–1702 [DOI] [PubMed] [Google Scholar]
- 17.Minaki Y., Mizuhara E., Morimoto K., Nakatani T., Sakamoto Y., Inoue Y., Satoh K., Imai T., Takai Y., Ono Y. (2005) Neurosci. Res. 52, 250–262 [DOI] [PubMed] [Google Scholar]
- 18.Serizawa S., Miyamichi K., Takeuchi H., Yamagishi Y., Suzuki M., Sakano H. (2006) Cell 127, 1057–1069 [DOI] [PubMed] [Google Scholar]
- 19.Khoshnoodi J., Sigmundsson K., Ofverstedt L. G., Skoglund U., Obrink B., Wartiovaara J., Tryggvason K. (2003) Am. J. Pathol. 163, 2337–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Putaala H., Soininen R., Kilpeläinen P., Wartiovaara J., Tryggvason K. (2001) Hum. Mol. Genet. 10, 1–8 [DOI] [PubMed] [Google Scholar]
- 21.Tamura S., Morikawa Y., Hisaoka T., Ueno H., Kitamura T., Senba E. (2005) Neuroscience 133, 615–624 [DOI] [PubMed] [Google Scholar]
- 22.Gerke P., Benzing T., Höhne M., Kispert A., Frotscher M., Walz G., Kretz O. (2006) J. Comp. Neurol. 498, 466–475 [DOI] [PubMed] [Google Scholar]
- 23.Shen K., Fetter R. D., Bargmann C. I. (2004) Cell 116, 869–881 [DOI] [PubMed] [Google Scholar]
- 24.Fischbach K. F., Linneweber G. A., Andlauer T. F., Hertenstein A., Bonengel B., Chaudhary K. (2009) J. Neurogenet. 23, 48–67 [DOI] [PubMed] [Google Scholar]
- 25.Niwa H., Yamamura K., Miyazaki J. (1991) Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 26.Hatanaka Y., Murakami F. (2002) J. Comp. Neurol. 454, 1–14 [DOI] [PubMed] [Google Scholar]
- 27.Tanaka D. H., Yanagida M., Zhu Y., Mikami S., Nagasawa T., Miyazaki J., Yanagawa Y., Obata K., Murakami F. (2009) J. Neurosci. 29, 1300–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y., Yamauchi J., Kaziro Y., Itoh H. (1999) J. Biochem. 125, 515–521 [DOI] [PubMed] [Google Scholar]
- 29.Zhu Y., Matsumoto T., Mikami S., Nagasawa T., Murakami F. (2009) Development 136, 1919–1928 [DOI] [PubMed] [Google Scholar]
- 30.Li H., Edlund H. (2001) Dev. Biol. 240, 247–253 [DOI] [PubMed] [Google Scholar]
- 31.Kawauchi D., Saito T. (2008) Dev. Biol. 322, 345–354 [DOI] [PubMed] [Google Scholar]
- 32.Andrews N. C., Faller D. V. (1991) Nucleic Acids Res. 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cockell M., Stevenson B. J., Strubin M., Hagenbüchle O., Wellauer P. K. (1989) Mol. Cell. Biol. 9, 2464–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masui T., Long Q., Beres T. M., Magnuson M. A., MacDonald R. J. (2007) Genes Dev. 21, 2629–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiebe P. O., Kormish J. D., Roper V. T., Fujitani Y., Alston N. I., Zaret K. S., Wright C. V., Stein R. W., Gannon M. (2007) Mol. Cell. Biol. 27, 4093–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beltcheva O., Kontusaari S., Fetissov S., Putaala H., Kilpeläinen P., Hökfelt T., Tryggvason K. (2003) J. Am. Soc. Nephrol. 14, 352–358 [DOI] [PubMed] [Google Scholar]
- 37.Beres T. M., Masui T., Swift G. H., Shi L., Henke R. M., MacDonald R. J. (2006) Mol. Cell. Biol. 26, 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hori K., Cholewa-Waclaw J., Nakada Y., Glasgow S. M., Masui T., Henke R. M., Wildner H., Martarelli B., Beres T. M., Epstein J. A., Magnuson M. A., MacDonald R. J., Birchmeier C., Johnson J. E. (2008) Genes Dev. 22, 166–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henke R. M., Savage T. K., Meredith D. M., Glasgow S. M., Hori K., Dumas J., MacDonald R. J., Johnson J. E. (2009) Development 136, 2945–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helms A. W., Battiste J., Henke R. M., Nakada Y., Simplicio N., Guillemot F., Johnson J. E. (2005) Development 132, 2709–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmén T., Ahola H., Palgi J., Aaltonen P., Luimula P., Wang S., Jaakkola I., Knip M., Otonkoski T., Holthöfer H. (2001) Diabetologia 44, 1274–1280 [DOI] [PubMed] [Google Scholar]
- 42.Rose S. D., Swift G. H., Peyton M. J., Hammer R. E., MacDonald R. J. (2001) J. Biol. Chem. 276, 44018–44026 [DOI] [PubMed] [Google Scholar]
- 43.Sun C., Kilburn D., Lukashin A., Crowell T., Gardner H., Brundiers R., Diefenbach B., Carulli J. P. (2003) Genomics 82, 130–142 [DOI] [PubMed] [Google Scholar]
- 44.Wagner N., Wagner K. D., Xing Y., Scholz H., Schedl A. (2004) J Am. Soc. Nephrol. 15, 3044–3051 [DOI] [PubMed] [Google Scholar]
- 45.Ristola M., Arpiainen S., Saleem M. A., Mathieson P. W., Welsh G. I., Lehtonen S., Holthöfer H. (2009) BMC Mol. Biol. 10, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shih N. Y., Li J., Cotran R., Mundel P., Miner J. H., Shaw A. S. (2001) Am. J. Pathol. 159, 2303–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyata T., Kawaguchi A., Okano H., Ogawa M. (2001) Neuron 31, 727–741 [DOI] [PubMed] [Google Scholar]
- 48.Huber T. B., Schmidts M., Gerke P., Schermer B., Zahn A., Hartleben B., Sellin L., Walz G., Benzing T. (2003) J. Biol. Chem. 278, 13417–13421 [DOI] [PubMed] [Google Scholar]
- 49.Garg P., Verma R., Nihalani D., Johnstone D. B., Holzman L. B. (2007) Mol. Cell. Biol. 27, 8698–8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.