Abstract
In Saccharomyces cerevisiae, lysine 4 on histone H3 (H3K4) is methylated by the Set1 complex (Set1C or COMPASS). Besides the catalytic Set1 subunit, several proteins that form the Set1C (Swd1, Swd2, Swd3, Spp1, Bre2, and Sdc1) are also needed to mediate proper H3K4 methylation. Until this study, it has been unclear how individual Set1C members interact and how this interaction may impact histone methylation and gene expression. In this study, Bre2 and Sdc1 are shown to directly interact, and it is shown that the association of this heteromeric complex is needed for proper H3K4 methylation and gene expression to occur. Interestingly, mutational and biochemical analysis identified the C terminus of Bre2 as a critical protein-protein interaction domain that binds to the Dpy-30 domain of Sdc1. Using the human homologs of Bre2 and Sdc1, ASH2L and DPY-30, respectively, we demonstrate that the C terminus of ASH2L also interacts with the Dpy-30 domain of DPY-30, suggesting that this protein-protein interaction is maintained from yeast to humans. Because of the functionally conserved nature of the C terminus of Bre2 and ASH2L, this region was named the SDI (Sdc1 Dpy-30 interaction) domain. Finally, we show that the SDI-Dpy-30 domain interaction is physiologically important for the function of Set1 in vivo, because specific disruption of this interaction prevents Bre2 and Sdc1 association with Set1, resulting in H3K4 methylation defects and decreases in gene expression. Overall, these and other mechanistic studies on how H3K4 methyltransferase complexes function will likely provide insights into how human MLL and SET1-like complexes or overexpression of ASH2L leads to oncogenesis.
Introduction
In Saccharomyces cerevisiae, histone H3 lysine 4 (H3K4)3 mono-, di-, and trimethylation is mediated by the Set1 histone methyltransferase (1, 2). Set1 and H3K4 methylation were first identified to be required for silencing at rDNA, telomeres, and mating-type loci (3–6). Set1 and H3K4 trimethylation are also found to be located at euchromatin and enriched in the 5′-ends of coding regions of transcriptionally active genes (7, 8). In addition, Set1 and its human homologs, MLL1, MLL2, and hSET1, associate with the phosphorylated C-terminal domain of RNA polymerase II (7, 9–11). More recently, it has been shown that various chromodomain and PHD zinc finger-containing proteins can bind to histones and H3K4 di- and trimethylated histone peptides and histones (12–14). Therefore, these and other studies suggest that Set1-mediated histone methylation plays a positive role in mediating gene expression through recruitment of effector proteins.
Unlike other yeast histone methyltransferases, Set1 requires a multiprotein complex for its activity in vitro and in vivo (1, 15–18). The yeast Set1 complex, also called Set1C or COMPASS (complex associated with Set1), consists of eight subunits (Set1, Bre2, Sdc1, Swd1, Swd2, Swd3, Spp1, and Shg1) (15, 18, 19). After the discovery of yeast Set1C, it was determined that the human H3K4 methyltransferases MLL1 through 4 and human SET1 (SET1A and SET1B) form yeast Set1-like complexes (2, 9, 20–26). The human MLL and SET1 complexes share most of their core subunits, and these are homologous to the subunits found in the yeast Set1 complex. The core subunits consist of ASH2/ASH2L (Bre2), DPY30 (Sdc1), RBBP5 (Swd1), WDR82 (Swd2), WDR5 (Swd3), and CFP1/CGBP (Spp1) (9, 24–27). To date, WDR82 has only been identified in the human SET1 complexes and not in the MLL complexes (2, 11, 23, 24, 28). In yeast, deletion of the individual Set1-associated genes BRE2, SDC1, SWD1, SWD3, and SPP1 disrupts, to varying degrees, the levels of H3K4 mono-, di-, or trimethylation in cells (15–17, 29). In addition, disruption of the human MLL and SET1 complex members WDR5, RBBP5, and ASH2L leads to decreases in H3K4 trimethylation, whereas embryonic stem cells lacking CFP1/CGBP exhibit an increase in H3K4 trimethylation (20, 21, 30, 31). These results suggest that the conserved Set1C subunits in addition to the SET domain catalytic subunit are essential for maintaining proper H3K4 methylation.
Several of the yeast and human complex members have been shown to interact with each other. In yeast lacking Set1, the only two heteromeric Set1C subunit interactions that remain intact are a Swd1-Swd3 heteromer and a Bre2-Sdc1 heteromer (18, 32). Bre2 and Sdc1 have also been shown to interact via bacterial two-hybrid analysis, and a Bre2-Sdc1 heteromeric association is needed for them to interact with Set1 (18, 32). Interestingly, deletion of either of the Set1C heteromer subunits displays similar defects in H3K4 methylation (15–17, 29). Deletion of SWD1 or SWD3 leads to global loss of H3K4 mono-, di-, and trimethylation similar to a SET1 deletion, whereas deletion of BRE2 or SDC1 results in global loss of H3K4 trimethylation and reductions in mono- and dimethylation (15–17, 29). However, how individual yeast or human Set1 complex members interact with each other mechanistically and how their interaction may impact histone methylation and gene expression has not been thoroughly investigated.
In this study, we focus on defining the interaction of Bre2 and Sdc1 and how this interaction impacts histone methylation and gene expression. We demonstrate that Bre2 and Sdc1 can directly interact with each other without additional yeast proteins. In addition, we identify that the C termini of Bre2 and ASH2L are required to bind to the Dpy-30 domain of Sdc1 and DPY-30, respectively. Because this domain appears to be functionally conserved from yeast to humans, we named this new domain the SDI (Sdc1 Dpy-30 interaction) domain. More importantly, disrupting the interaction between the SDI and Dpy-30 domains prevents Bre2 and Sdc1 association with Set1 and results in defective histone H3K4 methylation and gene expression. These results show the fundamental importance of this new and conserved protein-protein interaction domain.
EXPERIMENTAL PROCEDURES
Plasmids and Strains
All yeast strains and plasmids used in this study are described in the supplemental material (see supplemental Tables S1 and S2). Bacterial and yeast plasmids for full-length BRE2 and SDC1 were made by PCR amplification of yeast genomic DNA. PCR-amplified BRE2 was subcloned into a modified pET28b vector (pET28b-9) and a pGEX-KG vector to generate pPFS25 and pPFS52. SDC1 was subcloned into a pGEX-2T and pET28b-9 vector to generate pPFS15 and pPFS40, respectively. Constructs of BRE2 and SDC1 for in vivo methylation studies were engineered with a 5′-hemagglutinin (HA) sequence and subcloned into the yeast expression vector pRS415 containing an ADH1 promoter (pRS415/ADH1p) to make pPFS1 and pPFS2, respectively. Bre2 and Sdc1 constructs for co-immunoprecipitation studies were engineered with an N-terminal HA tag or FLAG tag by PCR and subcloned into a yeast expression vector under the control of the ADH1 promoter with a URA3- or LEU2-selectable marker to make pPFS45, pPFS46, or pPFS43. Plasmid constructs, pPFS44 and pPFS47, were generated by site-directed mutagenesis using pPFS43 as a template. ASH2L was subcloned into the pET28b-9 vector to generate pPFS36, and DPY-30 was subcloned into the pGEX2T vector to generate pPFS40. Full-length human ASH2L and DPY-30 were PCR-amplified from a human cDNA library. All bre2, ash2l, sdc1, and dpy-30 deletions and point mutations were generated either by PCR amplification or by using the Stratagene QuikChange site-directed mutagenesis method using BRE2-, ASH2L-, SDC1-, and DPY-30-containing plasmids as templates.
In Vitro Binding Analysis
GST, GST-Sdc1, His6-Bre2, and His6-Sdc1 constructs were bacterially expressed as described previously (33, 34). GST pull-down assays were done as described previously (33, 34). Briefly, GST and GST fusion proteins were bound to glutathione-agarose beads and washed with Buffer A (300 mm NaCl, 50 mm Tris-HCl pH 8.0, 1 mm phenylmethylsufonyl fluoride, and 1 μg/ml leupeptin, aprotinin, and pepstatin A). After washing, 4 μg of immobilized GST fusion proteins were incubated with 400 μl of bacterial whole cell extract containing His6-tagged proteins. GST fusion protein binding reactions were performed in Buffer A and incubated for 1 h at 4 °C with rotation. After 1 h, the beads were then washed three times in 1 ml of Buffer A and then resuspended and boiled in 10 μl of 5× SDS sample buffer with 10% β-mercaptoethanol. All 10 μl of each sample was loaded per lane on a 12% SDS-polyacrylamide gel, transferred to PVDF membrane, and immunoblotted using an α-His antibody (catalog number sc-8036, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)). After immunoblot analysis, the PVDF membrane was stained with Coomassie Blue stain to visualize GST and GST fusion proteins. GST pull-down assays using recombinant GST-DPY-30 and His6-ASH2L were performed as described above with the following modification. 2 μg of GST-DPY-30 bound to glutathione-agarose beads and 150 μl of bacteria whole cell extract containing His6-ASH2L or ASH2L mutants were used in the binding reaction.
Sdc1 Dimerization and Binding Assays
Bacteria whole cell extract containing His6-Sdc1 or His6-Sdc1 C47A was prepared as described above. His6-Sdc1 or His6-Sdc1 C47A was purified using nickel-agarose beads (Ni2+-NTA-agarose, Qiagen) from whole cell extracts according to the manufacturer's protocol. 2× SDS sample buffer with or without β-mercaptoethanol was added to Ni2+-NTA-agarose beads containing His6-Sdc1 or His6-Sdc1 C47A. The eluted purified samples were analyzed by SDS-PAGE and transferred to a PVDF membrane for immunoblot analysis using α-His antibody (catalog number sc-8036, Santa Cruz Biotechnology, Inc.) or stained with Coomassie Blue. Nickel-agarose binding assays were performed similarly to the GST fusion binding assays. His6-Sdc1 or His6-Sdc1 C47A was immobilized on Ni2+-NTA beads (Qiagen) and washed with buffer A (300 mm NaCl, 50 mm Tris-HCl, pH 8.0, 1 mm phenylmethylsufonyl fluoride, and 1 μg/ml leupeptin, aprotinin, and pepstatin A). Bacteria whole cell extract containing GST-Bre2 was incubated with the Ni2+-NTA beads for 1 h at 4 °C with rotation. After three washes in buffer A, pelleted samples were boiled in 10 μl of 5× SDS sample buffer with 10% β-mercaptoethanol and run on 12% SDS-polyacrylamide gels. For detection of His6- and GST-tagged Sdc1 and Bre2, SDS-polyacrylamide gels were transferred to PVDF and immunoblotted with α-His antibody (catalog number sc-8036, Santa Cruz Biotechnology, Inc.) or α-GST antibody (catalog number sc-138, Santa Cruz Biotechnology, Inc.).
Yeast Extraction and Immunoblot Analysis
Yeast extraction and immunoblot analysis to detect modified histones were performed as previously described (5). α-H3K4me1 (catalog number 07-436), α-H3K4me2 (catalog number 07-030), and α-H3K4me3 (catalog number 07-473) were obtained from Millipore and were used at 1:2500, 1:5000, and 1:5000 dilutions, respectively. The H3 (catalog number Ab1791, Abcam)-specific antibody at a 1:5000 dilution was used as a loading control. HA.11 (catalog number MMS-101R, Covance) and 12CA5 (catalog number 1666606, Roche Applied Science) antibodies were used at a 1:5000 dilution to detect HA-tagged Bre2 and Sdc1, respectively.
Co-immunoprecipitation Analysis
For co-immunoprecipitation of HA-Bre2 and FLAG-Sdc1, 50-ml yeast cultures were grown in select media to mid-log phase. Yeast whole cell extracts were prepared by bead beating in 400 μl of lysis buffer (40 mm HEPES-NaOH, pH 7.5; 350 mm NaCl; 0.1% Tween 20; 10% glycerol; 1 μg of leupeptin, aprotinin, and pepstatin A; 1 mm phenylmethylsulfonyl fluoride), and protein levels were normalized using a Bradford assay as described previously (35). To each sample, 10 μl of M2 α-FLAG resin (catalog number A2220; Sigma) was added to normalized extracts and incubated for 3 h at 4 °C with rotation. The immunoprecipitated samples were washed three times in 1 ml of lysis buffer. The remaining beads from the final wash were resuspended in 2× SDS sample buffer with 10% β-mercaptoethanol and boiled. Each immunoprecipitated sample was split in half, and 5 μl of sample was loaded per lane on a 12% SDS-polyacrylamide gel, transferred to PVDF membrane, and immunoblotted using α-HA polyclonal antibody (catalog number sc-805, Santa Cruz Biotechnology, Inc.) or α-FLAG polyclonal antibody (catalog number F7425, Sigma). Yeast cell extracts for loading inputs were detected using α-HA (HA.11, catalog number MMS-101R, Covance) and α-FLAG (catalog number F7425; Sigma) antibodies.
For immunoprecipitation of Myc3-Set1, the above procedure was performed with the following modifications. To each sample, 2 μg of α-Myc antibody (9E10, catalog number 11667203001, Roche Applied Science) was added to normalized extracts and incubated at 4 °C for 2 h. After 2 h, 12 μl of Protein G-Sepharose (catalog number 17-0618-01, GE Healthcare) was added to each sample and incubated for 1 h at 4 °C. The immunoprecipitated samples were washed three times with 1 ml of lysis buffer, and the remaining beads were resuspended in 10 μl of 2× SDS sample buffer with 10% β-mercaptoethanol and boiled. Samples were analyzed on an 8% SDS-polyacrylamide gel, transferred to a PVDF membrane, and immunoblotted with α-Myc antibody (9E10, catalog number 11 667 203 001, Roche Applied Science). For co-immunoprecipitation of HA-Bre2 and FLAG-Sdc1 by Myc3-Set1, Myc3-Set1 was immunoprecipitated, as described, and analyzed on a 12% SDS-polyacrylamide gel, transferred to PVDF membrane, and immunoblotted using α-HA polyclonal antibody (catalog number sc-805, Santa Cruz Biotechnology, Inc.) or α-FLAG polyclonal antibody (catalog number F7425, Sigma).
Gene Expression Analysis
Quantitative real-time PCRs were performed as previously described (35). Briefly, 6-ml cultures of yeast cells were grown to mid-log phase in selective media. Total RNA was extracted using the RNeasy kit (Qiagen) following the manufacturer's protocol, and 1 μg of RNA was reverse transcribed into cDNA using the Quantitec reverse transcription kit (Qiagen) according to the manufacturer's protocol. The real-time PCR mix consisted of 6.25 μl of SYBR green master mix (Applied Biosystems), 0.125 μl of each forward and reverse primer from a 5 μm stock, 0.5 μl of cDNA, and 5.5 μl of sterile water. DNA was amplified in a StepOne real-time PCR system (Applied Biosystems) with the following program: an initial hold of 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Three biological repeats, each including three technical repeats, were performed for all samples. Data were analyzed using the ΔΔCt method in which actin (ACT1) was used as the endogenous control, and relative quantity of MDH2 and GUA1 transcript was determined for each deletion strain compared with MDH2 and GUA1 transcript in a wild-type strain. Primer sequences used for PCR amplification are described in supplemental Table S3.
Chromatin Immunoprecipitation Analysis
Chromatin immunoprecipitation (ChIP) was performed as previously described (33, 35). Briefly, 100-ml cultures were grown to mid-log phase in selective media overnight and split in half. Cells were harvested and cross-linked with 1% formaldehyde. ChIP was performed using either H3K4 di- or trimethyl-specific antibodies (catalog numbers 07-030 and 07-473, Millipore) or an antibody specific to H3 (catalog number Ab1791, Abcam) and Protein G-Sepharose (GE Healthcare). Immunoprecipitated DNA was amplified by standard PCR methods using primer sets to analyze the 5′- and 3′-regions of the MDH2 and GUA1 open reading frames. Amplified PCR products were analyzed by agarose gel and ethidium bromide staining. Primers used in ChIP analysis are described in supplemental Table S4.
RESULTS
The Dpy-30 Domain of Sdc1 Is Required for Interaction with Bre2
In yeast lacking Set1, the Set1 complex is no longer intact, and only two heteromeric subunit associations remain. These include an association between Bre2 and Sdc1 and an association between Swd1 and Swd3 (18). More recently, Bre2 and Sdc1 have also been suggested to interact by bacterial two-hybrid analysis (32). Together, these studies indicate that the Bre2 and Sdc1 subunits directly interact. Until now, how Bre2 and Sdc1 or Swd1 and Swd3 interact has not been established. In this study, we focus on Bre2 and Sdc1 interaction and the functional significance of this interaction on histone methylation and gene expression.
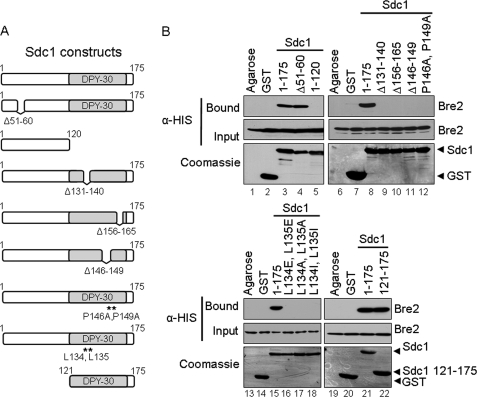
To investigate if Bre2 and Sdc1 directly interact, in vitro GST fusion binding assays were performed in which Escherichia coli extracts expressing recombinant His6-Bre2 were incubated with purified GST-Sdc1 fusion protein, GST, or glutathione-agarose beads. We observed that GST-Sdc1 could pull down His6-Bre2, indicating direct interaction (Fig. 1B, lane 3). This binding was specific to a Bre2 and Sdc1 protein-protein interaction because Bre2 was not detected when incubated with glutathione-agarose- or GST-containing beads (Fig. 1B, lanes 1 and 2). To determine the region of interaction, mutational analysis and GST fusion binding assays were used to determine which regions in Sdc1 were important for interacting with Bre2 (Fig. 1A). We initially hypothesized that the SPRY domain of Bre2 would be important for this interaction. The SPRY domain was first identified in the tyrosine kinase spore lysis A (SplA) and in the mammalian ryanodine receptor (RyR) from which it was named (36). Because SPRY domain-containing proteins have recently been shown to interact with proteins containing asparagine-rich sequences, we looked at the primary sequence of Sdc1 for asparagine residues (37, 38). Upon sequence analysis of Sdc1, we identified an N-terminal asparagine-rich region from amino acids 51–60 containing six asparagines. In addition, the Dpy-30 domain of Sdc1, which is a domain that was initially found in DPY-30 proteins required for sex determination and dosage compensation in Caenorhabditis elegans, has also been predicted to be a protein-protein interaction domain (39, 40). Dpy-30 domains are similar to RIIa domains found in cAMP protein kinase A regulatory subunits that are known to dimerize (41–43). Therefore, the Dpy-30 domain of Sdc1 could be facilitating dimerization or interactions with Bre2. To determine if the Sdc1 asparagine-rich sequence or the Dpy-30 domain was responsible for binding to Bre2, GST-Sdc1 deletion constructs were generated lacking these regions (Sdc1 Δ51–60 and Sdc1 1–120) (Fig. 1A). To our surprise, GST-Sdc1 Δ51–60 lacking the asparagine-rich sequence was still able to bind Bre2, similar to when full-length Sdc1 is used (Fig. 1B, compare lanes 3 and 4). However, GST-Sdc1 1–120 lacking the Dpy-30 domain was unable to bind to Bre2, suggesting that the Dpy-30 domain is required for Bre2 interaction (Fig. 1B, lane 5).
FIGURE 1.
The Dpy-30 domain of Sdc1 is required for interaction with Bre2 in vitro. A, schematic representation of Sdc1 and Sdc1 mutants used. The position of the Dpy-30 domain is indicated. B, GST fusion binding assays were performed to determine the region within Sdc1 responsible for binding Bre2. Purified GST-Sdc1 or GST-Sdc1 mutants were used to pull down bacterially expressed His6-Bre2 from whole cell extracts. GST and glutathione-agarose beads (Agarose) were used as controls for nonspecific binding. Input and bound fractions of His6-Bre2 were detected by immunoblotting using an α-His monoclonal antibody. Immunoblots were stained with Coomassie Blue to indicate the amount of GST, GST-Sdc1, and GST-Sdc1 mutant proteins used.
Modeling of the yeast Dpy-30 domain suggests that it forms a structure with an α-helix followed by a turn and then another α-helix (44). Interestingly, GST fusion binding assays using Sdc1 deletions (Sdc1 Δ131–140 and Sdc1 Δ156–165) showed that disrupting either predicted α-helix abolishes binding to Bre2 (Fig. 1, A and B, lanes 9 and 10). Because the deletions within the predicted α-helices of the Dpy-30 domain abolished binding to Bre2, we wanted to mutate conserved residues located in one of the predicted α-helices. Based on sequence alignment between various organisms, we identified two highly conserved leucine residues, Leu-134 and Leu-135 (Fig. 1A and supplemental Fig. S1A). These residues are located in the first predicted α-helix of the Dpy-30 domain of Sdc1. Interestingly, based on the x-ray crystal structure of the human Dpy-30 domain of DPY-30, it has been suggested that these two conserved leucine residues are probably forming a putative binding pocket (45). Similar to deletions within the predicted α-helix, all Sdc1 Leu-134 and Leu-135 mutations (Sdc1 L134E,L135E, Sdc1 L134A,L135A, and Sdc1 L134I, L135I) disrupted binding to Bre2, suggesting that the Dpy-30 domain of Sdc1 might be sufficient to mediate binding to Bre2 (Fig. 1B, lanes 16–18). In addition, another highly conserved region found in the Dpy-30 domain of Sdc1 is a PXXP motif, which is likely to form a turn and link the two predicted α-helices. Similar to deleting the conserved leucine residues, a PXXP deletion (Sdc1 Δ146–149) is also unable to bind to Bre2 (Fig. 1, A and B, lane 11). Furthermore, Sdc1 point mutations (Sdc1 P146A and P149A) converting the PXXP motif to an AXXA motif also completely disrupt binding of Bre2 to Sdc1, indicating that the PXXP motif is another important conserved region of the Dpy-30 domain (Fig. 1, A and B, lane 12). Because mutations in conserved amino acids in the predicted turn region and α-helix region of the Dpy-30 domain of Sdc1 disrupted binding to Bre2, we wanted to determine if the Dpy-30 domain in itself was sufficient in binding to Bre2 in vitro. Two sizes for the yeast Dpy-30 domain have been reported based on sequence alignments with conserved residues, with one being 41 amino acids in size by Pfam and the other 50 amino acids long published by Dong et al. (46) (supplemental Fig. S1A). Therefore, a GST-Sdc1 fusion construct representing the Dpy-30 domain of Sdc1 and the remaining C terminus of Sdc1 (Sdc1 121–175) was generated and used in a GST binding assay (Fig. 1A). Our results indicated that the Dpy-30 domain of Sdc1 is sufficient to bind full-length Bre2 (Fig. 1B, lane 22).
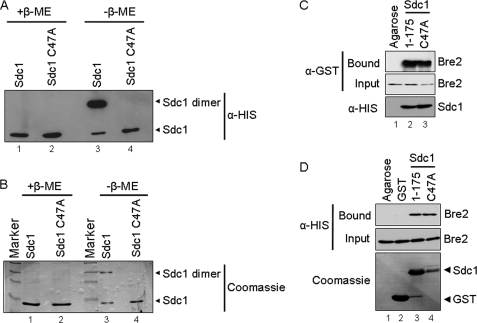
Previous bacterial two-hybrid results suggest that Sdc1 might dimerize (32). In addition, the DPY-30 crystal structure was solved as a dimer, and RIIa domains have also been shown to dimerize; therefore, we sought to determine if bacterially expressed Sdc1 can dimerize in vitro (41, 43, 45). Interestingly, Sdc1 dimers were detected on non-reducing polyacrylamide gels and disrupted in the presence of the sulfhydryl-reducing agent β-mercaptoethanol, suggesting that a cysteine residue is mediating this event (Fig. 2, A and B). Because only one cysteine is present in Sdc1, we mutated it to an alanine (Sdc1 C47A); as expected, this point mutation disrupted dimerization when run on non-reducing gels (Fig. 2, A and B). To determine if Sdc1 disulfide-mediated dimerization was important for binding to Bre2, a GST-Bre2 fusion protein was incubated with a His6-Sdc1 C47A point mutation (Fig. 2C). In addition, GST-Sdc1 and GST-Sdc1 C47A were incubated with His6-Bre2 (Fig. 2D). In both cases, the Sdc1 C47A point mutation, which failed to dimerize in vitro, was still capable of binding Bre2, suggesting that disulfide-mediated dimerization does not facilitate interaction between Sdc1 and Bre2 (Fig. 2, C and D). We were also unable to detect Sdc1 cysteine-mediated dimers when isolated from yeast (data not shown), suggesting that cysteine-mediated dimerization of Sdc1 is probably an artifact of bacterial expression. Altogether, our results show that Bre2 and Sdc1 can directly interact in vitro and indicate that the Dpy-30 domain of Sdc1 is functionally necessary and sufficient for interaction with Bre2.
FIGURE 2.
Sdc1 forms a dimer by a disulfide interaction. A, immunoblot analysis of purified His6-Sdc1 and His6-Sdc1 C47A in the presence and absence of the reducing agent β-mercaptoethanol (β-ME). B, Coomassie Blue stain of purified His6-Sdc1 and His6-Sdc1 C47A in the presence and absence of the reducing agent β-mercaptoethanol. Bands representing Sdc1 monomer and dimer are labeled accordingly. C, nickel-agarose binding assays were performed to determine if Sdc1 dimerization affected interaction with Bre2. Purified His6-Sdc1 and His6-Sdc1 C47A were used to pull down bacterially expressed GST-Bre2 from whole cell extracts. Ni2+-NTA-agarose beads were used as a control for nonspecific binding. Input and bound fraction of GST-Bre2, His6-Sdc1, and His6-Sdc1 C47A were detected by immunoblotting with α-GST or α-His monoclonal antibodies. D, GST fusion binding assays were performed to determine if the GST-Sdc1 C47A mutation affected binding of His6-Bre2. Purified GST-Sdc1 or GST-Sdc1 C47A was used to pull down bacterially expressed His6-Bre2 from whole cell extracts. Glutathione-agarose- or GST-bound beads were used as a control for nonspecific binding. Input and bound fractions of His6-Bre2, GST, GST-Sdc1, and GST-Sdc1 C47A were detected by immunoblotting with α-His monoclonal antibodies and Coomassie Blue staining of the immunoblot.
The C Terminus of Bre2 Is Required for Binding Sdc1
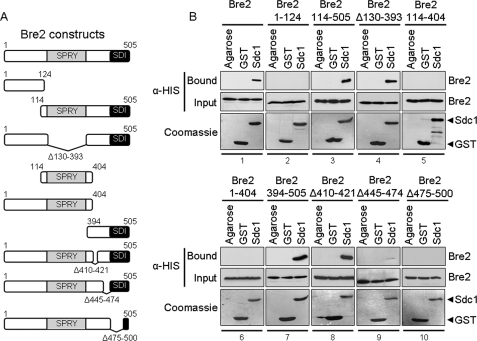
Because we determined that the Dpy-30 domain was necessary for Sdc1 interaction with Bre2, we wanted to determine the region of Bre2 important for interacting with Sdc1. The only known putative protein-protein interaction domain found within Bre2 is the SPRY domain (15, 18). Therefore, we predicted that the conserved SPRY domain of Bre2 would be the site of Sdc1 binding. Utilizing the GST fusion binding assay, GST-Sdc1, GST, and glutathione-agarose beads were incubated with extracts expressing either full-length His6-Bre2 or various His6-Bre2 truncation and deletion mutants (Fig. 3A). Again, GST-Sdc1 was able to associate with full-length Bre2 but not with GST or glutathione-agarose (Fig. 3B, panel 1). However, a Bre2 C-terminal truncation mutant (Bre2 1–124) lacking the SPRY domain and the C terminus was unable to bind to Sdc1 (Fig. 3B, panel 2). In addition, a Bre2 truncation mutant (Bre2 114–505) lacking the N terminus of Bre2 but retaining the SPRY domain and C terminus of Bre2 was able to bind to Sdc1 (Fig. 3B, panel 3). These results suggest the possibility that the SPRY domain or the C terminus of Bre2 is the domain necessary for binding Sdc1.
FIGURE 3.
The C terminus of Bre2 is required for interaction with Sdc1 in vitro. A, schematic representation of Bre2 and Bre2 mutants used. The positions of the SPRY domain and SDI domain are indicated. B, GST fusion binding assays were performed to determine the region within Bre2 responsible for binding Sdc1. Purified full-length GST-Sdc1 was used to pull down bacterially expressed His6-Bre2 or His6-Bre2 mutants (shown in panels 1–10) from whole cell extracts. GST and glutathione-agarose beads (Agarose) were used as controls for nonspecific binding. Input and bound fractions of His6-Bre2 were detected by immunoblotting using anti-His monoclonal antibody. Immunoblots were stained with Coomassie Blue to indicate the amount of GST and GST-Sdc1 proteins used.
To determine if the SPRY domain of Bre2 was responsible for binding to Sdc1, a Bre2 deletion mutant (Bre2 Δ130–393) lacking the SPRY domain and a Bre2 truncation mutant (Bre2 114–404) lacking the N and C terminus but containing the SPRY domain and 10 residues of surrounding flanking sequences were generated and tested for interaction with Sdc1 (Fig. 3A). Surprisingly, Sdc1 could still bind the Bre2 deletion mutant lacking the SPRY domain (Bre2 Δ130–393) but was not able to bind the Bre2 N- and C-terminal truncation mutant containing an intact SPRY domain (Bre2 114–404) (Fig. 3B, compare panels 4 and 5). These results clearly indicated that that the SPRY domain is not necessary for interaction with Sdc1 and suggest that an unknown domain in the C terminus of Bre2 is needed for the Bre2 and Sdc1 interaction.
To confirm that the C terminus of Bre2 is the region to which Sdc1 binds, a Bre2 truncation mutant lacking the C terminus (Bre2 1–404) and a Bre2 mutant lacking the N terminus (Bre2 394–505) were generated and tested for binding to Sdc1 (Fig. 3A). Again, a Bre2 truncation mutant lacking its C terminus (Bre2 1–404) was unable to bind Sdc1, whereas the Bre2 mutant lacking the N terminus (Bre2 394–505) could still bind Sdc1 (Fig. 3B, panels 6 and 7). This suggests that the C terminus of Bre2 is important for Sdc1 interaction. To further define the C-terminal region in Bre2 responsible for binding Sdc1, three Bre2 deletion constructs (Bre2 Δ410–421, Bre2 Δ445–474, and Bre2 Δ475–500) were generated (Fig. 3A). Although the Bre2 deletion mutant (Bre2 Δ410–421) could bind to Sdc1, similar to wild-type Bre2, Bre2 Δ445–474 had a significant defect in its ability to bind Sdc1 (Fig. 3B, panels 8 and 9). However, Sdc1 binding was completely abolished when using the Bre2 Δ475–500 deletion mutant, indicating that these amino acids of Bre2 are key determinants for interacting with Sdc1 (Fig. 3B, panel 10).
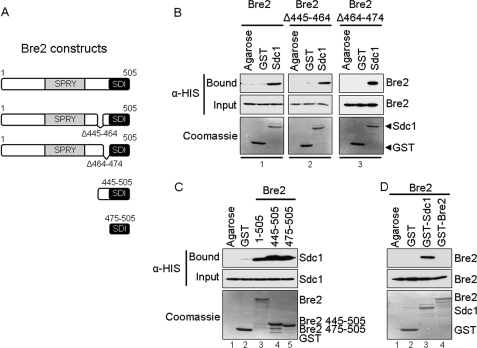
Because the Bre2 Δ445–474 deletion mutant almost abolishes binding to Sdc1, we wanted to further define the region of Bre2 needed for binding Sdc1. Therefore, two smaller Bre2 C-terminal deletions were generated (Bre2 Δ445–464 and Bre2 Δ464–474) and tested for their ability to bind Sdc1 (Fig. 4A). Interestingly, both Bre2 deletions (Bre2 Δ445–464 and Bre2 Δ464–474) were capable of binding to Sdc1 similar to full-length Bre2 (Fig. 4B, compare panels 1–3). Therefore, the Bre2 Δ445–474 deletion mutant that nearly abolishes interaction with Sdc1 is likely due to the size of the deletion and is probably disrupting the ability of the C-terminal residues 475–505 of Bre2 to interact with Sdc1.
FIGURE 4.
The SDI domain of Bre2 is sufficient for interaction with Sdc1 in vitro. A, schematic representation of Bre2 and Bre2 mutants used. Domains are indicated as in Fig. 3. B, GST fusion binding assays were performed to determine regions necessary for interaction with Sdc1. Purified full-length GST-Sdc1 was used to pull down bacterially expressed His6-Bre2 or His6-Bre2 mutants from whole cell extracts. C, GST fusion binding assays were performed to determine the Bre2 region sufficient for interacting with Sdc1. Purified full-length GST-Bre2 or GST-Bre2 truncation mutants were used to pull down bacterially expressed His6-Sdc1 from whole cell extracts. D, a GST fusion binding assay was performed to determine if Bre2 is able to form a homodimer in vitro. Purified full-length GST-Sdc1 or GST-Bre2 was used to pull down bacterially expressed His6-Bre2 from whole cell extracts. Controls for B, C, and D were similar. GST and glutathione-agarose beads (Agarose) were used as controls for nonspecific binding. Input and bound fractions were detected by immunoblotting using α-His monoclonal antibody. Immunoblots were stained with Coomassie Blue to indicate the amount of GST and GST fusion proteins used.
Our data show that C-terminal residues 475–500 of Bre2 are necessary for interaction with Sdc1 (Fig. 3B, panel 10). However, to determine if this region is sufficient for interacting with Sdc1, two Bre2 truncation mutants (Bre2 445–505 and Bre2 475–505) were generated and tested for binding to Sdc1 (Fig. 4A). Our results show that Bre2 445–505 and Bre2 475–505 can bind Sdc1 similar to full-length Bre2 (Fig. 4C, compare lanes 3–5). Therefore, the C-terminal region of Bre2 encompassing amino acid residues 475–505 is not only necessary but also sufficient for interaction with the Dpy-30 domain of Sdc1. In summary, our in vitro results demonstrate that the C terminus of Bre2 and the Dpy-30 domain of Sdc1 are both necessary and sufficient for binding to Sdc1 and Bre2, respectively. Furthermore, we have defined a novel protein-protein interaction domain in Bre2 that probably consists of amino acids 475–505, which we have designated the SDI domain.
Previous work has suggested that the Set1 complex may contain two copies of Bre2 by a Bre2 dimer (1, 32). To test if Bre2 does interact with itself, we performed an in vitro pull-down assay with GST-Bre2 and His6-Bre2. Our data show that we can pull down Bre2 with Sdc1, but we are unable to detect a direct interaction of Bre2 with itself, suggesting that Bre2 does not dimerize (Fig. 4D, compare lanes 3 and 4). This result indicates that if there are multiple Bre2 subunits in the Set1 complex, they may not be interacting directly with each other as a dimer. However, additional quantitative experiments will be needed to determine the accurate stoichiometry of Bre2 within the Set1 complex.
Interaction between Human ASH2L and DPY-30 Occurs through the C Terminus of ASH2L and the Dpy-30 Domain of DPY-30
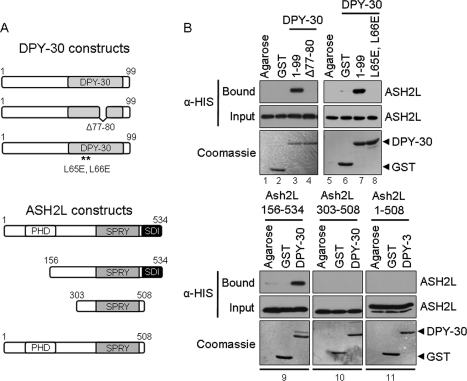
Consistent with the previous findings for the Bre2 and Sdc1 interaction, the human homolog of Bre2, ASH2L, has been shown to interact with the human homolog of Sdc1, DPY-30 (23). Similar to Bre2 and Sdc1, the regions important for how ASH2L and DPY-30 interact have not been established. In addition, no protein-protein interaction domains that mediate the interaction between ASH2L and DPY-30 have been identified. Although ASH2L has a conserved SPRY domain, it is distinct from Bre2 because it also contains a PHD zinc finger (Figs. 3A and 5A). DPY-30 and Sdc1 appear to be more similar because they both contain a Dpy-30 domain, with each being 41 amino acids in size as reported by Pfam with a 35.7% identity and 73.8% similarity (Figs. 1A and 5A and supplemental Fig. S1A) (44). GST fusion binding assays were performed to determine direct interaction between ASH2L and DPY-30. GST-DPY-30, GST, and glutathione-agarose beads were incubated with extracts expressing full-length His6-ASH2L (Fig. 5B, lanes 1–3). Our results show that ASH2L and DPY-30 can directly interact without the addition of other MLL- and SET1-associated protein factors. Similar to Sdc1, deletion of the highly conserved PXXP (Dpy-30 Δ77–80) motif within the Dpy-30 domain of DPY-30 disrupted binding of ASH2L (Fig. 5B, lane 4). Because the two highly conserved leucine residues Leu-134 and Leu-135 of the Dpy-30 domain of Sdc1 are needed for interacting with Bre2, we wanted to determine if the same conserved leucine residues Leu-65 and Leu-66 found within the Dpy-30 domain of DPY-30 are needed for interacting with ASH2L. Similar to the Sdc1 leucine mutations, a DPY-30 mutant (DPY-30 L65E,L66E) was unable to interact with ASH2L (Fig. 5B, lane 8). These results show the first functional importance of the conserved PXXP motif and leucine residues in Sdc1 and DPY-30.
FIGURE 5.
ASH2L and DPY-30 interact through their SDI and Dpy-30 domains. A, schematic representation of ASH2L and DPY-30 and corresponding mutants used. Positions of annotated domains are depicted. B, GST fusion binding assays were performed to determine critical regions for ASH2L and DPY-30 interaction. Purified full-length GST-DPY-30, GST-Dpy-30 Δ77–80, or GST-Dpy-30 L65E,L66E was used to pull down bacterially expressed His6-ASH2L or His6-Ash2L mutants from whole cell extracts. GST and glutathione-agarose beads (Agarose) were used as controls for nonspecific binding. Input and bound fractions of His6-ASH2L and His6-Ash2L mutants were detected by immunoblotting using anti-His monoclonal antibody. Immunoblots were stained with Coomassie Blue to indicate the amount of GST, GST-DPY-30, GST-Dpy-30 Δ77–80, and GST-Dpy-30 L65E,L66E proteins used.
To determine if the PHD zinc finger was mediating interaction with the Dpy-30 domain of DPY-30, the N terminus of ASH2L, which contains a PHD zinc finger, was deleted (Ash2l 156–534) and tested for binding to DPY-30. The PHD zinc finger deletion mutant could still bind to DPY-30 (Fig. 5B, panel 9), suggesting that the PHD zinc finger has other functions in the MLL and SET H3K4 methyltransferase complexes. Because the C terminus of Bre2 containing amino acids 475–500 and the last 26 amino acids of ASH2L are 25.9% identical and are 46.2% similar, we wanted to determine if the SDI domain in Bre2 was functionally conserved in ASH2L (supplemental Fig. S1B) (44). Consistent with our previous results with Bre2, binding of ASH2L with full-length DPY-30 was abolished only when the C terminus of ASH2L (Ash2L 303–508 and Ash2L 1–508) was missing (Fig. 5B, panels 10 and 11). These results suggest that the C terminus of ASH2L likely represents the SDI domain that we defined in the C terminus of Bre2. Altogether, our study defines, for the first time, the domains of interaction between the yeast and human Bre2/ASH2L and Sdc1/DPY-30 subunits. Furthermore, we define the SDI domain as a novel protein-protein interaction domain that interacts with the Dpy-30 domain and show that this interaction is functionally conserved in yeast and humans.
The Binding of Bre2 and Sdc1 in Yeast Requires the SDI-Dpy30 Domain Interaction
It has previously been reported that Bre2 and Sdc1 will co-immunoprecipitate from yeast whole cell extracts (18, 19). Therefore, we wanted to determine if the interaction between Bre2 and Sdc1 that occurs in vivo was also mediated by the SDI and Dpy-30 domain interaction that occurs in vitro. To determine this, we generated yeast expression constructs containing FLAG-Sdc1, FLAG-Sdc1 PXXP deletion mutant (FLAG-Sdc1 Δ146–149), HA-Bre2, or HA-Bre2 truncation mutant (HA-Bre2 1–404) lacking the SDI domain. All constructs were expressed from a CEN plasmid containing a constitutive ADH1 promoter (ADH1p) with either a LEU2- or URA3-selectable marker.
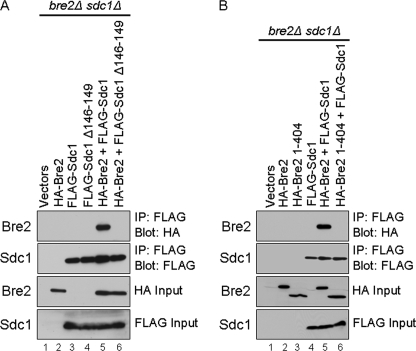
To determine if the Dpy-30 domain was responsible for mediating Sdc1 and Bre2 interactions in vivo, vector (LEU2), FLAG-Sdc1, or FLAG-Sdc1 PXXP deletion mutant (FLAG-Sdc1 Δ146–149) was coexpressed with full-length HA-Bre2 or vector (URA3) in a bre2Δsdc1Δ double deletion strain. Whole cell yeast extracts were generated, and co-IP analysis was preformed from the bre2Δsdc1Δ strain expressing these constructs. As expected, full-length HA-Bre2 was able to be co-immunoprecipitated with FLAG-Sdc1 (Fig. 6A, lane 5). This co-IP was specific for an Sdc1 and Bre2 interaction because nonspecific interactions were not detected in lanes expressing only vectors, HA-Bre2, FLAG-Sdc1, or FLAG-Sdc1 Δ146–149 (Fig. 6A, lanes 1–4, top panel). However, an Sdc1 mutant lacking its PXXP motif (FLAG-Sdc1 Δ146–149) was unable to associate with HA-Bre2 in vivo, although equivalent amounts of full-length Sdc1 and Sdc1 PXXP deletion mutant were immunoprecipitated (Fig. 6A, compare lanes 5 and 6, top two panels). These results are consistent with and support our in vitro GST fusion binding assays (Fig. 1). The inability of the Sdc1 PXXP deletion mutant (FLAG-Sdc1 Δ146–149) to co-immunoprecipitate Bre2 was not due to varying expression levels of Sdc1, Sdc1 Δ146–149 mutant, or Bre2 because nearly equivalent levels were detected (Fig. 6A, lanes 2–6, bottom two panels).
FIGURE 6.
The SDI and Dpy-30 domains are required for Bre2 and Sdc1 protein-protein interaction in vivo. A and B, whole cell extracts expressing vector or the indicated Sdc1 or Bre2 constructs in bre2Δsdc1Δ strains were immunoprecipitated (IP) with M2 α-FLAG resin. Detection of FLAG-Sdc1, FLAG-Sdc1 mutants, HA-Bre2, or HA-Bre2 mutants were determined by immunoblotting with α-FLAG or α-HA antibodies.
To determine if the SDI domain was also needed for in vivo interaction between Bre2 and Sdc1, vector (LEU2) or FLAG-Sdc1 was coexpressed with vector (URA3), full-length HA- Bre2, or HA-Bre2 C-terminal truncation mutant (HA-Bre2 1–404) lacking the SDI domain in a bre2Δsdc1Δ double deletion strain. Similar to Fig. 6A, co-immunoprecipitation analysis determined that full-length Sdc1 and Bre2 interact, whereas FLAG-Sdc1 was unable to co-immunoprecipitate HA-Bre2 1–404 lacking the SDI domain although equal amounts of Sdc1 was immunoprecipitated (Fig. 6B, compare lanes 5 and 6, top two panel). In addition, the co-IP was specific for an Sdc1 and Bre2 interaction because nonspecific interactions were not detected in lanes expressing vectors, HA-Bre2, FLAG-Sdc1, or HA-Bre2 1–404 (Fig. 6B, lanes 1–4, top panel). Again, these results are consistent with and support our in vitro binding assays (Figs. 3 and 4). The inability of Sdc1 to co-immunoprecipitate with the Bre2 mutant (Bre2 1–404) lacking the SDI domain was not due to varying expression levels of Sdc1, Bre2, or Bre2 1–404 because nearly equivalent levels were detected (Fig. 6B, lanes 2–6, bottom two panels). Altogether, our results show that the SDI-Dpy-30 protein-protein domain interaction is critical for Bre2 and Sdc1 interaction to occur in vitro and in vivo. In addition, our data would suggest that this interaction is probably important for in vivo histone H3K4 methylation.
Interaction between the Bre2 SDI and Sdc1 Dpy-30 Domains Is Required for Global H3K4 Methylation
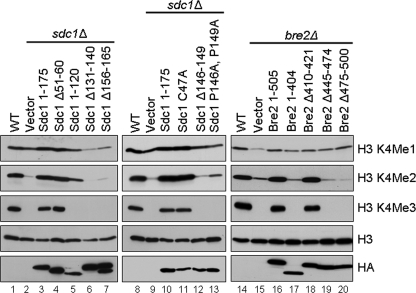
Although we have defined how Sdc1 and Bre2 interact, we wanted to determine if the interaction between the SDI and Dpy-30 domain was physiologically relevant for global H3K4 methylation. To determine if the Dpy-30 domain of Sdc1 was necessary for proper H3K4 methylation, HA-Sdc1 full-length (Sdc1 1–175), HA-Sdc1 deletion mutant (Sdc1 Δ51–60) lacking the asparagine-rich region, HA-Sdc1 C-terminal truncation mutant (Sdc1 1–120) lacking the entire Dpy-30 domain, HA-Sdc1 mutants (Sdc1 Δ131–140, Sdc1 Δ156–165, Sdc1 Δ146–149, and Sdc1 P146A,P149A) with deletions or point mutations in the Dpy-30 domain, and a HA-Sdc1 (Sdc1 C47A) point mutation were generated and expressed in the sdc1Δ strain. Immunoblot analysis of whole cell extracts using H3K4 mono-, di-, and trimethyl-specific antibodies were used to determine if the various Sdc1 constructs could rescue the H3K4 methylation defects in an sdc1Δ strain. As expected, full-length Sdc1 restored the H3K4 methylation in an sdc1Δ strain to a level similar to what is seen in wild-type (WT) cells (Fig. 7, compare lanes 1 and 3, top three panels). Interestingly, an Sdc1 deletion mutant (Sdc1 1–120) lacking the Dpy-30 domain and the ability to bind Bre2 in vitro and in vivo was unable to rescue H3K4 methylation and had H3K4 methylation levels similar to an sdc1Δ strain (Fig. 7, lanes 2 and 5). In addition, all mutations within the Dpy-30 domain of Sdc1 (Sdc1 Δ131–140, Sdc1 Δ156–165, Sdc1 Δ146–149, and Sdc1 P146A,P149A) that fail to interact with Bre2 also fail to rescue H3K4 methylation in sdc1Δ cells (Fig. 7, lanes 6, 7, 12, and 13). However, all of the Sdc1 mutants that were still able to interact with Bre2, such as the Sdc1 Δ51–60 lacking the asparagine-rich sequence or the Sdc1 C47A point mutation, were fully capable of rescuing H3K4 methylation (Fig. 7, lanes 4 and 11). These differences in H3K4 methylation patterns did not appear to be due to differences in histone H3 loading or expression of Sdc1 or Sdc1 mutants because nearly equivalent levels of histone H3 and Sdc1 expression were detected (Fig. 7, lanes 1–13, bottom two panels).
FIGURE 7.
The SDI and Dpy-30 domains are required for proper levels of global histone H3K4 methylation. Yeast whole cell extracts generated from wild-type (WT), sdc1Δ, and bre2Δ strains expressing vector, Sdc1, or Bre2 constructs were immunoblotted with histone H3K4 mono-, di-, and trimethyl-specific antibodies. Histone H3 and HA immunoblots were used for a loading control or detection of Sdc1 and Bre2 protein levels, respectively.
To investigate if the SDI domain of Bre2 was necessary for proper H3K4 methylation, full-length HA-Bre2 (Bre2 1–505), HA-Bre2 mutant (Bre2 1–404) lacking the C terminus containing the SDI domain, and HA-Bre2 deletion mutant (Bre2 Δ410–421, Bre2 Δ445–474, and Bre2 Δ474–500) constructs were generated and expressed in a bre2Δ strain. Immunoblot analysis of whole cell extracts using H3K4 mono-, di-, and trimethyl-specific antibodies were used to determine if the various Bre2 constructs could rescue the H3K4 methylation defects in a bre2Δ strain. As expected, full-length Bre2 could rescue H3K4 methylation in a bre2Δ strain similar to wild-type levels (Fig. 7, compare lanes 14 and 16). Interestingly, all Bre2 mutants (Bre 1–404, Bre2 Δ445–474, and Bre2 Δ474–500) that are compromised in their ability to bind Sdc1 were unable to rescue the H3K4 methylation defects of a bre2Δ strain (Fig. 7, compare lanes 15, 17, 19, and 20). In contrast, a Bre2 deletion mutant (Bre2 Δ410–421) retaining the SDI domain and ability to bind Sdc1 was able to rescue H3K4 methylation (Fig. 7, lane 18). The differences in ability to rescue H3K4 methylation levels in strains expressing Bre2 or the Bre2 mutants were not due to varying levels of protein expression or histone H3 levels because α-HA and H3 immunoblots detected nearly equivalent amounts of Bre2, Bre2 mutants, and histone H3 protein (Fig. 7, lanes 14–20, bottom two panels).
Interaction between the Bre2 SDI and Sdc1 Dpy-30 Domains Is Required for Association with Set1 and Proper H3K4 Methylation
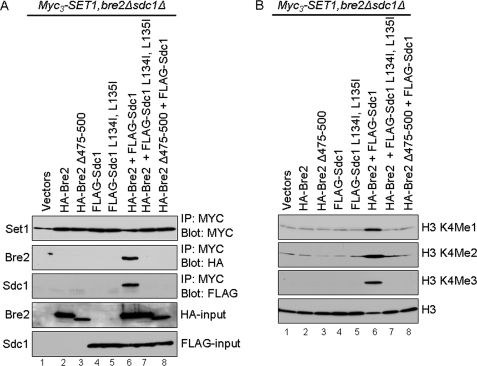
Prior work has suggested that the Bre2 and Sdc1 heteromer must be formed prior to incorporation into the Set1 complex, because deletion of either Bre2 or Sdc1 will prevent the other protein from interacting with the Set1 complex (18, 32). Furthermore, our data show that disrupting the binding between the SDI domain of Bre2 and Dpy-30 domain of Sdc1 in vivo leads to defects in global levels of H3K4 methylation similar to what is seen in bre2 and sdc1 deletion strains (Fig. 7). To determine if mutations in the Dpy-30 domain of Sdc1 and/or deletion of the SDI domain of Bre2 prevented interaction with Set1, BRE2 and SDC1 were both deleted in a yeast strain where Set1 was N-terminally Myc3-tagged at its endogenous locus (Myc3-Set1). Vector (LEU2), HA-Bre2, HA-Bre2 Δ475–500, FLAG-Sdc1, and FLAG-Sdc1 L134I,L135I constructs were coexpressed with vector (URA3), HA-Bre2, or FLAG-Sdc1 in the Myc3-SET1, bre2Δ, sdc1Δ strain. As expected, immunoprecipitated Set1 could co-immunoprecipitate full-length HA-Bre2 and FLAG-Sdc1 when cells were co-expressing wild-type Bre2 and Sdc1 (Fig. 8A, lane 6). The co-IP of Bre2 and Sdc1 by Set1 is specific because we do not detect nonspecific interactions in lanes expressing only vectors (Fig. 8A, lane 1). In contrast, Set1 could not co-immunoprecipitate HA-Bre2 or HA-Bre2 Δ475–500 in the absence of Sdc1 or co-immunoprecipitate FLAG-Sdc1 or FLAG-Sdc1 L134I,L135I in the absence of Bre2 (Fig. 8A, lanes 2, 3, 4, and 5). Furthermore, Set1 failed to co-immunoprecipitate HA-Bre2 Δ475–500 or FLAG-Sdc1 L134I,L135I in the presence of wild-type Sdc1 or Bre2, respectively (Fig. 8A, lanes 7 and 8). The inability of HA-Bre2, FLAG-Sdc1, HA-Bre2 Δ475–500, or FLAG-Sdc1 L134I,L135I to co-immunoprecipitate with Set1 was not due to significant change in protein expression levels because nearly equivalent levels were detected (Fig. 8A, lanes 2–8, bottom two panels). In addition, nearly equivalent levels of Set1 were immunoprecipitated in all samples (Fig. 8A, lanes 1–8, top panel). In agreement with the co-immunoprecipitation data, global H3K4 mono-, di-, and trimethylation are only restored when Bre2 and Sdc1 can interact and associate with Set1 (Fig. 8B, lane 6). In contrast, when Bre2 and Sdc1 cannot interact and thus cannot associate with Set1, global H3K4 methylation remains defective (Fig. 8B, lanes 2–5, 7, and 8). Taken together, our results show that the SDI-Dpy-30 interaction is not only critical for mediating a Bre2-Sdc1 heteromeric complex but also necessary for Bre2 and Sdc1 to associate with Set1. These results also help explain why disrupting the SDI-Dpy-30 interaction leads to defects in global H3K4 mono-, di-, and trimethylation. Further investigation will be needed to determine if the SDI and Dpy-30 domains are directly or indirectly associating with Set1 or if other regions within Bre2 and Sdc1 are responsible for direct or indirect contact with Set1.
FIGURE 8.
The SDI-Dpy-30 domain interaction is required for the Bre2 and Sdc1 heteromeric complex to associate with Set1. A, co-immunoprecipitation analysis from Myc3-SET1, bre2Δsdc1Δ strains expressing vector or the indicated Sdc1 or Bre2 constructs. Detection of Myc3-Set1, FLAG-Sdc1, FLAG-Sdc1 mutants, HA-Bre2, or HA-Bre2 mutants was determined by immunoblotting with α-Myc, α-FLAG, or α-HA antibodies. B, whole cell extracts generated from Myc3-SET1, bre2Δ, and sdc1Δ strains expressing vector or the indicated Sdc1 or Bre2 constructs were immunoblotted with histone H3K4 mono-, di-, and trimethyl-specific antibodies. Histone H3 immunoblots were used for a loading control.
The Bre2 SDI and Sdc1 Dpy-30 Domains Are Required for Gene-specific Methylation and Proper Gene Expression
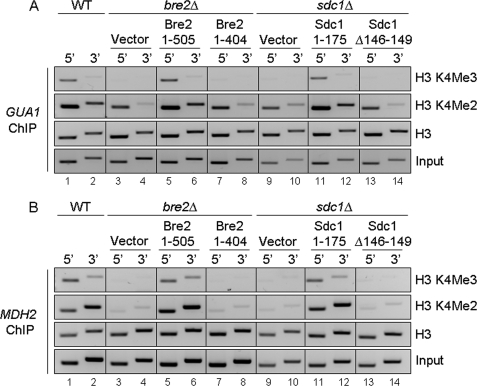
To verify that defects in global H3K4 methylation observed by Bre2 and Sdc1 mutants lacking the C-terminal SDI domain or the Dpy-30 domain, respectively, also occur on genes, ChIP analysis was performed on the GMP synthetase, GUA1, and malate dehydrogenase, MDH2, open reading frames using H3K4 di- and trimethyl-specific antibodies. ChIP inputs and histone H3 antibodies were used as controls (Fig. 9, A and B, bottom two panels). Chromatin immunoprecipitation eluates were analyzed by standard PCR using primers designed to analyze the 5′- and 3′-ends of GUA1 and MDH2 (supplemental Table S4). We chose to examine H3K4 di- and trimethylation levels at GUA1 and MDH2 because in a recently published microarray analysis, GUA1 and MDH2 were shown to be putative targets of the E3 ubiquitin ligase Not4 (47). In addition, it has recently been shown that Set1, Not4, and Jhd2 can modulate H3K4 methylation levels at GUA1 (35). Similar to global histone methylation levels, a loss of H3K4 trimethylation and a decrease in dimethylation at the 5′- and 3′-ends of GUA1 and MDH2 were detected in bre2Δ and sdc1Δ strains (Fig. 9, A and B, lanes 3, 4, 9, and 10). However, H3K4 tri- and dimethylation was restored to wild-type levels when full-length BRE2 and SDC1 were ectopically expressed (Fig. 9, A and B, lanes 5, 6, 11, and 12). Importantly, the Bre2 and Sdc1 mutants that are defective in binding to each other, Bre2 1–404 lacking the SDI domain or Sdc1 Δ146–149 lacking the PXXP motif within the Dpy-30 domain, had gene-specific H3K4 di- and trimethylation defects similar to what is observed in bre2Δ and sdc1Δ strain (Fig. 9, A and B, lanes 7, 8, 13, and 14).
FIGURE 9.
The SDI and Dpy-30 domains are required for proper gene-specific H3K4 methylation. A and B, ChIP analysis from the indicated strains was performed using antibodies specific to H3K4 di- and trimethylation and histone H3. ChIP eluates were PCR-amplified using primer sets directed to the 5′- and 3′-ends of the open reading frames of GUA1 and MDH2 and analyzed by agarose gel electrophoresis and ethidium bromide staining. Input and histone H3 were used as loading controls. The depicted ChIP analysis is a representation of three biological repeats. WT, wild type.
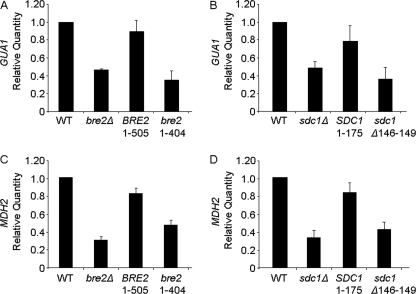
Set1-mediated H3K4 methylation is important for proper gene expression (3–6, 8, 35, 48–50). However, until now, it has not been demonstrated if Bre2 or Sdc1, two Set1-associated factors required for proper H3K4 methylation, are also needed for appropriate levels of expression at constitutively active genes. Because our chromatin immunoprecipitation analysis determined that a Bre2 and Sdc1 interaction is critical for modulating H3K4 di- and trimethylation levels at GUA1 and MDH2 open reading frames, we wanted to determine if GUA1 and MDH2 expression levels were also being modulated. Using quantitative real-time PCR analysis, we examined gene expression of GUA1 and MDH2. We determined that defects in H3K4 histone methylation that occur in bre2Δ and sdc1Δ strains coincided with an about 2-fold decrease in GUA1 expression relative to wild-type cells (Fig. 10, A and B, and supplemental Table S5). This decrease in GUA1 expression was similar to what we have observed for set1Δ and not4Δ strains (35). Furthermore, GUA1 gene expression levels in bre2Δ and sdc1Δ expressing full-length Bre2 and Sdc1, respectively, were rescued at levels similar to those of wild type (Fig. 10, A and B, and supplemental Table S5). More importantly, Bre2 and Sdc1 mutants (Bre2 1–404, and Sdc1 Δ146–149) that were defective in binding and H3K4 methylation showed a decrease in GUA1 expression levels (Fig. 10, A and B, and supplemental Table S5).
FIGURE 10.
The SDI and Dpy-30 domains are required for appropriate levels of gene expression. A–D, expression of GUA1 or MDH2 was determined in the indicated strains by quantitative real-time PCR analysis. All expression analysis was relative to wild-type (WT) cells using actin (ACT1) as an internal control to normalize expression levels. Data in A–D were analyzed from four biological repeats with three technical repeats each.
To confirm that these changes in GUA1 transcript levels were not specific only to the GUA1 locus, the MDH2 locus was also analyzed. Similar to GUA1, MDH2 transcript levels were decreased about 3-fold in bre2Δ and sdc1Δ strains when compared with wild-type cells (Fig. 10, C and D, and supplemental Table S5). Furthermore, MDH2 gene expression levels in bre2Δ and sdc1Δ expressing full-length Bre2 and Sdc1, respectively, were rescued to nearly wild-type levels (Fig. 10, C and D, and supplemental Table S5). More importantly, Bre2 and Sdc1 mutants (Bre2 1–404 and Sdc1 Δ146–149) that were defective in binding and H3K4 methylation also showed a defect in MDH2 expression levels (Fig. 10, A and B, and supplemental Table S5). Altogether, our results show that when interactions between Bre2 and Sdc1 are disrupted, Bre2 and Sdc1 no longer associate with each other or Set1; consequently, this leads to defects in global and gene-specific histone H3K4 methylation and decreases in gene expression.
DISCUSSION
In this study, we determined for the first time how two Set1-associated subunits, Bre2 and Sdc1, form a heteromeric complex. We show that the Dpy-30 domain of Sdc1 is critical for mediating interactions with Bre2. We identified a new protein-protein interaction domain located at the C terminus of Bre2, which we have named the SDI domain because it is needed to interact with the Dpy-30 domain of Sdc1. More importantly, we show that the SDI-Dpy-30 domain interaction is required for formation of the heteromeric Bre2 and Sdc1 complex and association with Set1. Without this interaction, histone H3K4 methylation and gene expression is disrupted. Finally, we determine that the human homologs of Bre2 and Sdc1, ASH2L and DPY-30, respectively, also associate in a similar manner involving the C terminus of ASH2L and the Dpy-30 domain of DPY-30. Our findings would suggest that the SDI-Dpy-30 domain interaction is a conserved interaction for yeast Set1 and human MLL and SET1-like complexes. The ASH2L and DPY-30 interaction is also likely to impact histone H3K4 methylation and gene expression in humans and other organisms.
In this study, we determined that the Dpy-30 domains of Sdc1 and human DPY-30 are required for interacting with Bre2 and ASH2L, respectively. Because all mutations within the Dpy-30 domain of Sdc1 disrupted interaction with Bre2, it is likely that the overall structure of the Dpy-30 domain is important for interacting with the C terminus of Bre2. Interestingly, during preparation of this manuscript, the x-ray crystal structure of the Dpy-30 domain of human DPY-30 was solved (45). The structure indicates that the human DPY-30 domain can form a four-helix bundle. Based on the human crystal structure and structure prediction analysis of yeast Sdc1, it appears that deletions of the PXXP motif or mutations that change the prolines to alanines are within a loop region connecting two α-helices (45, 51). Therefore, it is likely that this loop is important for the overall structure or position of the two α-helices. In addition, this might explain why Sdc1 deletion mutations within the predicted helices on either side of the PXXP loop would disrupt binding to Bre2 (Fig. 1B). From the crystal structure, it was also predicted that two highly conserved leucine residues, Leu-65 and Leu-66, form a putative binding pocket for interaction with ASH2L (45). Our data support this idea and show that mutations of the conserved leucines in Sdc1 and DPY-30 disrupt binding to Bre2 and ASH2L, respectively (Figs. 1B and 5B).
It has been predicted that Dpy-30 dimerization is important for Sdc1 and Bre2 association with Set1 (1). This idea is based on bacterial two-hybrid analysis of Sdc1 indicating that the Dpy-30 domain can dimerize and on the predicted stoichiometry of the individual Set1-associated proteins within the Set1 complex (18, 32). In addition, the recently solved x-ray crystal structure of the Dpy-30 domain of human DPY-30 also forms a dimer (45). Although we also show that Sdc1 could form disulfide-mediated dimers from bacterially expressed Sdc1, we were unable to determine if there is any physiological relevance for Sdc1 dimerization in yeast. For example, we were unable to detect disulfide-mediated dimerization of Sdc1 from yeast (data not shown), and a mutation of Sdc1 (Sdc1 C47A) that abolished dimerization in vitro had no effect on H3K4 methylation in vivo or binding to Bre2 (Figs. 2, C and D, and 7). More work will be needed to determine if the disulfide-dependent or independent dimerization of Sdc1 is physiologically relevant or an artifact of bacterial expression and/or crystallization conditions. It is also possible that the Sdc1 or DPY-30 dimerization observed in vitro occurs because a physiologically relevant binding partner, such as the SDI domain of Bre2 or ASH2L, is not present. Therefore, future studies with the co-crystal of the SDI domain of Bre2 or ASH2L with Sdc1 or DPY-30, respectively, would be useful in clarifying the importance of Sdc1 and DPY-30 dimerization.
A surprising result from our mutational analysis and binding studies indicated that the C termini of Bre2 and ASH2L are important for interacting with Sdc1 and DPY-30, respectively. This result was quite interesting because the only domain common to both Bre2 and ASH2L that was annotated as a protein-protein interaction domain is the SPRY domain. The function of the SPRY domain is currently unknown, but it is likely to be important for interacting with either Set1, Set1-associated subunits within the yeast and human MLL and SET1 complexes, or other transcription factors (15, 52). In this study, we have identified a new protein-protein interaction domain within Bre2 and ASH2L that we named the SDI domain. By homology searching, the SDI domain of Bre2 and ASH2L showed no significant matches to any other known domain. However, secondary structure prediction of the SDI domain of Bre2 and ASH2L suggests that the C terminus forms an α-helix structure (44). Interestingly, a domain that shares similarity to the Dyp-30 domain is the RIIa domain. Structural analysis has indicated that the Dpy-30 and RIIa domains can both form four-helix bundles and can dimerize. RIIa domains are commonly found within the regulatory subunit of cyclic AMP-dependent protein kinase A and are required for dimerization (41, 43). Recently, the structure of the RIIa domain and its binding partner A kinase-anchoring protein has been solved, and it was determined that an A kinase-anchoring protein α-helix was bound to the RIIa domain (41, 43). Intriguingly, when we compared the C terminus of Bre2 and mouse A kinase-anchoring protein α-helix that binds the RIIa domain, there was a 22.1% identity (44). Currently, it is unclear if the A kinase-anchoring protein α-helix is functionally similar to the SDI domain or structurally similar but with specificity toward the RIIa domain. Because of the functional conservation of the SDI predicted α-helical domain from yeast to humans, the SDI domain or domains of structural similarity may be important for interaction with other Dpy-30 or RIIa domains containing partners. Based on our data, the crystal structure of the human Dpy-30 domain, and the conserved amino acids within the SDI and Dpy-30 domains, it is likely that the SDI-Dpy-30 interaction is mediated by the conserved hydrophobic and/or charged amino acids (supplemental Fig. S1). However, additional biochemical and structural studies will be needed to determine the precise mode of interaction.
In this study, we also show that an interaction between the SDI domain of Bre2 and the Dpy-30 domain of Sdc1 is functionally important for Bre2 and Sdc1 to form a heteromeric complex and association with Set1 so that proper histone methylation and gene expression can occur. However, more work will be needed to establish the regions within Bre2 and Sdc1 required for association with Set1 or other Set1 complex members. Based on our work and recent data suggesting that Bre2, Sdc1, and ASH2L bind to the SET domain of yeast Set1 or human MLL1, it is likely that the Bre2 and Sdc1 heteromeric complex permits interaction with the SET domain of Set1, allowing for full methyltransferase activity (18, 32, 53–55). Because we also showed that human ASH2L and DPY-30 interact in a similar manner, we would expect this interaction could enhance the in vitro catalytic activity of the human MLL- and SET1-like complex or be required for in vivo trimethylation. Although it is still unclear if an SDI-Dpy-30 domain interaction is required, RNA interference knockdown of either human ASH2L or C. elegans DPY-30 can lead to defects in H3K4 methylation (20, 30, 56). In addition, it has recently been shown that recombinant ASH2L and DPY-30 heteromeric dimer and RbBP5 by itself can stimulate the catalytic activity of recombinant MLL1 by directly binding to the SET domain (53–55). However, the addition of both heteromeric dimers, ASH2L-DPY-30 and Wdr5-RbBP5, to recombinant MLL results in a further enhancement of catalytic activity (53–55). It has been proposed that the heteromeric dimers help stabilize the SET domains of MLL and SET1-like methyltransferases, so the SET domain is in the right conformation for full methyltransferase activity (53). Future studies will be required to determine if the SDI-Dpy-30 domain interaction is required for stabilizing the SET domain conformation and for H3K4 methylation in other organisms.
Although bre2Δ and sdc1Δ strains have been shown to have histone methylation defects, the basic observation that disruption of histone methylation in these strains coincides with defects in gene expression within euchromatin has not been reported. In this study, we show that loss of BRE2 or SDC1 or disruption of their SDI-Dpy-30 domain interaction results in expression defects at constitutively active genes. In addition, the decrease in gene expression observed at two loci strongly coincides with H3K4 methylation defects and is also consistent with microarray analysis of a set1 deletion strain showing a general 2-fold defect in global gene expression (8, 48, 50). Because RNA interference knockdown of ASH2L in mammalian cells results in histone methylation and gene expression defects, we would expect that disruption of the SDI-Dpy-30 interaction would also result in gene expression defects in human and other organisms (20, 30). Recently, ASH2L has also been found overexpressed in cancer cells and is thought to function as an oncoprotein (57). Although the molecular mechanism by which ASH2L mediates oncogenesis is unknown, it could be due to a consequence of either too little or too much of an association with DPY-30, resulting in reduced or overactive MLL or SET1 methyltransferase activity and consequently leading to aberrant gene expression. Therefore, our study and others that determine the interaction and function of yeast and human Set1-associated proteins are important and will probably impact human health and diseases and provide insight into novel therapeutic targets.
Supplementary Material
Acknowledgment
We thank Kayla M. Harmeyer for review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM74183 (to S. D. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1–S5.
- H3K4
- histone H3 lysine 4
- Set1C
- Set1 complex
- ChIP
- chromatin immunoprecipitation
- PHD
- plant homeodomain
- co-IP
- co-immunoprecipitation
- HA
- hemagglutinin
- PVDF
- polyvinylidene difluoride
- NTA
- nitrilotriacetic acid
- GST
- glutathione S-transferase.
REFERENCES
- 1.Dehé P. M., V Géli. (2006) Biochem. Cell Biol. 84, 536–548 [DOI] [PubMed] [Google Scholar]
- 2.Shilatifard A. (2008) Curr. Opin. Cell Biol. 20, 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggs S. D., Bryk M., Strahl B. D., Cheung W. L., Davie J. K., Dent S. Y., Winston F., Allis C. D. (2001) Genes Dev. 15, 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryk M., Briggs S. D., Strahl B. D., Curcio M. J., Allis C. D., Winston F. (2002) Curr. Biol. 12, 165–170 [DOI] [PubMed] [Google Scholar]
- 5.Fingerman I. M., Wu C. L., Wilson B. D., Briggs S. D. (2005) J. Biol. Chem. 280, 28761–28765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nislow C., Ray E., Pillus L. (1997) Mol. Biol. Cell 8, 2421–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng H. H., Robert F., Young R. A., Struhl K. (2003) Mol. Cell 11, 709–719 [DOI] [PubMed] [Google Scholar]
- 8.Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. (2002) Nature 419, 407–411 [DOI] [PubMed] [Google Scholar]
- 9.Hughes C. M., Rozenblatt-Rosen O., Milne T. A., Copeland T. D., Levine S. S., Lee J. C., Hayes D. N., Shanmugam K. S., Bhattacharjee A., Biondi C. A., Kay G. F., Hayward N. K., Hess J. L., Meyerson M. (2004) Mol. Cell 13, 587–597 [DOI] [PubMed] [Google Scholar]
- 10.Krogan N. J., Dover J., Wood A., Schneider J., Heidt J., Boateng M. A., Dean K., Ryan O. W., Golshani A., Johnston M., Greenblatt J. F., Shilatifard A. (2003) Mol. Cell 11, 721–729 [DOI] [PubMed] [Google Scholar]
- 11.Lee J. H., Skalnik D. G. (2008) Mol. Cell. Biol. 28, 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellor J. (2006) Cell 126, 22–24 [DOI] [PubMed] [Google Scholar]
- 13.Brehm A., Tufteland K. R., Aasland R., Becker P. B. (2004) BioEssays 26, 133–140 [DOI] [PubMed] [Google Scholar]
- 14.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy P. L., Griesenbeck J., Kornberg R. D., Cleary M. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krogan N. J., Dover J., Khorrami S., Greenblatt J. F., Schneider J., Johnston M., Shilatifard A. (2002) J. Biol. Chem. 277, 10753–10755 [DOI] [PubMed] [Google Scholar]
- 17.Mueller J. E., Canze M., Bryk M. (2006) Genetics 173, 557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roguev A., Schaft D., Shevchenko A., Pijnappel W. W., Wilm M., Aasland R., Stewart A. F. (2001) EMBO J. 20, 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller T., Krogan N. J., Dover J., Erdjument-Bromage H., Tempst P., Johnston M., Greenblatt J. F., Shilatifard A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dou Y., Milne T. A., Ruthenburg A. J., Lee S., Lee J. W., Verdine G. L., Allis C. D., Roeder R. G. (2006) Nat. Struct. Mol. Biol. 13, 713–719 [DOI] [PubMed] [Google Scholar]
- 21.Wysocka J., Myers M. P., Laherty C. D., Eisenman R. N., Herr W. (2003) Genes Dev. 17, 896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura T., Mori T., Tada S., Krajewski W., Rozovskaia T., Wassell R., Dubois G., Mazo A., Croce C. M., Canaani E. (2002) Mol. Cell 10, 1119–1128 [DOI] [PubMed] [Google Scholar]
- 23.Cho Y. W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G. R., Copeland T. D., Kalkum M., Ge K. (2007) J. Biol. Chem. 282, 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J. H., Tate C. M., You J. S., Skalnik D. G. (2007) J. Biol. Chem. 282, 13419–13428 [DOI] [PubMed] [Google Scholar]
- 25.Lee J. H., Skalnik D. G. (2005) J. Biol. Chem. 280, 41725–41731 [DOI] [PubMed] [Google Scholar]
- 26.Yokoyama A., Wang Z., Wysocka J., Sanyal M., Aufiero D. J., Kitabayashi I., Herr W., Cleary M. L. (2004) Mol. Cell. Biol. 24, 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dou Y., Milne T. A., Tackett A. J., Smith E. R., Fukuda A., Wysocka J., Allis C. D., Chait B. T., Hess J. L., Roeder R. G. (2005) Cell 121, 873–885 [DOI] [PubMed] [Google Scholar]
- 28.Wu M., Wang P. F., Lee J. S., Martin-Brown S., Florens L., Washburn M., Shilatifard A. (2008) Mol. Cell. Biol. 28, 7337–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider J., Wood A., Lee J. S., Schuster R., Dueker J., Maguire C., Swanson S. K., Florens L., Washburn M. P., Shilatifard A. (2005) Mol. Cell 19, 849–856 [DOI] [PubMed] [Google Scholar]
- 30.Steward M. M., Lee J. S., O'Donovan A., Wyatt M., Bernstein B. E., Shilatifard A. (2006) Nat. Struct. Mol. Biol. 13, 852–854 [DOI] [PubMed] [Google Scholar]
- 31.Tate C. M., Lee J. H., Skalnik D. G. (2009) Mol. Cell. Biol. 29, 3817–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dehé P. M., Dichtl B., Schaft D., Roguev A., Pamblanco M., Lebrun R., Rodríguez-Gil A., Mkandawire M., Landsberg K., Shevchenko A., Shevchenko A., Rosaleny L. E., Tordera V., Chávez S., Stewart A. F., Géli V. (2006) J. Biol. Chem. 281, 35404–35412 [DOI] [PubMed] [Google Scholar]
- 33.Du H. N., Fingerman I. M., Briggs S. D. (2008) Genes Dev. 22, 2786–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fingerman I. M., Li H. C., Briggs S. D. (2007) Genes Dev. 21, 2018–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mersman D. P., Du H. N., Fingerman I. M., South P. F., Briggs S. D. (2009) Genes Dev. 23, 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ponting C., Schultz J., Bork P. (1997) Trends Biochem. Sci. 22, 193–194 [DOI] [PubMed] [Google Scholar]
- 37.Woo J. S., Imm J. H., Min C. K., Kim K. J., Cha S. S., Oh B. H. (2006) EMBO J. 25, 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woo J. S., Suh H. Y., Park S. Y., Oh B. H. (2006) Mol. Cell 24, 967–976 [DOI] [PubMed] [Google Scholar]
- 39.Hsu D. R., Meyer B. J. (1994) Genetics 137, 999–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu D. R., Chuang P. T., Meyer B. J. (1995) Development 121, 3323–3334 [DOI] [PubMed] [Google Scholar]
- 41.Gold M. G., Lygren B., Dokurno P., Hoshi N., McConnachie G., Taskén K., Carlson C. R., Scott J. D., Barford D. (2006) Mol. Cell 24, 383–395 [DOI] [PubMed] [Google Scholar]
- 42.Morikis D., Roy M., Newlon M. G., Scott J. D., Jennings P. A. (2002) Eur. J. Biochem. 269, 2040–2051 [DOI] [PubMed] [Google Scholar]
- 43.Kinderman F. S., Kim C., von Daake S., Ma Y., Pham B. Q., Spraggon G., Xuong N. H., Jennings P. A., Taylor S. S. (2006) Mol. Cell 24, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramaniam S. (1998) Proteins 32, 1–2 [PubMed] [Google Scholar]
- 45.Wang X., Lou Z., Dong X., Yang W., Peng Y., Yin B., Gong Y., Yuan J., Zhou W., Bartlam M., Peng X., Rao Z. (2009) J. Mol. Biol. 390, 530–537 [DOI] [PubMed] [Google Scholar]
- 46.Dong X., Peng Y., Peng Y., Xu F., He X., Wang F., Peng X., Qiang B., Yuan J., Rao Z. (2005) Biochim. Biophys. Acta 1753, 257–262 [DOI] [PubMed] [Google Scholar]
- 47.Cui Y., Ramnarain D. B., Chiang Y. C., Ding L. H., McMahon J. S., Denis C. L. (2008) Mol. Genet. Genomics 279, 323–337 [DOI] [PubMed] [Google Scholar]
- 48.Boa S., Coert C., Patterton H. G. (2003) Yeast 20, 827–835 [DOI] [PubMed] [Google Scholar]
- 49.Carvin C. D., Kladde M. P. (2004) J. Biol. Chem. 279, 33057–33062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venkatasubrahmanyam S., Hwang W. W., Meneghini M. D., Tong A. H., Madhani H. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16609–16614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelley L. A., Sternberg M. J. (2009) Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 52.Demers C., Chaturvedi C. P., Ranish J. A., Juban G., Lai P., Morle F., Aebersold R., Dilworth F. J., Groudine M., Brand M. (2007) Mol. Cell 27, 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Southall S. M., Wong P. S., Odho Z., Roe S. M., Wilson J. R. (2009) Mol. Cell 33, 181–191 [DOI] [PubMed] [Google Scholar]
- 54.Patel A., Dharmarajan V., Cosgrove M. S. (2008) J. Biol. Chem. 283, 32158–32161 [DOI] [PubMed] [Google Scholar]
- 55.Patel A., Vought V. E., Dharmarajan V., Cosgrove M. S. (2008) J. Biol. Chem. 283, 32162–32175 [DOI] [PubMed] [Google Scholar]
- 56.Simonet T., Dulermo R., Schott S., Palladino F. (2007) Dev. Biol. 312, 367–383 [DOI] [PubMed] [Google Scholar]
- 57.Lüscher-Firzlaff J., Gawlista I., Vervoorts J., Kapelle K., Braunschweig T., Walsemann G., Rodgarkia-Schamberger C., Schuchlautz H., Dreschers S., Kremmer E., Lilischkis R., Cerni C., Wellmann A., Lüscher B. (2008) Cancer Res. 68, 749–758 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.