Abstract
The incorporation of histone variant H2A.Z into nucleosomes plays essential roles in regulating chromatin structure and gene expression. A multisubunit complex containing chromatin remodeling protein Swr1 is responsible for the deposition of H2A.Z in budding yeast and mammals. Here, we show that the JmjC domain protein Msc1 is a novel component of the fission yeast Swr1 complex and is required for Swr1-mediated incorporation of H2A.Z into nucleosomes at gene promoters. Loss of Msc1, Swr1, or H2A.Z results in loss of silencing at centromeres and defective chromosome segregation, although centromeric levels of CENP-A, a centromere-specific histone H3 variant that is required for setting up the chromatin structure at centromeres, remain unchanged. Intriguingly, H2A.Z is required for the expression of another centromere protein, CENP-C, and overexpression of CENP-C rescues centromere silencing defects associated with H2A.Z loss. These results demonstrate the importance of H2A.Z and CENP-C in maintaining a silenced chromatin state at centromeres.
Keywords: Chromatin/Epigenetics, Chromosomes/Centromeres, Genetics/Yeast, Histones, Centromeres, Chromatin Histone Modification, CENP-C, H2A.Z
Introduction
Centromeres are specialized regions of eukaryotic chromosomes that direct the assembly of kinetochores and are essential for the equal segregation of chromosomes during mitosis and meiosis (1–3). Despite significant differences in size and sequence composition, general features of centromeres are highly conserved, such as the enrichment of centromere-specific proteins and the existence of pericentric heterochromatin structures. However, with the exception of Saccharomyces cerevisiae, whose sites of centromere formation are determined by a short stretch of DNA, most eukaryotes establish and propagate active centromeres through chromatin-based epigenetic mechanisms, which are independent of DNA sequences (1–3).
As genomic DNA is folded with histone proteins in the form of chromatin, epigenetic mechanisms that regulate the genome include covalent modifications of histones and DNA, chromatin remodeling, and exchange of histone variants (4). The centromere differs fundamentally from the remainder of the genome due to its enrichment for a variant form of histone H3, CENP-A (also known as CID in Drosophila, Cse4 in budding yeast, and Cnp1 in fission yeast) (1–3). CENP-A serves as the foundation for the assembly of other kinetochore proteins and is essential for chromosome segregation in diverse organisms. Biochemical analysis has demonstrated that CENP-A-containing nucleosomes are more compact and assume a more rigid conformation than those containing histone H3, which might create a specialized chromatin environment at centromeres (5, 6). Consistent with this, centromere chromatin in fission yeast compacts differently from the remainder of the genome (7, 8), and reporter genes inserted within the centromeres are usually silenced (9). Several proteins that are required for chromosome segregation, including CENP-ACnp1, were identified in genetic screens for mutations that affect centromere silencing (10). However, whether the formation of CENP-ACnp1 chromatin directly contributes to centromere silencing is not known.
From fission yeast to mammals, centromere regions are usually surrounded by repetitive DNA elements that form heterochromatin structures (1, 2). The formation of heterochromatin requires the concerted actions of a diverse array of enzymatic activities that lead to the methylation of histone H3 at lysine 9 (H3K9me) and the recruitment of heterochromatin proteins such as HP1/Swi6 to ensure transcriptional silencing across the entire heterochromatin domain (11). Pericentric heterochromatin also contributes to the fidelity of chromosome segregation as it recruits cohesin proteins, which are required for proper chromosome cohesion (12, 13).
Recent studies in fission yeast demonstrate that a JmjC domain-containing protein, Msc1, is required for normal chromosome segregation and that overexpression of Msc1 rescues the lethality associated with a CENP-Acnp1 mutation (14). Msc1 also regulates the dynamics of pericentric heterochromatin, but whether this contributes to the regulation of centromere domain and chromosome segregation is not known (15). Msc1 shares strong sequence homology with the JARID1 family of proteins (16–18), which all use the JmjC domain to demethylate histones that are methylated at H3 lysine 4 (19–27). However, the JmjC domain of Msc1 lacks critical residues for catalysis (17), suggesting that Msc1 might function independently of histone demethylation. Interestingly, Msc1 overexpression suppresses a CENP-Acnp1 mutation only in the presence of the H2A variant H2A.ZPht1 (14). Whole genome genetic interaction analysis indicates that Msc1 functions together with the Swr1 complex (28), a multisubunit complex that catalyzes the incorporation of H2A.Z into chromatin in both budding yeast and mammals (29–32).
Using biochemical purification, we found that Msc1 is an integral component of the fission yeast Swr1 complex, as has been shown recently (33, 34). Chromatin immunoprecipitation (ChIP)3 coupled with DNA microarray (ChIP-chip) analysis demonstrated that both Msc1 and Swr1 are required for H2A.ZPht1 incorporation into chromatin, which shows a preference for gene promoters. Although H2A.ZPht1 is not enriched at centromeres, loss of H2A.ZPht1, as well as Msc1 and Swr1, results in loss of silencing at centromeres and defects in chromosome segregation. Interestingly, CENP-Acnp1 levels at centromeres are normal in the absence of H2A.ZPht1, suggesting that CENP-Acnp1 is not sufficient to impose silencing at centromere regions. Instead, H2A.ZPht1 regulates the expression of CENP-CCnp3, a centromere protein required for centromere silencing. These results demonstrate that H2A.ZPht1 maintains the silenced chromatin state that is critical for the fidelity of chromosome segregation.
EXPERIMENTAL PROCEDURES
Fission Yeast Strains
Msc1-FLAG, Swr1-FLAG, Swr1-Myc, Pht1-Myc, Cnp1-FLAG, Cnp1-GFP, Cnp3-Myc, swr1Δ, pht1Δ, and cnp3Δ strains were constructed using a PCR-based module method (35). Msc1 mutants were generated by first inserting a ura4+ gene in the coding region of msc1+ and subsequent replacement with mutated DNA sequences. Genetic crosses were used to construct all other strains. For serial dilution plating assays, 10-fold dilutions of a log-phase culture were plated on the indicated medium and grown for 3 days at 30 °C.
Chromosome Segregation Assay
Assaying the rate of chromosome loss was performed as described previously (36). The minichromosome Ch16 contains an ade6-M216 allele that complements an ade6-M210 allele at its normal chromosome location. Cells from Ade+ colonies were plated on adenine-limiting medium and incubated at 30 °C for 4 days. If chromosome loss occurs in the first division, half of the resultant colony carrying Ch16 will be white, whereas the half without Ch16 will be red. The number of half-sectored red/white colonies was determined, and the rate of chromosome loss per cell division was calculated by dividing the number of half-sectored colonies by the total number of white colonies plus half-sectored colonies.
Protein Purification and Mass Spectrometry Analysis
Affinity purification of Msc1-FLAG and Swr1-FLAG complexes and MudPIT mass spectrometry analysis were performed as described previously (37).
Western Blots and Antibodies
Protein extracts were prepared by lysis of cells with glass beads, followed by sonication to dissolve chromatin (37). The following antibodies were used for Western blot analyses: FLAG (Sigma, F7425 and F3165) and Myc (Covance, MMP-150).
Chromatin Immunoprecipitation
ChIP analysis was performed as described previously (36). Immunoprecipitation was performed with Myc or FLAG antibodies. ChIP-chip analysis was performed according to the “Agilent Yeast ChIP-on-chip Analysis” protocol. Blunt-end DNA was generated from immunoprecipitated chromatin fractions (ChIP) or from whole cell extract (WCE) with T4 DNA polymerase and then ligated to a linker. ChIP and WCE DNAs were amplified from the blunt-end DNA samples with primers annealing to the linker and were labeled with Cy5- or Cy3-dUTP, respectively, by random priming PCR (Invitrogen comparative genomic hybridization kit). 2.5–5 μg of Cy5-labeled ChIP DNA and corresponding Cy3-labeled WCE DNA were hybridized to an Agilent Schizosaccharomyces pombe whole genome ChIP-on-chip microarray (G4810A). The slides were washed and processed in accordance with Agilent protocols and scanned with an Agilent scanner. Data were collected with the Agilent Feature Extraction program. The enrichment value for each probe was calculated by dividing normalized ChIP signal by WCE signal.
For PCR-based quantification, DNA isolated from ChIP or WCE was quantitatively analyzed by competitive PCR in which one primer pair amplifies a region of interest, whereas the other primer pair amplifies control fbp1 (located in euchromatin) or cox3 (located in the mitochondrial genome). The ratios of intensities of the region of interest to control signals in the ChIP and WCE lanes were used to calculate the relative -fold enrichment. Values <1 indicate no enrichment. Quantitative real-time PCR was performed with Maxima SYBR Green qPCR Master Mix (Fermentas) in an ABI 7300 real-time PCR system. DNA serial dilutions were used as templates to generate a standard curve of amplification for each pair of primers, and the relative concentration of target sequence was calculated accordingly. A cox3 fragment was used as a reference to calculate the enrichment of ChIP over WCE for each target sequence.
RNA Extraction and Reverse Transcription (RT)-PCR
Total cellular RNA was isolated from log-phase cells using a MasterPure yeast RNA purification kit (EPICENTRE Biotechnologies) according to the manufacturer's protocol. Quantitative RT-PCR was performed with 250 ng of RNA as a template with the OneStep RT-PCR kit (Qiagen). Reactions in which the RT step was omitted were performed in parallel. Quantification with real-time RT-PCR was performed with Power SYBR Green RNA-to-CT 1-Step kit (Applied Biosystems). RNA serial dilutions were used as templates to generate the standard curve of amplification for each pair of primers, and the relative concentration of target sequence was calculated accordingly. An act1 fragment served as a reference to normalize the concentration of samples. The concentration of each target gene in the wild type was arbitrarily set to 1 and served as a reference for other samples.
RESULTS
Msc1 Associates with the Swr1 Complex
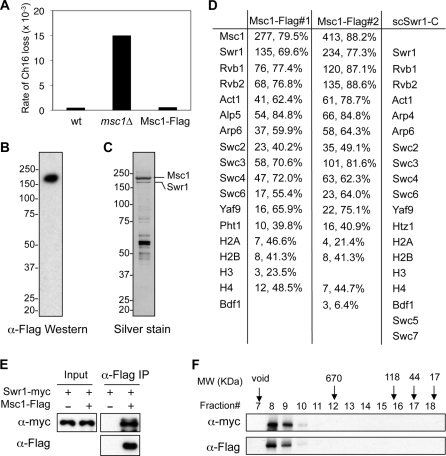
Msc1 has been shown to be required for proper chromosome segregation (14). To further understand the mechanism that regulates Msc1 function, we generated a yeast strain expressing Msc1 fused with a 3×FLAG epitope (Msc1-FLAG) at the endogenous msc1+ locus. Transcription of the modified gene is driven by its natural promoter from the endogenous chromatin environment to ensure wild-type expression levels. The fusion protein is functional, as no defect in the segregation of a nonessential minichromosome was observed (Fig. 1A). Expression of Msc1 was confirmed by Western blot analysis with a anti-FLAG antibody (Fig. 1B). Immunoaffinity-purified Msc1 complex contains a number of proteins in addition to Msc1 (Fig. 1C) and was subjected to MudPIT mass spectrometry analysis (38). Among the proteins identified were Swr1, Rvb1, Rvb2, Act1, Alp5, Arp6, Swc2, Swc3, Swc4, Swc6, Yaf9, and H2A.ZPht1, as well as core histones (Fig. 1D). Interestingly, their budding yeast homologs all belong to the Swr1 complex that catalyzes the exchange of H2A.ZHtz1 with H2A. The budding yeast Swr1 complex also contains three additional subunits, Bdf1, Swc5, and Swc7 (29–31). However, we did not detect any Swc5 or Swc7 homologs in either of two independent purifications of Msc1 and only low levels of Bdf1 in one purification (Fig. 1D).
FIGURE 1.
Msc1 is an integral component of the fission yeast Swr1 complex. A, Msc1-FLAG is functional. The loss rates of a nonessential chromosome (Ch16) in the indicated strains were measured. wt, wild type. B, cell extract prepared from an Msc1-FLAG strain was immunoprecipitated with anti-FLAG resin and subjected to Western blot analysis with anti-FLAG antibody. C, cell extract prepared from an Msc1-FLAG strain was used to perform affinity purification with anti-FLAG resin, resolved by SDS-PAGE, and silver-stained. D, shown are the results from MudPIT mass spectrometry analysis of proteins associated with Msc1-FLAG. The numbers of peptides, as well as the percentages of each protein these peptides cover, are indicated. The corresponding proteins of the S. cerevisiae Swr1 complex were also shown for comparison. E, protein extracts prepared from the indicated strains were immunoprecipitated (IP) with anti-FLAG resin and analyzed by Western blot analysis with anti-FLAG and anti-Myc antibodies. F, an affinity-purified complex containing Msc1-FLAG and Swr1-Myc was subjected gel filtration analysis with a Superose 6 column. Fractions were trichloroacetic acid-precipitated and subjected to Western blot analysis with anti-FLAG and anti-Myc antibodies. The elution volumes of protein standards (in kilodaltons) are also indicated.
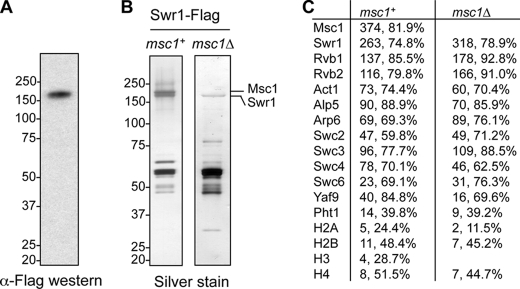
To confirm that Msc1 associates with the fission yeast Swr1 complex, we generated a yeast strain that expresses both Msc1-FLAG and Swr1-Myc at their native chromosomal loci. We found that Msc1-FLAG co-immunoprecipitated with Swr1-Myc (Fig. 1E), and gel filtration analysis of the purified complex showed that Msc1-FLAG and Swr1-Myc migrated in the same fraction (Fig. 1F). We caution that the molecular mass of the complex is very high, and the complex fractionates in a position very close to the resolution limit of the column. To further confirm that Msc1 is part of the Swr1 complex, we also generated a FLAG-tagged version of Swr1 at its endogenous chromosomal location (Fig. 2A). Silver staining of the purified Swr1 complex indicated that the components are similar to those of the Msc1 complex (Fig. 2B), which is corroborated by MudPIT analysis (Fig. 2C). These results demonstrate that Msc1 is an integral component of the Swr1 complex. Furthermore, Msc1 is not required for Swr1 complex assembly, as the Swr1 complex purified from an msc1Δ background shows the selective loss of Msc1, but not any other component of the Swr1 complex (Fig. 2, B and C).
FIGURE 2.
Msc1 is not required for the integrity of the Swr1 complex. A, cell extract prepared from an Swr1-FLAG strain was immunoprecipitated with anti-FLAG resin and subjected to Western blot analysis with anti-FLAG antibody. B, extracts prepared from Swr1-FLAG strains in the presence or absence of Msc1 were used to perform affinity purification with anti-FLAG resin, resolved by SDS-PAGE, and silver-stained. C, shown are the results from mass spectrometry analysis of proteins associated with Swr1-FLAG. The numbers of peptides, as well as the percentages of each protein these peptides cover, are indicated.
Msc1 and Swr1 Are Required for H2A.ZPht1 Incorporation at Gene Promoters
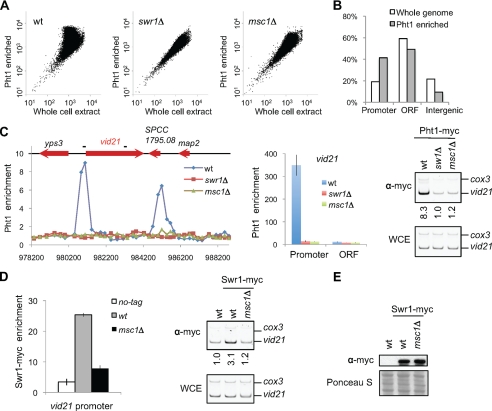
To examine whether Msc1 and Swr1 are required for H2A.ZPht1 incorporation in vivo, we performed ChIP of a Myc-tagged version of H2A.ZPht1 and examined its genome-wide occupancy using an Agilent whole genome ChIP-on-chip microarray. H2A.ZPht1 is not uniformly distributed across the genome, as indicated by scatter plot analysis (Fig. 3A), but is instead preferentially enriched at gene promoters (Fig. 3B). To confirm this observation, we randomly picked several genes that showed both high-level enrichment of H2A.ZPht1 in ChIP-chip analysis and are longer than 1.5 kb so that we could easily distinguish between promoter and coding regions. ChIP followed by quantification with PCR-based analyses showed that H2A.ZPht1 indeed preferentially localizes to gene promoters as compared with coding regions (Fig. 3C and supplemental Fig. S1). This pattern is similar to the distribution of H2A.Z in diverse organisms, suggesting that this is a highly conserved feature of H2A.Z distribution (39–47).
FIGURE 3.
Msc1 and Swr1 are required for H2A.ZPht1 incorporation into chromatin. A, scatter plots of Pht1-Myc ChIP-chip analysis. Log values of Pht1 ChIP and WCE signals for each probe were plotted against each other. wt, wild type. B, H2A.ZPht1 is enriched at gene promoters. The 42,708 probes are divided into three categories: open reading frame (ORF; coding region), promoter (within 500 bp upstream of the coding region), and intergenic (noncoding regions that do not belong to the promoter category). Pht1-enriched represents the top 5% (2,134) of probes that are enriched for Pht1, all of which have enrichment values of >3-fold. C, H2A.ZPht1 is enriched at gene promoters. Left panel, example of H2A.ZPht1 ChIP-chip analysis across a 10-kb region that includes vid21+. Gene coding regions are indicated on top, and the positions of PCR fragments used in the calculation are shown as short bars. Middle panel, quantification of H2A.ZPht1 enrichment at vid21+ by real-time PCR. Right panels, quantification of H2A.ZPht1 enrichment at the vid21+ promoter by competitive PCR. -Fold enrichment is shown below each lane. D, Msc1 is required for targeting of Swr1 to the vid21+ promoter. Quantification by both real-time PCR and competitive PCR is shown. E, Msc1 does not affect Swr1 protein levels. Cell extracts prepared from the indicated strains were subjected to Western blot analysis with anti-Myc antibody.
The loss of either Swr1 or Msc1 resulted in the loss of H2A.ZPht1 across the genome as well as at specific genes tested (Fig. 3, A and C, and supplemental Fig. S1), demonstrating that the fission yeast Swr1 complex indeed catalyzes the exchange of H2A.ZPht1 with H2A in vivo. Moreover, the loss of Msc1 resulted in delocalization of Swr1-Myc from the promoter of vid21+, indicating that Msc1 regulates the association of Swr1 with chromatin (Fig. 3D). Meanwhile, loss of Msc1 had no effect on the stability of Swr1 as indicated by Western blot analysis (Fig. 3E). Taken together, these results demonstrate that Msc1 is an essential component of the Swr1 complex that is required for targeting the complex to correct genomic locations.
Both Msc1 and Swr1 Are Required for Proper Chromosome Segregation
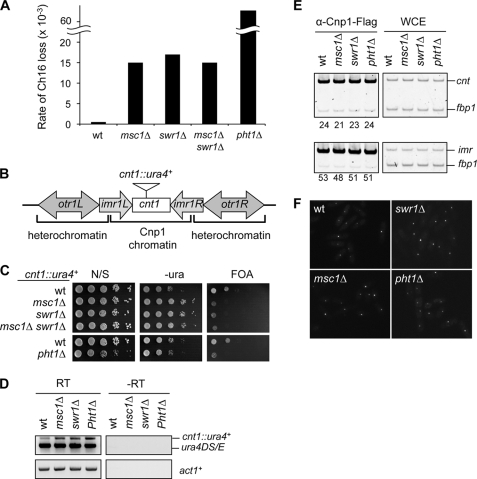
Because deletion of msc1+ or pht1+ results in chromosome segregation defects (14, 48), we investigated the role of the Msc1-Swr1 complex in regulating proper segregation of chromosomes during mitosis. Similar to msc1Δ cells, swr1Δ cells also showed elevated rates of chromosome loss (Fig. 4A). Consistent with the idea that Msc1 and Swr1 function in the same pathway genetically, an msc1Δ swr1Δ double mutant showed a rate of minichromosome loss similar to that of each single mutant (Fig. 4A). The pht1Δ mutant showed an even higher rate of chromosome loss, and the minichromosome was quickly lost from cell populations in the absence of selection pressure (Fig. 4A and data not shown) (48). This difference might be attributed to the existence of histone chaperones that mediate random incorporation of H2A.Z and compensate to some extent for the loss of Msc1 or Swr1 (49, 50).
FIGURE 4.
Msc1, Swr1, and H2A.ZPht1 are required for proper chromosome segregation. A, Msc1 and Swr1 are required for the maintenance of a nonessential minichromosome. wt, wild type. B, shown is a diagram of the fission yeast centromere 1. The insertion site of the cnt1::ura4+ reporter gene is indicated. C, serial dilution plating analysis of the indicated strains were performed to measure the expression of ura4+. FOA, 5-fluoroorotic acid; N/S, non-selective medium. D, RT-PCR analyses were performed to measure the RNA levels of ura4+ and act1+. −RT indicates the reverse transcription step was omitted. E, ChIP analysis was performed to measure the levels of Cnp1-FLAG at centromere regions. -Fold enrichment is shown below each lane. F, shown are the results from live cell imaging analysis of the indicated yeast strains expressing Cnp1-GFP.
Msc1 and Swr1 Are Required for Centromere Silencing
Centromeres form a unique chromatin environment due to the incorporation of the histone H3 variant CENP-A (5, 6). Reporter genes inserted at centromere regions are usually silenced, and defects in centromere chromatin structure are frequently associated with loss of silencing at these regions (9, 10). Loss of msc1+, swr1+, or pht1+ resulted in defective silencing of a ura4+ reporter gene inserted at CENP-A chromatin of chromosome I (cnt1::ura4+) as indicated by greater relative growth of cells in medium lacking uracil and reduced growth on medium containing 5-fluoroorotic acid, which is toxic to cells expressing Ura4 (Fig. 4B). In addition, an msc1Δ swr1Δ double mutant also alleviated silencing to a degree similar to each single mutant, further corroborating that Msc1 and Swr1 function in the same pathway (Fig. 4C). The slow growth rate of pht1Δ cells makes it difficult to interpret the silencing assay; however, RT-PCR analysis showed increased transcription of cnt1::ura4+ in pht1Δ as well as msc1Δ and swr1Δ cells, indicative of loss of silencing (Fig. 4D).
As pericentric heterochromatin is also required for chromosome segregation by regulating cohesin recruitment (12, 13), we examined the effect of the Swr1 complex on silencing of reporter genes inserted at pericentric heterochromatin of chromosome I (otr1::ura4+ and imr1::ura4+) (supplemental Fig. S2A). Both msc1Δ and swr1Δ showed better silencing at otr1::ura4+ than wild-type cells and little effect on silencing at imr1::ura4+ (supplemental Fig. S2B) (15). In contrast, loss of Clr4, which is a histone H3K9 methyltransferase essential for the integrity of pericentric heterochromatin, resulted in complete loss of silencing at both otr1::ura4+ and imr1::ura4+ (supplemental Fig. S2B). Thus, it seems that the effect of the Msc1-Swr1 complex on chromosome segregation is exerted through effects on centromere chromatin structure, but not the surrounding pericentric heterochromatin.
H2A.ZPht1 Regulates CENP-CCnp3 Expression
Although Msc1 has been previously reported to be required for CENP-ACnp1 incorporation at centromeres (14), we found that the localization of Cnp1-FLAG at centromeres (both cnt and imr) was not affected by msc1Δ, swr1Δ, or pht1Δ (Fig. 4E). In addition, live cell imaging of cells expressing Cnp1-GFP indicated that Cnp1 levels and localization were intact in msc1Δ, swr1Δ, and pht1Δ cells as well (Fig. 4F). The reason for this discrepancy is not clear, but our results suggest that centromere proteins other than CENP-ACnp1 enforce silencing at centromeres.
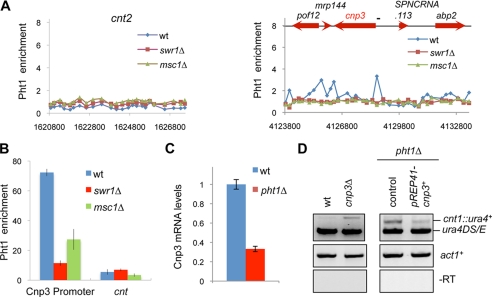
Our ChIP-chip analysis showed no enrichment of H2A.ZPht1 at any centromere positions (Fig. 5A), which was confirmed by quantification with PCR-based analysis (Fig. 5B). This suggests that the effect of Msc1-Swr1 on centromere silencing might be an indirect effect of H2A.ZPht1 on gene expression. Previous expression microarray analysis indicated that there are few genes whose expression levels change significantly in both pht1Δ and swr1Δ (34, 51). Among them, only Cnp3 has been implicated in centromere function (51). Cnp3 is a centromere protein homologous to mammalian CENP-C and is required for chromosome segregation (52–54). Our ChIP-chip results showed that H2A.ZPht1 is enriched at the promoter region of CENP-CCnp3, which was confirmed by PCR-based quantification (Fig. 5, A and B). Confirming previous microarray analysis, we found that CENP-CCnp3 mRNA levels were reduced in pht1Δ cells (Fig. 5C). These results suggest that H2A.ZPht1 directly regulates the expression of CENP-CCnp3, which might explain the centromere silencing defects in pht1Δ cells. Consistent with this idea, cnp3Δ cells were defective in silencing of cnt1::ura4+, and overexpression of CENP-CCnp3 rescued centromere silencing defects of pht1Δ cells (Fig. 5D). CENP-CCnp3 overexpression resulted in severe growth defects, preventing us from analyzing its effects on minichromosome maintenance in pht1Δ cells (data not shown). Nonetheless, these results demonstrate that H2A.Z regulates the expression of CENP-CCnp3 to establish a silenced chromatin environment at centromeres, although we caution that it is possible that H2A.ZPht1 could also regulate other centromere proteins whose functions contribute to chromosome segregation.
FIGURE 5.
H2A.ZPht1 regulates CENP-CCnp3 expression. A, ChIP-chip analysis of H2A.ZPht1 enrichment at the centromere of chromosome 2 (cnt2) and a 10-kb region that includes cnp3+. wt, wild type. B, ChIP analysis of H2A.ZPht1 at centromeres (cnt) and the cnp3 promoter. C, RT-PCR analysis of Cnp3 mRNA levels. D, CENP-CCnp3 is required for silencing at centromeres. RT-PCR analysis were performed to measure the expression of cnt1::ura4+.
Plant Homeodomains (PHDs) Are Required for Msc1 Function
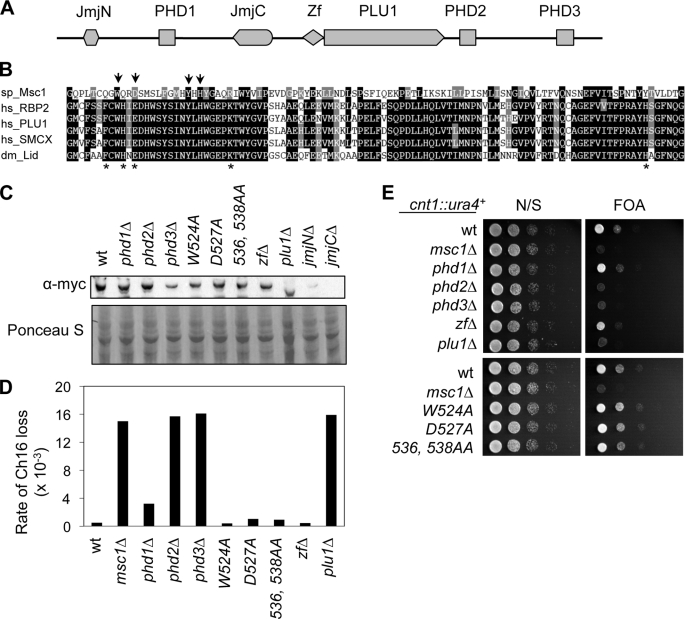
Msc1 belongs to the JARID1 family of histone demethylases, which use the JmjC domain to specifically demethylate histone H3K4 (19–27). Whether Msc1 functions as a histone demethylase is unknown, but it seems to lack some residues critical for catalysis (17). Msc1 also contains three plant homeodomains (PHD1–3) that have ubiquitin ligase activity, although the substrates are yet unknown (55). PHDs in other proteins have also been demonstrated to bind methylated histones (56–59). In addition, Msc1 also contains a JmjN domain, which usually associates with JmjC, as well as a zinc finger and a PLU1 domain of unknown function (Fig. 6A). To test whether these domains are required for Msc1 function, we systematically deleted each domain and examined the effect on chromosome segregation and centromere silencing. We found that loss of PHD2 and PHD3 resulted in elevated rates of chromosome loss and defects in centromere silencing, indicating that the E3 ubiquitin ligase activity or ability to bind methylated histones might be essential for Msc1 function (Fig. 6, D and E). Deletion of either JmjC or JmjN severely affected Msc1 stability (Fig. 6C). To circumvent this problem, we generated point mutations of several highly conserved residues in the JmjC domain, including one residue that is critical for histone demethylase activity (Fig. 6B) (17, 60). However, none of these mutations had any effect on chromosome segregation or centromere silencing, indicating that Msc1 might not exert its effect through histone demethylation.
FIGURE 6.
PHDs are required for Msc1 function. A, shown is the domain architecture of Msc1. Zf, zinc finger. B, shown is sequence alignment of the JmjC domains of the JARID1 family proteins. Arrows indicate amino acid residues mutated in Msc1. Asterisks represent residues that are essential for histone demethylase activity (17, 60). sp, S. pombe; hs, Homo sapiens; dm, Drosophila melanogaster. C, cell extracts prepared from the indicated strains were subjected to Western blot analysis with anti-Myc antibody. Ponceau S staining of part of the membrane is shown below as a control for loading. wt, wild type. D, shown is the maintenance of a nonessential minichromosome in Msc1 mutants. E, serial dilution plating analysis of the indicated strains was performed to measure the expression of cnt1::ura4+. FOA, 5-fluoroorotic acid; N/S, non-selective medium.
DISCUSSION
Centromeres are essential for the segregation of chromosomes during mitosis and meiosis, ensuring the faithful inheritance of genetic material. In this study, we characterized the function of the JmjC domain protein Msc1 in fission yeast, which is required for proper chromosome segregation (14). We found that Msc1 is an integral component of the Swr1 complex and that both Msc1 and Swr1 are required for the preferential incorporation of H2A.ZPht1 at gene promoters. Furthermore, loss of H2A.ZPht1 results in defects in centromere silencing and chromosome segregation without affecting CENP-ACnp1 levels at centromeres. Instead, H2A.ZPht1 is required for the expression of CENP-CCnp3, a centromere protein required for maintaining centromere silencing.
The Swr1 complex was originally characterized in S. cerevisiae, and it is required for the incorporation of H2A.ZHtz1 both in vitro and in vivo (29–31). With the exception of Msc1, the Swr1 complex of fission yeast is very similar to that of S. cerevisiae in composition (Figs. 1C and 2C) (33, 34). Msc1 does not seem to have a homolog in budding yeast, but it is essential for the localization of Swr1 to chromatin and H2A.ZPht1 incorporation at gene promoters (Fig. 3). Given the similar enrichment of H2A.Z at gene promoters in both organisms (Fig. 3) (34, 39–43), it is surprising that Msc1 is required for the function of the fission yeast Swr1 complex, as the budding yeast Swr1 complex could function without an Msc1 homolog.
One possibility is that Msc1 could replace the function of S. cerevisiae-specific subunits of the Swr1 complex, such as Bdf1, Swc5, and Swc7. Budding yeast Bdf1 contains two bromodomains that bind to acetylated histones (61, 62) and regulates H2A.ZHtz1 chromatin incorporation (41, 42), but whether fission yeast Bdf1 is required for H2A.Z incorporation has not been characterized. Budding yeast Swc5 is essential for Swr1-mediated histone H2A.Z exchange in vitro (63). The fission yeast genome contains a homolog of Swc5, and genome-wide genetic interaction analysis suggests that it is involved in Swr1 function as well (28). Although a separate study indicated that both Swc5 and Bdf1 are part of the fission yeast Swr1 complex (33), we detected only low levels of Bdf1 in some of our purifications and did not detect any Swc5 peptides. This discrepancy is possibly due to more stringent conditions used in our purifications and indicates that Bdf1 and Swc5 transiently interact with the Swr1 complex. Swc7 is a protein of unknown function, and no homolog of Swc7 is found in fission yeast. Although it is not required for Swr1 activity in vitro (64), whether it is required for H2A.ZHtz1 incorporation in vivo is not known, and Msc1 might functionally substitute for Swc7 in the fission yeast Swr1 complex.
Another possibility is that the chromatin environment of the fission yeast is quite different from that of the budding yeast, which necessitates the presence of Msc1 in fission yeast. For example, H2A.Z in fission yeast genetically interacts with heterochromatin machinery and the RNA interference pathway, which are absent in S. cerevisiae, to suppress antisense transcription (34). In addition, budding yeast does not seem to have some histone methylation marks, such as H3K9me and H4K20me (65–67). In that sense, the PHDs of JARID1 family proteins, such as mammalian RBP2 and fission yeast Lid2, bind to methylated histones to target these enzymes to correct genomic locations (27, 68). Thus, the PHD2 and PHD3 domains might recruit Msc1 to chromatin through histone lysine methylation, analogous to the function of budding yeast Bdf1 in recruiting the Swr1 complex through histone acetylation (41, 42). It is unlikely that Msc1 exerts its effect through histone demethylation, as Msc1 lacks some critical residues for catalysis (17), and mutations of critical residues within the JmjC domain do not affect Msc1 function (Fig. 6, D and E).
The centromeres of fission yeast maintain a unique chromatin environment as indicated by a diffuse micrococcal nuclease digestion pattern (7, 8) and silencing of reporter genes inserted within centromeres (9), both of which depend on the presence of histone H3 variant CENP-ACnp1 (10, 69). As nucleosomes reconstituted with CENP-A are more compact compared with those formed with histone H3 (5, 6), it seems that CENP-A could directly maintain the silenced chromatin environment. However, the fact that loss of Msc1, Swr1, or H2A.ZPht1 affected silencing at centromeres without affecting CENP-ACnp1 levels demonstrates that silencing is not imposed by CENP-ACnp1 itself (Fig. 4).
Our ChIP assays indicated that H2A.ZPht1 is not enriched at the centromere region (Fig. 5, A and B), thus ruling out the possibility that H2A.ZPht1 directly impacts centromere chromatin structure. In mammals, H2A.Z is enriched at pericentric heterochromatin in addition to centromeres, and H2A.Z is required for heterochromatin integrity (70, 71, 93). However, our results suggest that in fission yeast H2A.ZPht1 does not exert its effect on centromere function through heterochromatin (supplemental Fig. S2). Because H2A.Z has been shown to control gene expression (29–31, 72), it is plausible that H2A.ZPht1 affects the levels of other centromere components that function downstream of CENP-ACnp1 to regulate centromere silencing. Our results suggest that H2A.ZPht1 directly regulates the expression of CENP-CCnp3 (Fig. 5C), which is essential for centromere silencing and chromosome segregation (Fig. 5D) (54). However, given the pleiotropic defects associated with pht1Δ cells, it is possible that H2A.ZPht1 also regulates the expression of other proteins involved in centromere function, which were not detected in previous microarray analyses (51).
CENP-C was originally identified as a kinetochore protein from analysis of scleroderma patient autoantibody (73), and it is essential for kinetochore assembly (74–78). It is a member of the constitutive centromere-associated network that copurifies with CENP-A and colocalizes with CENP-A throughout the cell cycle (79–82). Kinetochore localization of CENP-C is downstream of CENP-A but upstream of most other kinetochore proteins (77, 78, 83, 84). Unlike other proteins of the constitutive centromere-associated network, CENP-C has been identified in all eukaryotes (52, 74, 85–87). Fission yeast CENP-CCnp3 also serves as a scaffold for the recruitment of other kinetochore proteins (54). However, its molecular function is not clear.
We found that CENP-CCnp3 is required for maintaining a silenced chromatin state at centromeres (Fig. 5D). The biological function of centromere silencing is not understood, although mutations that affect silencing of centromere reporter genes usually result in defects in chromosome segregation (10). It is possible that silencing is a byproduct of kinetochore assembly. However, the detection of transcripts derived from the centromere region in mammals and plants suggests that silencing at centromeres might have significant biological consequences (2), and it fits into a broad body of evidence implicating RNA in chromatin-related functions (88, 89). Whether centromeric sequences are transcribed in fission yeast is currently unknown, but the region is enriched for low levels of RNA polymerase II and methylated histone H3 lysine 4, a modification usually associated with gene transcription (90, 91). Interestingly, CENP-C binds to transcripts derived from human α-satellite DNA, and the transcripts are required for CENP-C localization (92). Thus, examining whether fission yeast centromeres are transcribed and elucidating the role this transcription might have in centromere function are promising new directions in centromere chromatin research.
Supplementary Material
Acknowledgments
We thank Kenichi Shimada for help with microarray analysis, Han Kim and Jie Zhang for assistance with strain construction, Shiv Grewal and Nancy Walworth for strains, Michael Keogh and Shiv Grewal for helpful discussions, and Bharat Reddy for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM085145 (to S. J.) and Grant P41 RR11823 from the National Center for Research Resources (to T. N. Davis). This work was also supported by a Basil O'Connor Starter Scholar Award from the March of Dimes Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- ChIP
- chromatin immunoprecipitation
- GFP
- green fluorescent protein
- WCE
- whole cell extract; reverse transcription
- PHD
- plant homeodomain.
REFERENCES
- 1.Cleveland D. W., Mao Y., Sullivan K. F. (2003) Cell 112, 407–421 [DOI] [PubMed] [Google Scholar]
- 2.Allshire R. C., Karpen G. H. (2008) Nat. Rev. Genet. 9, 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malik H. S., Henikoff S. (2009) Cell 138, 1067–1082 [DOI] [PubMed] [Google Scholar]
- 4.Allis C., Jenuwein T., Reinberg D. (2006) Epigenetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 5.Black B. E., Foltz D. R., Chakravarthy S., Luger K., Woods V. L., Jr., Cleveland D. W. (2004) Nature 430, 578–582 [DOI] [PubMed] [Google Scholar]
- 6.Black B. E., Brock M. A., Bédard S., Woods V. L., Jr., Cleveland D. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5008–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polizzi C., Clarke L. (1991) J. Cell Biol. 112, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi K., Murakami S., Chikashige Y., Funabiki H., Niwa O., Yanagida M. (1992) Mol. Biol. Cell 3, 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allshire R. C., Javerzat J. P., Redhead N. J., Cranston G. (1994) Cell 76, 157–169 [DOI] [PubMed] [Google Scholar]
- 10.Pidoux A. L., Richardson W., Allshire R. C. (2003) J. Cell Biol. 161, 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grewal S. I., Jia S. (2007) Nat. Rev. Genet. 8, 35–46 [DOI] [PubMed] [Google Scholar]
- 12.Bernard P., Maure J. F., Partridge J. F., Genier S., Javerzat J. P., Allshire R. C. (2001) Science 294, 2539–2542 [DOI] [PubMed] [Google Scholar]
- 13.Nonaka N., Kitajima T., Yokobayashi S., Xiao G., Yamamoto M., Grewal S. I., Watanabe Y. (2002) Nat. Cell Biol. 4, 89–93 [DOI] [PubMed] [Google Scholar]
- 14.Ahmed S., Dul B., Qiu X., Walworth N. C. (2007) Genetics 177, 1487–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrence R. J., Volpe T. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed S., Palermo C., Wan S., Walworth N. C. (2004) Mol. Cell. Biol. 24, 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klose R. J., Kallin E. M., Zhang Y. (2006) Nat. Rev. Genet. 7, 715–727 [DOI] [PubMed] [Google Scholar]
- 18.Shi Y., Whetstine J. R. (2007) Mol. Cell 25, 1–14 [DOI] [PubMed] [Google Scholar]
- 19.Eissenberg J. C., Lee M. G., Schneider J., Ilvarsonn A., Shiekhattar R., Shilatifard A. (2007) Nat. Struct. Mol. Biol. 14, 344–346 [DOI] [PubMed] [Google Scholar]
- 20.Huarte M., Lan F., Kim T., Vaughn M. W., Zaratiegui M., Martienssen R. A., Buratowski S., Shi Y. (2007) J. Biol. Chem. 282, 21662–21670 [DOI] [PubMed] [Google Scholar]
- 21.Iwase S., Lan F., Bayliss P., de la Torre-Ubieta L., Huarte M., Qi H. H., Whetstine J. R., Bonni A., Roberts T. M., Shi Y. (2007) Cell 128, 1077–1088 [DOI] [PubMed] [Google Scholar]
- 22.Klose R. J., Yan Q., Tothova Z., Yamane K., Erdjument-Bromage H., Tempst P., Gilliland D. G., Zhang Y., Kaelin W. G., Jr. (2007) Cell 128, 889–900 [DOI] [PubMed] [Google Scholar]
- 23.Lee M. G., Norman J., Shilatifard A., Shiekhattar R. (2007) Cell 128, 877–887 [DOI] [PubMed] [Google Scholar]
- 24.Lee N., Zhang J., Klose R. J., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2007) Nat. Struct. Mol. Biol. 14, 341–343 [DOI] [PubMed] [Google Scholar]
- 25.Secombe J., Li L., Carlos L., Eisenman R. N. (2007) Genes Dev. 21, 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamane K., Tateishi K., Klose R. J., Fang J., Fabrizio L. A., Erdjument-Bromage H., Taylor-Papadimitriou J., Tempst P., Zhang Y. (2007) Mol. Cell 25, 801–812 [DOI] [PubMed] [Google Scholar]
- 27.Li F., Huarte M., Zaratiegui M., Vaughn M. W., Shi Y., Martienssen R., Cande W. Z. (2008) Cell 135, 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roguev A., Bandyopadhyay S., Zofall M., Zhang K., Fischer T., Collins S. R., Qu H., Shales M., Park H. O., Hayles J., Hoe K. L., Kim D. U., Ideker T., Grewal S. I., Weissman J. S., Krogan N. J. (2008) Science 322, 405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krogan N. J., Keogh M. C., Datta N., Sawa C., Ryan O. W., Ding H., Haw R. A., Pootoolal J., Tong A., Canadien V., Richards D. P., Wu X., Emili A., Hughes T. R., Buratowski S., Greenblatt J. F. (2003) Mol. Cell 12, 1565–1576 [DOI] [PubMed] [Google Scholar]
- 30.Kobor M. S., Venkatasubrahmanyam S., Meneghini M. D., Gin J. W., Jennings J. L., Link A. J., Madhani H. D., Rine J. (2004) PLoS Biol. 2, E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizuguchi G., Shen X., Landry J., Wu W. H., Sen S., Wu C. (2004) Science 303, 343–348 [DOI] [PubMed] [Google Scholar]
- 32.Ruhl D. D., Jin J., Cai Y., Swanson S., Florens L., Washburn M. P., Conaway R. C., Conaway J. W., Chrivia J. C. (2006) Biochemistry 45, 5671–5677 [DOI] [PubMed] [Google Scholar]
- 33.Shevchenko A., Roguev A., Schaft D., Buchanan L., Habermann B., Sakalar C., Thomas H., Krogan N. J., Shevchenko A., Stewart A. F. (2008) Genome Biol. 9, R167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zofall M., Fischer T., Zhang K., Zhou M., Cui B., Veenstra T. D., Grewal S. I. (2009) Nature 461, 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998) Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 36.Jia S., Kobayashi R., Grewal S. I. (2005) Nat. Cell Biol. 7, 1007–1013 [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Reddy B., Thompson J., Wang H., Noma K., Yates J. R., 3rd, Jia S. (2009) Mol. Cell 33, 428–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 39.Guillemette B., Bataille A. R., Gévry N., Adam M., Blanchette M., Robert F., Gaudreau L. (2005) PLoS Biol. 3, e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li B., Pattenden S. G., Lee D., Gutiérrez J., Chen J., Seidel C., Gerton J., Workman J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18385–18390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raisner R. M., Hartley P. D., Meneghini M. D., Bao M. Z., Liu C. L., Schreiber S. L., Rando O. J., Madhani H. D. (2005) Cell 123, 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H., Roberts D. N., Cairns B. R. (2005) Cell 123, 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millar C. B., Xu F., Zhang K., Grunstein M. (2006) Genes Dev. 20, 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 45.Mavrich T. N., Jiang C., Ioshikhes I. P., Li X., Venters B. J., Zanton S. J., Tomsho L. P., Qi J., Glaser R. L., Schuster S. C., Gilmour D. S., Albert I., Pugh B. F. (2008) Nature 453, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whittle C. M., McClinic K. N., Ercan S., Zhang X., Green R. D., Kelly W. G., Lieb J. D. (2008) PLoS Genet. 4, e1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zilberman D., Coleman-Derr D., Ballinger T., Henikoff S. (2008) Nature 456, 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carr A. M., Dorrington S. M., Hindley J., Phear G. A., Aves S. J., Nurse P. (1994) Mol. Gen. Genet. 245, 628–635 [DOI] [PubMed] [Google Scholar]
- 49.Park Y. J., Chodaparambil J. V., Bao Y., McBryant S. J., Luger K. (2005) J. Biol. Chem. 280, 1817–1825 [DOI] [PubMed] [Google Scholar]
- 50.Luk E., Vu N. D., Patteson K., Mizuguchi G., Wu W. H., Ranjan A., Backus J., Sen S., Lewis M., Bai Y., Wu C. (2007) Mol. Cell 25, 357–368 [DOI] [PubMed] [Google Scholar]
- 51.Anders A., Watt S., Bähler J., Sawin K. E. (2008) Yeast 25, 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holland S., Ioannou D., Haines S., Brown W. R. (2005) Chromosome Res. 13, 73–83 [DOI] [PubMed] [Google Scholar]
- 53.Hayashi A., Asakawa H., Haraguchi T., Hiraoka Y. (2006) Mol. Biol. Cell 17, 5173–5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka K., Chang H. L., Kagami A., Watanabe Y. (2009) Dev. Cell 17, 334–343 [DOI] [PubMed] [Google Scholar]
- 55.Dul B. E., Walworth N. C. (2007) J. Biol. Chem. 282, 18397–18406 [DOI] [PubMed] [Google Scholar]
- 56.Li H., Ilin S., Wang W., Duncan E. M., Wysocka J., Allis C. D., Patel D. J. (2006) Nature 442, 91–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peña P. V., Davrazou F., Shi X., Walter K. L., Verkhusha V. V., Gozani O., Zhao R., Kutateladze T. G. (2006) Nature 442, 100–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi X., Hong T., Walter K. L., Ewalt M., Michishita E., Hung T., Carney D., Peña P., Lan F., Kaadige M. R., Lacoste N., Cayrou C., Davrazou F., Saha A., Cairns B. R., Ayer D. E., Kutateladze T. G., Shi Y., Côté J., Chua K. F., Gozani O. (2006) Nature 442, 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wysocka J., Swigut T., Xiao H., Milne T. A., Kwon S. Y., Landry J., Kauer M., Tackett A. J., Chait B. T., Badenhorst P., Wu C., Allis C. D. (2006) Nature 442, 86–90 [DOI] [PubMed] [Google Scholar]
- 60.Chen Z., Zang J., Whetstine J., Hong X., Davrazou F., Kutateladze T. G., Simpson M., Mao Q., Pan C. H., Dai S., Hagman J., Hansen K., Shi Y., Zhang G. (2006) Cell 125, 691–702 [DOI] [PubMed] [Google Scholar]
- 61.Ladurner A. G., Inouye C., Jain R., Tjian R. (2003) Mol. Cell 11, 365–376 [DOI] [PubMed] [Google Scholar]
- 62.Matangkasombut O., Buratowski S. (2003) Mol. Cell 11, 353–363 [DOI] [PubMed] [Google Scholar]
- 63.Wu W. H., Alami S., Luk E., Wu C. H., Sen S., Mizuguchi G., Wei D., Wu C. (2005) Nat. Struct. Mol. Biol. 12, 1064–1071 [DOI] [PubMed] [Google Scholar]
- 64.Wu W. H., Wu C. H., Ladurner A., Mizuguchi G., Wei D., Xiao H., Luk E., Ranjan A., Wu C. (2009) J. Biol. Chem. 284, 6200–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fang J., Feng Q., Ketel C. S., Wang H., Cao R., Xia L., Erdjument-Bromage H., Tempst P., Simon J. A., Zhang Y. (2002) Curr. Biol. 12, 1086–1099 [DOI] [PubMed] [Google Scholar]
- 66.Nishioka K., Rice J. C., Sarma K., Erdjument-Bromage H., Werner J., Wang Y., Chuikov S., Valenzuela P., Tempst P., Steward R., Lis J. T., Allis C. D., Reinberg D. (2002) Mol. Cell 9, 1201–1213 [DOI] [PubMed] [Google Scholar]
- 67.Garcia B. A., Hake S. B., Diaz R. L., Kauer M., Morris S. A., Recht J., Shabanowitz J., Mishra N., Strahl B. D., Allis C. D., Hunt D. F. (2007) J. Biol. Chem. 282, 7641–7655 [DOI] [PubMed] [Google Scholar]
- 68.Wang G. G., Song J., Wang Z., Dormann H. L., Casadio F., Li H., Luo J. L., Patel D. J., Allis C. D. (2009) Nature 459, 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takahashi K., Chen E. S., Yanagida M. (2000) Science 288, 2215–2219 [DOI] [PubMed] [Google Scholar]
- 70.Rangasamy D., Berven L., Ridgway P., Tremethick D. J. (2003) EMBO J. 22, 1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rangasamy D., Greaves I., Tremethick D. J. (2004) Nat. Struct. Mol. Biol. 11, 650–655 [DOI] [PubMed] [Google Scholar]
- 72.Meneghini M. D., Wu M., Madhani H. D. (2003) Cell 112, 725–736 [DOI] [PubMed] [Google Scholar]
- 73.Earnshaw W. C., Rothfield N. (1985) Chromosoma 91, 313–321 [DOI] [PubMed] [Google Scholar]
- 74.Saitoh H., Tomkiel J., Cooke C. A., Ratrie H., 3rd, Maurer M., Rothfield N. F., Earnshaw W. C. (1992) Cell 70, 115–125 [DOI] [PubMed] [Google Scholar]
- 75.Tomkiel J., Cooke C. A., Saitoh H., Bernat R. L., Earnshaw W. C. (1994) J. Cell Biol. 125, 531–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fukagawa T., Pendon C., Morris J., Brown W. (1999) EMBO J. 18, 4196–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwon M. S., Hori T., Okada M., Fukagawa T. (2007) Mol. Biol. Cell 18, 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hori T., Amano M., Suzuki A., Backer C. B., Welburn J. P., Dong Y., McEwen B. F., Shang W. H., Suzuki E., Okawa K., Cheeseman I. M., Fukagawa T. (2008) Cell 135, 1039–1052 [DOI] [PubMed] [Google Scholar]
- 79.Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., Yoda K. (2004) Genes Cells 9, 105–120 [DOI] [PubMed] [Google Scholar]
- 80.Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., 3rd, Cleveland D. W. (2006) Nat. Cell Biol. 8, 458–469 [DOI] [PubMed] [Google Scholar]
- 81.Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J. R., 3rd, Desai A., Fukagawa T. (2006) Nat. Cell Biol. 8, 446–457 [DOI] [PubMed] [Google Scholar]
- 82.Cheeseman I. M., Desai A. (2008) Nat. Rev. Mol. Cell Biol. 9, 33–46 [DOI] [PubMed] [Google Scholar]
- 83.Régnier V., Vagnarelli P., Fukagawa T., Zerjal T., Burns E., Trouche D., Earnshaw W., Brown W. (2005) Mol. Cell. Biol. 25, 3967–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu S. T., Rattner J. B., Jablonski S. A., Yen T. J. (2006) J. Cell Biol. 175, 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meluh P. B., Koshland D. (1995) Mol. Biol. Cell 6, 793–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moore L. L., Roth M. B. (2001) J. Cell Biol. 153, 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heeger S., Leismann O., Schittenhelm R., Schraidt O., Heidmann S., Lehner C. F. (2005) Genes Dev. 19, 2041–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bernstein E., Allis C. D. (2005) Genes Dev. 19, 1635–1655 [DOI] [PubMed] [Google Scholar]
- 89.Cam H. P., Chen E. S., Grewal S. I. (2009) Cell 136, 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cam H. P., Sugiyama T., Chen E. S., Chen X., FitzGerald P. C., Grewal S. I. (2005) Nat. Genet. 37, 809–819 [DOI] [PubMed] [Google Scholar]
- 91.Chen E. S., Zhang K., Nicolas E., Cam H. P., Zofall M., Grewal S. I. (2008) Nature 451, 734–737 [DOI] [PubMed] [Google Scholar]
- 92.Wong L. H., Brettingham-Moore K. H., Chan L., Quach J. M., Anderson M. A., Northrop E. L., Hannan R., Saffery R., Shaw M. L., Williams E., Choo K. H. (2007) Genome Res. 17, 1146–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Greaves I. K., Rangasamy D., Ridgway P., Tremethick D. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.