Abstract
The crystal structure of Escherichia coli NhaA determined at pH 4 has provided insights into the mechanism of activity of a pH-regulated Na+/H+ antiporter. However, because NhaA is activated at physiological pH (pH 5.5–8.5), many questions related to the active state of NhaA have remained elusive. Our experimental results at physiological pH and computational analyses reveal that amino acid residues in transmembrane segment II contribute to the cation pathway of NhaA and its pH regulation: 1) transmembrane segment II is a highly conserved helix and the conserved amino acid residues are located on one side of the helix facing either the cytoplasmic or periplasmic funnels of NhaA structure. 2) Cys replacements of the conserved residues and measuring their antiporter activity in everted membrane vesicles showed that D65C, L67C, E78C, and E82C increased the apparent Km to Na+ and Li+ and changed the pH response of the antiporter. 3) Introduced Cys replacements, L60C, N64C, F71C, F72C, and E78C, were significantly alkylated by [14C]N-ethylmaleimide implying the presence of water-filled cavities in NhaA. 4) Several Cys replacements were modified by MTSES and/or MTSET, membrane impermeant, negatively and positively charged reagents, respectively, that could reach Cys replacements from the periplasm only via water-filled funnel(s). Remarkably, the reactivity of D65C to MTSES increased with increasing pH and chemical modification by MTSES but not by MTSET, decreased the apparent Km of the antiporter at pH 7.5 (10-fold) but not at pH 8.5, implying the importance of Asp65 negative charge for pH activation of the antiporter.
Keywords: Membrane, Na+/H+ Antiporter, NhaA, NhaA pH-activation, Crystal Structure-based Functional Study, Transport Protein
Introduction
Sodium proton antiporters are ubiquitous membrane proteins found in the cytoplasmic and organelle membranes of cells of many different origins, including plants, animals, and microorganisms. They are involved in cell energetics and play primary roles in the regulation of intracellular pH, cellular Na+ content, and cell volume (see reviews in Refs. 1–3).
Ec-NhaA,2 the main Na+/H+ antiporter of Escherichia coli (henceforth NhaA), is indispensable for adaptation to high salinity, for challenging Li+ toxicity, and for growth at alkaline pH (in the presence of Na+ (2, 4)). It is widely spread in enterobacteria (1) and has orthologs throughout the biological kingdoms including humans (5).
NhaA is an electrogenic antiporter with a stoichiometry of 2H+/Na+ (2, 6). Similar to many other prokaryotic (2) and eukaryotic (7–11) Na+/H+ antiporters, the activity of NhaA is strongly dependent on pH; it changes over 3 orders of magnitude between pH 7.0 and 8.5 (2, 12, 13). This pH activation is accompanied by a conformational change as probed by a monoclonal antibody (monoclonal antibody 1F6 (14)) and by accessibility of NhaA to trypsin (15) or 2-(4′-maleimidylanilino)-naphthalene-6-sulfonic acid, a fluorescent probe (13).
The recently determined crystal structure of the acid pH down-regulated NhaA (16) has provided the first structural insight into the mechanism of antiport and pH regulation of a Na+/H+ antiporter (3). The structure shows that NhaA consists of 12 TMS with the N and C termini pointing into the cytoplasm, in line with previous biochemical and genetic results (17, 18). NhaA contains two negatively charged ion funnels; a cytoplasmic funnel, lined by TMSs II, IX, IVc, and V, opens to the cytoplasm and ends in the middle of the membrane at the putative ion binding site, where Asp164 of helix V is located (see Ref. 16 and Fig. 1a); a periplasmic funnel, lined by TMSs II, VIII, and XIp, opens to the periplasm and is separated from the cytoplasmic funnel by a group of densely packed hydrophobic residues.
FIGURE 1.
a, TMS II and the cytoplasmic and periplasmic cation funnels of NhaA. The crystal structure of helices comprising the cytoplasmic (blue) and periplasmic (green) funnels (black lines) of NhaA in addition to TMS II (maroon) are shown in ribbon representation viewed parallel to the membrane (gray broken lines). The figure was generated using the PyMOL program. TMS II is colored according to its evolutionary conservation using Consurf program. The color-coding bar, with turquoise through maroon indicates variable through conserved residues (the most variable, score 1 and the most conserved, score 9). The Cys-replaced residues on TMS II are marked in black and shown (ball and stick). For orientation, amino acids Lys57 and Glu252 are shown in yellow. b, helical wheel presentation of TMS II. A black background indicates conserved residues. Residues of which Cys replacements were alkylated by NEM are encircled in red (35–100%) or green (20–35%).
Hence, based on the crystal structure obtained at pH 4, TMS II is the only TMS that lines both the cytoplasmic and the periplasmic funnels of the protein (Fig. 1a), suggesting an important role in the ion translocation by NhaA. However, because the three-dimensional crystals were obtained at pH 4, at which NhaA is down-regulated, the structure of the active conformation(s) and the process(es) leading to it have remained open questions. To answer these questions, it is important to conduct experiments, both structurally and functionally oriented, at physiological pH when NhaA is active.
The present work explores the structural and functional role of TMS II in NhaA at physiological pH. Our results reveal that amino acid residues in TMS II participate in the cation passage of NhaA and its pH regulation. Furthermore, Asp65, which is located at the rim of the periplasmic funnel (16) changes its conformation upon increasing the pH and its negative charge is critical for the activation of the antiporter at alkaline pH.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
EP432 is an E. coli K-12 derivative, which is melBLid, ΔnhaA1::kan, ΔnhaB1::cat, ΔlacZY, thr1 (4). TA16 is nhaA+nhaB+lacIQ (TA15lacIQ) and otherwise isogenic to EP432 (12). Cells were grown either in L broth (LB) or in modified L broth (LBK (19)) in which NaCl was replaced by 87 mm KCl. Where indicated, the medium was buffered with 60 mm 1,3-bis-[tris(hydroxymethyl)-methylamino]propane. Cells were also grown in minimal medium A without sodium citrate (20) with 0.5% glycerol, 0.01% MgSO4·7H2O, and thiamine (2.5 μg/ml). For plates, 1.5% agar was used. Antibiotics were 100 μg/ml of ampicillin and/or 50 μg/ml of kanamycin. To test resistance to Li+ and Na+, EP432 cells transformed with the respective plasmids were grown on LBK to A600 of 0.5. Samples (2 μl) of serial 10-fold dilutions of the cultures were spotted onto agar plates containing the indicated concentrations of NaCl or LiCl at the various pH values and incubated for 2 days.
Plasmids
Plasmid pGM36 encodes NhaA (21). pCL-GMAR100, a derivative of pGM36 encodes Cys-less NhaA (CL-NhaA (17)). pECO, a derivative of pGMAR100 (with an EcoRI site at position 5319) encodes NhaA (22). Plasmid pAXH (previously called pYG10), a pET20b (Novagen) derivative encodes His-tagged NhaA (17). pCL-AXH, a derivative of pAXH encodes a His-tagged CL-NhaA (17). pCL-AXH2, a derivative of pCL-AXH lacks the BglII site at position 3382 (23). pCL-AXH3, a derivative of pCL-AXH2 contains the BstXI silent site at position 248 in nhaA. pCL-HAH4, a derivative of pCL-HAH3 bears the silent BstXI mutation at codon 248 in nhaA and encodes His-tagged CL-NhaA (22). All plasmids carrying mutations are designated by the name of the plasmid followed by the mutation.
Site-directed Mutagenesis
Site-directed mutagenesis was conducted following a polymerase chain reaction-based protocol (24) or DpnI-mediated site-directed mutagenesis (25). For Cys replacements W62C, F71C, F72C, and G76C, pCL-HAH4 was used as a template. For Cys replacements L60C, N64C, L67C, and M68C, pCL-AXH3 was used as a template. For Cys replacement E82C, pECO was used as a template. E78C was constructed as described in Ref. 26.
Mutation D65C was constructed on plasmid pAXH. pCL-HAH4D65C was obtained by ligating the EcoRI-BglII fragment (465 bp) from pAXHD65C with an EcoRI-BglII fragment (4.28 kb) of the plasmid pCL-HAH4. Mutants W62C, D65C F71C, F72C, G76C, and E82C on pCL-AXH3 background were obtained by ligating the EcoRI-BglII fragment (465 bp) with the EcoRI-BglII fragment (4.3 kb) of the plasmid pCL-AXH3. Mutants A118C and S246C were constructed as described in Ref. 22 and mutant H225C was previously described (27). All mutations were verified by DNA sequencing of the entire gene, through the ligation junction with the vector plasmid.
Isolation of Membrane Vesicles, Assay of Na+/H+ Antiporter Activity
EP432 cells transformed with the respective plasmids were grown and everted vesicles were prepared and used to determine the Na+/H+ or Li+/H+ antiporter activity as described (28, 29). The assay of antiporter activity was based upon the measurement of Na+- or Li+-induced changes in the ΔpH as measured by acridine orange, a fluorescent probe of ΔpH. The fluorescence assay was performed with 2.5 ml of reaction mixture containing 50–100 μg of membrane protein, 0.5 μm acridine orange, 150 mm KCl, 50 mm 1,3-bis-[tris(hydroxymethyl)-methylamino]propane, 5 mm MgCl2, and the pH was titrated with HCl. After energization with either ATP (2 mm) or d-lactate (2 mm), quenching of the fluorescence was allowed to achieve a steady state, and then either Na+ or Li+ was added. A reversal of the fluorescence level (dequenching) indicates that protons are exiting the vesicles in antiport with either Na+ or Li+. As shown previously the end level of dequenching is a good estimate of the antiporter activity (30) and the concentration of the ion that gives half-maximal dequenching is a good estimate of the apparent Km of the antiporter (30, 31). The concentration range of the cations tested was 0.01–100 mm at the indicated pH values and the apparent Km values were calculated by linear regression of a Lineweaver-Burk plot.
Detection and Quantitation of NhaA and Its Mutated Derivatives in the Membrane
Total membrane protein was determined according to Ref. 32. The expression level of His-tagged NhaA mutants was determined by resolving the Ni-NTA purified proteins on SDS-PAGE, staining the gels by Coomassie Blue, and quantification of the band densities by Image Gauge (Fuji) software (17).
Determination of Accessibility to NEM
TA16 cells were transformed with nhaA mutant-expressing plasmids. Preparation of membrane vesicles was carried out as described above, but with no dithiothreitol in the resuspension medium. The membrane vesicles (200 μg) were resuspended in 500 μl of buffer containing 100 mm potassium phosphate, pH 7.5, and 5 mm MgSO4. Membranes were treated with 0.5 mm [14C]NEM (specific activity of 4 μCi/μmol) and the NEM-alkylated residues were quantified as a percentage of the control after normalization by the amount of the protein, as described (17). The standard deviation was between 5 and 10%.
Accessibility to Membrane Impermeant Sulfhydryl Reagent MTSES/MTSET
The accessibility test in intact cells was performed as described (33) but the Tris buffer was replaced by 100 mm potassium phosphate and 5 mm MgSO4 at different pH values as indicated, and the incubation time with the SH reagents was 20 min at room temperature. The reaction was stopped by dilution into 40 ml of potassium Pi buffer at the respective pH values and centrifuged (10,000 × g). The cells were washed twice, resuspended in 1 ml of the respective potassium Pi buffer and sonicated (three times for 20 s at 4 °C, Vebra Cell Model VCX 750). Unbroken cells were removed by centrifugation (10,000 × g) and the membranes collected by centrifugation (Beckman, TLA 100.4, 265,000 × g for 30 min at 4 °C) were resuspended in 0.5 ml of TSC buffer (10 mm Tris, pH 7.5, 250 mm sucrose, and 140 mm choline chloride). The protein was extracted and affinity purified on Ni2+-NTA (Qiagen) and left bound on the beads. The beads were washed twice in binding buffer (17) at pH 7.4, washed in 500 μl of SDS-urea buffer (containing 6 m urea, 20 mm Tris, pH 7.5, 2% SDS, and 500 mm NaCl), and resuspended in 100 μl of the SDS-urea buffer containing 0.5 mm fluorescein 5-maleimide (Molecular Probes) and further incubated for 30 min at 23 °C to determine the free Cys following MTSES/MTSET treatment. Then, the beads were washed with 500 μl of the SDS-urea buffer, and the protein was eluted in a sample buffer containing 300 mm imidazole and separated on SDS-PAGE. For evaluation of the fluorescence intensity, the gels were photographed under UV light (260 nm) as described (22). For quantifying the amount of the protein, the gels were Coomassie Blue-stained and the density of the bands was determined. After normalization of the fluorescence intensity to the amount of protein in the band, the accessibility to MTSES/MTSET was determined from the difference in the fluorescence of the reagent-treated and untreated samples (100% fluorescence = 0% accessibility (22)). The standard deviation was between 5 and 10%.
The Effect of SH Reagents on the Antiporter Activity
Everted membrane vesicles were isolated from EP432 cells transformed with the indicated plasmids. Membranes (0.5 mg of membrane protein) were resuspended in a reaction mixture (0.5 ml) containing 5 mm MgSO4, 100 mm potassium Pi, pH 7.5, and one of the following reagents: 10 mm MTSES (Anatrace) or 10 mm MTSET, (Anatrace), and incubated for 20 min at room temperature with gentle shaking. The reaction was stopped by addition of 3 ml of TSC and then membranes were centrifuged (Beckman, TLA 100.4, 265,000 × g, 20 min, 4 °C). For measurement of Na+/H+ antiporter activity, the treated everted membranes were resuspended in TSC (5–10 mg of membrane protein/ml).
Computational Methods and Coordinates
The initial atomic model of the wild type enzyme was derived from crystallographic data obtained under cryo-conditions of NhaA from E. coli at 3.45-Å resolution (Protein Data Bank entry 1ZCD). NhaA consists of 388 amino acid residues with N and C termini exposed to the cytoplasm. The atomic model comprises residues 9–384, which form 12 TMSs (16).
Continuum Electrostatics
Multiconformation continuum electrostatic calculations (MCCE version 2.0) have been used to investigate the pH dependence of the D65C variant electrostatics (34–36). The interior dielectric constant of the protein was set to 4, and the solvent dielectric constant was set to 80 with an ionic strength of 150 mm. The molecular surface of the protein as the boundary between high and low dielectric medium was defined by a solvent probe of radius 1.4 Å (37). The protein backbone and nonpolar side chains were fixed. Hydrogens were added to the structure after creating side chain rotamers. The program DELPHI (38) was used with PARSE (parameters of solvation energies) atomic charges and radii (39). Monte Carlo sampling yielded the conformer occupancies according to a Boltzmann distribution of states as a function of pH. Conformation and ionization degrees of freedom were simultaneously relaxed to low-energy states.
RESULTS
Cys Replacement Mutations of Conserved Residues in TMS II, Expression in the Membrane and Growth Phenotypes
The structure of the acid-locked down-regulated conformation of NhaA has revealed that TMS II lines both the cytoplasmic and periplasmic cation funnels; the former funnel ends at the cation binding site (Asp164) and is separated from the latter funnel by a hydrophobic barrier (see Ref. 16 and Fig. 1a). Alignment of the NhaA protein sequence with the updated available sequences of putative prokaryotic Na+/H+ antiporters (UniProt/TrEMBL databases (40)) shows that the residues of TMS II are highly conserved (Fig. 1a). Furthermore, the conserved amino acid residues are located on one face of helix II, which according to the crystal structure lines the cytoplasmic or periplasmic funnels (Fig. 1, a and b). We therefore considered the possibility that amino acid residues in TMS II line the cation passage of NhaA at physiological pH, when NhaA is active, and are important for the functionality of NhaA. To test this suggestion, we constructed Cys-replacement mutations of the most conserved amino acid residues in TMS II (Fig. 1a) in Cys-less NhaA (CL-NhaA), a variant expressed and active as the native NhaA (17).
To characterize the mutations with respect to expression, growth, and antiporter activity (Table 1 and Fig. 2), the mutated plasmids were transformed into EP432, an E. coli strain that lacks the two Na+-specific antiporters (NhaA and NhaB). This strain, neither grows on selective medium, (0.6 m NaCl at pH 7 or pH 8.3 or 0.1 m LiCl at pH 7.0) nor exhibits any Na+/H+ antiporter activity in isolated everted membrane vesicles, unless transformed with a plasmid encoding an active antiporter (reviewed in Ref. 2).
TABLE 1.
Single Cys replacements of conserved residues in TMS II of NhaA
For characterization of the mutations, EP432 cells transformed with the plasmids carrying the indicated mutations were used. The expression level was expressed as percentage of the control cells (EP432/pCL-AXH3). Na+/H+ and Li+/H+ antiporter activity at pH 8.5 was determined with 10 mm NaCl or LiCl. The activity is expressed as percentage of dequenching. EP432/pBR322 served as a negative control. The apparent Km for the ions was determined at pH 8.5, as described under “Experimental Procedures.”
| Mutation in TMS II | Expression | Growth |
Activity |
Apparent Km |
||||
|---|---|---|---|---|---|---|---|---|
| Na+ (7) | Na+ (8.3) | Li+ (7) | Na+ | Li+ | Na+ | Li+ | ||
| % | mm | |||||||
| L60C | 165 | +++ | ++ | +++ | 83 | 91 | 0.15 | 0.02 |
| W62C | 100 | +++ | ++ | +++ | 73 | 90 | 1.0 | 0.02 |
| N64C | 190 | +++ | ++ | +++ | 93 | 85 | 0.12 | 0.02 |
| D65C | 52 | +++ | ++ | +++ | 56 | 65 | 2.26 | 0.7 |
| L67C | 100 | +++ | ++ | +++ | 62 | 84 | 1.8 | 0.67 |
| M68C | 150 | +++ | ++ | +++ | 58 | 52 | 0.41 | 0.02 |
| F71C | 100 | + | – | +++ | 83 | 55 | 0.83 | 0.14 |
| F72C | 76 | +++ | ++ | +++ | 67 | 43 | 0.54 | 0.02 |
| G76C | 82 | +++ | ++ | +++ | 80 | 73 | 0.18 | 0.02 |
| E78C | 22 | +++ | ++ | +++ | 40 | 24 | 1.12 | 0.68 |
| E82C | 17 | +++ | ++ | +++ | 33 | 12 | 2.61 | NDa |
| Controls | ||||||||
| pCL-AXH3 | 100 | +++ | ++ | +++ | 88 | 75 | 0.19 | 0.07 |
| pBR322 | – | – | – | – | – | – | – | |
a ND, not determined.
FIGURE 2.
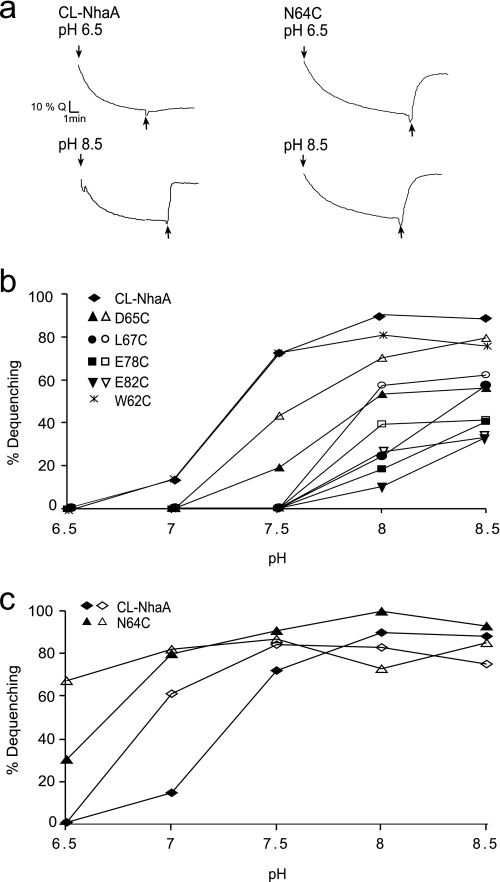
Li+/H+ and Na+/H+ antiporter activity in everted membrane vesicles of NhaA mutants in TMS II. Everted membrane vesicles were prepared from EP432 cells expressing the indicated NhaA mutants and grown in LBK, pH 7. The Na+/H+ antiporter activity was determined using acridine orange fluorescence to monitor ΔpH. a, the data of typical experiments are shown. At the onset of the reaction, ATP or d-lactate (2 mm) were added (↓) and the fluorescence quenching (Q) was recorded until a steady-state level of ΔpH (100% quenching) was reached. LiCl (10 mm) was then added (↑), and the new steady state of fluorescence obtained (dequenching) was monitored. b, the Na+/H+ antiporter activity was determined at the indicated pH values in the presence of 10 mm NaCl (closed symbols) or 100 mm NaCl (open symbols). The results are as the end level of dequenching (%). c, the Na+/H+ and Li+/H+ antiporter activity were determined as in b in the presence of 10 mm NaCl (closed symbols) or 10 mm LiCl (open symbols) at the indicated pH values. All experiments were repeated at least three times with practically identical results.
Although most variants were significantly (≥52%) expressed as compared with the level of expression of the wild type (100%), Cys-replacement mutants E78C and E82C were expressed to about 20% of the control level (Table 1). Notably, because all variants are expressed from multicopy plasmids, even the low level of expression is way above the level expressed from a single chromosomal gene, which confers a Na+ resistance phenotype (41).
Except for mutant F71C, all variants grew similar to the wild type on the selective agar plates. For unknown reasons, mutant F71C grew slower than the wild type on the NaCl selective medium at neutral pH and did not grow at all at alkaline pH (Table 1).
The Effect of Cys-replacement Mutations in TMS II on the Na+/H+ and Li+/H+ Antiporter Activity
The Na+/H+ and Li+/H+ antiport activity was measured in everted membrane vesicles isolated from EP432 transformed with plasmids encoding the various mutations. The activity was estimated from the change caused by either Na+ or Li+ to the ΔpH maintained across the membrane, as measured by acridine orange, a fluorescent probe of ΔpH. After energization (Fig. 2a, down arrow) with either ATP or d-lactate, quenching of the fluorescence achieved a steady state level and then Li+ (Fig. 2a, up arrow) or Na+ (data not shown) was added (further details under “Experimental Procedures”). EP432 transformed with plasmid pCL-AXH3 encoding CL-NhaA or the vector plasmid, pBR322, served as positive and negative controls, respectively (Fig. 2a and Table 1). The apparent Km values for Na+ and Li+ at pH 8.5 and the extent of activity (maximal dequenching) at pH 8.5 were determined for each mutant (Table 1 and data not shown). Mutations D65C, L67C, E78C, and E82C significantly increased the apparent Km of the antiporter to Na+ (6–10-fold) and mutations D65C, L67C, and E78C caused a similar increase of the apparent Km of the antiporter to Li+ (10-fold for all three mutants, Table 1). The apparent Km of the antiporter to Li+ of variant E82C was not determined due to very low activity (12% in the presence of 100 mm Li+). The other mutants did not affect the apparent Km for either Na+ or Li+ (Table 1).
We have previously shown (23, 42) that, when measured at saturating concentrations of Na+, mutations that affect the apparent Km but not the pH dependence of NhaA, show a pH dependence very similar to that of the wild type at saturating concentration of the substrate. In contrast, mutations that affect both the apparent Km and the pH dependence of the antiporter retain the abnormal pH dependence even at saturating concentrations of the ions.
We therefore measured the effect of Na+ concentrations (10 versus 100 mm) on the pH dependence of the activity of the variants. Variants D65C, L67C, E78C, and E82C were of the second type; both at low and saturating concentrations of Na+ they showed an alkaline pH shift in their activity profile (Fig. 2b). Whereas, only a small shift was caused by Cys replacement D65C (about 0.5 of a pH unit), a shift of 1 pH unit was caused by the other three Cys replacement mutants. These mutations also caused an alkaline shift of the pH dependence of the Li+/H+ antiporter activity (data not shown). Mutation N64C caused an acidic shift in the pH dependence of the Na+-dependent antiport and lost the pH dependence of the Li+/H+ antiporter activity; it was as active with Li+ at pH 6.5 as at 8.5 (Fig. 2c). The rest of the mutants showed Na+/H+ and Li+/H+ antiport activity with a pH profile similar to that of the wild type (for example, see Fig. 2b shows W62C). Taken together, our experiments carried out at physiological pH found novel mutants in TMS II that markedly increase the apparent Km of the antiporter to the respective ions (D65C, L67C, E78C, and E82C) and change the pH dependence of NhaA (N64C, D65C, L67C, E78C, and E82C).
Accessibility to [14C]NEM of the Cys-replacement Mutations in TMS II
At the present resolution (3.45 Å), water molecules cannot be detected in the crystal structure of NhaA (16). We therefore tested whether Cys replacements in TMS II are accessible to [14C]NEM, a membrane-permeable SH-reagent. Modification of Cys by this reagent depends upon the ionization of the Cys sulfhydryl to its thiolate form, which occurs in the presence of water. Therefore, chemical modification by [14C]NEM may reflect the presence of water.
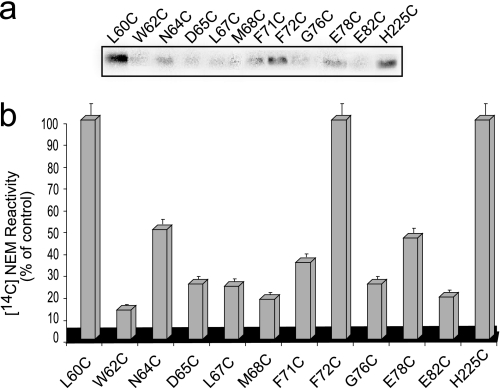
Membrane vesicles, expressing the various Cys-replacement variants, constructed in CL-NhaA were incubated with [14C]NEM at pH 7.5. Then, the His-tagged antiporters were affinity purified, separated on SDS-PAGE, and the radioactivity of the proteins was measured by autoradiography (Fig. 3a) and normalized to the amount of protein (see Figs. 1b and 3b). As a positive control (100% labeling), we used H225C, which was previously shown to be efficiently labeled by [14C]NEM (17). As a negative control we used CL-NhaA, which is not labeled by [14C]NEM (0% labeling; data not shown and see Ref. 17). We also verified that all inserted cysteines can theoretically react with NEM (supplemental Fig. S1). The standard deviation of this experiment is between 5 and 10%. Therefore, [14C]NEM labeling above 20% was considered significant. Variants W62C, M68C, and E82C were not accessible to NEM. Variants D65C, L67C, and G76C were moderately labeled (20–35%) (see Figs. 1b and 3b) and variants L60C, N64C, F71C, F72C, and E78C were strongly labeled (35–100%) (see Figs. 1b and 3b). The results highly suggest that many of the Cys-replaced residues in TMS II line a water-filled passage.
FIGURE 3.
Accessibility of Cys-replacements in TMS II to [14C]NEM. a, membrane vesicles were prepared from cells expressing the Cys-replacement mutants in TMS II of Cys-less NhaA. Membrane vesicles (200 μg of protein) were incubated with [14C]NEM as described under “Experimental Procedures.” The membranes were washed, solubilized, and the protein was affinity purified. Equal volumes of the purified protein were resolved on SDS-PAGE. The dried gel was exposed to a phosphorimager (Fuji Bas 1000). b, for quantification, the radioactivity was expressed as % of the control after normalization by the amount of the protein and presented graphically. The standard deviation (5–10%) is indicated (error bars).
Accessibility of the Cys-replacement Mutations in TMS II to MTSES/MTSET
We next tested whether MTSES and MTSET, membrane impermeant and water-soluble reagents, can penetrate the funnels and reach the Cys replacements lining the funnels. MTSES and MTSET are negatively and positively charged (43, 44), respectively, with diameters quite similar to the diameter of hydrated Na+ (7.2 Å diameter); MTSET is about 11.5 Å long and 6 Å wide.
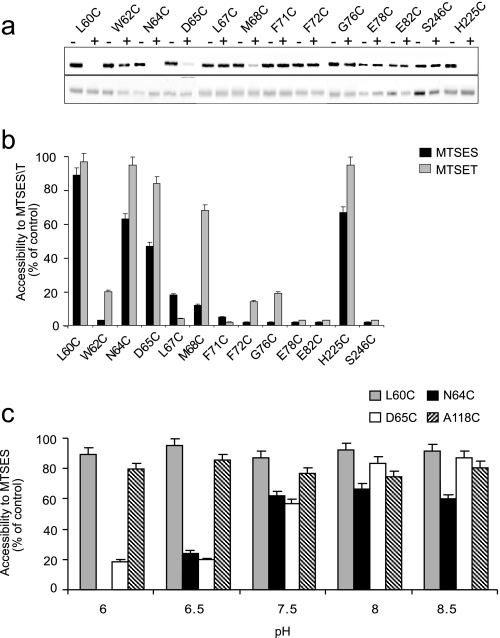
Test of accessibility of the Cys replacements to MTSES (Fig. 4, a and b) and MTSET (Fig. 4b) from the periplasmic side of the membrane was conducted at physiological pH when NhaA is active. Intact cells were incubated with MTSES or MTSET at pH 7.5. Then, NhaA was affinity purified on Ni2+-NTA beads, left bound on the beads, and treated with fluorescein maleimide, a fluorescent SH-reagent. The latter has previously been shown to bind any free Cys in purified NhaA protein and not to bind to CL-NhaA (22). Following separation of the proteins on SDS-PAGE, the level of fluorescence (Fig. 4a, upper panel) and the amount of the proteins on the gels (Fig. 4a, lower panel) were monitored and the fluorescent intensity was normalized to the amount of the proteins. For each variant, an untreated control was processed and its fluorescent level was used as 100% (0% accessibility) to express the MTSES/MTSET accessibility in percent. Variant H225C (17), located at the periplasmic end of TMS VIII and A118C (22), located at loop III-IV, were previously shown to be exposed to the periplasm and served as positive controls (Fig. 4, a–c). Variant S246C located on cytoplasmic loop VIII–IX (22) served as a negative control (Fig. 4, a and b).
FIGURE 4.
Accessibility of Cys replacements in TMS II to MTSES/MTSET from the periplasm. Intact cells expressing the Cys-replacement mutants in TMS II of Cys-less NhaA were incubated with MTSES or MTSET as described under “Experimental Procedures” at pH 7.5. Proteins were purified by Ni2+-NTA affinity chromatography and labeled on the beads with fluorescein maleimide for estimating the percentage of free cysteines remaining. The eluted proteins were resolved on SDS-PAGE, and the fluorescence level was monitored and photographed under UV light. Then, the gel was stained with Coomassie Blue and photographed under ordinary light to assess the protein concentration. a, MTSES (top panel, fluorescence of fluorescein-maleimide; bottom panel, protein level). b, the fluorescence intensity was normalized by the respective protein concentration and expressed in % of the fluorescence intensity of the untreated control. Accessibility to MTSES/T = 100% − % fluorescent intensity of the treated sample, and is presented by bars. The standard deviation was between 5 and 10% as indicated. Black bars, accessibility to MTSES; gray bars, accessibility to MTSET. c, intact cells expressing the indicated variant were incubated with MTSES at different pH values (pH 6–8.5) and processed as above. The standard deviation was between 5 and 10% as indicated (error bars).
The cysteines of variants L60C, N64C, and D65C were accessible both to MTSES and MTSET in intact cells (i.e. from the periplasmic side), about 90, 60, and 50% of the control for MTSES, and about 100, 95, and 85% of the control for MTSET, respectively. Remarkably, the cysteine of variant M68C was accessible to about 70% to MTSET but not to MTSES (Fig. 4b). Hence, the accessibility to both reagents decreased with the depth in the membrane but the positively charged MTSET was more reactive than the negatively charged MTSES and reached a deeper depth inside the funnel.
The Effect of pH on Accessibility of Cys Replacements to MTSES from the Periplasmic Side of the Membrane
In the crystal structure determined at pH 4, Asp65 is located at the rim of the periplasmic funnel (16), in close proximity to the hydrophobic barrier separating the periplasmic and cytoplasmic funnels (Fig. 1a). We therefore, considered the possibility that D65C changes its conformation with pH and used intact cells to test whether D65C and N64C change their accessibility to MTSES at different pH values (pH 6.5–8.5) as compared with other residues in TMS II. The rate of the chemical modification itself is pH sensitive. Therefore, for control we used two variants: A118C, which is located in loop III–IV on the periplasmic side of the membrane (22) and variant L60C, which is located on TMS II at the periplasmic side (Fig. 1a). The control variants were accessible to MTSES to the same level throughout the tested pH range (80–90% for both mutants, Fig. 4c). As a control for nonspecific binding of MTSES, we used CL-NhaA and no labeling was obtained throughout the pH range (data not shown). Variants D65C and N64C changed their reactivity to MTSES, with pH; whereas at pH 6.5 they had a very low reactivity, it increased as the pH increased to pH 8.5 (Fig. 4c). The other Cys replacements in TMS II did not change their MTSES accessibility with pH (Table 2).
TABLE 2.
MTSES accessibility of Cys replacements in TMS II at pH 6.5 and pH 8.5 from the periplasm
Accessibility test to MTSES (10 mm) in intact cells was conducted as described in Fig. 4 at pH 6.5 and pH 8.5.
| Residue | pH 6.5 | pH 8.5 |
|---|---|---|
| % of control | ||
| L60C | 94 | 97 |
| W62C | 3 | 5 |
| N64C | 18 | 50 |
| D65C | 14 | 97 |
| L67C | 8 | 10 |
| M68C | 4 | 6 |
| F71C | 3 | 6 |
| F72C | 2 | 3 |
| G76C | 2 | 2 |
| E78C | 2 | 5 |
| E82C | 1 | 5 |
The Effect of MTSES/MTSET on the Activity and pH Profile of D65C
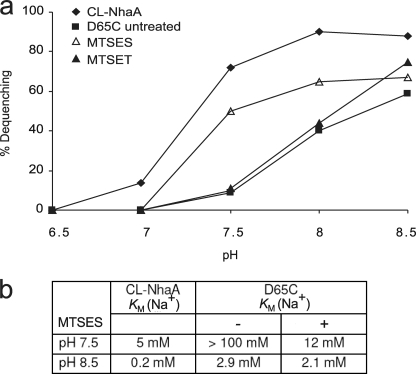
To test the effect of chemical modification by MTSES and MTSET on the Na+/H+ antiport activity of D65C, everted membrane vesicles were treated with the reagents at pH 7.5, and then the Na+/H+ antiporter activity and its dependence on pH was determined. For a control we used untreated membranes (Fig. 5a). At pH 7.5, the apparent Km for Na+ of CL-NhaA was 5 mm and at pH 8.5 it was 0.2 mm (Fig. 5b). The pH effect on the apparent Km for Na+ was slightly larger than that of wild type NhaA (15). This result suggests that there are two steps in pH activation of NhaA between pH 7.5 and 8.5: one involves a pH effect on the apparent Km; the second involves a more drastic pH effect on the Vmax of NhaA, which increases by 3 orders of magnitude between pH 7.5 and 8.5 (2, 15).
FIGURE 5.
Treatment with MTSES drastically reduces the apparent Km of the mutant D65C. a, everted membrane vesicles isolated from EP432 cells expressing D65C or the control CL-NhaA were treated or not with MTSES or MTSET and tested for antiporter activity at various pH values. Each experiment was repeated at least three times, and the results were essentially identical. b, the apparent Km for Na+ of the MTSES-treated and untreated variants are shown.
Cys replacement D65C increased by more than 10-fold the apparent Km for Na+ at pH 7.5 and 8.5 as compared with the CL-NhaA control (see Table 1 and Fig. 5b). Whereas chemical modification by the positively charged MTSET had no effect (see Fig. 5 and data not shown) on the apparent Km for Na+ of D65C membranes, the negatively charged MTSES decreased (by more than 10-fold) the apparent Km for Na+ of the variant at pH 7.5 (Fig. 5b) bringing it almost to that of the CL-NhaA control. However, at pH 8.5 there was no effect of MTSES on the apparent Km for Na+ of D65C. The chemical modification had no effect on the pH dependence of D65C; the MTSES modified D65C had a pH dependence (at saturating Na+ concentration) similar to that of D65C (compare Figs. 2b and 5a). These results imply that a negative charge at position Asp65 of TMS II is critical for the apparent Km for Na+ of D65C at pH 7.5 but not at pH 8.5.
Computational Analysis Predicts a pH-induced Conformational Change of Mutant D65C
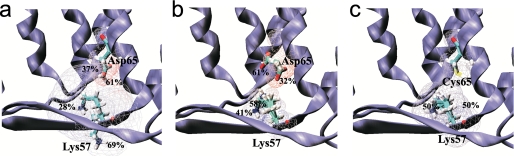
Our MTSES accessibility tests imply that at physiological pH the conformation of both N64C and D65C changes with respect to that in the crystal structure at pH 4. Although, as yet, we do not have the crystal structure of NhaA at physiological pH, we can use the available crystal structure at pH 4 to compute the possible conformations that Asp65 can acquire at physiological pH values. For this purpose we applied MCCE calculations. In the crystal structure at pH 4 (see Ref. 16 and Fig. 1a), Asp65 is located 3.1 Å away from the nitrogen atom of Lys57 (in the periplasmic β sheet), 16 Å away from Asp164 at the bottom of the cytoplasmic funnel (part of the putative cation binding site), and about 18 Å away from Glu252 (TMS IX) of the “pH sensor” at the orifice of the cytoplasmic funnel. Our previous MCCE calculations (45) demonstrated that the pK of Asp65 is about 6.7 so that at pH 4 of most side chains of Asp65 are protonated and have a conformation similar to that in the crystal structure (facing the periplasm) (Fig. 6a). In contrast, at pH 8, deprotonation of Asp65 triggers a pH-dependent structural change of its side chain (Fig. 6b). At alkaline pH, according to the results of our calculations 32% of Asp65 side chains adopt a conformation similar to that in the crystal structure, whereas 61% adopt a conformation in the direction of the ion binding site/cytoplasm. The carbonyl oxygen faces a cavity at the rim of the periplasmic funnel and is likely in contact with a water molecule. Indeed, MD simulation at pH 8 confirmed the reorganization of the Asp65 side chain into the nonpolar barrier associated with penetration of water into the hydrophobic region (46). The remaining 7% of Asp65 side chains occupy an intermediate conformation. Therefore, conformational reorientation of Asp65 side chains is pH-dependent and influences the local polarizability of the antiporter. These calculations support our experimental results implying a pH-induced conformational change at Asp65 of NhaA. Most probably due to its vicinity to Asp65, Asn64 behaves similar to Asp65.
FIGURE 6.
Most occupied conformers of Asp65 and Lys57 of NhaA for the wild type and D65C mutant. a, MCCE analysis shows that in the wild type NhaA, Asp65 has two conformations almost identical to that in the crystal structure at pH 4 and Lys57 side chain adopts two conformations with 69 and 28% occupancy. b, at pH 8, deprotonation of the Asp65 side chain with occupancy of 61% orients into the direction of the ion binding site; side chain with occupancy of 32% corresponds to the conformation similar to that of the crystal structure. c, two different conformations of Lys57 side chains in the D65C variant at pH 8 with equal occupancies (50%). Electrostatic potential maps were calculated using GRASP software. Zones of negative and positive potentials are colored red and blue, respectively.
To explore the influence of the negative charge of Asp65 on the electrostatic properties of NhaA, we performed MCCE calculations for mutant D65C. The most significant shift of a pKa value was observed for residue Lys57. Its pKa decreases from 12.2 (45) in the wild type to 8.0 in variant D65C, indicating that at pH 8, about 50% of Lys57 side chains are deprotonated and adopt a conformation very similar to that in the crystal structure (Fig. 6c). The other 50% of the Lys57 side chains are protonated and show reorientation (Fig. 6c). Therefore, Lys57 becomes pH sensitive at the pH range between pH 7.5 and 8.5 in variant D65C.
DISCUSSION
The three-dimensional crystal structure of the acidic pH down-regulated NhaA has provided the basis for understanding the mechanism of Na+/H+ exchange and its unique regulation by pH (3, 16). Furthermore, it paved the way to conduct structure-based studies aimed to answer questions that have remained open. These questions, in particular, are related to the open active conformation of NhaA at physiological pH. In contrast to the crystal structure obtained at pH 4, activation of NhaA occurs above pH 6.5 and the cation exchange rate is maximal at pH 8.5 (2, 12). Here, we conducted a structure-based biochemical study at physiological pH combined with computational analysis and show that TMS II of NhaA is functionally important at physiological pH.
TMS II Bears Conserved Residues of Which Cys Replacement Change the NhaA Antiport Kinetics
Evolutionary conservation analysis showed that residues of TMS II are highly conserved and the conserved amino acid residues are located on one side of the helix facing either the cytoplasmic or periplasmic funnels observed in the crystal structure at pH 4 (Fig. 1, a and b). Cys scanning mutagenesis of the conserved residues and measuring the antiporter activity of the Cys replacements at physiological pH values identified new amino acid residues of which Cys replacements increased the apparent Km to Na+ and Li+; Mutations D65C, L67C, E78C, and E82C increased the apparent Km of the antiporter to Na+ by 6–10-fold and mutations D65C, L67C, and E78C increased the apparent Km of the antiporter to Li+ by 10-fold (see Table 1 and Fig. 2). Cys replacements N64C, D65C, L67C, E78C, and E82C changed the pH dependence of NhaA (Fig. 2, b and c).
It is apparent that our experimental study presented here has been conducted on Cys-replacement variants that are normally expressed and exhibit full or at least a partial antiport activity in the membrane (see Table 1). Therefore, the results reflect the wild type NhaA active conformation.
TMS II Participates in the NhaA Cation Passage to the Active Site at Physiological pH
The crystal structure at pH 4 shows that TMS II is the only TMS that lines both the cytoplasmic and periplasmic funnels of NhaA and crosses the hydrophobic barrier separating the funnels (see Ref. 16 and Fig. 1a). This structural insight has raised the suggestion that at physiological pH, both funnels exist and TMS II lines the cation passage of NhaA. Indeed, our experimental results, obtained at physiological pH, revealed that amino acid residues in TMS II participate in the cation translocation pathway of NhaA. 1) As described above and later, Cys replacement of many TMS II conserved residues increase the apparent Km of NhaA to either Na+ or Li+ and change the pH response of the antiporter. 2) Several of the conserved Cys replacements were alkylated significantly (above 20%) by [14C]NEM implying the presence of water needed for Cys ionization. The Cys replacements that are most strongly labeled by [14C]NEM cluster on one face of TMS II that according to the crystal structure lines the cytoplasmic (F71C, F72C, and E78C) or the periplasmic (L60C and N64C) funnels (see Figs. 1b and 3, a and b). A lower but significant level of NEM accessibility was observed with D65C and L67C, which are located near the hydrophobic barrier and with G76C that most probably faces the membrane lipids (Fig. 1). Residues either in the barrier (M68C) or facing the lipids (W62C and E82C) were not significantly labeled. 3) Many of the Cys residues replacing the conserved residues were modified by MTSES and/or MTSET, membrane impermeant water-soluble charged reagents that can reach the introduced Cys from the periplasm only via water-filled funnels.
Notably, other parameters such as stereochemistry can also affect the reactivity of the Cys replacements to any of the reagents. Therefore, to test whether each Cys replacement can theoretically react, we purified and denatured each protein variant and tested its reactivity to fluorescein maleimide, NEM, MTSES, and MTSET. We found that each denatured protein fully reacted with each of the reagents (supplemental Fig. S1).
The NhaA Funnels at Physiological pH as Compared with pH 4
Because ionized MTSES and MTSET reach the Cys replacements in TMS II, only via water-filled funnels either from the cytoplasm or the periplasm can they be used to trace the funnels at physiological pH as compared with the funnels observed in the crystal structure at pH 4. Our results conducted on intact cells strongly suggest that a periplasmic funnel exists at physiological pH. Thus, in line with the acidic pH crystal structure (Fig. 1), at pH 7.5, Cys replacements at the cytoplasmic side of TMS II (F71C, F72C, G76C, E78C, and E82C) were not accessible to MTSES from the periplasm in contrast to L60C, N64C, and D65C that were labeled by both MTSES and MTSET (about 90, 60, and 50% of the control for MTSES, and about 100, 95, and 85% of the control for MTSET, respectively) (see Fig. 4 and Table 2). Interestingly, accessibility from the periplasm to both reagents decreased with depth in the membrane, but the positively charged MTSET was more reactive than the negatively charged MTSES, and reached a deeper depth; variant M68C was accessible to about 70% of MTSET but not MTSES (see Fig. 4b). These results suggest that at physiological pH a negatively charged periplasmic funnel exists and narrows with depth in the membrane.
The NhaA Hydrophobic Barrier That Separates the Funnels at Acidic pH Changes at Physiological pH
In line with our computational analysis (46), our presented experimental results imply that, the barrier between the cytoplasmic and periplasmic funnels change at physiological pH; in the crystal structure at pH 4, the barrier length (normal to the membrane) is about 16 Å (see Fig. 1a) and is comprised of many hydrophobic residues (including Phe71, Phe72, and Met68). In marked contrast to the pattern observed at acidic pH, our experimental results at physiological pH show that both F71C and F72C were NEM alkylated (Fig. 3), a reaction that is dependent on the presence of water (in the absence of other proton acceptor). This result suggests a structural change in the barrier and water penetration into this area. In further contrast to acidic pH crystal structure, where the periplasmic funnel ends at Asp65 (Fig. 1a), at the physiological pH, the funnel penetrates much deeper into the membrane; M68C is 70% accessible from the periplasm at least to MTSET, which is similar in charge and dimensions to hydrated Na+ (see above and Fig. 4b). Our previous MTSET accessibility test of Cys replacements in TMS IX also showed that the cytoplasmic funnel deepens into the membrane at physiological pH as compared with acidic pH (48).
TMS II Participates in the pH Response of NhaA
Our results show that TMS II contains amino acid residues that participate in the pH regulation of the protein. Cys replacements in TMS II caused a drastic alkaline shift of one pH unit (L67C, E78C, and E82C) or one-half a pH unit (D65C) in the pH profile of NhaA both at low and saturating concentrations of Na+ (see Fig. 2b) and Li+. As compared with the wild type, mutant N64C had an acidic shift of 0.5 pH unit of the Na+/H+ antiporter activity but lost the pH dependence of the Li+/H+ antiporter activity; its activity with Li+ at pH 6.5 and 8.5 was very similar (Fig. 2c). An effect on the pH dependence of NhaA can occur by mutations of residues that: (i) are involved in the pH sensor. In line with our results, Glu78 and Glu82 have previously been suggested most important in the “pH sensor” of NhaA on the basis of MCCE and MD simulation (45, 46). (ii) Change their conformation with pH for transduction of the pH signal from the “pH sensor” to the active site. We believe that Leu67, Asn64, and Asp65 belong to this category and as described below Asp65 has a unique role.
N64C and D65C Change Their Conformation with pH
Most intriguing results were obtained with N64C and D65C in the MTSES-accessibility test. Whereas, MTSES accessibility of most Cys replacements in TMS II was not affected by pH, MTSES accessibility of D65C was dependent on pH (see Fig. 4c and Table 2) and N64C showed a more modest similar pattern. Thus, D65C was not or hardly accessible at pH 6 and 6.5, whereas above pH 6.5 its accessibility progressively increased and reached a maximum at about pH 8.5 (Fig. 4c). This result suggests that Asp65 changes its conformation with pH and this change might be involved in the pH activation of NhaA.
Our experimental results are in line with our previous (45) and present MCCE computation analysis. Thus, at pH 8, deprotonation of Asp65 triggers a pH-dependent structural change affecting the charge pair Asp65–Lys57 (compare Fig. 6, a and b).
MD simulation at pH 8 confirmed the reorganization of Asp65 side chains into the non-polar barrier associated with penetration of water (46). Therefore, conformational reorientation of Asp65 side chains is pH-dependent and influences the local polarizability of the antiporter. In particular, a change in one of the partners of the charge pair Asp65–Lys57 is expected to affect the other partner and cause a change that may also affect antiporter functionality.
Our MCCE calculations for variant D65C indeed show that the pKa value of Lys57 is drastically changed in the mutant; whereas, the pKa of Lys57 in the wild type is 12.2, it is reduced to 8 in the variant. It can be argued that in the absence of a titratable residue at position D65C, Lys57 mediates an almost normal pH response of NhaA as shown here experimentally. The pH profile of D65C is alkaline shifted by only one-half a pH unit as compared with the pH profile of the wild type (Fig. 2b).
Lys57 is located in the β-sheet in the loop connecting TMS I and II of NhaA (Fig. 1a). The growth phenotype and exchange activity of K57C have recently been described (51). Its growth phenotype is normal and apparent Km for Li+ is very similar to that of the wild type, whereas the apparent Km for Na+ is 4-fold higher than that of the wild type. Whereas, the pH profile of K57C was drastically changed by an acidic shift of one pH unit, the pH profile of the other Cys replacements in the β-sheet exhibited normal pH profile. In contrast to the wild type, which is inactive at pH 6.5 and 7 in Na+/H+ exchange, K57C shows 30 or 50% dequenching in the respective pH values. Taken together, the charge pair Asp65–Lys57 is important for the functionality and pH response of NhaA.
The Negative Charge on Asp65 Is Critical for a pH-induced Conformational Change of Asp65
Strikingly, the negative charge of Asp65 was found critical for pH activation of NhaA. Thus, the impaired Na+/H+ antiporter activity of D65C, reflected in a very high Km, was dramatically improved after treatment with MTSES, the negatively charged SH-reagent, but not by the positively charged SH-reagent, MTSET (Fig. 5, a and b). Furthermore, the repair effect of MTSES was most pronounced around pH 7.5 and vanished at pH 8.5 (see Fig. 5b). These results reveal that a negative charge at position Asp65 is crucial for the activation of the antiporter at pH 7.5, whereas it has no influence when the antiporter is already active, pH 8.5. MTSES treatment did not completely cure the impaired phenotype of D65C; the apparent Km for Na+ of the MTSES-treated D65C at pH 8.5 was still 10-fold higher than that of the control (Fig. 5b). Hence, following the conformational change at pH 7.5, an additional conformational change must take place during the pH-induced activation of NhaA. In this respect it is remarkable that cryo-electron microscopy of two-dimensional crystals has recently identified a pH-induced conformational change of NhaA between pH 4 and 7 and a second conformational change at alkaline pH (49).
The importance of TMS II has recently been highlighted in a study modeling the human NHE1 on the basis of NhaA. Although NhaA and NHE1 share very low sequence identity (about 10%), they share a similar fold, and similar cation and inhibitor binding sites. As shown computationally and validated experimentally, the binding site of the inhibitor 2-aminoperimidine of Ec-NhaA involves N64C and F71C of TMS II (50).
It is clear that crystal structure(s) of the open active conformation(s) of NhaA at alkaline pH is required to answer how the barrier between the cytoplasmic and periplasmic funnels change during activation of the protein, and how residues of TMS II become exposed to either sides of the membrane in the open state of the protein. Nevertheless, our biochemical results presented here reveal that TMS II is most important in the activity of NhaA and its regulation by pH.
Supplementary Material
This work was supported by USA-Israel Binational Science Foundation Grant 501/03-16.2 and European Union European Drug Initiative on Channels and Transporters Grant FP 7 (to E. P.) and a grant from the Max-Planck-Gesellschaft (to E. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- Ec-NhaA
- E. coli NhaA
- TMS
- transmembrane segment
- CL-NhaA
- cysteine-less NhaA
- NEM
- N-ethylmaleimide
- MTSES
- 2-sulfonatoethyl methanethiosulfonate
- MTSET
- [2-(trimethyammonium)ethyl]methanethiosulfonate bromide
- Ni-NTA
- Ni2+-nitrilotriacetic acid
- MCCE
- multiconformation continuum electrostatic.
REFERENCES
- 1.Padan E., Venturi M., Gerchman Y., Dover N. (2001) Biochim. Biophys. Acta 1505, 144–157 [DOI] [PubMed] [Google Scholar]
- 2.Padan E., Bibi E., Ito M., Krulwich T. A. (2005) Biochim. Biophys. Acta 1717, 67–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Padan E. (2008) Trends Biochem. Sci. 33, 435–443 [DOI] [PubMed] [Google Scholar]
- 4.Pinner E., Kotler Y., Padan E., Schuldiner S. (1993) J. Biol. Chem. 268, 1729–1734 [PubMed] [Google Scholar]
- 5.Brett C. L., Donowitz M., Rao R. (2005) Am. J. Physiol. Cell Physiol. 288, C223–C239 [DOI] [PubMed] [Google Scholar]
- 6.Taglicht D., Padan E., Schuldiner S. (1993) J. Biol. Chem. 268, 5382–5387 [PubMed] [Google Scholar]
- 7.Orlowski J., Grinstein S. (2003) in The Sodium Hydrogen Exchange, from Molecule to its Role in Disease (Karmazin M., Avkiran N., Fliegel L. eds) pp. 17–34, Kluwer Academic Publishers, Boston [Google Scholar]
- 8.Orlowski J., Grinstein S. (2004) Pflugers Arch. 447, 549–565 [DOI] [PubMed] [Google Scholar]
- 9.Putney L. K., Denker S. P., Barber D. L. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 527–552 [DOI] [PubMed] [Google Scholar]
- 10.Wakabayashi S., Hisamitsu T., Pang T., Shigekawa M. (2003) J. Biol. Chem. 278, 43580–43585 [DOI] [PubMed] [Google Scholar]
- 11.Fliegel L. (2008) J. Mol. Cell Cardiol. 44, 228–237 [DOI] [PubMed] [Google Scholar]
- 12.Taglicht D., Padan E., Schuldiner S. (1991) J. Biol. Chem. 266, 11289–11294 [PubMed] [Google Scholar]
- 13.Tzubery T., Rimon A., Padan E. (2004) J. Biol. Chem. 279, 3265–3272 [DOI] [PubMed] [Google Scholar]
- 14.Venturi M., Rimon A., Gerchman Y., Hunte C., Padan E., Michel H. (2000) J. Biol. Chem. 275, 4734–4742 [DOI] [PubMed] [Google Scholar]
- 15.Gerchman Y., Rimon A., Padan E. (1999) J. Biol. Chem. 274, 24617–24624 [DOI] [PubMed] [Google Scholar]
- 16.Hunte C., Screpanti M., Venturi M., Rimon A., Padan E., Michel H. (2005) Nature 435, 1197–1202 [DOI] [PubMed] [Google Scholar]
- 17.Olami Y., Rimon A., Gerchman Y., Rothman A., Padan E. (1997) J. Biol. Chem. 272, 1761–1768 [DOI] [PubMed] [Google Scholar]
- 18.Rothman A., Padan E., Schuldiner S. (1996) J. Biol. Chem. 271, 32288–32292 [DOI] [PubMed] [Google Scholar]
- 19.Padan E., Maisler N., Taglicht D., Karpel R., Schuldiner S. (1989) J. Biol. Chem. 264, 20297–20302 [PubMed] [Google Scholar]
- 20.Davis B. D., Mingioli E. S. (1950) J. Bacteriol. 60, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karpel R., Olami Y., Taglicht D., Schuldiner S., Padan E. (1988) J. Biol. Chem. 263, 10408–10414 [PubMed] [Google Scholar]
- 22.Rimon A., Tzubery T., Galili L., Padan E. (2002) Biochemistry 41, 14897–14905 [DOI] [PubMed] [Google Scholar]
- 23.Galili L., Rothman A., Kozachkov L., Rimon A., Padan E. (2002) Biochemistry 41, 609–617 [DOI] [PubMed] [Google Scholar]
- 24.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 25.Fisher C. L., Pei G. K. (1997) BioTechniques 23, 570–574 [DOI] [PubMed] [Google Scholar]
- 26.Kozachkov L., Herz K., Padan E. (2007) Biochemistry 46, 2419–2430 [DOI] [PubMed] [Google Scholar]
- 27.Gerchman Y., Olami Y., Rimon A., Taglicht D., Schuldiner S., Padan E. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 1212–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosen B. P. (1986) Methods Enzymol. 125, 328–336 [DOI] [PubMed] [Google Scholar]
- 29.Goldberg E. B., Arbel T., Chen J., Karpel R., Mackie G. A., Schuldiner S., Padan E. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 2615–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuldiner S., Fishkes H. (1978) Biochemistry 17, 706–711 [DOI] [PubMed] [Google Scholar]
- 31.Tsuboi Y., Inoue H., Nakamura N., Kanazawa H. (2003) J. Biol. Chem. 278, 21467–21473 [DOI] [PubMed] [Google Scholar]
- 32.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 33.Ninio S., Elbaz Y., Schuldiner S. (2004) FEBS Lett. 562, 193–196 [DOI] [PubMed] [Google Scholar]
- 34.Alexov E. G., Gunner M. R. (1997) Biophys. J. 72, 2075–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexov E. G., Gunner M. R. (1999) Biochemistry 38, 8253–8270 [DOI] [PubMed] [Google Scholar]
- 36.Gunner M. R., Alexov E. (2000) Biochim. Biophys. Acta 1458, 63–87 [DOI] [PubMed] [Google Scholar]
- 37.Beroza P., Fredkin D. R., Okamura M. Y., Feher G. (1995) Biophys. J. 68, 2233–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholls A. H., Honig B. (1991) J. Comput. Chem. 12, 435–445 [Google Scholar]
- 39.Sitkoff D., Lockhart D. J., Sharp K. A., Honig B. (1994) Biophys. J. 67, 2251–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Combet C., Blanchet C., Geourjon C., Deléage G. (2000) Trends Biochem. Sci. 25, 147–150 [DOI] [PubMed] [Google Scholar]
- 41.Rimon A., Gerchman Y., Kariv Z., Padan E. (1998) J. Biol. Chem. 273, 26470–26476 [DOI] [PubMed] [Google Scholar]
- 42.Galili L., Herz K., Dym O., Padan E. (2004) J. Biol. Chem. 279, 23104–23113 [DOI] [PubMed] [Google Scholar]
- 43.Akabas M. H., Kaufmann C., Archdeacon P., Karlin A. (1994) Neuron 13, 919–927 [DOI] [PubMed] [Google Scholar]
- 44.Akabas M. H., Stauffer D. A., Xu M., Karlin A. (1992) Science 258, 307–310 [DOI] [PubMed] [Google Scholar]
- 45.Olkhova E., Hunte C., Screpanti E., Padan E., Michel H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olkhova E., Padan E., Michel H. (2007) Biophys. J. 92, 3784–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deleted in proof
- 48.Tzubery T., Rimon A., Padan E. (2008) J. Biol. Chem. 283, 15975–15987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Appel M., Hizlan D., Vinothkumar K. R., Ziegler C., Kühlbrandt W. (2009) J. Mol. Biol. 386, 351–365 [DOI] [PubMed] [Google Scholar]
- 50.Landau M., Herz K., Padan E., Ben-Tal N. (2007) J. Biol. Chem. 282, 37854–37863 [DOI] [PubMed] [Google Scholar]
- 51.Herz K., Rimon A., Jeschke G., Padan E. (2009) J. Biol. Chem. 284, 6337–6347 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.