Abstract
In most mammals, the expression of SRY (sex-determining region on the Y chromosome) initiates the development of testes, and thus determines the sex of the individual. However, despite the pivotal role of SRY, its mechanism of action remains elusive. One important missing piece of the puzzle is the identification of genes regulated by SRY. In this study we used chromatin immunoprecipitation to identify direct SRY target genes. Anti-mouse SRY antibody precipitated a region 7.5 kb upstream of the transcriptional start site of cerebellin 4 precursor (Cbln4), which encodes a secreted protein. Cbln4 is expressed in Sertoli cells in the developing gonad, with a profile mimicking that of the testis-determining gene SRY-box containing gene 9 (Sox9). In transgenic XY mouse embryos with reduced Sox9 expression, Cbln4 expression also was reduced, whereas overexpression of Sox9 in XX mice caused an upregulation of Cbln4 expression. Finally, ectopic upregulation of SRY in vivo resulted in ectopic expression of Cbln4. Our findings suggest that both SRY and SOX9 contribute to the male-specific upregulation of Cbln4 in the developing testis, and they identified a direct in vivo target gene of SRY.
Keywords: Cbln4, developmental biology, gene regulation, mouse, Sertoli cells, sex determination, SOX9, SRY, testis, transcriptional regulation
Cbln4 expression is upregulated by SRY and maintained by SOX9 during early testis development.
INTRODUCTION
A pivotal point in the growth of all sexually dimorphic organisms is the initiation of either the male or the female pathway of development. The gene that initiates the male pathway in almost all mammals, including humans, is SRY. The requirement for SRY in testis development was indicated by the discovery of mutations in this gene in two sex-reversed XY women but not their fathers [1, 2]. Male development of transgenic XX mice expressing Sry confirmed the significance of this gene [3]. SRY protein has been characterized as a DNA-binding transcription factor, but the molecular mechanisms of its action remain a mystery (reviewed in Polanco and Koopman [4]).
SRY belongs to the SOX family of transcription factors that are distinguished by the presence of a high mobility group domain. SRY has been shown to bind and bend DNA in vitro [5, 6]. However, whether SRY acts as a transcriptional activator, repressor, or architectural protein remains unclear. In vitro biochemical assays showed that SRY activated transcription of a reporter gene driven by multiple copies of the SRY-binding motif [7]. In addition, it has been shown recently that mouse SRY binds to a SRY-box containing gene 9 (Sox9) enhancer region in vivo and activates Sox9 expression [8]. Conversely, analysis of more than 100 human XX males supports a model in which SRY acts by repressing a negative regulator of male sex determination [9]. Other in vitro experiments have demonstrated that SRY can act as a repressor, depending on its phosphorylation status [10].
SRY is expressed in the supporting cell lineage within the developing XY genital ridge. Its expression in mice resembles a wave starting in the central portions of the genital ridge, expanding to the anterior part, and finally encompassing the posterior region. Expression levels then subside in a similar center-anterior-posterior wave, suggesting that Sry is active for only a few hours in each single cell [11–13]. Closely following the onset of Sry expression, another gene of the Sox family, Sox9, is expressed in the developing testis. Sox9 expression follows a wave similar to that of Sry, but rather than being rapidly extinguished, its expression continues in the supporting cell lineage of the testis for the remainder of embryonic development [14]. SOX9-positive cells become Sertoli cells that not only form cords surrounding germ cells but also are thought to orchestrate the differentiation of the other cell types in the testis. SOX9 protein has been shown to bind to the same DNA motifs as SRY [15, 16], and a number of direct target genes expressed during testis differentiation have been identified, including anti-Müllerian hormone (Amh) [17, 18], prostaglandin-d-synthetase (Ptgds) [19], and vanin 1 (Vnn1) [20].
Like Sry, Sox9 is essential for male development, and its ectopic expression in mice leads to XX sex reversal [21, 22]. However, in these cases of sex reversal, it is unclear whether SOX9 is regulating its normal targets or, as a consequence of its early expression, is recapitulating the function of SRY. Given that normal testis development requires SRY to be expressed within a narrow time window [23, 24] and that SRY and SOX9 recognize similar or identical DNA-binding sites, it may be that SOX9, expressed at the right time, can fulfill the early functions of SRY. This possibility raises the question of whether Sox9 is the single gene through which SRY influences male determination, or whether SRY regulates multiple targets, one of which is Sox9.
To understand the early events in sex determination, and particularly to disentangle the relationship between SRY and SOX9, we sought here to identify direct target genes of SRY. We performed chromatin immunoprecipitation (ChIP) with an anti-SRY antibody and identified a noncoding region 7.5 kb upstream of the transcriptional start site of the gene cerebellin 4 precursor (Cbln4). Cbln4 is expressed in Sertoli cells of the developing gonad, with a profile mimicking that of Sox9. Using transgenic mouse models, we show that both SRY and SOX9 are sufficient to upregulate Cbln4 in vivo and that SOX9 is necessary for the maintenance of Cbln4 expression. Together, these data suggest that Cbln4 is directly regulated by SRY and SOX9, allowing us to propose a model in which SRY normally activates multiple targets, whose expression is subsequently maintained by SOX9.
MATERIALS AND METHODS
Animal Strains
Embryos were collected from timed matings of CD1 outbred and We mutant mice [25], with noon of the day on which the mating plug was observed designated as 0.5 days postcoitum (dpc). For more accurate staging, the tail somite (ts) stage of the embryo was determined by counting the number of somites posterior to the hind limb [26]. Using this method, 10.5 dpc corresponds to approximately 8 ts, 11.5 dpc to 18 ts, and 12.5 dpc to 30 ts. Embryos at 11.5 dpc or younger were sexed by PCR using Zfy gene-specific primers Zfy-F, 5′-CCTATTGCATGGACTGCAGCTTATG-3′; and Zfy-R, 5′-GACTAGACATGTCTTAACATCTGTCC-3′. Older embryos were sexed by morphological criteria. Protocols and use of animals in these experiments were approved by the Animal Welfare Unit of the University of Queensland, registered as an institution that uses animals for scientific purposes under the Queensland Animal Care and Protection Act (2001).
ChIP Assay
Male and female genital ridges (gonad and mesonephros) were dissected from approximately 550 staged mouse embryos, and ChIP was performed as described previously [19]. Antibodies used in the ChIP assays were anti-mouse SRY [13, 27] and anti-mouse SOX9 [19]. To identify novel targets of SRY, eluted fragments were cloned into pPCR-Script Amp SK(+) according to the manufacturer's instructions (Stratagene). DNA was extracted from cultures using a FastPlasmid mini kit (Eppendorf) and was sequenced at the Australian Genome Research Facility (Brisbane, Australia). Sequences were aligned to regions in the mouse genome using an Ensembl BLAST [28] search. To confirm enrichment of the binding fragments, a second ChIP (αSRY, αSOX9) was performed as above using eluted fragments as template in PCR. Primers used were ChIPPCR1 F, 5′-ATAGAGTGCCTGGTGCACAAGCAT-3′; ChIPPCR1 R, 5′-GCAGCCACAACTCAGTCTCCATACT-3′; ChIPPCR2 F, 5′-ACTTAGCATTGCCCTCTGGCTTTG-3′; and ChIPPCR2 R, 5′-AAGTTACAAACCCTCTTTGAGCCG-3′.
Western Blot Analysis
Western blot analysis was performed as described previously [29], with an anti-HA antibody (HA-7; Sigma) at a 1:1000 dilution.
Cell Culture and Transfections
MCF-7 cells (catalogue no. HTB-22; American Type Culture Collection) were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions and were analyzed 24–48 h after transfection. Plasmids used were isoprenylcysteine-O-carboxyl methyltransferase-GFP (IGMT-GFP) based on the vector pEGFP-C1 (Clontech), Transgolgi network 38 (TGN38-YFP [30]) based on the vector pYFP-C1 (Clontech), and Cbln4-myc, containing the open reading frame of Cbln4 with a 3′ myc tag cloned into pcDNA3 (Invitrogen).
Immunofluorescence
For immunofluorescence, cells were fixed in 4% paraformaldehyde (PFA) for 10 min on ice and then permeabilized with 0.25% Triton X-100 for 5 min at room temperature prior to labeling. Antibodies used were a rabbit polyclonal antibody to GFP (Molecular Probes) and a mouse monoclonal antibody to the MYC-tag (9B11; Cell Signaling Technology). As a nuclear stain, 4′,6′-diamidino-2-phenylindole (DAPI; Roche) was used.
Epi-illumination fluorescence microscopy of fixed specimens was performed using an IX81 microscope with a 60×, 1.40 numerical aperture objective (Olympus), and imaging was performed with Orca-1 ER cameras (Hamamatsu) driven by Metamorph imaging software (Universal Imaging). Background correction and contrast adjustment of raw data images were performed with ImageJ (National Institutes of Health) or Adobe Photoshop (Adobe).
Secretion and Deglycosylation Assays
MCF-7 cells were transfected with the Cbln4-myc construct for 6 h prior to PBS) wash and addition of fresh medium. Medium was removed following 24 h of incubation and was spun for 5 min at 5000 × g at 4°C to remove cell debris.
For deglycosylation, medium was heated to 100°C for 5 min in PBS containing 1% SDS and 1 M dithiothreitol (Sigma). Samples were then incubated with 1% Triton X-100 in PBS in the presence or absence of peptide-N-glycosydase (Roche) for 3 h at 37°C. Proteins in untreated and treated media, as well as cell lysates, were separated by SDS-PAGE; CBLN4-MYC was detected by Western blot analysis using a mouse monoclonal antibody to the MYC tag (9B11; Cell Signaling Technology), as described previously [29].
In Situ Hybridization
Probes for Amh and Oct4 were made as described previously [31, 32]. A 792-bp Cbln4 fragment was cloned from cDNA from 13.5 dpc mouse testes. Primers used were Cbln4-F, 5′-ATAGAACCCGACTTCTCCGTGATG-3′; and Cbln4-R, 5′-ACCAAGGAGAGGTACTTTGCCAAG-3′. Embryos and dissected gonads/mesonephroi were fixed in 4% PFA in PBS for several hours at 4°C. Whole-mount in situ hybridization (ISH) with digoxygenin-labeled RNA probes was carried out as described by Hargrave et al. [33]. Section ISH was performed on 7-μm sagittal sections from paraffin-embedded embryos as described previously [19].
RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR
Fetal gonads (pooled) or gonad and mesonephros (individual) were collected and frozen on dry ice. Total RNA was isolated using an RNeasy Micro kit (Qiagen) according to the manufacturer's instructions, including the optional DNaseI step. cDNA was synthesized from 1 μg of RNA from pooled gonads or 400 ng of RNA from individual gonads and mesonephroi by reverse transcriptase (Superscript III; Invitrogen) using random primers (Invitrogen) according to the manufacturer's instructions. Relative cDNA levels were analyzed by quantitative real-time RT-PCR using an Applied Biosystems Inc. Prism-7000 Sequence Detector System or Light Cycler Instrument (Roche).
Quantitative real-time RT-PCR was carried out on pooled samples in triplicate of three independent biological samples and is represented as mean ± SEM of the three individual experiments. Samples were analyzed in 25-μl reactions as described previously [34]. Reactions lacking reverse transcriptase were carried out in parallel with each cDNA sample to identify genomic DNA contamination. Primers used were rtSRY-F, 5′-CAGCCTGCAGTTGCCTCAA-3′ and rtSRY-R, 5′-GGTGTGCAGCTCTACTCCAGTCT-3′; rtSOX9-F, 5′-AGTACCCGCATCTGCACAAC-3′ and rtSOX9-R, 5′-TACTTGTAATCGGGGTGGTCT-3′; rtAMH-F, 5′-CGAGCTCTTGCTGAAGTTCCA-3′ and rtAMH-R, 5′-GAAGTCCACGGTTAGCACCAA-3′; rtMVH-F, 5′-CAGGAATGCCATCAAAGGAACAAC-3′ and rtMVH-R, 5′-CCCAACAGCGACAAACAAGTAACTG-3′; rtCBLN4-F, 5′-TGTCTTTGTGGCACCGAGGAA-3′ and rtCBLN4-R, 5′-GCGGCTTCACGGGTCACATCT-3′; rt18S-F, 5′-GATCCATTGGAGGGCAAGTCT-3′ and rt18S-R, 5′-CCAAGATCCAACTACGAGCTTTTT-3′.
Real-time RT-PCR performed on cDNA from individual gonad/mesonephros pairs was done either in triplicate and represented as individual data points with standard deviation or as individual reactions. Reactions prepared in triplicate included Taqman PCR master mix (Applied Biosystems Inc.) and 1× Taqman gene expression sets (Applied Biosystems Inc.) with 200 nM each forward and reverse primers. Taqman gene expression sets (Applied Biosystems Inc.) were used for rt18S (4319413E), rtSRY (Mm00441712_s1), rtSOX9 (Mm00448840_m1), rtAMH (Mm00431795_g1), and rtCBLN4 (Mm0055863_m1).
Quantitative real-time RT-PCR was performed on Wt1:Sox9 and Sox9-KO mice as described previously [35]. Primers were Gapdh, Sry, Sox9 [35], and Cbln4 (5′-GCACCGAGGAAAGGAATCTA-3′ and 5′-TGCAGAGATGACTGGTTTTCC-3′ and universal probe library probe no. 21).
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay analysis was performed as described previously [36] using recombinant, bacterially expressed GST fusion proteins of full-length SRY, SOX9, SF1, and SOX7. Recombinant plasmids were constructed using the vector pGEX-KG [37].
RESULTS
Identification of Putative SRY Target Genes
To identify direct target genes of SRY, we performed in vivo ChIP using approximately 550 pairs of mouse genital ridges at 11.5 dpc as starting material. For the immunoprecipitation, we used a recently described antibody to mouse SRY [13, 27]. We showed that this antibody can precipitate mouse SRY protein by using extracts of cells transfected with an Sry expression construct (Supplemental Fig. S1, available online at www.biolreprod.org). Genomic fragments, presumably bound to endogenous SRY, were isolated and cloned into plasmid vector. In all, 162 cloned fragments were retrieved and sequenced. Of these, 53 mapped to single regions of the mouse genome with a low probability of false alignment and were larger than 200 bp (Supplemental Table S1, available online at www.biolreprod.org). Fragments with poor alignment, multiple alignments with similar probability, or those smaller than 200 bp were discarded. We examined these fragments bioinformatically to determine if they contained putative SRY consensus binding sites (Supplemental Table S1). Any genes close to these regions or containing these fragments in an intron were considered as candidate SRY target genes.
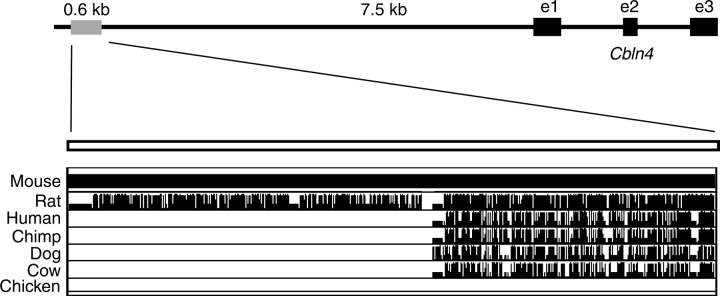
A bona fide SRY target gene ought to be either expressed at the same time and in the same cells as Sry—that is, in the supporting cell lineage of XY genital ridges at 11.5 dpc in mice—or downregulated in these cells at the onset of Sry expression. We first assessed candidate target genes by analyzing their expression in sorted gonadal cell populations at 11.5 dpc using Affymetrix microarray data (data not shown and Beverdam and Koopman [38]). Based on this analysis, we chose one candidate target gene, Cbln4, for further investigations. The immunoprecipitated genomic fragment mapped 7.5 kb upstream of the Cbln4 transcription start site within the qH3 region of mouse chromosome 2 (Fig. 1). Using the University of California, Santa Cruz genome browser [39], we confirmed that this genomic fragment is a noncoding region and is highly conserved between mouse and rat and, to a lesser extent, human, dog, cow, and chimpanzee genomes (Fig. 1), suggesting that it may be important in the regulation of Cbln4 gene expression.
FIG. 1.
Sequence homology of the ChIP fragment. Location of the 600-bp ChIP fragment relative to the mouse Cbln4 mRNA (top). Sequence comparison of the mouse fragment with the respective sequences of rat, human, chimp, dog, cow, and chicken showed that approximately 300 bp were conserved between mouse, dog, cow, chimp, and human; approximately 600 bp were conserved between mouse and rat. Black boxes indicate exons 1–3. Pairwise alignments of each species to the mouse genome are displayed, with the height of the vertical bars indicating alignment quality. e1, exon 1; e2, exon 2; e3, exon 3.
Cbln4 Expression in Developing Mouse Testes
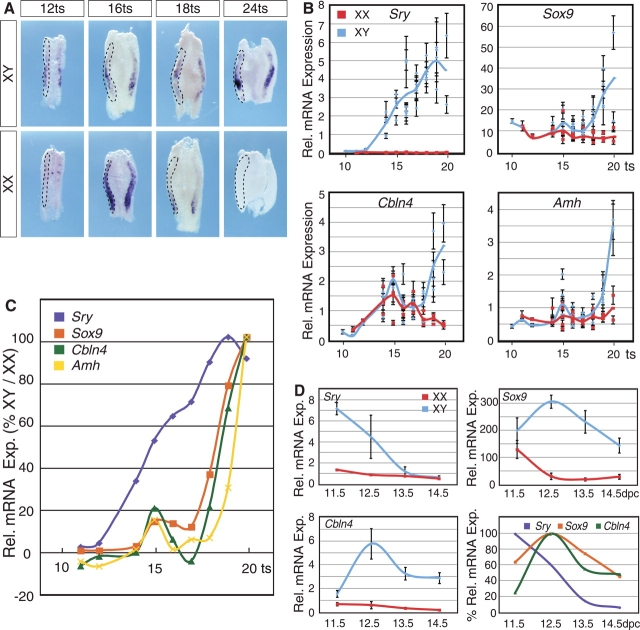
We next investigated in detail the expression of Cbln4 in relation to the known schedule of events in sex determination and testis differentiation. We used whole-mount ISH of mouse fetal gonad samples from 12 to 24 ts as a qualitative assay to assess the spatiotemporal expression pattern. This analysis showed that Cbln4 was expressed in XY and XX genital ridges until 16 ts. Approximately 4 h later, at 18 ts, the stage at which Sry expression reaches its maximum [11, 13], Cbln4 was downregulated in ovaries, whereas its expression seemed to increase in the developing testis (Fig. 2A). By 24 ts, Cbln4 was male-specifically expressed, with no detectable levels in ovaries (Fig. 2A).
FIG. 2.
Comparative expression profiling of Cbln4, Sry, Sox9, and Amh. A) Whole-mount ISH for Cbln4 was performed on XX and XY genital ridges collected from embryos between 12 and 24 ts. Expression of Cbln4 is present in both male and female gonads at 12 ts. By 18 ts, expression is downregulated in the ovary, and at 24 ts it is present in the testis only. The left gonad of each pair is outlined by dotted lines. Original magnification ×10. B) Real-time RT-PCR for Sry, Sox9, Cbln4, and Amh was performed on RNA extracted from gonad and mesonephros from embryos between 10 and 20 ts. Individual data points represent single experiments performed in triplicate (mean ± SD). Input RNA was normalized against 18S RNA, with y-axis denoting relative (Rel.) mRNA expression of individual genes. Blue and red curves denote best fit of mRNA expression over time in XY and XX gonads, respectively. C) Expression (Exp.) of Sry, Sox9, Cbln4, and Amh in XY gonads relative to XX gonads. A relative expression value above zero indicates a higher expression in XY gonads relative to XX gonads, whereas a negative value represents a higher expression in XX gonads relative to XY gonads. D) Real-time RT-PCR analysis for Sry, Sox9, and Cbln4 on RNA extracted from pooled gonads at 11.5, 12.5, 13.5, and 14.5 dpc. Data sets represent mRNA expression relative to 18S (mean ± SEM of three biologically independent experiments performed in triplicates). Bottom right graph shows relative expression of Sry, Sox9, and Cbln4 represented as percentage of maximal expression in XY gonads.
To quantify and directly compare the mRNA expression of Cbln4 with that of Sry, we employed quantitative real-time RT-PCR. Sox9 was included for comparison as an SRY target gene, and Amh as a SOX9 target gene [17, 18]. As expected, Sry expression increased over time and reached a maximum around 18 ts before declining in XY gonads, whereas it was absent in XX (Fig. 2B, top left). Sox9 and Amh, which showed similar expression profiles, were expressed in XX gonads at low levels at 10–14 ts, with a small, transient increase at 15 ts. In XY gonads, both genes were strongly upregulated after 18 ts (Fig. 2B, top right and bottom right). Although Cbln4 expression appeared the highest in XX gonads at 16 ts (Fig. 2A) in the ISH, quantitative RT-PCR showed that Cbln4 expression increased in both XY and XX gonads until approximately 15 ts before decreasing over the next 4 h. Around 18 ts, Cbln4 expression was strongly upregulated in XY and downregulated in XX gonads (Fig. 2B, bottom left).
To determine at which developmental stage the expression of each gene in males diverged from that in females, we normalized the levels of gene expression in XY gonads to those measured in XX gonads. Using this method, positive values were obtained when a given gene was expressed at higher levels in XY compared with XX. These data showed that except for a brief increase at 15 ts, Amh, Sox9, and Cbln4 were maintained at similar levels of expression until 18 ts, when both Sox9 and Cbln4 mRNA rapidly increased in XY compared with XX (Fig. 2C). One tail somite (approximately 2 h) later in development, Amh expression increased in XY genital ridges (Fig. 2C).
To analyze the expression of Cbln4 in comparison with Sry and Sox9 after 11.5 dpc, we employed quantitative real-time RT-PCR of embryonic gonads from 11.5 to 14.5 dpc. As expected, expression of Sry was detected only in XY genital ridges, declining after 11.5 dpc (Fig. 2D, top left). In contrast, Sox9 and Cbln4 expression levels increased in testes after 11.5 dpc and decreased in ovaries (Fig. 2D, top right and bottom left). Combining the expression profiles of all three genes within XY gonads demonstrated that Sox9 and Cbln4 follow similar kinetics (Fig. 2D, bottom right), supporting the hypothesis that Cbln4 might be regulated by SRY.
We next sought to determine whether Cbln4 is expressed in the same cell type as Sry. We first preformed quantitative real-time RT-PCR analysis on We/We mice for Cbln4 in parallel with Sox9 as a Sertoli cell marker and mouse vasa homologue (Mvh) as a germ cell marker. We/We mice harbor a mutation in the Kit gene that prevents the migration of germ cells into the genital ridge, resulting in a lack of this cell type within the gonad [14]. As expected, Sox9 was expressed at approximately the same level in wild-type and We/We mice (Fig. 3A, left), whereas expression of the germ cell marker Mvh was lost in We/We mutants (Fig. 3A, middle). Cbln4 was expressed at similar levels in We/We compared with wild-type mice (Fig. 3A, right), demonstrating that Cbln4 expression is neither in germ cells nor dependent on the presence of germ cells. To further identify the cell type that expresses Cbln4, section ISH was carried out on whole embryos at 13.5 dpc. For comparison, we performed section ISH with the germ cell marker Pou5f1 (Oct4, octamer 4) and the Sertoli cell marker Amh on adjacent sections of the same embryo (Fig. 3B). Cbln4 expression resembled the expression of Amh within the testis cords, suggesting Cbln4 is expressed by Sertoli cells (Fig. 3B, left and right columns), whereas Pou5f1 showed the expected expression in germ cells only (Fig. 3B, middle column).
FIG. 3.
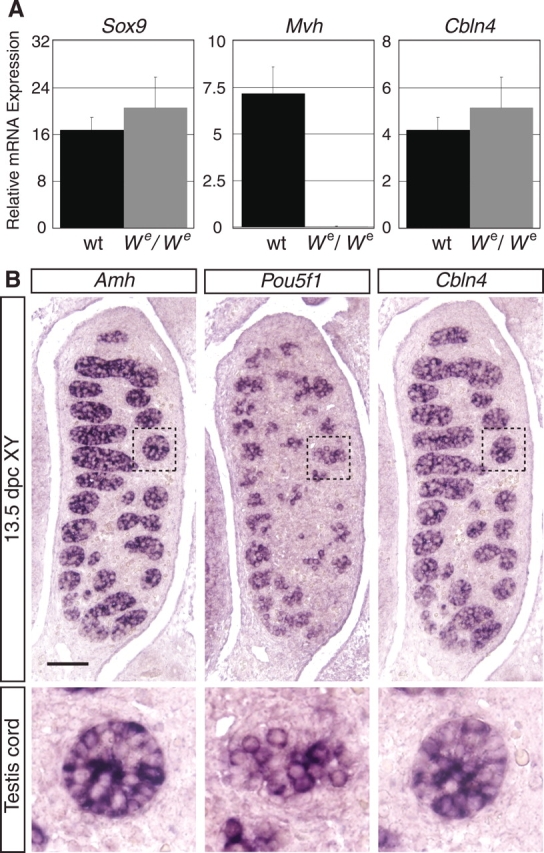
Sertoli cell-specific expression of Cbln4. A) Real-time RT-PCR analysis of wild-type (wt) and homozygous We/We littermate gonads for Cbln4, Sox9 (Sertoli cell marker), and Mvh (germ cell marker) relative to 18S RNA (mean ± SEM of three independent experiments). Individual experiments were performed in triplicate on RNA obtained from pooled gonads from three to four littermates. Cbln4 showed expression similar to Sox9 and is maintained in the germ cell-depleted gonads. In comparison, Mvh expression was lost in We/We samples. B) In situ hybridization on sagittal paraffin sections showed Cbln4 (right column) and Amh (left column) expression specifically in Sertoli cells at 13.5 dpc, whereas the germ cell marker Pou5f1 was expressed by germ cells only (middle column). The bottom row shows enlargements (magnified ×4) of the regions marked with rectangles in the top row. Images are oriented so that the anterior pole is at the top. Bar = 200 μm.
CBLN4 Is a Secreted Protein
Having established that Cbln4 is expressed by the same cell type and at the same time as Sry, we wanted to identify the type of protein that is encoded by Cbln4. Using the Web-based protein prediction tool TMHMM [40], CBLN4 has a predicted transmembrane helix domain from amino acids 7 to 24. SignalP 3.0 analysis [41] predicted a signal peptide with a cleavage site between amino acids 24 and 25, suggesting that it is a secreted factor or a membrane-bound protein (Fig. 4A). Using SMART [42], an online domain architecture tool, we found that CBLN4 also contains a C1Q domain (Fig. 4A), a protein-protein association domain present in a wide variety of secreted proteins [43, 44].
FIG. 4.
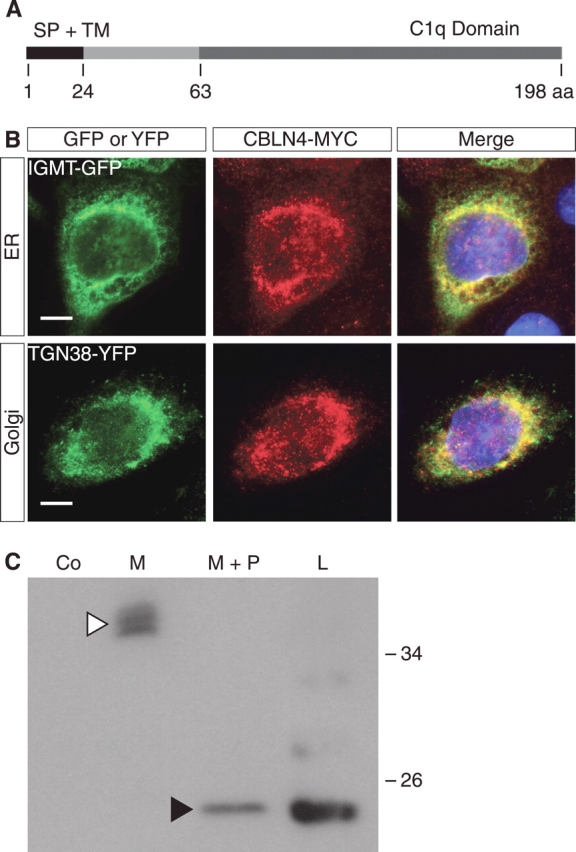
CBLN4 is a secreted protein. A) Schematic representation of CBLN4 protein with predicted signal peptide cleavage site at position 24 and C-terminal C1q domain. B) Coimmunofluorescence of MCF-7 cells transfected with the Cbln4-Myc construct and either isoprenylcysteine-O-carboxyl methyltransferase-GFP (IGMT-GFP, an endoplasmic reticulum [ER] marker; top row) or TGN38-YFP (a golgi marker; bottom row) constructs demonstrates colocalization of CBLN4-MYC with both IGMT-GFP and TGN38-YFP. DAPI staining (blue) marks cell nuclei. Bar = 10 μm. C) Secretion and deglycosylation assays performed on MCF-7 cells transfected with Cbln4-Myc construct. Western blot analysis shows that CBLN4-MYC is detected in the cell medium at approximately 38 kDa (white arrowhead). Deglycosylation results in a decrease of the molecular weight to approximately 20 kDa, similar to CBLN4-MYC from cell lysates (black arrowhead), suggesting that the difference in size of intracellular and secreted CBLN4 is the result of posttranslational glycosylation. aa, amino acid; Co, mock-transfected control; M, medium; M+P, medium with peptide-N-glycosydase; L, cell lysate; SP, signal peptide; TM, transmembrane domain.
To confirm experimentally the intracellular location of CBLN4, we transfected MCF-7 cells with a construct expressing CBLN4 with an MYC-tag at the C-terminus, and we performed coimmunofluorescence using an antibody specific for the MYC-tag together with antibodies detecting marker proteins for the secretory pathway. CBLN4-MYC was localized to the endoplasmic reticulum (Fig. 4B, top row) and the golgi apparatus (Fig. 4B, bottom row), suggesting that it is trafficked through the secretory pathway. To test whether CBLN4 is indeed secreted, we performed Western blot analysis on isolated compartments of cultures of Cbln4-Myc-transfected cells. We detected CBLN4-MYC in both the cell lysate and the medium of transfected cells, but not in control cells transfected with empty vector (Fig. 4C), confirming the bioinformatic predictions that CBLN4 is a secreted factor. CBLN4-MYC within the medium was approximately 20 kDa larger than that found in the cell lysate. Deglycosylation with peptide-N-glycosydase was able to reduce the secreted protein size to that of the lysate, indicating that secreted CBLN4 is posttranslationally glycosylated (Fig. 4C).
SRY and SOX9 Bind to the ChIP Fragment In Vitro
We next aimed to confirm that SRY regulates the expression of Cbln4 via the genomic fragment identified by ChIP analysis. To this end, we first examined bioinformatically the sequence of the ChIP fragment for the presence of SRY/SOX, steroidogenic factor 1 (SF1), and Wilms tumor suppressor 1 (WT1) binding motifs. We included SF1 and WT1 because both proteins have been shown to be expressed and to be important for the formation of the genital ridges [45, 46]. This approach identified three potential SRY/SOX sites (here referred to as SOX sites) and one SF1 and one WT1 binding site within the ChIP fragment (Fig. 5A). To determine whether SRY, SOX9, and SF1 can bind to the ChIP fragment, we performed electrophoretic mobility shift assays. All three factors—SRY, SOX9, and SF1—bound in vitro to the Cbln4 enhancer fragment. SRY bound to the fragment to either one (Fig. 5B, white arrowhead) or two (Fig. 5B, black arrowhead) of the three sites, whereas SOX9 seemed to be bound to at least two of the SOX sites. The related factor SOX7, as well as the GST-only control, did not bind under these conditions.
FIG. 5.
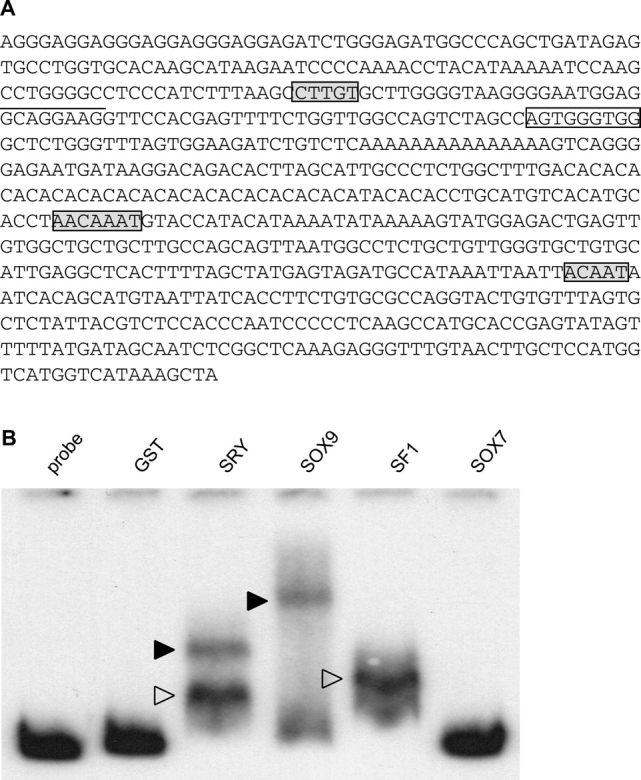
SRY, SOX9, and SF1 bind the Cbln4 ChIP fragment. A) Sequence of the Cbln4 ChIP fragment. Putative SRY/SOX binding sites are marked with gray boxes, potential SF1 binding site is underlined, and WT1 site is indicated with a box. B) Electrophoretic mobility shift assay with radioactive-labeled ChIP fragment and bacterially expressed, purified GST fusion proteins. GST-SRY binds to one (open arrowhead) or two (black arrowhead) of the three SRY/SOX sites, whereas GST-SOX9 (black arrowhead) binds at least two of the three sites simultaneously. In addition, SF1 binds to the ChIP fragment in contrast to GST-SOX7 and GST-only, which do not bind.
Cbln4 Expression Is Regulated by SRY and SOX9 In Vivo
Both SRY and SOX9 bind to essentially the same binding motif [47, 48], and both Sry and Sox9 are expressed in the same cells in overlapping time windows during gonad development, making it difficult to determine which of these factors might be responsible for the male-specific expression of Cbln4 in vivo. To solve this problem, we made use of three mouse models in which the expression of Sry and Sox9 is uncoupled.
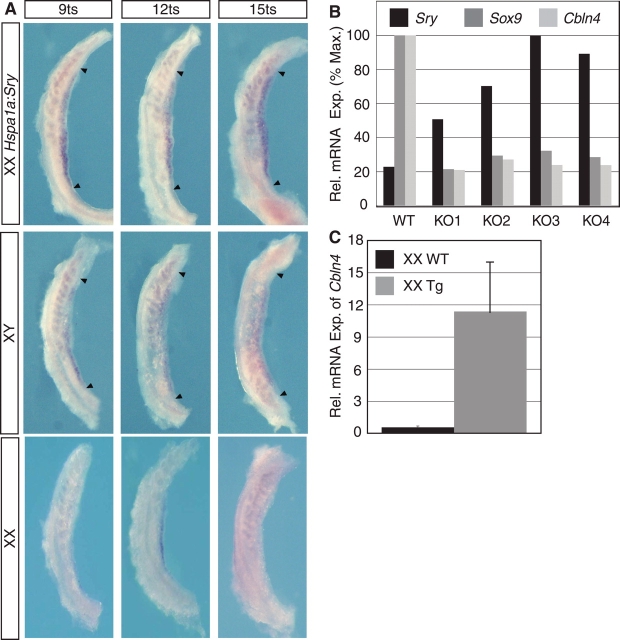
We first employed a mouse model in which SRY is expressed under the control of the Hspa1a (Hsp70.3) promoter (Hspa1a:Sry; Kidokoro et al. [49]). These mice express Sry in XX and XY genital ridges from a very early stage along the whole length of the gonad. However, although Sry is expressed at least 8 h earlier than normal, the onset of Sox9 expression is not altered [49]. Therefore, these mice provide a useful mouse model with a longer than normal time window in which Sry is expressed prior to the onset of Sox9 expression. Using this mouse model, we investigated whether Cbln4 expression is altered by the ectopic expression of Sry. Whole-mount ISH revealed that Cbln4 was expressed robustly along the whole length of the gonad by 9 ts, whereas in wild-type XX and XY gonads, Cbln4 expression was weak (Fig. 6A). These results demonstrate that ectopic SRY expression was able to elicit a corresponding ectopic upregulation of Cbln4 expression.
FIG. 6.
Both SRY and SOX9 affect Cbln4 expression in vivo. A) Cbln4 whole-mount ISH of 9–15 ts XX wild-type (bottom row), XY wild-type (middle row), and sex-reversed Sry-transgenic (XXHspa1a:Sry; top row) genital ridges. Cbln4 expression is detectable at all investigated time points in XXHspa1a:Sry genital ridges, but it is only weakly or not expressed in XX and XY wild-type genital at these time points, showing that Cbln4 expression is upregulated by the ectopic Sry expression in these mice. Arrowheads denote gonad poles. Original magnification ×10. B) Real-time RT-PCR for Sry, Sox9, and Cbln4 was performed on RNA extracted from gonad pairs at 13.5 dpc from wild-type (WT) and Sf1:Cre;Sox9flox/Sox9flox (KO1 to KO4) embryos. Variation in input RNA was normalized against Gapdh. Experiments were performed in triplicates, and relative mRNA expression is represented as percentage of maximum (% Max.) expression (Exp.). Loss of Sox9 results in a loss of Cbln4 expression at 13.5 dpc. The y-axis shows percentage of relative maximal expression. C) Real-time RT-PCR for Cbln4 was performed on RNA extracted from gonad pairs at 12.5 dpc from Wt1:Sox9 embryos. Data represent fold expression as mean ± SEM of three independent experiments normalized against 18S. Tg, transgenic.
Next, we used quantitative real-time RT-PCR to measure the expression of Cbln4 in mice harboring a null mutation of Sox9 [35]. In these mice, Sox9 was conditionally deleted by CRE recombinase driven by the Sf1 promoter (Sf1:Cre;Sox9flox/Sox9flox), here referred to as Sox9-KO [35]. Expression of Sox9 mRNA in Sox9-KO embryos is variable from almost normal to a complete absence of transcription at 13.5 dpc, with the result that, unlike wild-type mice, Sry expression in the Sox9-KO gonads remains until at least 13.5 dpc [35]. This aberrant expression profile provides a time window of gonad development in which Sry is expressed but Sox9 is absent. We found that at 13.5 dpc Cbln4 expression was reduced in XY Sox9-KO mice compared with wild-type littermates (Fig. 6B), suggesting that, at least at this late developmental stage, SOX9 is necessary for Cbln4 expression in vivo.
Having established that SOX9 is necessary for Cbln4 expression in vivo, we next sought to determine whether SOX9 also was sufficient to initiate the expression of Cbln4. To answer this question, we used quantitative real-time RT-PCR analysis to examine the expression of Cbln4 in mice (Wt1:Sox9) that express Sox9 driven by the Wt1 regulatory region of the XX gonads [22]. Ectopic expression of Sox9 was sufficient to upregulate Cbln4 in transgenic XX gonads at 13.5 dpc (Fig. 6C). Hence, SOX9 is not only necessary but also sufficient to activate Cbln4 expression in vivo in the absence of SRY. In summary, we showed that SRY and SOX9 are both able to upregulate Cbln4 expression and that SOX9 is essential for the maintenance of Cbln4 expression.
SRY and SOX9 Bind to the ChIP Fragment In Vivo
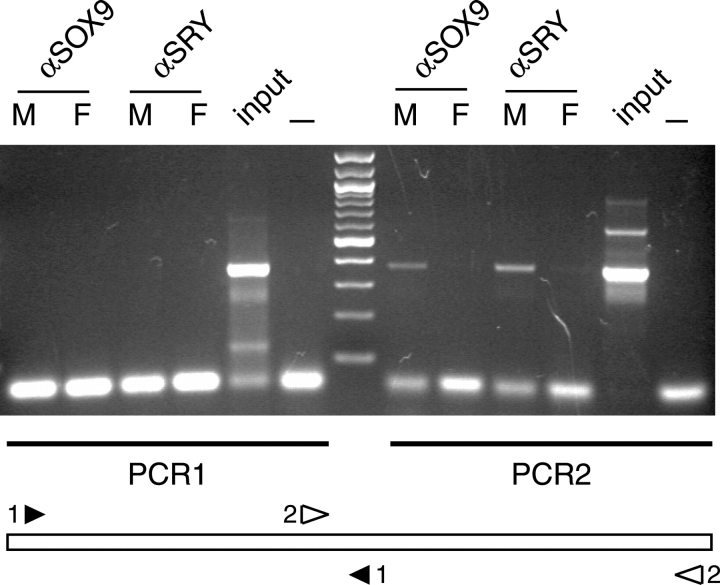
Induced ectopic expression in vivo does not necessarily mean that SRY and SOX9 bind directly and regulate the identified Cbln4 enhancer during normal development. To investigate whether endogenous SRY and SOX9 bind to the identified enhancer fragment in vivo, we performed PCR on ChIP extracts of isolated 11.5 dpc XX and XY genital ridges that have been used previously [19]. Any transcription factor that functions as a direct regulator of Cbln4 in vivo would be expected to associate with the promoter or enhancer fragment in Sertoli cell nuclei, which can be measured by an enrichment of these fragments in the ChIP extract. Two primer sets, together covering the 667-bp fragment, were used in a PCR, with the ChIP extract as template (Fig. 7). No PCR product was detected using PCR primer set 1 (Fig. 7). In contrast, using PCR primer set 2 we detected clear bands with the XY but not the XX genital ridge extract when using the SRY- and the SOX9-specific antibodies (Fig. 7). These results confirmed that both SRY and SOX9 can occupy the Cbln4 enhancer fragment in vivo in XY genital ridges at 11.5 dpc, and therefore are likely to be jointly responsible for the regulation of its male-specific expression.
FIG. 7.
SRY and SOX9 bind to putative Cbln4 enhancer region in vivo. Chromatin immunoprecipitation of XY (M) and XX (F) genital ridges at 11.5 dpc with antibodies specific to SOX9 (αSOX9) and SRY (αSRY), respectively. Polymerase chain reaction analysis with primer pair 1 (PCR1; black arrowhead 1) and primer pair 2 (PCR2; open arrowhead 2) shows an enrichment of fragment 2 in the SRY and SOX9 ChIP extracts of XY genital ridges. Genomic DNA before immunoprecipitation (input) was used as positive control, and PCR without DNA template (—) was used as negative control. The PCR fragment sizes are 361 bp (PCR1) and 367 bp (PCR2).
DISCUSSION
The male-determining gene Sry was discovered more than 15 years ago, yet very little is known about its molecular mode of action. In this study, we identified Cbln4 as a direct target gene of SRY by employing in vivo ChIP analysis. In vivo ChIP is a powerful tool for studying protein-DNA interactions [50, 51], offering the unique possibility of identifying novel target genes and their potential regulatory regions [52, 53]. However, because of the occurrence of random protein-DNA cross-linking, nonspecific DNA fragments are likely to be precipitated together with the DNA genuinely bound by the protein of interest. Therefore, to separate nonspecific fragments from genuine in vivo SRY-binding sites, we combined ChIP with detailed expression analyses and a number of in vivo studies to confirm Cbln4 as a target of the SRY transcription factor.
The Role of Cbln4
CBLN4 is a member of the C1q and tumor necrosis factor superfamily. Precerebellin 1, the first identified family member, was originally described as a precursor of the Purkinje cell-specific peptide cerebellin [54]. More recently, cerebellins 1 to 4 have been hypothesized to act as signal molecules themselves [55]. Our results here support this hypothesis by showing that Cbln4 is secreted as a glycoprotein.
One of the early roles of the supporting cell lineage in the testis is to initiate cell migration from the mesonephros into the differentiating gonad [56], cell proliferation within the gonad and the coelomic epithelium [57], and the formation of a male-specific vasculature [58]. For Sertoli cells to initiate these events, they must use some form of paracrine signaling. The expression of Cbln4 is upregulated and male specific at 18 ts, approximately the time when these processes are set in motion. It is therefore feasible that CBLN4 may be a paracrine factor produced and secreted by Sertoli cells to initiate one or several of these early testis-specific events.
In addition, Cbln4's early expression in both male and female genital ridges suggests that it might also play a sex-independent role in the formation of the early bipotential gonad. It is known for other genes involved in gonad development to have multiple roles. For example, a dual role during gonad development has been described for WT1. Mice carrying homozygous null mutations in Wt1 lack gonads entirely in males and females [45], suggesting an early role for Wt1 in the formation of the primordial genital ridge. Other studies in which Wt1 was conditionally ablated in Sertoli cells by 14.5 dpc showed that Wt1 is essential also for the maintenance of Sox9 expression and tubular architecture in the developing testis [59]. Cbln4 null mice have not been generated to date. However, future functional analyses of Cbln4 could employ similar conditional knockout strategies to identify its role in the development of the bipotential gonad as well as during testis differentiation.
SRY as a Transcriptional Activator
A longstanding question in the biology of SRY is whether SRY acts as a transcriptional activator or a repressor. Our analyses showed an increase in Cbln4 expression in the Hspa1a:Sry transgenic mice. This upregulation could be due to a direct activation by SRY, or indirectly via repression of a repressor. In vitro luciferase assays may provide further information about the transcriptional control of Cbln4 by SRY, but extrapolation of in vitro results to the in vivo setting must be treated cautiously. However, the observed in vivo binding of SRY to the putative Cbln4 enhancer in the ChIP experiments confirmed a direct interaction, and it suggests that SRY acts as a transcriptional activator of Cbln4. This is in agreement with a recent study showing that SRY binds to and activates transcription from a gonad-specific enhancer of Sox9 [8]. In contrast, other studies have suggested that SRY-negative XX maleness occurs at too high a frequency to be explained by gain of function of a male-determining gene, and is more likely a loss of function of a repressor of male development, an argument that has been used to suggest that SRY might act as a transcriptional repressor [9]. Available in vitro data show that SRY might act as both a transcriptional activator and a repressor, depending on its phosphorylation state [10]. A similar scenario has been described for a number of transcription factors, such as the thyroid hormone receptor POU1F1 (pituitary-specific transcription factor 1 [PIT-1]) and members of the homeobox gene (HOX) family, which can both activate and repress gene expression [60, 61]. Further studies are required to determine whether SRY is able to repress the expression of a specific target gene or genes during gonadal development.
Cbln4 Expression Is Regulated by SRY and SOX9
One of the complications in identifying a direct target of SRY has been the differentiation of a potential target of SRY from a potential target of SOX9. Based on the known, almost identical DNA binding and bending abilities of SRY and SOX9, any gene activated by one factor could theoretically also be activated by the other. Given that early ectopic expression of Sox9 is sufficient to induce XX sex reversal, it has been proposed that Sox9 is the only gene that is induced by SRY and that, once activated, Sox9 initiates all pathways necessary for male development. If this were the case, SRY would simply function as a switch that is present in most mammals but not in any other vertebrate class [4, 62].
However, published data do not exclude the possibility that the expression of multiple genes might be upregulated initially by SRY and subsequently maintained by SOX9. Indeed, it has been shown that Sox9 expression in the testis is initially SRY dependent. Thereafter, Sox9 transcription is maintained by SOX9 protein in a self-regulatory fashion [8]. We did not isolate the Sox9 enhancer region in our ChIP; however, the amount of starting material used is very small, and therefore it is not surprising to not have pulled down this regulatory region. Our results using the three different mouse models indicate a similar regulatory mechanism for Cbln4. The analysis of the Hspa1a:Sry mice showed that SRY might play a role in the early, male-specific upregulation of Cbln4. On the other hand, analysis of Sox9-KO mice demonstrated that SRY is not sufficient to maintain Cbln4 expression at later stages. This may be because of an absence of specific cofactors required for SRY to transactivate Cbln4 at 13.5 dpc or because of insufficient levels of SRY expression. However, we demonstrated that SOX9 is necessary and sufficient for the expression of Cbln4, which would explain why during normal development, Cbln4 is expressed in the Sertoli cells long after Sry expression has extinguished. Moreover, the second ChIP analysis proved that both SRY and SOX9 are bound to the identified Cbln4 enhancer in vivo at the right time of development, supporting the hypothesis of direct Cbln4 regulation by both SRY and SOX9. Further experiments are needed to establish the regulatory network that is needed to initiate Cbln4 expression in the bipotential genital ridges of both sexes.
A Multiple-Target Model for SRY
Based on our data, we suggest a model for the male-specific expression of Cbln4 in which SRY initiates male-specific upregulation, and SOX9 further upregulates, and then maintains, Cbln4 expression in the developing gonad (Fig. 8). This model is similar to a feed-forward network, which has been described previously, where a given transcription factor controls the expression of another transcription factor, and together the two factors regulate the transcription of a shared target gene [50, 63]. It is possible that a cooperative interaction between SRY and SOX9 may operate to transactivate Cbln4, although no direct interaction between SRY and SOX9 has been shown to date. However, SOX9 has been shown previously to interact with other SOX proteins to cooperatively activate gene expression. For example, the L-SOX5/SOX6 protein complex and SOX9 cooperate in activating the expression of collagen type 2 alpha 1 (Col2a1) by binding to a 48-bp chondrocyte-specific enhancer [64, 65]. This coupling of transcription factors accommodates switchlike transcriptional responses that are important in development [63] and may help reinforce the molecular commitment down the male pathway.
FIG. 8.
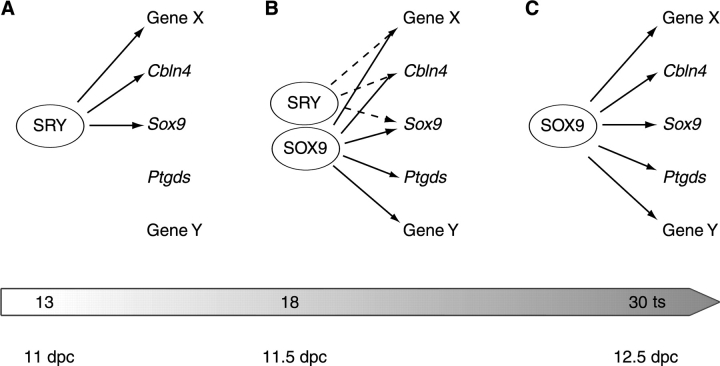
Model of SRY and SOX9 function during mammalian sex determination. A) SRY activates the transcription of a number of male-specific genes, including Sox9 and Cbln4. B) Once SOX9 is active, it contributes to the activation of SRY target genes, in addition to activating and/or repressing other, non-SRY targets. C) With SRY expression extinguished, SOX9 maintains the expression of these genes throughout testis development.
Although our data support a model in which both SRY and SOX9 regulate Cbln4 expression, we suspect that this mode of regulation may be more broadly applicable in early sex determination. According to this hypothesis, SRY is responsible for the male-specific expression of a number of genes, including Cbln4 and Sox9 (Fig. 8A). However, because the expression of SRY is transient, it is not sufficient for the maintenance of gene expression throughout testis development. Consequently, SOX9 replaces SRY (Fig. 8B) to maintain the expression of genes required for testis differentiation, and it possibly represses genes of the female pathway (Fig. 8C).
If a multiple-target model for SRY is correct, then the male-specific increase in expression of Sox9, Cbln4, and Amh apparent at 15 ts might be due to transactivation by SRY. This early upregulation of Sox9, Cbln4, and Amh by SRY might then be counteracted by genes involved in the female pathway. Once SOX9 reaches a threshold level, it replaces SRY and upregulates testis-specific genes, and it represses ovary-specific genes. A detailed expression analysis, along with in vitro studies, is necessary to determine whether this mechanism is also the case for other genes that are regulated by SOX9. One SOX9 target gene, Ptgds, has been excluded already from being initially upregulated by SRY because its promoter was found to be regulated by SOX9 but not by SRY [19].
In summary, our data show that Cbln4 is upregulated in the same spatiotemporal pattern as Sox9, making it one of the first genes to be upregulated in early testis determination. The increased expression in the testis is mediated by both SRY and SOX9. CBLN4 is likely to work in a paracrine fashion and be part of the signaling mechanisms of early Sertoli cells to direct the differentiation of other cell types within the developing testis. Our data identify Cbln4 as a likely direct target gene of SRY and support a transcriptional model in which SRY has multiple targets and cooperates with SOX9 to regulate early gene expression in the developing testis.
Supplementary Material
Acknowledgments
We thank the staff of the Institute for Molecular Bioscience Animal Facility for breeding mice used in this study; Marcel Dinger and Martin Frith for assistance with bioinformatics; Annemiek Beverdam, Fred Martinson, Jo Bowles, Cassy Spiller, Fiona Simpson, Rajith Aturaliya, Markus Kerr, and Andrew Jackson for technical help; and Annemiek Beverdam and Terje Svingen for comments on the manuscript.
Footnotes
1Supported by research grants from the National Institutes of Health (R03-HD049431), the National Health and Medical Research Council of Australia, the Australian Research Council, and the Agence Nationale de la Recherche (ANR-07-BLAN-0044).
REFERENCES
- Berta P, Hawkins JR, Sinclair AH, Taylor A, Griffiths BL, Goodfellow PN, Fellous M.Genetic evidence equating SRY and the male sex determining gene. Nature 1990; 348: 448–450. [DOI] [PubMed] [Google Scholar]
- Jäger RJ, Anvret M, Hall K, Scherer G.A human XY female with a frame shift mutation in the candidate testis-determining gene SRY. Nature 1990; 348: 452–454. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R.Male development of chromosomally female mice transgenic for Sry. Nature 1991; 351: 117–121. [DOI] [PubMed] [Google Scholar]
- Polanco JC, Koopman P.Sry and the hesitant beginnings of male development. Dev Biol 2007; 302: 13–24. [DOI] [PubMed] [Google Scholar]
- Giese K, Pagel J, Grosschedl R.Distinct DNA-binding properties of the high mobility group domain of murine and human SRY sex-determining factors. Proc Natl Acad Sci U S A 1994; 91: 3368–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner MH, Bianchi ME, Gronenborn AM, Clore GM.NMR spectroscopic analysis of the DNA conformation induced by the human testis determining factor SRY. Biochemistry 1995; 34: 11998–12004. [DOI] [PubMed] [Google Scholar]
- Dubin RA, Ostrer H.Sry is a transcriptional activator. Mol Endocrinol 1994; 8: 1182–1192. [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R.Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 2008; 453: 930–934. [DOI] [PubMed] [Google Scholar]
- McElreavey K, Vilain E, Herskowitz I, Fellous M.A regulatory cascade hypothesis for mammalian sex determination: SRY represses a negative regulator of male development. Proc Natl Acad Sci U S A 1993; 90: 3368–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desclozeaux M, Poulat F, Barbara PD, Capony JP, Turowski P, Jay P, Mejean C, Moniot B, Boizet B, Berta P.Phosphorylation of an N-terminal motif enhances DNA-binding activity of the human SRY protein. J Biol Chem 1998; 273: 7988–7995. [DOI] [PubMed] [Google Scholar]
- Bullejos M, Koopman P.Spatially dynamic expression of Sry in mouse genital ridges. Dev Dyn 2001; 221: 201–205. [DOI] [PubMed] [Google Scholar]
- Sekido R, Bar I, Narvaez V, Penny G, Lovell-Badge R.SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev Biol 2004; 274: 271–279. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Martinson F, Bradford S, Wilson MJ, Combes AN, Beverdam A, Bowles J, Mizusaki H, Koopman P.Sertoli cell differentiation is induced both cell-autonomously and through prostaglandin signaling during mammalian sex determination. Dev Biol 2005; 287: 111–124. [DOI] [PubMed] [Google Scholar]
- Kent J, Wheatley SC, Andrews JE, Sinclair AH, Koopman P.A male-specific role for SOX9 in vertebrate sex determination. Development 1996; 122: 2813–2822. [DOI] [PubMed] [Google Scholar]
- Harley VR, Lovell-Badge R, Goodfellow PN.Definition of a consensus DNA binding site for SRY. Nucleic Acids Res 1994; 22: 1500–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Harley VR.Molecular mechanisms of SOX9 action. Mol Genet Metab 2000; 71: 455–462. [DOI] [PubMed] [Google Scholar]
- Arango N, Lovell-Badge R, Behringer R.Targeted mutagenesis of the endogenous mouse Mis gene promoter: in vivo definition of genetic pathways of vertebrate sexual development. Cell 1999; 99: 409–419. [DOI] [PubMed] [Google Scholar]
- de Santa Barbara P, Bonneaud N, Boizet B, Desclozeaux M, Moniot B, Südbeck P, Scherer G, Poulat F, Berta P.Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Müllerian hormone gene. Mol Cell Biol 1998; 18: 6653–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm D, Hiramatsu R, Mizusaki H, Widjaja L, Combes AN, Kanai Y, Koopman P.SOX9 regulates prostaglandin D synthase gene transcription in vivo to ensure testis development. J Biol Chem 2007; 282: 10553–10560. [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Jeyasuria P, Parker KL, Koopman P.The transcription factors steroidogenic factor-1 and SOX9 regulate expression of Vanin-1 during mouse testis development. J Biol Chem 2005; 280: 5917–5923. [DOI] [PubMed] [Google Scholar]
- Bishop CE, Whitworth DJ, Qin Y, Agoulnik AI, Agoulnik IU, Harrison WR, Behringer RR, Overbeek PA.A transgenic insertion upstream of Sox9 is associated with dominant XX sex reversal in the mouse. Nat Genet 2000; 26: 490–494. [DOI] [PubMed] [Google Scholar]
- Vidal V, Chaboissier M, de Rooij D, Schedl A.Sox9 induces testis development in XX transgenic mice. Nat Genet 2001; 28: 216–217. [DOI] [PubMed] [Google Scholar]
- Bullejos M, Koopman P.Delayed Sry and Sox9 expression in developing mouse gonads underlies B6-YDOM sex reversal. Dev Biol 2005; 278: 473–481. [DOI] [PubMed] [Google Scholar]
- Nagamine C, Morohashi K, Carlisle C, Chang D.Sex reversal caused by Mus musculus domesticus Y chromosomes linked to variant expression of the testis-determining gene Sry. Dev Biol 1999; 216: 182–194. [DOI] [PubMed] [Google Scholar]
- Buehr M, McLaren A, Bartley A, Darling S.Proliferation and migration of primordial germ cells in We/We mouse embryos. Dev Dyn 1993; 198: 182–189. [DOI] [PubMed] [Google Scholar]
- Hacker A, Capel B, Goodfellow P, Lovell-Badge R.Expression of Sry, the mouse sex determining gene. Development 1995; 121: 1603–1614. [DOI] [PubMed] [Google Scholar]
- Bradford ST, Wilhelm D, Koopman P.Comparative analysis of anti-mouse SRY antibodies. Sex Dev 2007; 1: 305–310. [DOI] [PubMed] [Google Scholar]
- Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, et al. Ensembl 2008. Nucleic Acids Res 2008; 36: D707–D714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radler-Pohl A, Sachsenmaier C, Gebel S, Auer HP, Bruder JT, Rapp U, Angel P, Rahmsdorf HJ, Herrlich P.UV-induced activation of AP-1 involves obligatory extranuclear steps including Raf-1 kinase. EMBO J 1993; 12: 1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P, Toomre D, Diaz E, White J, Simons K.Multicolour imaging of post-Golgi sorting and trafficking in live cells. Nat Cell Biol 2001; 3: 140–149. [DOI] [PubMed] [Google Scholar]
- Munsterberg A, Lovell-Badge R.Expression of the mouse anti-mullerian hormone gene suggests a role in both male and female sexual differentiation. Development 1991; 113: 613–624. [DOI] [PubMed] [Google Scholar]
- Schepers G, Wilson M, Wilhelm D, Koopman P.SOX8 is expressed during testis differentiation in mice and synergizes with SF1 to activate the Amh promoter in vitro. J Biol Chem 2003; 278: 28101–28108. [DOI] [PubMed] [Google Scholar]
- Hargrave M, Bowles J, Koopman P.In situ hybridization of whole-mount embryos. Methods Mol Biol 2006; 326: 103–113. [DOI] [PubMed] [Google Scholar]
- Svingen T, Beverdam A, Bernard P, McClive P, Harley VR, Sinclair AH, Koopman P.Sex-specific expression of a novel gene Tmem184a during mouse testis differentiation. Reproduction 2007; 133: 983–989. [DOI] [PubMed] [Google Scholar]
- Chaboissier MC, Kobayashi A, Vidal VI, Lutzkendorf S, van de Kant HJ, Wegner M, de Rooij DG, Behringer RR, Schedl A.Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development 2004; 131: 1891–1901. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Englert C.The Wilms tumor suppressor WT1 regulates early gonad development by activation of Sf1. Genes Dev 2002; 16: 1839–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Dixon JE.Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem 1991; 192: 262–267. [DOI] [PubMed] [Google Scholar]
- Beverdam A, Koopman P.Expression profiling of purified mouse gonadal somatic cells during the critical time window of sex determination reveals novel candidate genes for human sexual dysgenesis syndromes. Hum Mol Genet 2006; 15: 417–431. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D.The human genome browser at UCSC. Genome Res 2002; 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TMHMM server v. 2.0: prediction of transmembrane helices in proteins. Center for Biological Sequence Analysis, Technical University of Denmark, Lyngby, Denmark.; 2007. World Wide Web (URL: http://www.cbs.dtu.dk/services/TMHMM/). (June, 2007). [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H.Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2007; 2: 953–971. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P.SMART 6: recent updates and new developments. Nucleic Acids Res 2009; 37: D229–D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innamorati G, Bianchi E, Whang MI.An intracellular role for the C1q-globular domain. Cell Signal 2006; 18: 761–770. [DOI] [PubMed] [Google Scholar]
- Kishore U, Gaboriaud C, Waters P, Shrive AK, Greenhough TJ, Reid KB, Sim RB, Arlaud GJ.C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol 2004; 25: 551–561. [DOI] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R.WT-1 is required for early kidney development. Cell 1993; 74: 679–691. [DOI] [PubMed] [Google Scholar]
- Luo X, Ikeda Y, Parker KL.A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell 1994; 77: 481–490. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Harley VR, Pontiggia A, Goodfellow PN, Lovell-Badge R, Bianchi ME.SRY, like HMG1, recognizes sharp angles in DNA. EMBO J 1992; 11: 4497–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertin S, McDowall SG, Harley VR.The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res 1999; 27: 1359–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro T, Matoba S, Hiramatsu R, Fujisawa M, Kanai-Azuma M, Taya C, Kurohmaru M, Kawakami H, Hayashi Y, Kanai Y, Yonekawa H.Influence on spatiotemporal patterns of a male-specific Sox9 activation by ectopic Sry expression during early phases of testis differentiation in mice. Dev Biol 2005; 278: 511–525. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 2002; 298: 799–804. [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 2007; 4: 651–657. [DOI] [PubMed] [Google Scholar]
- Shimomura Y, Wajid M, Shapiro L, Christiano AM.P-cadherin is a p63 target gene with a crucial role in the developing human limb bud and hair follicle. Development 2008; 135: 743–753. [DOI] [PubMed] [Google Scholar]
- White RB, Ziman MR.Genome-wide discovery of Pax7 target genes during development. Physiol Genomics 2008; 33: 41–49. [DOI] [PubMed] [Google Scholar]
- Slemmon JR, Blacher R, Danho W, Hempstead JL, Morgan JI.Isolation and sequencing of two cerebellum-specific peptides. Proc Natl Acad Sci U S A 1984; 81: 6866–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima T, Miura E, Matsuda K, Kamekawa Y, Watanabe M, Yuzaki M.Characterization of a transneuronal cytokine family Cbln–regulation of secretion by heteromeric assembly. Eur J Neurosci 2007; 25: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Capel B, Albrecht KH, Washburn LL, Eicher EM.Migration of mesonephric cells into the mammalian gonad depends on Sry. Mech Dev 1999; 84: 127–131. [DOI] [PubMed] [Google Scholar]
- Schmahl J, Eicher E, Washburn L, Capel B.Sry induces cell proliferation in the mouse gonad. Development 2000; 127: 65–73. [DOI] [PubMed] [Google Scholar]
- Brennan J, Karl J, Capel B.Divergent vascular mechanisms downstream of Sry establish the arterial system in the XY gonad. Dev Biol 2002; 244: 418–428. [DOI] [PubMed] [Google Scholar]
- Gao F, Maiti S, Alam N, Zhang Z, Deng JM, Behringer RR, Lecureuil C, Guillou F, Huff V.The Wilms tumor gene, Wt1, is required for Sox9 expression and maintenance of tubular architecture in the developing testis. Proc Natl Acad Sci U S A 2006; 103: 11987–11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman DS.Transcription factors: bound to activate or repress. Trends Biochem Sci 2001; 26: 211–213. [DOI] [PubMed] [Google Scholar]
- Pinsonneault J, Florence B, Vaessin H, McGinnis W.A model for extradenticle function as a switch that changes HOX proteins from repressors to activators. EMBO J 1997; 16: 2032–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JA.From brain determination to testis determination: evolution of the mammalian sex-determining gene. Reprod Fertil Dev 2001; 13: 665–672. [DOI] [PubMed] [Google Scholar]
- Komili S, Silver PA.Coupling and coordination in gene expression processes: a systems biology view. Nat Rev Genet 2008; 9: 38–48. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Behringer RR, de Crombrugghe B.L-Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage 2001; 9(suppl A):S69–S75. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Li P, de Crombrugghe B.A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J 1998; 17: 5718–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.