Abstract
The involvement of egg integrins in mammalian sperm-egg interactions has been controversial, with data from integrin inhibitor studies contrasting with evidence from knockouts showing that specific integrin subunits are not essential for fertility. An alpha4/alpha9 (ITGA4/ITGA9) integrin subfamily member has been implicated in fertilization but not extensively examined, so we tested the following three hypotheses: 1) an ITGA4/ITGA9 integrin participates in sperm-egg interactions, 2) short-term acute knockdown by RNA interference of integrin subunits would result in a fertilization phenotype differing from that of chronic depletion via knockout, and 3) detection of a fertilization phenotype is sensitive to in vitro fertilization (IVF) assay conditions. We show that mouse and human eggs express the alpha9 integrin subunit (ITGA9). RNA interference-mediated knockdown resulted in reduced levels of Itga9 mRNA and surface protein in mouse eggs. RNA interference attempts to knockdown ITGA9′s likely beta partner, beta1 (ITGB1), resulted in reduced Itgb1 mRNA but no reduction in ITGB1 surface protein. Therefore, studies using a function-blocking anti-ITGB1 antibody tested the hypothesis that ITGB1 participates in gamete interactions. Analyses of sperm-egg interactions with Itga9-knockdown eggs and anti-ITGB1 antibody-treated eggs in IVF assays using specific sperm:egg ratios revealed the following: 1) a reduction, but not complete loss, of sperm-egg binding and fusion was observed and 2) the reduction of sperm-egg binding and fusion was not detected in inseminations with high sperm:egg ratios. These data demonstrate that ITGA9 and ITGB1 participate in sperm-egg interactions but clearly are not the only molecules involved. This also shows that careful design of IVF parameters allows detection of deficiencies in gamete interactions.
Keywords: ADAM, fertilization, integrin, in vitro fertilization, ovum, sperm-egg binding
Mouse and human eggs express the integrin ITGA9, and IVF assays reveal defects in sperm binding and fusion in mouse eggs in which integrin function has been disrupted by RNAi-mediated knockdown of Itga9 or by function-blocking antibodies to ITGB1.
INTRODUCTION
A crucial step of fertilization is the cell-cell interaction that allows the two gametes to fuse and create the zygote, although the mechanistic basis of this remains elusive. Despite concerted efforts to identify molecules that mediate mammalian gamete membrane interactions, only two key molecules have emerged, CD9 on the egg and IZUMO1 (also known as Izumo) on the sperm. IZUMO1 is a member of the immunoglobulin superfamily (IgSF), and Izumo1−/− male mice sire no pups in mating trial investigations, and sperm from Izumo1−/− males are unable to fertilize eggs in vitro [1]. CD9 is a member of the tetraspanin family of membrane proteins and is expressed in numerous tissues [2–4], but the knockout phenotype uniquely affects female reproduction. Only 60% of Cd9−/− females became pregnant, and the pregnancies in this subset of animals took longer to achieve and resulted in litters that were ∼25% the size of litters from control females [5]. Cd9−/− eggs have a severe defect in sperm-egg fusion [6–8] and abnormal microvillar structure on the egg surface [9]. These knockout studies are complemented by antibody studies [1, 5, 10, 11] showing that anti-IZUMO1 antibodies and anti-CD9 antibodies have inhibitory effects in in vitro fertilization (IVF) assays.
The role of CD9 in egg membrane organization [9] suggests that additional egg molecules are involved in gamete membrane interaction, although in contrast to the robust effects observed in investigations using CD9 knockout eggs and anti-CD9 antibodies, disruption of other molecules leads to more subtle effects. CD81, another tetraspanin, is an example of this; CD81−/− females have modest defects in various end points of female fertility [5], and anti-CD81 antibodies have moderate or no effect in IVF assays [5, 12]. Members of the integrin family of heterodimeric cell adhesion molecules are another group of egg candidate molecules. Mammalian eggs express at least three integrin β subunits and seven α subunits, with a few differences between species [13–25]. Egg integrins have been suggested to participate in gamete interactions based on inhibition of sperm-egg interactions in IVF assays using antibodies to specific integrin subunits or peptides blocking specific subfamilies of integrins [18, 21, 23, 24, 26–29]. However, no defect in fertility has been reported in the egg-expressed integrins for the knockouts that are viable (e.g., Itga1 [α1], Itga2 [α2], Itgb3 [β3], and Itgb5 [β5]). Other integrin subunit knockouts die during embryonic development or the neonatal period, although three of these subunits have been investigated by oocyte-specific conditional knockout (Itgb1, β1) or by performance of kidney capsule transplants of ovaries recovered from neonates and evaluation of the oocytes from these ovaries (Itga3 [α3] and Itga6 [α6]) [30, 31]. Eggs deficient in Itga3, Itga6, or Itgb1 can be fertilized in vitro, and female mice with Itgb1-deficient eggs have no deficits in fertility [30, 31].
Our study examines two issues regarding a putative role for egg integrins in fertilization. First, we performed short-term acute integrin subunit depletion in eggs by RNA interference (RNAi), reducing opportunity for compensatory changes occurring that could allow the egg membrane to be capable of supporting sperm interactions. Second, a possible explanation for why no deficit in egg function has been detected in knockout animals is that the defects are not apparent in conventional mating trials or IVF assays. A leader in the integrin field, Richard Hynes, and his colleague, have written, “Failure to observe a phenotype does not mean that one is not present; it may simply mean that the physiological analyses are not yet sufficiently refined to reveal it” [32, p. 2194]. In this work, we perform a series of systematic IVF assays using a range of specific sperm:egg ratios (25:1, 100:1, or 500:1), with these IVF assays used as much for cell-cell interaction as for reproductive function. This concept of varying the sperm number/concentration has been applied to assessments of sperm fertilizing ability, and it has been suggested that the sperm number used in an assay to characterize a deficiency in fertilizing ability needs to be below the sperm number at which the maximum in some fertility outcome (e.g., percentage of eggs fertilized) is achieved [33]. We apply this concept of varying the amount of sperm in IVF to examine the ability of experimentally manipulated eggs to support sperm binding and fusion and, indeed, are able to detect a phenotype in Itga9 (α9) integrin subunit knockdown eggs.
MATERIALS AND METHODS
Eggs and Oocytes
All work involving animals was conducted with approval from the Johns Hopkins University Animal Care and Use Committee. Ovulated mature metaphase II eggs were collected from 6- to 8-wk-old superovulated CF1 mice (Harlan, Indianapolis, IN) at ∼13 h after human chorionic gonadotropin injection. Cumulus cells were removed by brief incubation (<5 min) in Whitten medium containing 15 mM Hepes (109.5 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 5.5 mM glucose, 0.23 mM pyruvic acid, 4.8 mM lactic acid, 7 mM NaHCO3, 15 mM Hepes; hereafter referred to as Whitten-Hepes [34]), 30 mg/ml of bovine serum albumin (BSA) (Albumax I; Gibco-BRL, Gaithersburg, MD), and 0.02% type IV-S hyalouronidase (Sigma, St. Louis, MO). The zonae pellucidae (ZP) were then removed by a brief incubation (∼15 sec) in acidic culture medium-compatible buffer (10 mM Hepes, 1 mM NaH2PO4, 0.8 mM MgSO4, 5.4 mM KCl, 116.4 mM NaCl, pH 1.5) and then allowed to recover for 60 min in Whitten medium containing 22 mM NaHCO3 (109.5 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 5.5 mM glucose, 0.23 mM pyruvic acid, 4.8 mM lactic acid, 22 mM NaHCO3; hereafter referred to as Whitten-bicarbonate) supplemented with 15 mg/ml of Albumax BSA or with 0.05% polyvinyl alcohol (PVA), as indicated. All egg culture was performed in drops covered with mineral oil at 37°C in a humidified atmosphere of 5% CO2 in air.
Germinal vesicle (GV)-intact oocytes were collected from the ovaries of 6- to 9-wk-old female CF-1 mice (Harlan) as previously described [35] in Whitten-Hepes supplemented with 0.05% PVA (Sigma) and 2.5 μM milrinone (a phosphodiesterase 4 inhibitor to prevent GV breakdown; Sigma). Cumulus cells were removed by pipetting oocytes through a thin-bore pipette.
Discarded human oocytes (at various stages of maturation between GV intact and metaphase II) that were deemed unusable for IVF were obtained, with approval from the Johns Hopkins Bloomberg School of Public Health Institutional Review Board. They were used for immunofluorescence as described below.
Immunofluorescence
Antibodies used for immunofluorescent staining of live mouse eggs were dialyzed against Whitten-compatible dialysis buffer (109.5 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 7 mM NaHCO3, 15 mM Hepes, pH 7.4). ZP-free live mouse eggs were cultured with 50 μg/ml of chicken anti-ITGA9 IgY antibody (Genway Biotech, Inc., San Diego, CA) or nonimmune chicken IgY control (Genway Biotech, Inc.) in Whitten-bicarbonate supplemented with 0.05% PVA for 1 h at 37°C (10 eggs per 10-μl drops). For recombinant antigen crossabsorption experiments, 15 pM (78.75 μg/ml) bacterially expressed ITGA9 fusion protein was preincubated with 3 pM (50 μg/ml) anti-ITGA9 antibody in 10 μl of Whitten-bicarbonate with 0.05% PVA at 37°C for 1 h before eggs were added. After this incubation in primary antibody, the eggs were washed through three 100-μl drops of Whitten-bicarbonate with 0.05% PVA and then fixed with 4% paraformaldehyde in PBS (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.4) for 1 h at room temperature. After fixation, eggs were washed three times in PBS, then incubated in 10% cold fish gelatin in PBS for 1 h, then incubated in 15 μg/ml of fluorescein isothiocyanate (FITC)-conjugated goat anti-chicken IgY secondary antibody (Jackson Immunoresearch, West Grove, PA) in PBS containing 10% fish gelatin for 1 h, and finally washed three times in PBS containing 10% fish gelatin, once in PBS, once in 25% glycerol in PBS, once in 50% glycerol in PBS, and once in 75% glycerol in PBS.
ZP-free human oocytes were fixed in 3.7% paraformaldehyde in PBS, then permeabilized with 0.1% Triton X-100 in PBS, and then incubated in blocking solution (PBS containing 0.1% BSA and 0.01% Tween 20). Staining used 15 μg/ml of anti-ITGA9 Y9A2 (Chemicon, Temecula, CA) or, as a negative control, 15 μg/ml of mouse nonimmune IgG. This was followed by 5 μg/ml of biotin-conjugated goat anti-mouse IgG (Jackson Immunoresearch) and then 2.5 μg/ml of avidin-Texas Red or avidin-FITC (Vector Laboratories, Burlingame, CA). As a positive control, oocytes were stained with 10 μg/ml of anti-CD9 SYB-1 [36] (gift of Claude Boucheix and Eric Rubinstein), followed by 15 μg/ml of FITC-conjugated goat anti-mouse IgG.
Eggs were mounted in VectaShield mounting medium (Vector Laboratories) containing 1.5 μg/ml of 4′,6′-diamidino-2-phenylindole (DAPI; Sigma). They were viewed on a Nikon (Natick, MA) Eclipse 800 microscope, with images captured using a Princeton Instruments 5-MHz Cooled CCD camera and IPLab software (Scanalytics, Fairfax, VA).
Immunoprecipitation
ZP-free mouse eggs were surface biotinylated in Whitten-bicarbonate medium containing 0.05% PVA (WB-PVA) and 0.5 mg/ml of EZ-link-sulfo-LC-biotin (Pierce Chemical Company, Rockford, IL) for 30 min at 37°C, after which Tris-HCl (pH 7.5) was added to a final concentration of 10 mM. The eggs were then quickly washed through three drops of Whitten medium containing 0.05% PVA. The eggs were lysed in 150 mM NaCl, 50 mM Tris-HCl, 2 mM CaCl2, 1% Triton X-100 supplemented with 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (Sigma), 1 μg/ml of leupeptin (Sigma), and 1 μg/ml of pepstatin (Sigma) for 30 min on ice and then centrifuged at 18 000 × g for 15 min at 4°C. Before immunoprecipitation, egg lysates (prepared from 250–300 eggs) were precleared overnight at 4°C with 40 μl of packed protein G-sepharose beads (Upstate, Lake Placid, NY). The precleared lysates were then combined with 1 μg of the anti-ITGB1/β1 monoclonal Armenian hamster antibody Hmβ1-1 (BD Pharmingen, San Diego, CA) [37], the anti-ITGA9/α9 rabbit polyclonal antibody 1057 (gift of Dean Sheppard, University of California, San Francisco [38]), or nonimmune controls (Armenian hamster or rabbit IgG; Jackson Immunoresearch) and incubated for 4 h at 4°C, after which 20 μl of packed protein G beads was added; this mixture incubated overnight at 4°C. The beads were then washed five times with ice-cold lysis buffer, after which the beads were resuspended in SDS-PAGE sample buffer (2% SDS, 5.5% sucrose, 0.0006% bromophenol blue, 80 mM Tris-HCl, pH 6.8) and heated at 100°C for 5 min; the eluted proteins were separated by SDS-PAGE on a 7% nonreducing gel. Following electrophoresis, proteins were transferred to an Immobilon membrane (Millipore Corporation, Bedford, MA). The membrane was blocked with 10% cold-water fish gelatin (Sigma) in PBS for 60 min at room temperature, and then biotinylated proteins were detected using the ImmunoPure ABC Peroxidase staining kit (Pierce Chemical Company) according to the manufacturer's instructions.
Double-Stranded RNA Preparation
Templates for in vitro transcription of double-stranded RNA (dsRNA) were prepared by PCR using primers containing T7 promoter sequences. The Itgb1 forward primer was 5′-TAATACGACTCACTATAGGGTACTGGGCACACTGTCTGGAAACT-3′, and the Itgb1 reverse primer was 5′-TAATACGACTCACTATAGGGTACTCGCCATTGCCTCCACAAATT-3′, generating a 646-bp product corresponding to a portion of the mouse Itgb1 coding region (nucleotides 1071–1717, counting the A of the start ATG as nucleotide 1). The template used for the Itgb1 PCR was the plasmid pMINTβ-13A (from the laboratory of Richard Hynes; provided by Daniel Leahy, Johns Hopkins School of Medicine). The Itga9 forward primer was 5′-TAATACGACTCACTATAGGGACATCTACTACGAAGCAGACC-3′, and the Itga9 reverse primer was 5′-TAATACGACTCACTATAGGGACCATGATAGATGTAGACTGC-3′, generating an 801-bp product corresponding to a portion of the Itga9 coding region (nucleotides 669-1427) from PCR using the plasmid pGEX-4T-1-mα9bp as template. The resulting PCR products were checked on agarose gels and sequenced.
The Itga9 and Itgb1 PCR products were used as templates for in vitro transcription and dsRNA preparation according to the manufacturer's instructions (MEGAscript RNAi kit; Ambion, Austin, TX). Briefly, 2 μl of the PCR product, 2 μl of 10× T7 reaction buffer, 2 μl of 75 mM NTP solution, 2 μl of T7 RNA polymerase, and 20 μl of nuclease-free water were combined and incubated for 4 h at 37°C. DNA template and unannealed single-stranded RNA were then removed by treatment with DNase I and RNase for 1 h at 37°C. Double-stranded RNA was purified on a filter cartridge provided with the MEGAscript RNAi kit (Ambion) and eluted in sterile water. The resulting dsRNA was examined on an agarose gel. The dsRNA concentration was determined and then diluted to 100 μg/ml and stored at −80°C.
Mos dsRNA was used herein as a negative dsRNA control. It was prepared from pCRII-Mos (gift of Paula Stein) as previously described [39].
Oocyte Microinjection
ZP-intact oocytes were injected in groups of 10 in a 100-μl drop of Whitten-Hepes containing 0.05% PVA and 2.5 μM milrinone covered with mineral oil, using Eppendorf (Hamburg, Germany) FemtoJet on a Nikon Eclipse TE 2000–5 microscope. The concentration of dsRNA stocks was 100 μg/ml. Injection parameters were injection pressure of 270–350 hPa, injection time of 0.1–0.5 sec, and compensation pressure of 50 hPa. Injection was considered successful when recoiling in the oocyte cytoplasm was observed. Injected oocytes were cultured for 48 h in Whitten-bicarbonate supplemented with 0.05% PVA and 2.5 μM milrinone, transferring the oocytes to fresh medium every 24 h.
To assess mRNA knockdown by RT-PCR, injected oocytes were collected 48 h after injection. For luminometric assays to assess surface protein knockdown and IVF assays, oocytes were cultured for 48 h after injection and then washed through twelve 100-μl drops of Whitten-bicarbonate media containing 0.05% PVA without milrinone. Oocytes were then cultured overnight to allow meiotic maturation to metaphase II.
Isolation of Oocyte RNA and RT-PCR
Total RNA was isolated from 35 dsRNA-injected mouse oocytes for RT-PCR using either RNAqueous-Micro Isolation kit (Ambion) or Trizol reagent (Invitrogen). First-strand cDNA was synthesized from RNA of 35 oocyte equivalents using random hexamer primers and SuperScript Reverse Transcriptase III (Invitrogen). For semiquantitative PCR to assess RNA knockdown in dsRNA-injected oocytes, PCR was performed using Itga9- or Itgb1-specific primers or tissue plasminogen activator as a control (Plat forward primer, 5′-CATGGGCAAGCGTTACACAG-3′; reverse primer, 5′-CAGAGAAGAATGGAGACGAT-3′ [39]). For each set of gene-specific primers, preliminary experiments assessed the amount of PCR product as a function of cycle number to identify linear range. The PCR was conducted in 25-μl reactions with cDNA of two oocyte equivalents for Itgb1 and Plat or with cDNA of eight oocyte equivalents for Itga9. The PCR products were examined on agarose gels, viewed either using Typhoon 9200 Variable Mode Imager (Molecular Dynamics, Sunnyvale, CA) for samples prepared with QuantiTect SYBR Green PCR kit (Qiagen, Valencia, CA) or using UV light following ethidium bromide staining. Quantification of band intensities was performed using either ImageQuant version 5.0 software (Molecular Dynamics) or Quantity One version 4.4.1 software (BioRad, Hercules, CA).
Luminometric Immunoassay Detection of ITGA9 or ITGB1 Protein on the Egg Surface
To assess surface protein knockdown following dsRNA injection, microinjected oocytes were cultured for 48 h in Whitten-bicarbonate and the phosphodiesterase inhibitor milrinone and were then matured overnight (∼16 h) without milrinone to allow meiotic maturation to metaphase II. The ZP were removed, and ZP-free eggs were incubated in Whitten-bicarbonate with 0.05% PVA containing either 100 μg/ml of anti-ITGB1 antibody Hmβ1–1, 100 μg/ml of nonimmune Armenian hamster IgG control, 50 μg/ml of anti-ITGA9 antibody (Genway Biotech, Inc.), or 50 μg/ml of nonimmune chicken IgY control for 1 h. Eggs were washed through three 100-μl drops of Whitten-bicarbonate with 0.05% PVA, then fixed with 4% paraformaldehyde in PBS for 1 h, and then processed for luminometric detection of egg-bound antibody as previously described [27]. Eggs labeled with the hamster anti-ITGB1 antibody were incubated for 1 h in alkaline phosphatase (AP) blocking buffer (PBS containing 1% AP-free casein [I-Block; Tropix, Bedford, MA, or Applied Biosystems, Foster City, CA]), 0.01% Tween-20, and 0.02% sodium azide, and eggs labeled with the chicken anti-ITGA9 antibody were incubated in PBS containing 10% cold-water fish gelatin. Eggs were then incubated for 1 h in the species-appropriate AP-conjugated secondary antibody (6 μg/ml in PBS containing 10% fish gelatin; Jackson Immunoresearch). The eggs were then washed through ten 50-μl drops of AP blocking buffer and three drops of PBS (10 min per each wash). To detect egg-associated AP activity, individual eggs were transferred to 25 μl of AP assay buffer (20 mM Tris-HCl, pH 9.5, 1 mM MgCl2) in a 12 × 75-mm polystyrene tube. One hundred microliters of enhancer-substrate solution (AP assay buffer containing 25 mM Sapphire II or Sapphire III enhancer and 0.25 mM disodium 3-[4-methoxyspiro(1,2-dioxetane-3, 2′-[5′-chloro]tricyclo[3.3.1.13,7]decan)-4-yl]phenyl phosphate substrate [CSPD]; Tropix or Applied Biosystems) was added to each tube, and tubes were incubated for 20 min at room temperature. Photon emission was measured by raw light units (RLU) detected during 10 sec (RLU/10 sec) in a Monolight 3010 luminometer (Analytical Luminescence Laboratory, Sparks, MD).
In Vitro Fertilization
In vitro fertilization of ZP-free eggs was performed as described [40] with the following modifications. Cauda epididymal sperm were prepared from CD1 retired breeders (Harlan) by swim-up as previously described [41]. Sperm were cultured for a total of 2.5–3 h in Whitten-bicarbonate medium containing 15 mg/ml of BSA (Albumax I) to allow the sperm to undergo capacitation and spontaneous acrosome exocytosis. Double-stranded RNA-injected oocytes were cultured for 48 h and then matured in vitro for 16 h to metaphase II, and then ZP removal was performed as already described. Only mature metaphase II eggs (i.e., without a GV, with a first polar body) were used for IVF. For experiments examining the effects of the anti-ITGB1 antibody Hmβ1-1 [37], ovulated metaphase II eggs were collected from superovulated females as already described. After the ZP were removed, the eggs were incubated in Whitten- bicarbonate medium containing 15 mg/ml of BSA and either 100 μg/ml of anti-ITGB1 Hmβ1-1 antibody or 100 μg/ml of nonimmune Armenian hamster IgG for 1 h before insemination.
Inseminations were performed for 60 min with sperm:egg ratios of 25:1, 100:1, and 500:1 (i.e., 10 eggs per 10-μl drop with 25 000, 100 000, and 500 000 sperm/ml, respectively). In inseminations with antibody-treated eggs, the eggs were pretreated with antibody in 10-μl drops, and 10 μl of a 2× sperm suspension was added, so that the final sperm:egg ratio was 100:1 or 500:1, as indicated; thus, the antibody concentration during insemination was 50 μg/ml. After insemination, the eggs were washed through six 100-μl drops of WB-PVA using a thin-bore pipette to remove loosely attached sperm; the same person performed all washes, using the same pipette and the same washing pressure for all experimental groups. Eggs were then fixed in 4% paraformaldehyde in PBS for 1 h and then stained with DAPI to visualize sperm and assess the stage of maternal DNA [42, 43]. The number of sperm bound and fused per egg was determined. Error bars in the figures represent the SEM.
Statistical analyses were performed using Statview 5.0 (SAS Institute, Cary, NC). Chi-square analysis was used to examine the frequency distributions of sperm fused per egg shown in Figure 4. Data on the average number of sperm bound or fused per egg were analyzed using ANOVA. ANOVA P < 0.05 and Fisher protected least significant difference post hoc testing were then used to examine differences between groups.
RESULTS
Our previous investigations demonstrated that treatment of mouse eggs with a peptide inhibitor of ITGA4/ITGA9 (α4/α9) integrins containing the tripeptide sequence MLD reduced the binding of ADAM2 [29]. In vitro fertilization studies with this peptide inhibitor also revealed that this ITGA4/ITGA9 inhibitor reduced sperm-egg binding, sperm-egg fusion, and subsequent incidence of fertilization (untreated eggs, 93% fertilized; MLD-treated eggs, 48% fertilized; eggs treated with a control peptide [MAA instead of MLD], 85% fertilized). The known members of the ITGA4/ITGA9 integrin subfamily are α4β1, α4β7, and α9β1; therefore, we examined the expression of these integrin subunits in eggs. An anti-ITGA9 (α9) antibody immunoprecipitated two bands (molecular weight [Mr], ∼142 000 [ITGA9] and ∼115 000 [ITGB1]) from mouse egg lysates (Fig. 1A, lane 1); similar results were obtained using this anti-ITGA9 antibody with the mouse spleen positive control tissue (data not shown). Immunoprecipitations from mouse egg lysates with an anti-ITGB1 monoclonal antibody yielded a major band with Mr of ∼115 000 and several other bands, one of which comigrated with the band with Mr of ∼142 000 immunoprecipitated by the anti-ITGA9 antibody (Fig. 1A, lane 3), in agreement with other anti-ITGB1 immunoprecipitations from egg lysates [17, 19]. No bands were detected in immunoprecipitations with nonimmune rabbit or hamster IgGs (Fig. 1A, lanes 2 and 4). Immunofluorescence revealed that ITGA9 is present in the microvillar region of the egg plasma membrane (Fig. 1B, panels i and iii). This signal on the egg surface is reduced to undetectable levels when the anti-ITGA9 antibody is preincubated with a molar excess of the ITGA9 fusion protein used as the antigen (Fig. 1B, panel iv), and no signal is detected when eggs were labeled with a nonimmune antibody (Fig. 1B, panel ii). Furthermore, to determine if ITGA9 could be a candidate to mediate human gamete membrane interaction, we examined ITGA9 expression using an anti-human ITGA9 monoclonal antibody (Y9A2) with discarded human oocytes that were not used for IVF attempts. This antibody labeled the cell surface of discarded human oocytes; the anti-ITGA9 staining was typically weaker than that of the positive control, anti-CD9 staining (Fig. 2). We did not detect ITGA4 (α4) expression in mouse eggs by RT-PCR or by immunoprecipitation or immunofluorescence with two rat anti-mouse anti-ITGA4 monoclonal antibodies (PS/2 and R1–2; data not shown), nor did we detect Itgb7 (β7) expression by RT-PCR (data not shown). This focused our attention on the subfamily member α9β1.
FIG. 1.
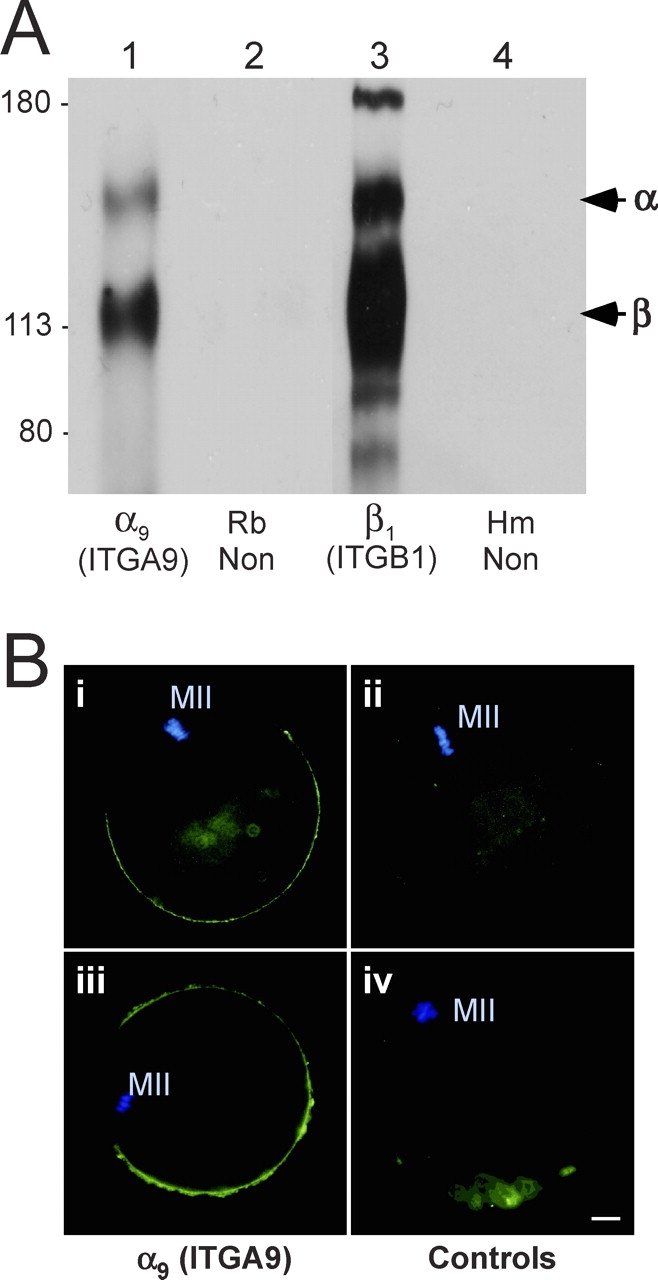
Expression of ITGA9 (α9) by mouse eggs. A) Immunoprecipitations from lysates of 300 surface-biotinylated eggs per lane. Lane 1: a purified rabbit anti-ITGA9 polyclonal IgG; lane 2: a nonimmune rabbit IgG (Rb Non); lane 3: an anti-ITGB1 monoclonal antibody (Hmβ1-1); lane 4: a nonimmune Armenian hamster IgG (Hm Non). Sizes shown are Mr × 10−3. B) Immunofluorescence studies of mouse eggs. Panels i and iii: anti-ITGA9 chicken IgY; panel ii: nonimmune chicken IgY; panel iv: anti-ITGA9 chicken IgY preincubated with a 5-fold molar excess of ITGA9 fusion protein. An FITC-conjugated goat anti-chicken IgY secondary antibody was used in all panels, shown in green. Blue staining shows DAPI labeling of the metaphase II DNA (MII). Bar = 10 μm.
FIG. 2.
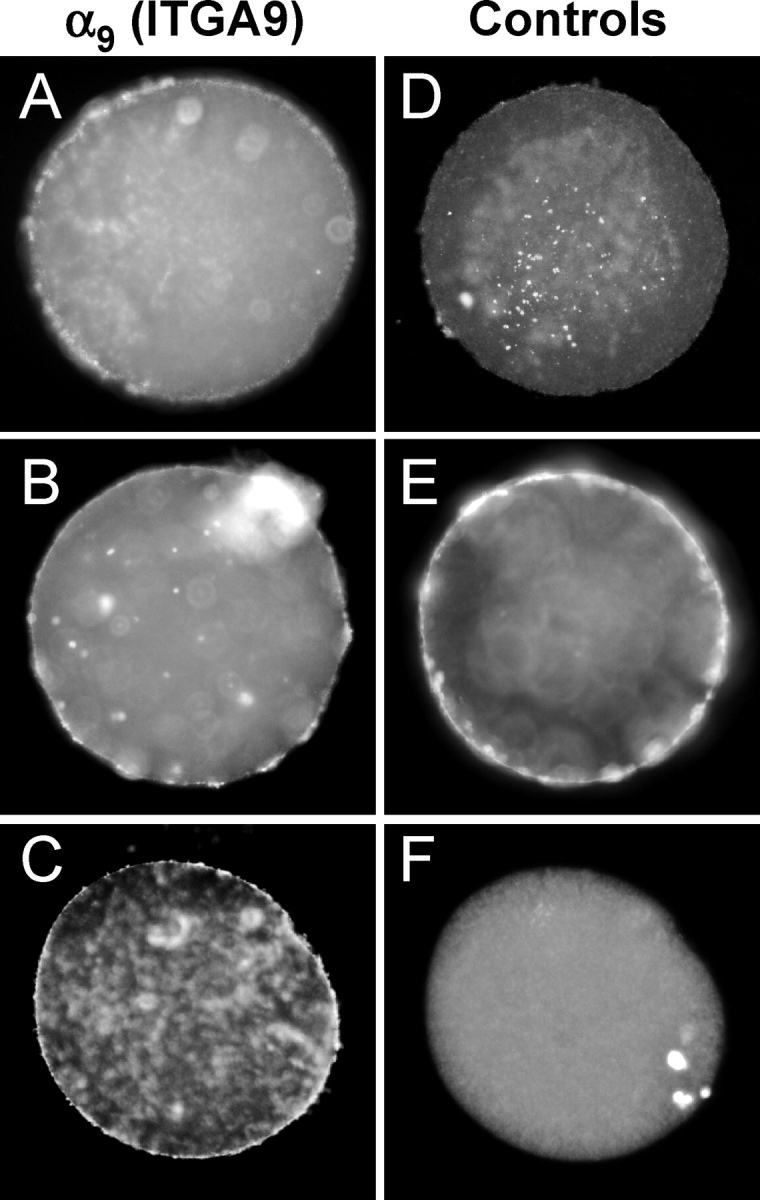
Expression of ITGA9 (α9) by human oocytes. Discarded human oocytes were labeled with the anti-ITGA9 antibody Y9A2 (A–C), negative control nonimmune mouse IgG (D), or positive control, the anti-CD9 antibody SYB-1 (E). F) DAPI staining corresponding to the image in C. In total, 12/14 oocytes stained positively with the anti-ITGA9 antibody, 7/7 oocytes stained positively with the anti-CD9 antibody, and 0/7 oocytes showed staining with the nonimmune IgG. Bar = 10 μm.
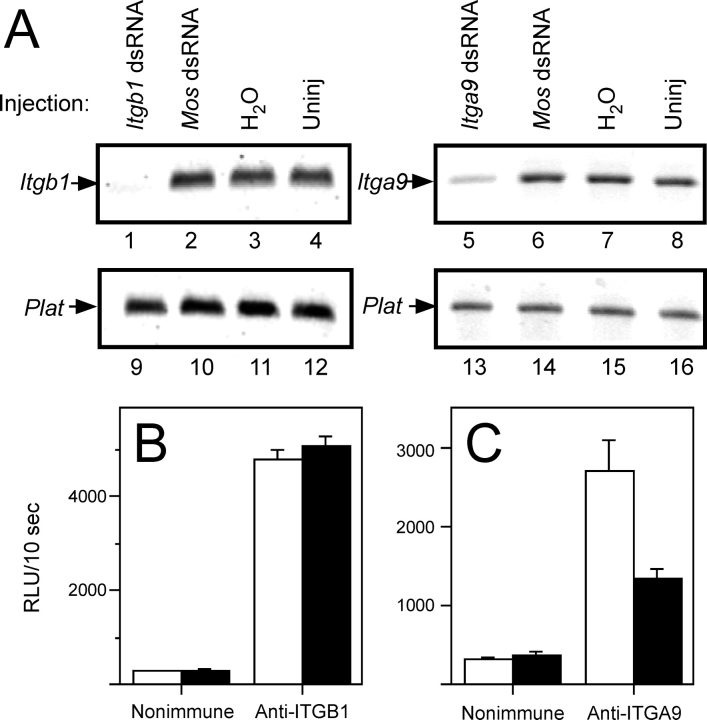
We sought to knockdown Itga9 and Itgb1 expression in eggs by RNAi through injection of GV-intact oocytes with dsRNA [39, 44]. Itgb1 mRNA levels were knocked down an average of 67.4% ± 10.8% relative to uninjected controls (Fig. 3A). However, this knockdown of Itgb1 mRNA levels by dsRNA injection did not lead to decreased ITGB1 surface protein levels; ITGB1 surface protein levels were an average of 96.4% ± 9.9% of the levels detected on uninjected control eggs (no reduction was observed in five experiments, and ∼35% reduction was observed in two experiments) (Fig. 3B). Itga9 mRNA levels in dsRNA-injected eggs were knocked down an average of 90.3% ± 0.9% relative to the Itga9 mRNA levels in uninjected control eggs (Fig. 3A). Itga9 RNA knockdown resulted in an ∼50% reduction in ITGA9 protein on the egg surface compared with uninjected control eggs (n = 4; 34%, 50%, 51%, and 66%; average, 50.0% ± 6.5%) (Fig. 3C).
FIG. 3.
The mRNA and surface protein levels of Itgb1 (β1) and Itga9 (α9) in dsRNA-injected eggs. Messenger RNA levels were assessed by RT-PCR. A) Data from one representative experiment. Lanes 1–4 show the Itgb1 PCR product for Itgb1 dsRNA-injected (lane 1), Mos dsRNA-injected (lane 2), water-injected (lane 3), and uninjected (Uninj) (lane 4) eggs. Lanes 5–8 show the Itga9 PCR product for Itga9 dsRNA-injected (lane 5), Mos dsRNA-injected (lane 6), water-injected (lane 7), and uninjected (lane 8) eggs. Lanes 9–16 show the control Plat (tissue plasminogen activator) PCR product for Itgb1 or Itga9 dsRNA-injected (lanes 9 and 13), Mos dsRNA-injected (lanes 10 and 14), water-injected (lanes 11 and 15), and uninjected (lanes 12 and 16) eggs. This experiment was repeated five times for Itgb1 dsRNA and three times for Itga9 dsRNA. B and C) Analysis of egg surface protein levels for ITGB1 (B) and ITGA9 (C) using a luminometric immunoassay with anti-ITGB1 and anti-ITGA9 antibodies, respectively (solid bars), or species-matched nonimmune negative controls (open bars). These panels show data from representative luminometric assays, expressed as RLU/10 sec. This analysis was performed seven times for Itgb1 dsRNA-injected eggs and four times for Itga9 dsRNA-injected eggs (with 8–16 eggs analyzed per experimental group per experiment). Error bars represent the SEM.
The IVF assays used ZP-free eggs inseminated with different sperm:egg ratios (25:1, 100:1, and 500:1). Figure 4 illustrates results with control eggs inseminated using these different sperm:egg ratios, showing the distribution of sperm numbers fused per egg and the resulting percentages of fertilized and polyspermic eggs in each of the three insemination conditions. The extents of sperm-egg binding also were affected in these different insemination conditions (3.1 ± 0.3 sperm bound/egg for 25:1; 7.9 ± 0.5 for 100:1; and 19.8 ± 0.8 for 500:1; P < 0.0001, ANOVA). These data show that the occurrence of sperm-egg interaction and fertilization varies in these different sperm:egg ratios, demonstrating that fertilization occurs readily in inseminations with a sperm:egg ratio of 500:1 and occurs less frequently when eggs are challenged with a sperm:egg ratio of 25:1.
FIG. 4.
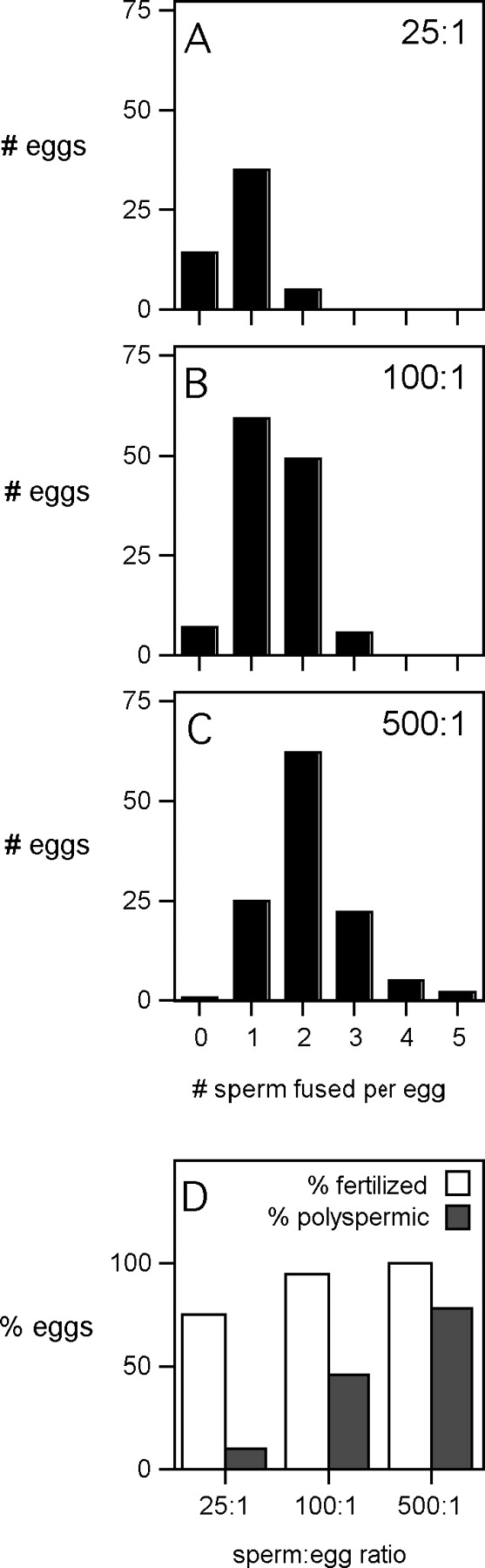
Results of inseminating ZP-free eggs with sperm:egg ratios of 25:1, 100:1, or 500:1. ZP-free eggs were inseminated for 60 min with sperm:egg ratios of 25:1, 100:1, or 500:1. A–C) Frequency distributions of the number of sperm fused per egg (0–5, as shown on the x-axis). D) Percentages of eggs fertilized (open bars) and polyspermic (solid bars) that result from these inseminations with these sperm:egg ratios. The differences between each of these three groups were statistically significant (P < 0.0001, chi-square analysis). Data are based on three or more experiments and 54–120 total eggs per insemination condition.
Using this experimental design, we determined that Itga9 dsRNA-injected eggs had reduced levels of sperm-egg binding and fusion compared with uninjected control eggs in inseminations in which the eggs were challenged with sperm:egg ratios of 25:1 and 100:1 (Fig. 5). Sperm-egg binding was reduced to 38% and 45% of control levels in inseminations with 25:1 and 100:1, respectively, and sperm-egg fusion was reduced to 45% and 58% of control levels in inseminations with 25:1 and 100:1, respectively. This resulted in a decrease in the percentage of eggs fertilized (36% for Itga9 dsRNA-injected eggs versus 74% for uninjected eggs in 25:1 inseminations; 69% for Itga9 dsRNA-injected eggs versus 92% for uninjected eggs in 100:1 inseminations). However, when the eggs were challenged with a sperm:egg ratio of 500:1, the levels of sperm-egg binding and fusion were similar in the Itga9 dsRNA-injected eggs and uninjected controls, resulting in similar percentages of fertilized eggs (98% for uninjected and 97% for Itga9 dsRNA injected).
FIG. 5.
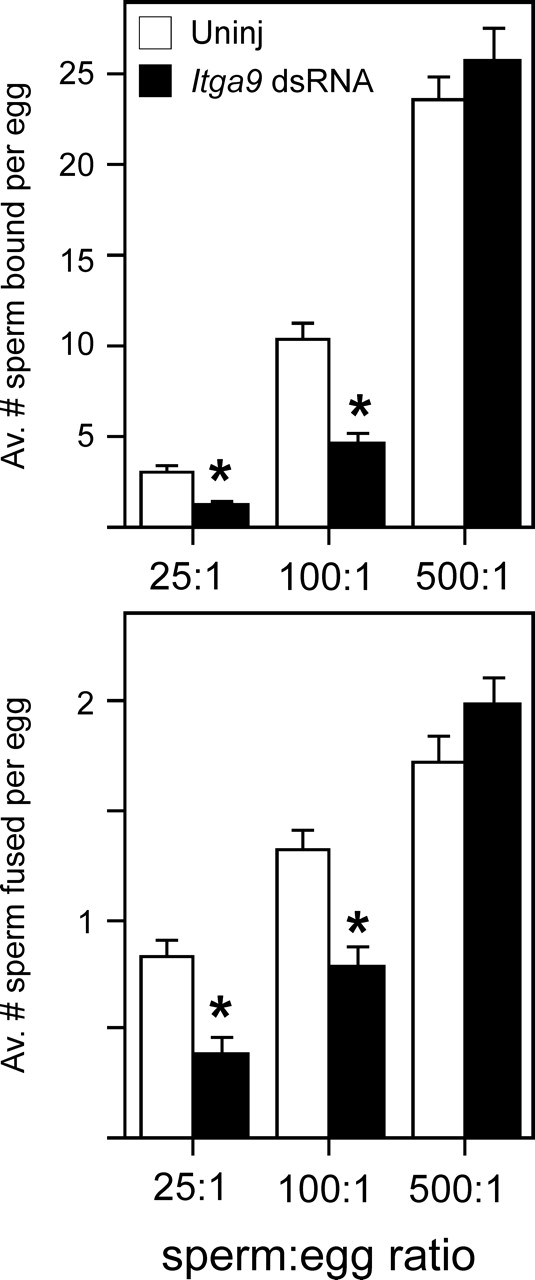
Effects of Itga9 knockdown on sperm-egg binding and fusion. Metaphase II eggs were matured from GV-intact oocytes that were injected with Itga9 dsRNA (Itga9 dsRNA, closed bars) or left uninjected (Uninj, open bars). Figure 2 shows that levels of Itga9 RNA were similar in all controls, including uninjected oocytes, water-injected oocytes, and oocytes injected with an irrelevant RNA. ZP-free eggs were inseminated with a sperm:egg ratio of 25:1, 100:1, or 500:1 for 60 min and then were fixed and assessed for sperm-egg binding (top panel) and fusion (bottom panel). Data are based on three experiments, with 41–54 total eggs per treatment group and insemination condition; the number of eggs per group in an individual experiment was in the range of 13–19. *Statistically significant differences between the Itga9 dsRNA-injected eggs and uninjected controls (P < 0.05, ANOVA with Fisher protected least significant difference post hoc testing).
Similar IVF investigations of the Itgb1 dsRNA-injected eggs showed, not surprisingly, that there were no differences in the numbers of sperm bound or fused with Itgb1 dsRNA-injected eggs compared with uninjected control eggs (data not shown), as we did not achieve a reduction of ITGB1 protein on the egg surface (Fig. 3B). Because RNAi-mediated knockdown of ITGB1 protein was not successful, we tested the hypothesis that ITGB1 on wild-type eggs had a role in sperm-egg interactions, using antibody blocking of the ITGB1 subunit on eggs with a function-blocking anti-mouse ITGB1 antibody, Hmβ1–1 [37]. Previous IVF studies [18, 23, 24, 27, 28] of the inhibitory effects of integrin-blocking antibodies or peptides have used a wide range of sperm concentrations (a few thousand to millions per milliliter) and insemination times (<1 h to 18 h), and different extents of inhibition have been detected, from ∼25% to ∼80% inhibition of binding and/or fusion. Herein, we systematically compared anti-ITGB1-treated eggs inseminated with sperm:egg ratios of 100:1 and 500:1. The anti-ITGB1-treated eggs showed reduced sperm-egg binding and fusion in inseminations in which eggs were challenged with a sperm:egg ratio of 100:1. Sperm-egg binding was reduced to 31% of the level observed in eggs treated with the nonimmune control antibody, and sperm-egg fusion was reduced to 73% of the level observed in nonimmune controls (Fig. 6). However, when eggs were challenged with a sperm:egg ratio of 500:1, no inhibitory effect of the anti-ITGB1 antibody was observed on sperm-egg binding or fusion (Fig. 6).
FIG. 6.
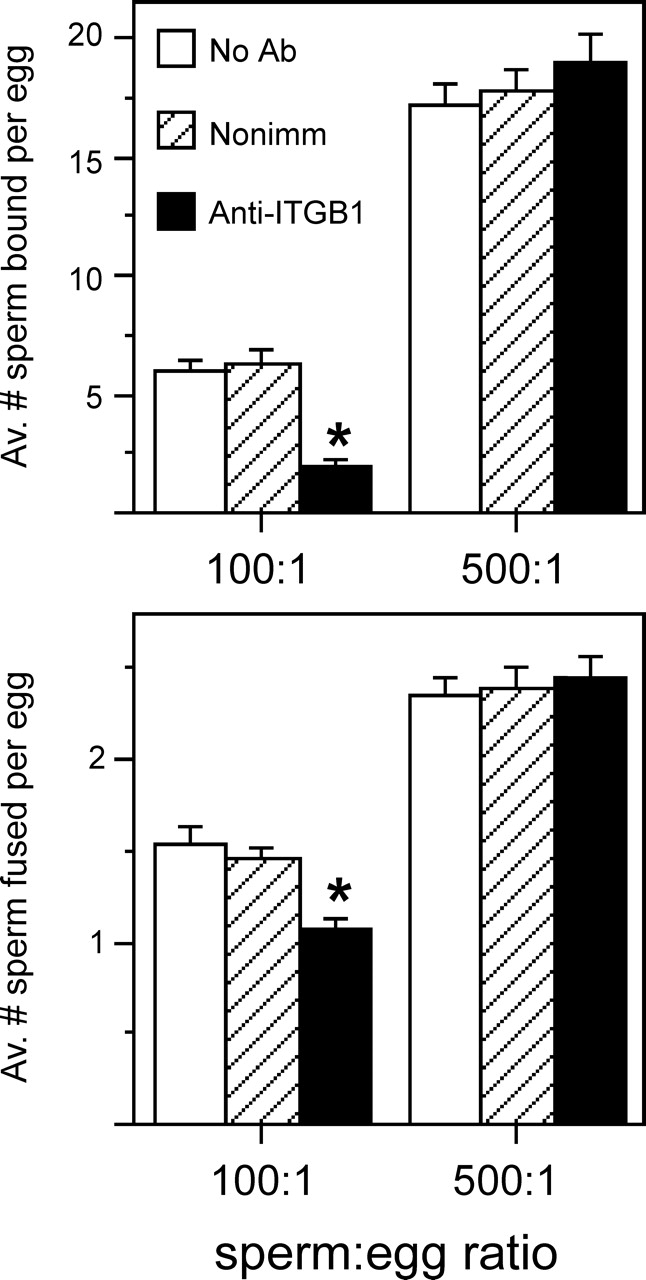
Effects of a function-blocking anti-ITGB1 antibody on sperm-egg binding and fusion. ZP-free eggs treated in medium containing no antibody (No Ab, open bars), 100 μg/ml of a nonimmune Armenian hamster IgG (Nonimm, hatched bars), or 100 μg/ml of Hmβ1-1, an anti-mouse ITGB1 function-blocking Armenian hamster monoclonal antibody (Anti-ITGB1, closed bars) were inseminated with a sperm:egg ratio of 100:1 or 500:1 for 60 min and then were fixed and assessed for sperm-egg binding (top panel) and fusion (bottom panel). Data are based on four experiments, with 67–77 total eggs per treatment group and insemination condition; the number of eggs per group in an individual experiment was in the range of 12–30. *Statistically significant differences between the anti-ITGB1 β1 antibody-treated eggs and the nonimmune antibody-treated eggs (P < 0.05, ANOVA with Fisher protected least significant difference post hoc testing).
DISCUSSION
The work shows that ITGA9 is expressed on the mouse and human egg surface and that reduction of ITGA9 on the mouse egg surface reduces fertilizability. Our detection of ITGA9 protein expression herein differs from a previous finding of an inability to detect ITGA9 in mouse eggs [45], although these varying results may be due to technical differences. We detected ITGA9 in mouse eggs using two different antibodies in two different techniques, although neither the anti-human ITGA9 Y9A2 monoclonal antibody nor the 1057 polyclonal antibody labeled mouse eggs by immunofluorescence (data not shown). We also detected ITGA9 on discarded human oocytes with the anti-ITGA9 Y9A2 monoclonal antibody; thus, ITGA9 may be a candidate molecule to mediate sperm-egg membrane interactions in human gametes.
A decrease of ∼50% in the amount of ITGA9 on the egg surface reduced sperm-egg binding and fusion and the percentage of eggs fertilized. This partial effect could be due to the incomplete knockdown (i.e., the remaining ITGA9 on the egg surface mediates sperm-egg interactions). It is also highly probable that other molecules on the egg surface, in addition to ITGA9, function in gamete membrane interaction. Based on what is known about integrin function, the most likely role that ITGA9 would have in fertilization is as a cell adhesion molecule binding partner for a ligand(s) on sperm to mediate sperm-egg binding. Members of the ADAM (a disintegrin and a metalloprotease) family on the sperm have been characterized as ligands for ITGA4/ITGA9 integrins, and sperm deficient in certain members of the ADAM family have reduced ability to bind to ZP-free eggs in IVF assays, and these knockouts have other phenotypes that affect male fertility [12, 29, 45–48]. Certain IgSF members can interact with different integrins, including members of the ITGA4/ITGA9 subfamily, suggesting that IgSFs on sperm such as IZUMO1 could interact with egg integrins.
This work demonstrates that the design of experimental assays for gamete function can be critical for detecting a phenotype. The incidence of fertilization and sperm-egg binding and fusion is higher when eggs are inseminated with a sperm:egg ratio of 500:1 than when eggs are inseminated with 100:1 or 25:1. An assay to assess sperm-egg interactions would be expected to be sensitive to sperm:egg ratios, just as other cell interaction assays are sensitive to similar variables such as the concentration of substrate or the number of cells used (e.g., more α9-expressing Chinese hamster ovary cells adhere 10 μg/ml of ADAM7 than to 2.5 μg/ml of ADAM7 [49]). We observed reduced sperm-egg interactions as a result of Itga9 knockdown or anti-ITGB1 antibody treatment in inseminations with a sperm:egg ratio of 100:1 but not in inseminations with 500:1. The concept of titrating the number of sperm is not a novel one [33, 50], but it is frequently overlooked that the concentration of sperm used in IVF or in artificial insemination can affect outcomes. Use of assays that take this experimental variable into account can reveal phenotypes that otherwise would be missed. There are genetic mouse models that are fertile in conventional housing and/or mating trials, but deficiencies in sperm function can be detected when fewer sperm are used in IVF or intrauterine insemination, whereas the deficiency is masked when more sperm are used [51, 52]. In a broader context, there are numerous examples of knockout mice that are not completely infertile but whose gametes have deficiencies that are revealed in specific assays of gamete function [5, 53–55].
Investigations of mouse eggs deficient in integrin subunits have failed to detect a phenotype, and there is no disputing that the ITGB1 subunit (the likely partner for ITGA9 in eggs) is not essential for eggs to be fertilized [31]. One possible explanation for this is that ITGB1 integrins have no functions in fertilization, although this is argued against by the findings that ITGB1 antibodies reduce sperm-egg interactions [18, 23, 24, 27, 28] (Fig. 5). It is likely that egg molecules other than ITGB1 integrins participate in gamete interactions. An additional explanation why knockouts of individual integrin subunits do not have obvious phenotypes is that these long-term chronic depletions of one integrin subunit possibly allow the remaining integrin subunits to dimerize with alternative subunit partners. A computational study [56] examining α-β transmembrane domain interactions found that models of possible α-β interaction were conserved for almost all α-β combinations, including the 24 known combinations and the other 120 possible combinations. Instead, the major factor affecting what α-β combinations are present on the cell surface seems to be the cell's integrin subunit expression profile, the types, and relative abundance. For example, manipulating the amount of an α subunit (e.g., by overexpression) affects the α-β combinations expressed by a cell [57–62]. In many cell types (including eggs [17, 19]), ITGB1 is a highly abundant β subunit, favoring ITGB1 dimerization with the available α subunits. However, in the absence of ITGB1, α subunits may be able to dimerize with other partners. Although we show herein that reduction of ITGA9 on the egg surface resulted in eggs with a fertilization phenotype, we unfortunately were unable to test the hypothesis that acute short-term knockdown of Itgb1 would result in a fertilization phenotype, as the RNAi-induced reduction of Itgb1 mRNA was not sufficient to reduce the levels of ITGB1 protein on the egg surface. This may be due to the abundance and/or stability of ITGB1 in eggs. Nevertheless, with our alternative approach of examining anti-ITGB1-treated eggs, reduced sperm-egg binding and fusion were detected in inseminations with a sperm:egg ratio of 100:1, as was observed with the Itga9 dsRNA-injected eggs.
One additional message from this work is that the lack of a strong phenotype merits cautious interpretation. No defect in fertility has been observed in conventional matings with knockout mice lacking in certain egg-expressed integrins (Itga1, Itga2, Itgb3, and Itgb5 [63]) or with female mice having Itgb1-deficient eggs [31], but such a lack of phenotype does not explicitly rule out a function for the molecule in question. This of course applies to more than reproductive function; for example, Hras1−/− and Hras1−/−/Nras−/− mice do not have an obvious phenotype, but careful phenotypic analysis and manipulation of Hras1 expression reveal important functions in carcinogenesis and tumor maintenance; thus, RAS pathway-related therapeutics are being pursued [64–67]. Certain gene products may not be essential for reproduction, but they could be advantageous for reproductive success; thus, these genes would be passed on and maintained in the genome as a result of conveying this reproductive advantage to individuals who have them. The multiple and possibly redundant molecules that mediate gamete interaction would likely be in this category. Finally, in considering the implications for human health that come from studies of mouse reproduction and gamete function, it is worth bearing in mind the differences between mouse and human female reproductive physiology. With mice ovulating multiple eggs in very short estrous cycles, while humans typically ovulate only one egg once every ∼28 days, a gamete function deficiency that has modest to minimal effects in the mouse could still translate to an effect on human fertility such as increased time for a woman to achieve a pregnancy. This is significant because the clinical definition of infertility is an inability to conceive after a year of unprotected intercourse. Thus, to have a full picture of what factors can affect human reproductive health, there is value in uncovering the dramatic effects such as infertility or subfertility detected in mating trials, as well as more subtle reproductive phenotypes.
Acknowledgments
We thank Paula Stein, Petr Svoboda, and Francesca Duncan for advice on dsRNA-mediated knockdown; Matt Marcello for advice on biostatistics; Peter Donovan for help with microinjection; and the anonymous reviewers for their comments.
Footnotes
1Supported by National Institutes of Health grant HD037696 to J.P.E.
REFERENCES
- Inoue N, Ikawa M, Isotani A, Okabe M.The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 2005; 434: 234–238. [DOI] [PubMed] [Google Scholar]
- Boucheix C, Rubinstein E.Tetraspanins. Cell Mol Life Sci 2001; 58: 1189–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler ME.Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Ann Rev Cell Dev Biol 2003; 19: 397–422. [DOI] [PubMed] [Google Scholar]
- Hemler ME.Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 2005; 6: 801–811. [DOI] [PubMed] [Google Scholar]
- Rubinstein E, Ziyyat A, Prenant M, Wrobel E, Wolf JP, Levy S, Le Naour F, Boucheix C.Reduced fertility of female mice lacking CD81. Dev Biol 2006; 290: 351–358. [DOI] [PubMed] [Google Scholar]
- Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, Tada N, Miyazaki S, Kudo A.The gamete fusion process is defective in eggs of CD9-deficient mice. Nat Genet 2000; 24: 279–282. [DOI] [PubMed] [Google Scholar]
- Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C.Severely reduced female fertility in CD9-deficient mice. Science 2000; 287: 319–321. [DOI] [PubMed] [Google Scholar]
- Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, Suzuki K, Kosai K, Inoue K, Ogura A, Okabe M, Mekada E.Requirement of CD9 on the egg plasma membrane for fertilization. Science 2000; 287: 321–324. [DOI] [PubMed] [Google Scholar]
- Runge KE, Evans JE, He ZY, Gupta S, McDonald KL, Stahlberg H, Primakoff P, Myles DG.Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Dev Biol 2007; 304: 317–325. [DOI] [PubMed] [Google Scholar]
- Okabe M, Yagasaki M, Oda H, Matzno S, Kohama Y, Mimura T.Effect of a monoclonal anti-mouse sperm antibody (OBF13) on the interaction of mouse sperm with zona-free mouse and hamster eggs. J Reprod Immunol 1988; 13: 211–219. [DOI] [PubMed] [Google Scholar]
- Chen MS, Tung KSK, Coonrod SA, Takahashi Y, Bigler D, Chang A, Yamashita Y, Kincade PW, Herr JC, White JM.Role of the integrin associated protein CD9 in binding between sperm ADAM 2 and the egg integrin α6β1: implications for murine fertilization. Proc Natl Acad Sci U S A 1999; 96: 11830–11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Bigler D, Ito Y, White JM.Sequence-specific interaction between the disintegrin domain of mouse ADAM3 and murine eggs: role of the b1 integrin-associated proteins CD9, CD81, and CD98. Mol Biol Cell 2001; 12: 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusi FM, Vigali M, Busacca M, Bronson RA.Evidence for the presence of an integrin cell adhesion receptor on the oolemma of unfertilized human oocytes. Mol Reprod Dev 1992; 31: 215–222. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Abbott AF, Jack RM.The role of complement component C3b and its receptor in sperm-oocyte interaction. Proc Natl Acad Sci U S A 1993; 90: 10051–10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusi FM, Vigali M, Gailit J, Bronson RA.Mammalian oocytes exhibit specific recognition of the RGD (Arg-Gly-Asp) tripeptide oolemmal integrins. Mol Reprod Dev 1993; 36: 212–219. [DOI] [PubMed] [Google Scholar]
- Sutherland AE, Calarco PG, Damsky CH.Developmental regulation of integrin expression at the time of implantation in the mouse embryo. Development 1993; 119: 1175–1186. [DOI] [PubMed] [Google Scholar]
- Tarone G, Russo MA, Hirsch E, Odorisio T, Altruda F, Silengo L, Siracusa G.Expression of β1 integrin complexes on the surfaces of unfertilized mouse oocyte. Development 1993; 117: 1369–1375. [DOI] [PubMed] [Google Scholar]
- Almeida EAC, Huovila APJ, Sutherland AE, Stephens LE, Calarco PG, Shaw LM, Mercurio AM, Sonnenberg A, Primakoff P, Myles DG, White JM.Mouse egg integrin α6β1 functions as a sperm receptor. Cell 1995; 81: 1095–1104. [DOI] [PubMed] [Google Scholar]
- Evans JP, Schultz RM, Kopf GS.Identification and localization of integrin subunits in oocytes and eggs of the mouse. Mol Reprod Dev 1995; 40: 211–220. [DOI] [PubMed] [Google Scholar]
- Campbell S, Swann HR, Seif MW, Kimber SJ, Aplin JD.Cell adhesion molecules on the oocyte and preimplantation human embryo. Hum Reprod 1995; 10: 1571–1578. [DOI] [PubMed] [Google Scholar]
- de Nadai C, Fenichel P, Donzeau M, Epel D, Ciapa B.Characterisation and role of integrins during gametic interaction and egg activation. Zygote 1996; 4: 31–40. [DOI] [PubMed] [Google Scholar]
- Capmany G, Mart M, Santalo J, Bolton VN.Distribution of α3, α5, and αv integrin subunits in mature and immature human oocytes. Mol Hum Reprod 1998; 4: 951–956. [DOI] [PubMed] [Google Scholar]
- Linfor J, Berger T.Potential role of αv and β1 integrins as oocyte adhesion molecules during fertilization in pigs. J Reprod Fertil 2000; 120: 65–72. [PubMed] [Google Scholar]
- Sengoku K, Takuma N, Miyamoto T, Horikawa M, Ishikawa M.Integrins are not involved in the process of human sperm-oolemmal fusion. Hum Reprod 2004; 19: 639–644. [DOI] [PubMed] [Google Scholar]
- Pate BJ, White KL, Winger QA, Rickords LF, Aston KI, Sessons BR, Li GP, Campbell KD, Weimer B, Bunch TD.Specific integrin subunits in bovine oocytes, including novel sequences for α6 and β3 subunits. Mol Reprod Dev 2007; 74: 600–607. [DOI] [PubMed] [Google Scholar]
- Bronson RA, Fusi F.Evidence that an Arg-Gly-Asp adhesion sequence plays a role in mammalian fertilization. Biol Reprod 1990; 43: 1019–1025. [DOI] [PubMed] [Google Scholar]
- Evans JP, Kopf GS, Schultz RM.Characterization of the binding of recombinant mouse sperm fertilin β subunit to mouse eggs: evidence for adhesive activity via an egg β1 integrin-mediated interaction. Dev Biol 1997; 187: 79–93. [DOI] [PubMed] [Google Scholar]
- Ji YZ, Wolf JP, Jouannet P, Bomsel M.Human gamete fusion can bypass β1 integrin requirement. Hum Reprod 1998; 13: 682–689. [DOI] [PubMed] [Google Scholar]
- Zhu X, Evans JP.Analysis of the roles of RGD-binding integrins, α4/α9 integrins, α6 integrins, and CD9 in the interaction of the fertilin β (ADAM2) disintegrin domain with the mouse egg membrane. Biol Reprod 2002; 66: 1193–1202. [DOI] [PubMed] [Google Scholar]
- Miller BJ, Georges-Labouesse E, Primakoff P, Myles DG.Normal fertilization occurs with eggs lacking the integrin α6β1 and is CD9-dependent. J Cell Biol 2000; 149: 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He ZY, Brakebusch C, Fassler R, Kreidberg JA, Primakoff P, Myles DG.None of the integrins known to be present on the mouse egg or to be ADAM receptors are essential for sperm-egg binding and fusion. Dev Biol 2003; 254: 226–237. [DOI] [PubMed] [Google Scholar]
- Hynes RO, Wagner DD.Genetic manipulation of vascular adhesion molecules in mice. J Clin Invest 1996; 98: 2193–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RP, Hammerstedt RH.Detection of differences in fertility. J Androl 2002; 23: 317–325. [PubMed] [Google Scholar]
- Whitten WK.Nutrient requirements for the culture of preimplantation embryos in vitro. Adv Biosci 1971; 6: 129–139. [Google Scholar]
- Evans JP, Foster JA, McAvey BA, Gerton GL, Kopf GS, Schultz RM.The effects of perturbation of cell polarity on molecular markers of sperm-egg binding sites on mouse eggs. Biol Reprod 2000; 62: 76–84. [DOI] [PubMed] [Google Scholar]
- Ziyyat A, Rubinstein E, Monier-Gavelle F, Barraud V, Kulski O, Prenant M, Boucheix C, Bomsel M, Wolf JP.CD9 controls the formation of clusters that contain tetraspanins and the integrin α6β1, which are involved in human and mouse gamete fusion. J Cell Sci 2006; 119: 416–424. [DOI] [PubMed] [Google Scholar]
- Noto K, Kato K, Okumura K, Yagita H.Identification and functional characterization of mouse CD29 with a mAb. Int Immunol 1995; 7: 835–842. [DOI] [PubMed] [Google Scholar]
- Palmer EL, Ruegg C, Ferrando R, Pytela R, Sheppard D.Sequence and tissue distribution of the integrin α9 subunit, a novel partner of β1 that is widely distributed in epithelia and muscle. J Cell Biol 1993; 123: 1289–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Hayashi H, Schultz RM.Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 2000; 127: 4147–4156. [DOI] [PubMed] [Google Scholar]
- McAvey BA, Wortzman GB, Williams CJ, Evans JP.Involvement of calcium signaling and the actin cytoskeleton in the membrane block to polyspermy in mouse eggs. Biol Reprod 2002; 67: 1342–1352. [DOI] [PubMed] [Google Scholar]
- Gardner AJ, Knott JG, Jones KT, Evans JP.CaMKII can participate in but is not sufficient for the establishment of the membrane block to polyspermy in mouse eggs. J Cell Physiol 2007; 212: 275–280. [DOI] [PubMed] [Google Scholar]
- Wortzman GB, Evans JP.Membrane and cortical abnormalities in post-ovulatory aged eggs: analysis of fertilizability and establishment of the membrane block to polyspermy. Mol Hum Reprod 2005; 11: 1–9. [DOI] [PubMed] [Google Scholar]
- Gardner AJ, Williams CJ, Evans JP.Establishment of the mammalian membrane block to polyspermy: evidence for calcium-dependent and -independent regulation. Reproduction 2007; 133: 383–393. [DOI] [PubMed] [Google Scholar]
- Svoboda P.Long dsRNA and silent genes strike back: RNAi in mouse oocytes and early embryos. Cytogenet Genome Res 2004; 105: 422–434. [DOI] [PubMed] [Google Scholar]
- Tomczuk M, Takahashi Y, Huang J, Murase S, Mistretta M, Klaffky E, Sutherland A, Bolling L, Coonrod S, Marcinkiewicz C, Sheppard D, Stepp MA, et al. Role of multiple β1 integrins in cell adhesion to the disintegrin domains of ADAMs 2 and 3. Exp Cell Res 2003; 290: 68–81. [DOI] [PubMed] [Google Scholar]
- Cho C, Bunch DO, Faure JE, Goulding EH, Eddy EM, Primakoff P, Myles DG.Fertilization defects in sperm from mice lacking fertilin b. Science 1998; 281: 1857–1859. [DOI] [PubMed] [Google Scholar]
- Nishimura H, Cho C, Branciforte DR, Myles DG, Primakoff P.Analysis of loss of adhesive function in sperm lacking cyritestin or fertilin β. Dev Biol 2001; 233: 204–213. [DOI] [PubMed] [Google Scholar]
- Eto K, Huet C, Tarui T, Kupriyanov S, Liu HZ, Puzon-McLaughlin W, Zhang XP, Sheppard D, Engvall E, Takada Y.Functional classification of ADAMs based on a conserved motif for binding to integrin α9β1: implications for sperm-egg binding and other cell interactions. J Biol Chem 2002; 277: 17804–17810. [DOI] [PubMed] [Google Scholar]
- Bridges LC, Sheppard D, Bowditch RD.ADAM disintegrin-like domain recognition by the lymphocyte integrins α4β1 and α4β7. Biochem J 2005; 387: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey RP, Pyrzak R, Lu PY, Taylor SN, Rye PH.Comparison of the sperm quality necessary for successful intrauterine insemination with World Health Organization threshold values for normal sperm. Fertil Steril 1999; 71: 684–689. [DOI] [PubMed] [Google Scholar]
- Johnson LR, Pilder SH, Bailey JL, Olds-Clarke P.Sperm from mice carrying one or two t haplotypes are deficient in investment and oocyte penetration. Dev Biol 1995; 168: 138–149. [DOI] [PubMed] [Google Scholar]
- Sutton KA, Jungnickel MK, Florman HM.A polycystin-1 controls postcopulatory reproductive selection in mice. Proc Natl Acad Sci U S A 2008; 105: 8661–8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba D, Kashiwabara S, Honda A, Yamagata K, Wu Q, Ikawa M, Okabe M, Baba T.Mouse sperm lacking cell surface hyaluronidase PH-20 can pass through the layer of cumulus cells and fertilize the egg. J Biol Chem 2002; 277: 30310–30314. [DOI] [PubMed] [Google Scholar]
- Ensslin MA, Shur BD.Identification of mouse sperm SED1, a bimotif EGF repeat and discoidin-domain protein involved in sperm-egg binding. Cell 2003; 114: 405–417. [DOI] [PubMed] [Google Scholar]
- Da Ros VG, Maldera JA, Willis WD, Cohen DJ, Goulding EH, Gelman DM, Rubinstein M, Eddy EM, Cuasnicu PS.Impaired sperm fertilizing ability in mice lacking Cysteine-RIch Secretory Protein 1 (CRISP1). Dev Biol 2008; 320: 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Tan SM, Alex Law SK, Torres J.Unambiguous prediction of human integrin transmembrane heterodimer interactions using only homologous sequences. Proteins 2006; 65: 274–279. [DOI] [PubMed] [Google Scholar]
- Heino J, Ignotz RA, Hemler ME, Crouse C, Massague J.Regulation of cell adhesion receptors by transforming growth factor-β: concomitant regulation of integrins that share a common β1 subunit. J Biol Chem 1989; 264: 380–388. [PubMed] [Google Scholar]
- Jaspers M, de Meirsman C, Schollen E, Vekemans S, Cassiman JJ.Stable expression of VLA-4 and increased maturation of the β1-integrin precursor after transfection of CHO cells with α4m cDNA. FEBS Lett 1994; 353: 239–242. [DOI] [PubMed] [Google Scholar]
- Webb DL, Conrad PJ, Ma L, Blue ML.Induction of mouse β integrin expression following transfection with human α4 chain. J Cell Biochem 1996; 61: 127–138. [DOI] [PubMed] [Google Scholar]
- Retta SF, Cassara G, D'Amato M, Alessandro R, Pellegrino M, Degani S, De Leo G, Silengo L, Tarone G.Cross talk between β1 and αV integrins: β1 affects β3 mRNA stability. Mol Biol Cell 2001; 12: 3126–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koistinen P, Heino J.The selective regulation of αvβ1 integrin expression is based on the hierarchical formation of αv-containing heterodimers. J Biol Chem 2002; 277: 24835–24841. [DOI] [PubMed] [Google Scholar]
- Conesa M, Prat A, Mort JS, Marvaldi J, Lissitzky JC, Seidah NG.Down-regulation of αv/β3 integrin via misrouting to lysosomes by overexpression of a β3Lamp1 fusion protein. Biochem J 2003; 370: 703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO.Integrins: bidirectional, allosteric signaling machines. Cell 2002; 110: 673–687. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M.RAS oncogenes: the first 30 years. Nat Rev Cancer 2003; 3: 459–465. [DOI] [PubMed] [Google Scholar]
- Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O'Hagan R, Pantginis J, Zhou H, Horner JW, Cordon-Cardo C, et al. Essential role for oncogenic Ras in tumour maintenance. Nature 1999; 400: 468–472. [DOI] [PubMed] [Google Scholar]
- Ise K, Nakamura K, Nakao K, Shimizu S, Harada H, Ichise T, Miyoshi J, Gondo Y, Ishikawa T, Aiba A, Katsuki M.Targeted deletion of the H-ras gene decreases tumor formation in mouse skin carcinogenesis. Oncogene 2000; 19: 2951–2956. [DOI] [PubMed] [Google Scholar]
- Esteban LM, Vicario-Abejon C, Fernandez-Salguero P, Fernandez-Medarde A, Swaminathan N, Yienger K, Lopez E, Malumbres M, McKay R, Ward JM, Pellicer A, Santos E.Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol Cell Biol 2001; 21: 1444–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]