Abstract
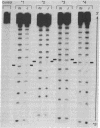
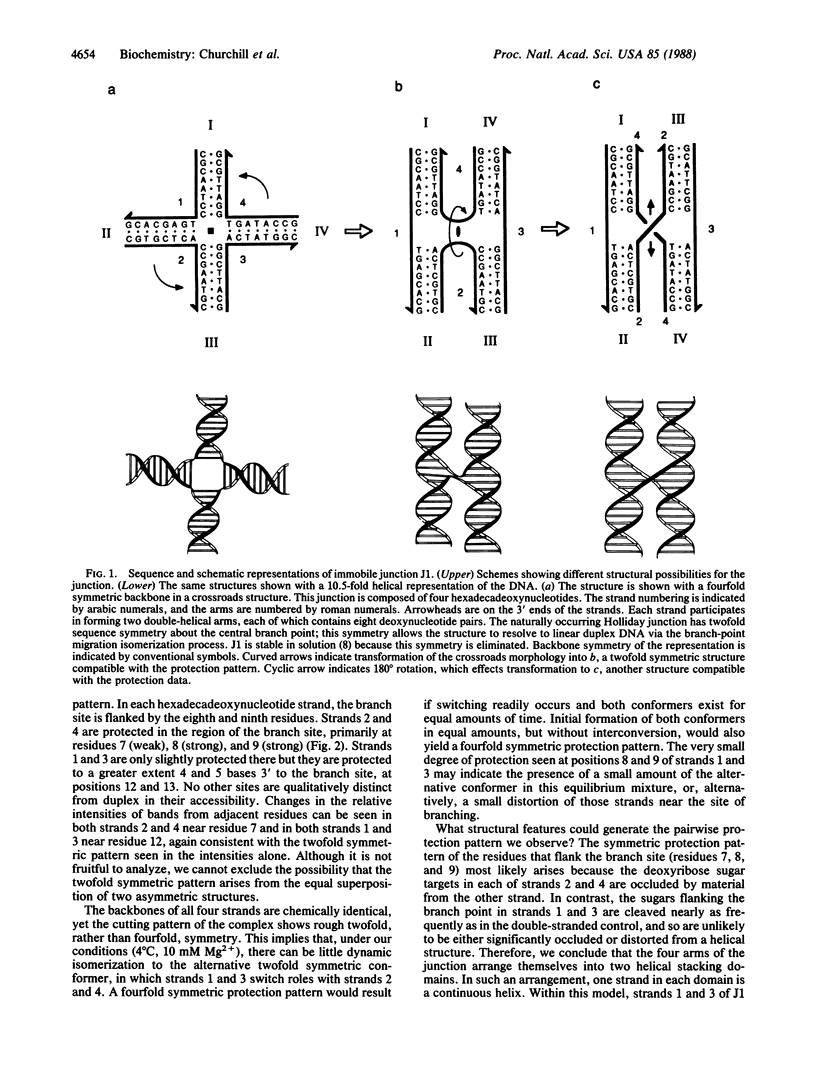
Four-arm Holliday structures are ephemeral intermediates in genetic recombination. We have used an oligodeoxynucleotide system to form immobile DNA junctions, which are stable analogs of Holliday structures. We have probed the equilibrium structure of a junction by means of hydroxyl radicals generated by the reaction of iron(II)EDTA with hydrogen peroxide. The hydroxyl radical cleavage pattern shows twofold symmetry throughout the molecule. Strong protection from hydroxyl radical attack is evident on two strands near the branch site, and weaker protection may be seen four or five residues 3' to the branch site on the other two strands. No other position appears significantly distinct from double-helical DNA controls. From these data, we conclude that the Holliday junction is a twofold symmetric complex whose four arms form two stacking domains.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Cooper J. P., Hagerman P. J. Gel electrophoretic analysis of the geometry of a DNA four-way junction. J Mol Biol. 1987 Dec 20;198(4):711–719. doi: 10.1016/0022-2836(87)90212-9. [DOI] [PubMed] [Google Scholar]
- Dressler D., Potter H. Molecular mechanisms in genetic recombination. Annu Rev Biochem. 1982;51:727–761. doi: 10.1146/annurev.bi.51.070182.003455. [DOI] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Hsu P. L., Landy A. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage lambda. Nature. 1984 Oct 25;311(5988):721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Homology-dependent interactions in phage lambda site-specific recombination. Nature. 1987 Sep 24;329(6137):346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Ma R. I., Kallenbach N. R., Sheardy R. D., Petrillo M. L., Seeman N. C. Three-arm nucleic acid junctions are flexible. Nucleic Acids Res. 1986 Dec 22;14(24):9745–9753. doi: 10.1093/nar/14.24.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marky L. A., Kallenbach N. R., McDonough K. A., Seeman N. C., Breslauer K. J. The melting behavior of a DNA junction structure: a calorimetric and spectroscopic study. Biopolymers. 1987 Sep;26(9):1621–1634. doi: 10.1002/bip.360260912. [DOI] [PubMed] [Google Scholar]
- McGavin S. A model for the specific pairing of homologous double-stranded nucleic acid molecules during genetic recombination. Heredity (Edinb) 1977 Aug;39(1):15–25. doi: 10.1038/hdy.1977.39. [DOI] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Matsumoto L., Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987 Aug 28;50(5):779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- Seeman N. C. Nucleic acid junctions and lattices. J Theor Biol. 1982 Nov 21;99(2):237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- Sigal N., Alberts B. Genetic recombination: the nature of a crossed strand-exchange between two homologous DNA molecules. J Mol Biol. 1972 Nov 28;71(3):789–793. doi: 10.1016/s0022-2836(72)80039-1. [DOI] [PubMed] [Google Scholar]
- Thompson B. J., Camien M. N., Warner R. C. Kinetics of branch migration in double-stranded DNA. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2299–2303. doi: 10.1073/pnas.73.7.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- Wemmer D. E., Wand A. J., Seeman N. C., Kallenbach N. R. NMR analysis of DNA junctions: imino proton NMR studies of individual arms and intact junction. Biochemistry. 1985 Oct 8;24(21):5745–5749. doi: 10.1021/bi00342a009. [DOI] [PubMed] [Google Scholar]
- Wilson J. H. Nick-free formation of reciprocal heteroduplexes: a simple solution to the topological problem. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3641–3645. doi: 10.1073/pnas.76.8.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]