Abstract
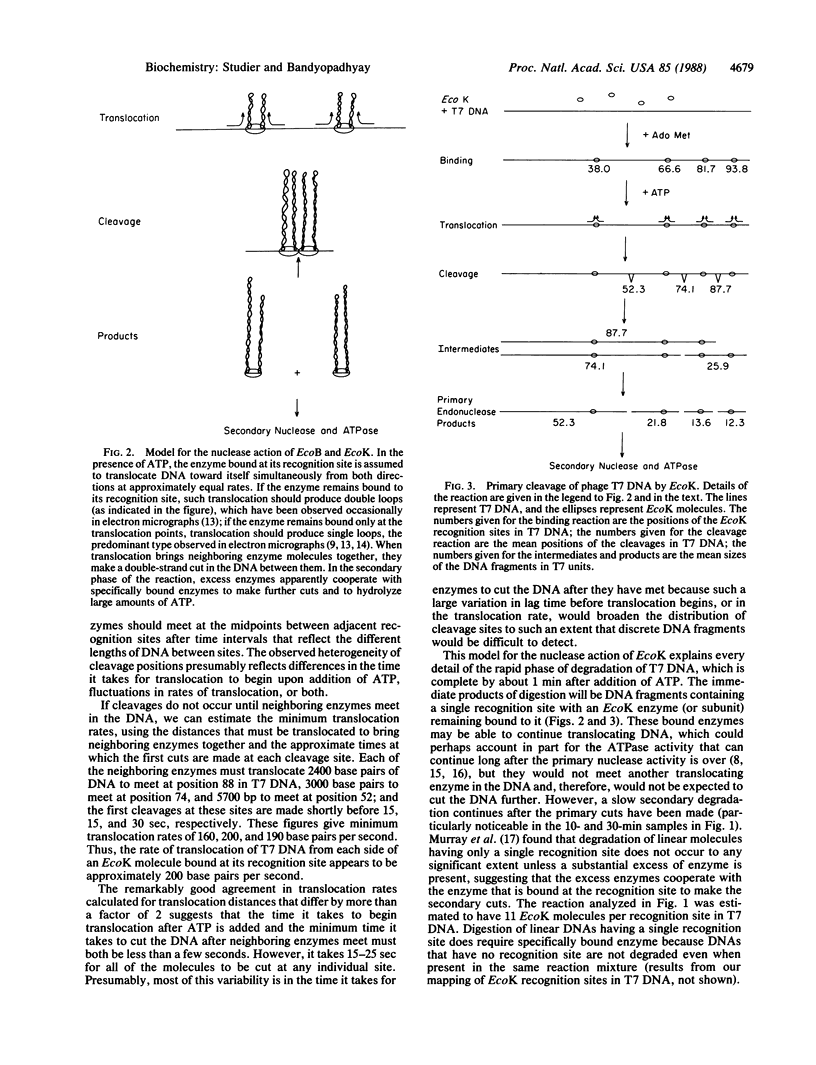
Under appropriate conditions, digestion of phage T7 DNA by the type I restriction enzyme EcoK produces an orderly progression of discrete DNA fragments. All details of the fragmentation pattern can be explained on the basis of the known properties of type I enzymes, together with two further assumptions: (i) in the ATP-stimulated translocation reaction, the enzyme bound at the recognition sequence translocates DNA toward itself from both directions simultaneously; and (ii) when translocation causes neighboring enzymes to meet, they cut the DNA between them. The kinetics of digestion at 37 degrees C indicates that the rate of translocation of DNA from each side of a bound enzyme is about 200 base pairs per second, and the cuts are completed within 15-25 sec of the time neighboring enzymes meet. The resulting DNA fragments each contain a single recognition site with an enzyme (or subunit) remaining bound to it. At high enzyme concentrations, such fragments can be further degraded, apparently by cooperation between the specifically bound and excess enzymes. This model is consistent with a substantial body of previous work on the nuclease activity of EcoB and EcoK, and it explains in a simple way how cleavage sites are selected.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler S. P., Nathans D. Studies of SV 40 DNA. V. Conversion of circular to linear SV 40 DNA by restriction endonuclease from Escherichia coli B. Biochim Biophys Acta. 1973 Mar 19;299(2):177–188. doi: 10.1016/0005-2787(73)90340-7. [DOI] [PubMed] [Google Scholar]
- BOYER H. GENETIC CONTROL OF RESTRICTION AND MODIFICATION IN ESCHERICHIA COLI. J Bacteriol. 1964 Dec;88:1652–1660. doi: 10.1128/jb.88.6.1652-1660.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay P. K., Studier F. W., Hamilton D. L., Yuan R. Inhibition of the type I restriction-modification enzymes EcoB and EcoK by the gene 0.3 protein of bacteriophage T7. J Mol Biol. 1985 Apr 20;182(4):567–578. doi: 10.1016/0022-2836(85)90242-6. [DOI] [PubMed] [Google Scholar]
- Bickle T. A., Brack C., Yuan R. ATP-induced conformational changes in the restriction endonuclease from Escherichia coli K-12. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3099–3103. doi: 10.1073/pnas.75.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack C., Eberle H., Bickle T. A., Yuan R. Mapping of recognition sites for the restriction endonuclease from Escherichia coli K12 on bacteriophage PM2 DNA. J Mol Biol. 1976 Dec 15;108(3):583–593. doi: 10.1016/s0022-2836(76)80138-6. [DOI] [PubMed] [Google Scholar]
- Brammar W. J., Murray N. E., Winton S. Restriction of lambda trp bacteriophages by Escherichia coli K. J Mol Biol. 1974 Dec 25;90(4):633–647. doi: 10.1016/0022-2836(74)90529-4. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Endlich B., Linn S. The DNA restriction endonuclease of Escherichia coli B. I. Studies of the DNA translocation and the ATPase activities. J Biol Chem. 1985 May 10;260(9):5720–5728. [PubMed] [Google Scholar]
- Endlich B., Linn S. The DNA restriction endonuclease of Escherichia coli B. II. Further studies of the structure of DNA intermediates and products. J Biol Chem. 1985 May 10;260(9):5729–5738. [PubMed] [Google Scholar]
- Eskin B., Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. II. Purification, subunit structure, and catalytic properties of the restriction endonuclease. J Biol Chem. 1972 Oct 10;247(19):6183–6191. [PubMed] [Google Scholar]
- Eskin B., Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. J Biol Chem. 1972 Oct 10;247(19):6192–6196. [PubMed] [Google Scholar]
- Horiuchi K., Vovis G. F., Zinder N. D. Effect of deoxyribonucleic acid length on the adenosine triphosphatase activity of Escherichia coli restriction endonuclease B. J Biol Chem. 1974 Jan 25;249(2):543–552. [PubMed] [Google Scholar]
- Horiuchi K., Zinder N. D. Cleavage of bacteriophage fl DNA by the restriction enzyme of Escherichia coli B. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3220–3224. doi: 10.1073/pnas.69.11.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn S., Lautenberger J. A., Eskin B., Lackey D. Host-controlled restriction and modification enzymes of Escherichia coli B. Fed Proc. 1974 May;33(5):1128–1134. [PubMed] [Google Scholar]
- Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature. 1968 Mar 23;217(5134):1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Morrow J. F., Berg P. Cleavage of Simian virus 40 DNA at a unique site by a bacterial restriction enzyme. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3365–3369. doi: 10.1073/pnas.69.11.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Batten P. L., Murray K. Restriction of bacteriophage lambda by Escherichia coli K. J Mol Biol. 1973 Dec 15;81(3):395–407. doi: 10.1016/0022-2836(73)90149-6. [DOI] [PubMed] [Google Scholar]
- Rosamond J., Endlich B., Linn S. Electron microscopic studies of the mechanism of action of the restriction endonuclease of Escherichia coli B. J Mol Biol. 1979 Apr 25;129(4):619–635. doi: 10.1016/0022-2836(79)90472-8. [DOI] [PubMed] [Google Scholar]
- Roulland-Dussoix D., Boyer H. W. The Escherichia coli B restriction endonuclease. Biochim Biophys Acta. 1969 Nov 19;195(1):219–229. doi: 10.1016/0005-2787(69)90618-2. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Gene 0.3 of bacteriophage T7 acts to overcome the DNA restriction system of the host. J Mol Biol. 1975 May 15;94(2):283–295. doi: 10.1016/0022-2836(75)90083-2. [DOI] [PubMed] [Google Scholar]
- Yuan R., Bickle T. A., Ebbers W., Brack C. Multiple steps in DNA recognition by restriction endonuclease from E. coli K. Nature. 1975 Aug 14;256(5518):556–560. doi: 10.1038/256556a0. [DOI] [PubMed] [Google Scholar]
- Yuan R., Hamilton D. L., Burckhardt J. DNA translocation by the restriction enzyme from E. coli K. Cell. 1980 May;20(1):237–244. doi: 10.1016/0092-8674(80)90251-2. [DOI] [PubMed] [Google Scholar]
- Yuan R., Heywood J., Meselson M. ATP hydrolysis by restriction endonuclease from E. coli K. Nat New Biol. 1972 Nov 8;240(97):42–43. doi: 10.1038/newbio240042a0. [DOI] [PubMed] [Google Scholar]
- Yuan R. Structure and mechanism of multifunctional restriction endonucleases. Annu Rev Biochem. 1981;50:285–319. doi: 10.1146/annurev.bi.50.070181.001441. [DOI] [PubMed] [Google Scholar]