Abstract
MicroRNAs (miRNAs) are important gene regulators that could play a profound role in tumorigenesis. Our previous studies indicate that miR-145 is a tumor suppressor capable of inhibiting tumor cell growth both in vitro and in vivo. In this study, we show that miR-145 exerts its function in a cell-specific manner. Although miR-145 inhibits cell growth in MCF-7 and HCT-116 cells, it has no significant effect on cell growth in metastatic breast cancer cell lines. However, miR-145 significantly suppresses cell invasion in these cells; in contrast, the antisense oligo against miR-145 increases cell invasion. miR-145 is also able to suppress lung metastasis in an experimental metastasis animal model. This miR-145-mediated suppression of cell invasion is in part due to the silencing of the metastasis gene mucin 1 (MUC1). Using luciferase reporters carrying the 3'-untranslated region of MUC1 combined with western blot and immunofluorescence staining, we identify MUC1 as a direct target of miR-145. Moreover, ectopic expression of MUC1 enhances cell invasion, which can be blocked by miR-145. Of interest, suppression of MUC1 by miR-145 causes a reduction of β-catenin as well as the oncogenic cadherin 11. Finally, suppression of MUC1 by RNAi mimics the miR-145 action in suppression of invasion, which is associated with downregulation of β-catenin and cadherin 11. Taken together, these results suggest that as a tumor suppressor, miR-145 inhibits not only tumor growth, but also cell invasion and metastasis.
Keywords: breast cancer, invasion, metastasis, miRNA, miR-145, posttranscriptional regulation
Cell migration and invasion are the major features of metastatic tumor cells which are responsible for the most of cancer related death. It is well known that the potential of a tumor cell to metastasize depends on numerous factors. Accumulating evidence suggests that microRNAs (miRNAs) could be key players in regulation of tumor cell invasion and metastasis (1). miRNAs are small non-coding RNAs that serve as negative regulators of gene expression (2–5). Through interactions with the 3'-untranslated region (3'-UTR) of mRNA by partial sequence homology, miRNAs cause gene silencing either by mRNA degradation or translation repression (6). Given the unique feature of their targeting, each single miRNA could have over a hundred of targets (7) and thus a large number of protein-coding genes could be under control of miRNAs. As a result, miRNAs play a fundamential role in regulation of diverse cellular functions, and deregulation of miRNA expression is often associated with a variety of disorders including human malignancy.
Increasing evidence indicates miRNAs may function as either oncogenes or tumor suppressors (8). For example, miR-145 is a putative tumor suppressive miRNA that is underexpressed in several types of tumors (9–11) and causes cell growth inhibition by targeting c-Myc (12) and IRS-1 (13). In addition,, miR-145 is able to target the pluripotency factors OCT4, SOX2, and KLF4 and functions as a key regulator of human stem cells (14) or promotes differentiation and repressing proliferation of smooth muscle cells (15). We have previously shown that miR-145 plays an important role in p53-mediated repression of c-Myc (12). During the further characterization of miR-145 in different cancer cell lines, we found that miR-145 functions as a tumor suppressor in a cell type specific manner. We demonstrated that miR-145 is a tumor suppressor impacting invasion and metastasis in part by targeting mucin 1 (MUC1).
Materials and Methods
Reagents
Primary antibodies were purchased from following vendors: MUC1 (small isoform), β-catenin and cyclin D1 from Epitomics (Burlingame, CA); cadherin 11 from Invitrogen (Carlsbad, CA); and Myc-tag from Applied Biological Materials (Vancouver, British Columbia, Canada); the antibody specific to large isoforms of MUC1 from Santa Cruz (Santa Cruz, CA). Secondary antibodies conjugated with IRDye 800CW or IRDye 680 were purchased from LI-COR Biosciences (Lincoln, NE). PCR primers and anti-miR-145 LNA oligo were purchased from IDT (Coralville, IA). MUC1 siRNA was purchased from Open Biosystems (Huntsville, AL). Freshly frozen breast tumor specimens and their matching normal breast specimens were obtained from Cooperative Human Tissue Network (CHTN) (Midwestern Division, Columbus, OH).
Cell culture
All cell lines were purchased from American Tissue Culture Collection (ATCC) (Manassas, VA) except for LM2-4142 (16) which was a generous gift from Dr. Joan Massagué (Sloan-Kettering Institute). Breast cancer cell lines BT-549, MDA-MB-231 and LM2-4142 were grown in RPMI 1640 (Cambrex, Walkersville, MD) supplemented with 10% FBS (Sigma-Aldrich). HEK-293T cells were cultured in DMEM (Cambrex) supplemented with 10% FBS. All media contained 2 mM glutamine, 100 units of penicillin/ml, and 100 μg of streptomycin/ml. Cells were incubated at 37 °C and supplemented with 5% CO2 in the humidified chamber.
Transfection
MDA-MB-231, LM2-4142 or BT-549 cells were transfected with anti-miR-145 using RNAfectin reagent (Applied Biological Materials) following the manufacturer's protocol.
Plasmids
The plasmid expressing miR-145 in pCMV or in lentiviral vector pCDH-CMV-MCS-EF1-copGFP (System Biosciences) or a mutant miR-145 expression vector has been described previously (12). Expression of the mature miR-145 was verified by TaqMan real-time RT-PCR (17).
To ectopically express MUC1, we cloned the MUC1 coding region in pCMV-Myc. The PCR product for MUC1 without UTR was obtained by primers MUC1-R1-5.1 (5'GAATTCTGACACCGGGCACCCAGTCTC) and MUC1-Not1-3.1 (5'GCGGCCGCCTACAAGTTGGCAGAAGTGGC; for MUC1 with UTR we used primers MUC1-R1-5.1 and MUC1-UTR-Not1-3.1 (see below). The PCR product was first cloned into a PCR cloning vector (pCR8) and then subcloned into pCMV-Myc at EcoR1 and Not1 sites.
The luciferase-UTR reporter constructs were generated by introducing the MUC1 3'-UTR carrying a putative miR-145 binding site into pGL3 control vector (Promega, Madison, WI). We first amplified the MUC1 3′-UTR sequence by PCR using primers MUC1-UTR-5.1 (5'TCTGCCAACTTGTAGGGGCAC) and MUC1-UTR-Not1-3 . 1 (5 'GCGGCCGCTTTTTTGGCGCAGTGGGAGAC), and MCF10A cDNA as a template. The PCR product was also first cloned into a PCR cloning vector (pCR8) and then subcloned into a modified pGL3 control vector where EcoR1 and Not1 sites were introduced into the original Xba1 site. To delete the putative miR-145 binding site in the MUC1 3'-UTR, we amplified the UTR by using primers MUC1-UTR-5.1 and MUC1-UTR-Not13.2 (5'GCGGCCGCCAGGATCCCCGCTATCTCAGG), and then cloned into the modified pGL3 control vector at EcoR1 and Not1 sites. Site-directed mutagenesis of the miR-145 binding site in the MUC1 3'-UTR was carried out by the two-step PCR approach as described previously (12). All PCR products were verified by DNA sequencing.
Luciferase Assay
Luciferase assays were carried out in 293T cells. First, cells were transfected with appropriate plasmids in 12-well plates. Then the cells were harvested and lysed for luciferase assay 24 h after transfection. Luciferase assays were performed using a luciferase assay kit (Promega) according to the manufacturer's protocol. β-galactosidase or renilla luciferase was used for normalization.
PCR/RT-PCR and real-time RT-PCR
PCR reactions were performed to amplify the MUC1 with or without 3'-UTR according to the standard three-step procedure. To detect MUC1 mRNA, we used the SYBR Green method with primers MUC1-5.1 (5'ACAGCTACCACAGCCCCTAA) and MUC1-3.1 (5'CAGCTGCCCGTAGTTCTTTC); Average levels of 5s RNA and β-actin were used as an internal control.
Cell proliferation assay
Cell growth assays were carried out by MTT [3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assays according to standard methods, as previously described (18).
Invasion assay
Matrigel chambers (BD Biosciences, San Jose, CA) were used to determine the effect of miR-145 or MUC1 on invasiveness per the manufacturer's protocol. In brief, infected cells were harvested, resuspended in serum free medium and then transferred to the hydrated matrigel chambers (~25,000 cells/well). The chambers were then incubated for 24 h in culture medium with 10% FBS in the bottom chambers before examination. The cells on the upper surface were scraped and washed away while the invaded cells on the lower surface were fixed and stained with 0.05% crystal violet for 2 h. Finally, invaded cells were counted under a microscope and the relative number was calculated.
Western Blot
Cells were harvested and protein was extracted from transfected cells as previously described (12, 18).
Immunofluorescence microscopy
To determine the effect of miR-145 on the protein level of MUC1, we also performed immunofluorescence staining using the MUC1 antibody using the previously described procedure (19).
Immunohistochemistry (IHC)
Immunohistochemistry was used to detect MUC1 in paraffin-embedded breast tumor tissue or to detect cadherin 11 in cell culture. For paraffin-embedded breast tumor tissue, slides were pretreated at 65°C for 2 h, followed by deparaffinization using standard procedures. After antigen retrieval, MUC1 antibody was applied to slides, followed by the secondary antibody conjugated with horse radish peroxidase (HRP). Signals were revealed Histostain Plus kit (Invitrogen) according to the manufacturer's instruction. To detect cadherin 11 expression in stably transduced MDA-MB-231 cells, the cells were grown on the coated coverslips overnight before immunohistochemical staining.
Experimental metastasis assay
Female athymic nude (nu/nu) mice (4–5 weeks old) were purchased from Harlan Sprague Dawley (Indianapolis, IN) and were maintained in the Southern Illinois University School of Medicine's accredited animal facility. All animal studies were conducted in accordance with NIH animal use guidelines and a protocol approved by SIU Animal Care Committee. In brief, 1.5 × 106 exponentially growing MDA-MB-231 or LM2-4142 cells with ectopic expression of appropriate genes were injected into nude mice through tail vein. Four weeks after injection, the animals were sacrificed. The lungs were harvested, fixed in Bouin's solution and tumor nodules were counted.
Statistical analyses
Statistical significance of the studies was analyzed by Student's t test. Differences with p values less than 0.05 are considered significant. The linear correlation coefficient (Pearson's r) was calculated to estimate the correlation between miR-145 values and MUC1 levels in the matched breast tumor specimens.
Results
miR-145 suppresses cell invasion in metastatic breast cancer cell lines
We have previously shown that as a tumor suppressor, miR-145 inhibited cell growth in MCF-7 and HCT-116 cells (12). However, further characterization indicated that miR-145 had no significant effect on cell growth in other cell lines like MDA-MB-231 and LM2-4142 (S-Fig. 1), suggesting cell type-specific function of miR-145. Of note, we observed changes in morphology of the miR-145 infected cells which revealed a less invasive morphological phenotype, i.e., round and flat compared to vector control. These results suggest that miR-145 may play a suppressive role in cell invasion in these cells because both MDA-MB-231 and LM2- 4142 are metastatic breast cancer cell lines.
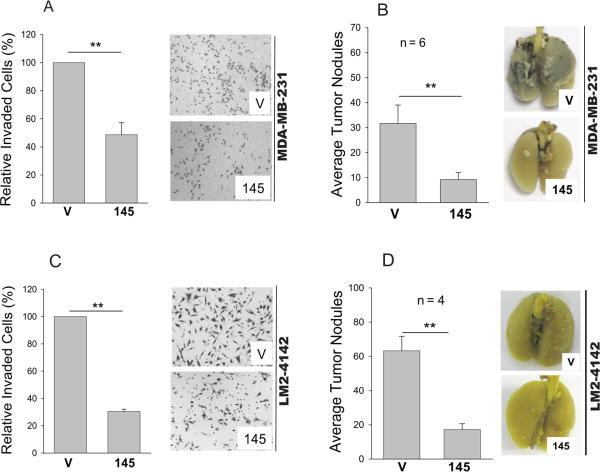
Therefore, we determined whether miR-145 impacts cell invasion by matrigel chamber assays. As expected, miR-145 significantly impaired invasion (Fig. 1A). For example, in MDA-MB-231 cells, miR-145 caused a reduction of invasive cells by over 50%. In contrast, anti-miR-145 enhanced cell invasion up to 50%, suggesting that this suppression is specific to miR-145. We found more reduction of invasion by miR-145 (by about 75%) in LM2-4142 whereas anti-miR-145 increased their invasiveness (Fig. 1C). To determine whether miR-145 suppresses cell invasion in vivo, we used an experimental metastasis model by tail vein injection. As shown in Fig 1B, while the average lung tumor nodules for the vector control were 32, there were only 9 nodules for miR-145. A similar result was obtained for LM2-4142 cells (Fig. 1D). These results further support the role of miR-145 in suppression of invasion and metastasis.
Fig. 1. Effect of miR-145 on cell invasion and lung metastasis.
A & B, Stably transduced MDA-MB-231 or LM2-4142 cells ectopically expressing miR-145 (145) or vector alone (V) were subject to matrigel chamber assay as detailed in Materials and Methods. Invaded cells on the membrane were counted as follows. Since the distribution of cells on the membrane was not always even, we first took a picture at a low magnification and then enlarged the image in a computer screen with grids so that all of the cells on the entire membrane were counted. B & D, Lung metastasis as revealed by the experimental metastasis animal model. Stably transduced MDA-MB-231 (B) or LM2-4142 (D) cells ectopically expressing miR-145 (145) or vector alone (V) were introduced into female nude mice through tail vein as described in Materials and Methods. Values A and C are means ± SE of three independent experiments. **, p < 0.01; SC, scrambled oliog; anti-145, anti-miR-145.
miR-145directly targets MUC1 by interaction with the 3'-UTR
To understand the mechanism of the miR-145-mediated inhibition of cell invasion, we first examined the effect of c-Myc on cell invasion because we previously demonstrated that miR-145 directly targets c-Myc (12). As a well known oncogene, c-Myc has been implicated in the involvement of cell invasion indirectly (20). However, we found that ectopic expression of c- Myc gave rise to a modest increase in cell invasion (not shown), which is not comparable to the suppressive effect of miR-145, suggesting that some other miR-145 targets may play a more important role in this aspect.
Hence, we searched for additional miR-145 targets using computer-aided miRNA target prediction programs, such as TargetScan4 (21) and miRBase targets and found several putative miR-145 target genes that might play a role in cell invasion, including ATF1, ATF3, MMP11, ANGPT2 and MUC1. Luciferase reporter assays showed that two of them, MUC1 and MMP11, met the arbitrary 35% reduction cutoff. We were particularly interested in MUC1 because MUC1 is a well known metastasis promoting gene which is upregulated in several types of tumors (22, 23). Therefore, we further characterized how miR-145 suppresses MUC1.
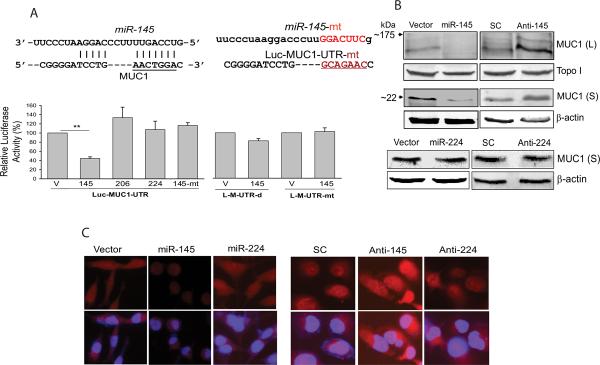
As shown in Fig.2A, miR-145 suppressed over 55% activity for Luc-MUC1-UTR compared to vector control; neither miR-206 nor miR-224 suppressed Luc-MUC1-UTR. Furthermore, the mutant miR-145 had no suppressive effect. We then deleted this site from the MUC1-UTR or made a site-directed mutant (Fig. 2A, top panels), and this suppression was abolished in the construct without this miR-145 binding site (Luc-MUC1-UTR-d) or with a mutant site (Luc-MUC1-UTR-mt) (Fig. 2A, bottom).
Fig. 2. miR-145 directly targets MUC1.
A, Effect of miR-145 on MUC1-3'-UTR luciferase reporters. Top panels: alignment of MUC1 3'-UTR and miR-145, along with a mutant miR-145 or mutant MUC1-3'-UTR in which the sequences in red were deleted or mutagenized. Bottom panels: a luciferase reporter carrying the 3'-UTR of MUC1 (Luc-MUC1-UTR), deletion of miR-145 binding site at the 3'-UTR (Luc-MUC1-UTR-d) or mutant that the miR-145 binding site was mutated (Luc-MUC1-UTR-mt), was introduced into 293T cells along with miR-145 (145), miR-206 (206), miR-224 (224), mutant miR-145 (145-mt) or vector control (V) and the cells were harvested 24 h later for luciferase assays. miR-206 and miR-224 serve as a negative control. B, miR-145 suppresses, whereas anti-miR-145 enhances the endogenous protein levels of MUC1, as detected by western blot. Stably transduced MDA-MB-231 cells ectopically expressing miR-145 or transiently transfected with anti-miR-145 were used for western blot analysis. Top panels: Effect miR-145 or anti-miR-145 on large (L) and small (S) isoforms of MUC1. Bottom panels: stably transduced MDA-MB-231 cells with miR-224 reveal no suppression of MUC1. Topoisomerase I (topo I) and β-actin serve as loading controls. C, Immunofluorescence staining further confirms suppression of MUC1 by miR-145 in stably transduced MDA-MB-231 cells. The cells were grown on coverslips overnight and then subject to immunostaining with anti-MUC1 antibody, as described in Materials and Methods. All images with red signals (MUC1) were taken at the same fixed time. Merged pictures are overlays of both MUC1 red signals and nuclear staining by Hoechst dye (blue). Values in A are means ± SE of three independent experiments. **, p < 0.01; SC, scrambled oligo; anti-145, anti-miR-145.
Next, we determined whether ectopic expression of miR-145 can suppress the endogenous MUC1 at the protein level by Western blot. It has been reported of several MUC1 isoforms in the lysates of MUC1-positive cultured cancer cells: high molecular weight isoforms (150–300 kDa) presumably due to glycosylation and a low molecular weight isoform (20–35 kDa) (24). As shown in Fig. 2B (top panels), miR-145 suppressed both large and small isoforms of MUC1. Since the small isoform is the predominant form in cancer cell lines and tumor specimens, and it mediates tumor cell growth (24), this study focused on this small isoform. We further showed that the level of small isoform of MUC1 in the miR-145 cells was less than 40% of vector control cells, whereas anti-miR-145 increased its protein level by over 200% (S-Fig. 2). In contrast, miR-224 or anti-miR-224 had no effect on MUC1 expression (Fig. 2B, bottom panels). We also tested two additional metastatic breast cancer cell lines LM2-4142 and BT-549. The relative expression levels of miR-145 in these cells were shown in S-Fig. 3. Similar to the result in MDA-MB-231 cells, while miR-145 suppressed, anti-miR-145 increased level of MUC1 in these cells (S-Fig. 4). Finally, we confirmed this miR-145-mediated repression of MUC1 by immnunofluoroscence staining. The red signal of (MUC1) in the miR-145 transduced MDA-MB-231 (Fig. 2C) and LM2-4142 cells (S-Fig. 5) were visibly low compared to that of the cells infected with vector control, especially compared to the cells transfected with anti-miR-145 (Fig. 2C, middle of right panels and S-Fig. 5), supporting the suppressive role of miR-145 in MUC1 expression.
Real-time RT-PCR analysis detected a slight reduction of MUC1 mRNA in miR-145 cells compared to the vector control in MDA-MB-231 or LM2-4142 cells, but this difference was not significant (S-Fig. 6), suggesting that miR-145 silences MUC1 mainly at the translational level.
Ectopic expression of MUC1 promotes invasiveness
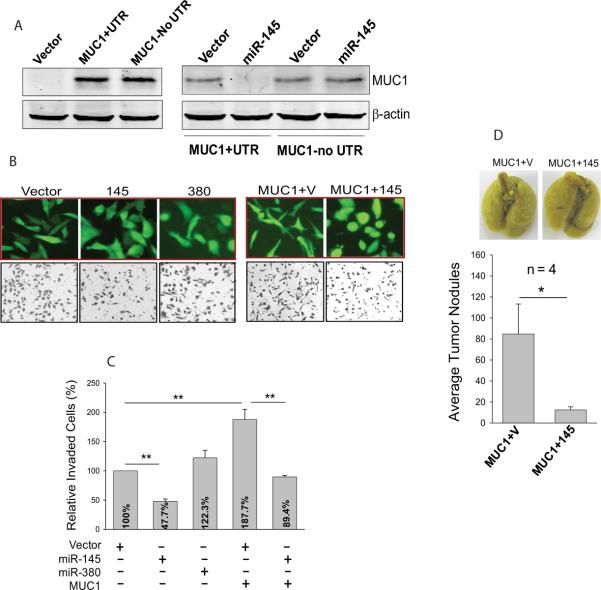
To confirm the role of miR-145-mediated MUC1 suppression in invasion, we cloned the coding region of MUC1 with or without the 3'-UTR and tagged them with Myc epitope. We first verified the ectopic expression for these two constructs. About the same amount of the exogenous MUC1 was expressed with or without the 3'-UTR (Fig. 3A, left panel). Co-transfection with miR-145 caused a marked reduction of MUC1 protein level for the clone carrying the 3'-UTR, but not with the construct carrying no 3'-UTR (Fig. 3A, right panel).
Fig. 3. Ectopically expressed MUC1 promotes cell invasion which can be reversed by miR-145.
A, Left panel: expression vector carrying Myc-tagged MUC1 with or without the 3'-UTR was introduced into 293T cells and western blot was performed 24 h after transfection. The membrane was probed with Myc tag antibody. Right panel: miR-145 suppresses MUC1 with the corresponding 3'-UTR. 293T cells were transfected with UTR (MUC1+UTR) or without UTR (MUC1 no UTR) along with vector alone or miR-145. The cells were harvested for extraction of protein 24 h after transfection. B, Top panels: Effect of miR-145 on morphology changes. Left three pictures: stably transduced MDA-MB-231 cells with vector pCDH, miR-145 (145) or miR-380 (380). We used miR-380 as a negative control here instead of miR-224 because our separate studies suggested that miR-224 is able to suppress invasion by targeting other metastatic genes (Zhu et al unpublished). Right two pictures: MDA-MB-231 cells infected with MUC1 along with vector control (V) or miR-145 (145). Images were taken before invasion assays. Bottom panels: Representative fields of invaded cells on the membranes after invasion assays. C, Quantitative analysis of invasion assays. D. Suppression of MUC1-mediated metastasis by miR-145 in experimental metastasis assays. MDA-MB-231 cells ectopically expressing MUC1 and vector (MUC1+V) or MUC1 and miR-145 (MUC1+145) were injected into female nude mice through tail vein as detailed in Materials and Methods. Values in C are means ± SE of three independent experiments. **, p < 0.01; *, p < 0.05.
Having demonstrated that MUC1 is functionally expressed, we next determined the invasive potential of MUC1. We found notable morphology changes in these cells; they appeared more elongated, a more invasive phenotype which can be reversed by miR-145 (Fig. 3B, top right panels). In contrast, about 30% miR-145 cells were round and more epithelial characteristic compared to <5% round cells for vector control (Fig. 3B, top left panels). Consistent with the morphological changes, MUC1 enhanced cell invasiveness (Fig. 3C), which was inhibited by miR-145. Experimental metastasis assays further supported the role of miR-145 in suppressing invasion and metastasis by targeting MUC1. For example, MUC1 plus vector produced an average of 85 tumor nodules compared to 13 nodules from the MUC1 plus miR-145 cells (Fig. 3D), similar to the invasion results.
Effect ofmiR-145on cadherin 11
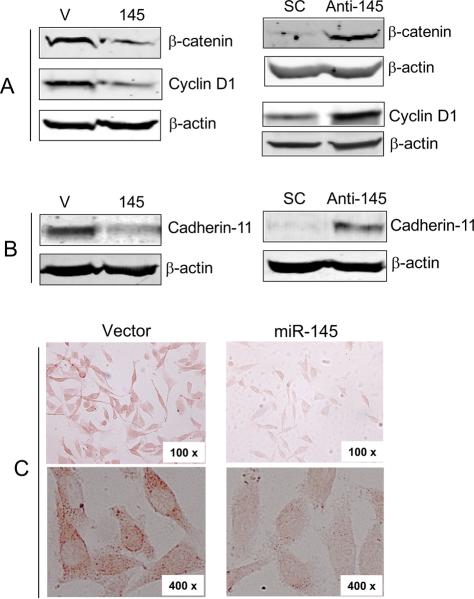
MUC1 promotes cell invasion and metastasis possibly through interactions with different cell signaling molecules, causing stabilization of β-catenin (25). Thus, suppression of MUC1 by miR-145 leading to the reduction of cell invasion is of particular interest because this may suggests that miR-145 also affects the level of β-catenin. Therefore, we first determined the effect of miR-145 on β-catenin. As expected, suppression of MUC1 by miR-145 decreased the level of β-catenin and its downstream gene cyclin D1 (Fig. 4A, left panels). In contrast, anti-miR-145 increased β-catenin and cyclin D1 (Fig. 4A, right panels). Since in silico analysis did not find any miR-145 binding site in the 3'-UTRs of β-catenin and cyclin D1; furthermore, luciferase reporters carrying the corresponding UTR revealed no effect by miR-145 (not shown), thus suppression of β-catenin and cyclin D1 by miR-145 is possibly an indirect effect through MUC1. It has been reported that β-catenin forms a complex with E-cadherin and down-regulation of MUC1 in cancer cells inhibits cell migration by promoting the E-cadherin/catenin complex formation (26). However, there is no detectable E-cadherin in MDA-MB-231 cells which, instead, express a high level of cadherin 11 (27). Evidence further suggests that cadherin 11 plays an oncogenic role and it also interacts with β-catenin (27–29). Therefore, we examined the effect of miR-145 on cadherin 11. While miR-145 suppressed, anti-miR-145 increased cadherin 11 (Fig. 4B). We further confirmed by immunohistochemistry that cadherin 11 was repressed in MDA-MB-231 cells expressing miR-145 (Fig. 4C). Therefore, suppression of cadherin 11 by miR-145 through targeting MUC1 is likely to contribute to the miR-145-mediated reduction of cell invasion.
Fig. 4. Effect of miR-145 on β-catenin and cadherin 11.
A and B, miR-145 suppresses, while anti-miR-145 enhances β-catenin, cyclin D1 and cadherin 11. Stably transduced MDA-MB-231 cells ectopically expressing vector alone (V) or miR-145 (145); and MDA-MB-231 cells transfected with scrambled olig (SC) or anti-miR-145 were used for western blot analysis. C, Suppression of cadherin 11 by miR-145 in MDA-MB-231 cells as detected by immunohistochemistry. Stably transduced MDA-MB-231 cells ectopically expressing vector alone or miR-145 were used for immunohistochemistry analysis as detailed in Materials and Methods.
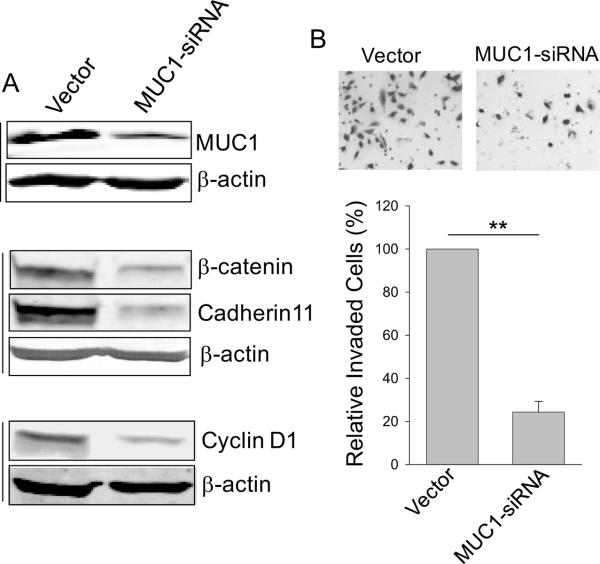
To further determine the role of MUC1 in miR-145-mediated suppression of invasion and metastasis, we suppressed MUC1 by RNAi (Fig. 5A, top panels). This suppression of MUC1 significantly reduced invasiveness (Fig. 5B). At the same time, MUC1 siRNA also caused downregulation of β-catenin and cadherin 11 as well as cyclin D1 (Fig. 5A), similar to the effect of miR-145 on these proteins (Fig. 4 A and B). These results suggest that miR-145 targets MUC1, which in turn causes downregulation of β-catenin, cyclin D1 and cadherin 11, leading to suppression of invasion and metastasis.
Fig. 5. Suppression of MUC1 by RNAi reduces levels of β-catenin, cyclin D1 and cadherin 11 as well as invasiveness.
A, Effect of MUC1 siRNA on MUC1 protein as well as β-catenin, cadherin 11 and cyclin D1. B, Suppression of cell invasion by MUC1 siRNA. MDA-MB-231 cells were transfected with MUC1 siRNA, followed by invasion assay as detailed in Materials and Methods. Values are means ± SE of three separate experiments. **, p < 0.01.
MUC1 is upregulated in breast cancer specimens
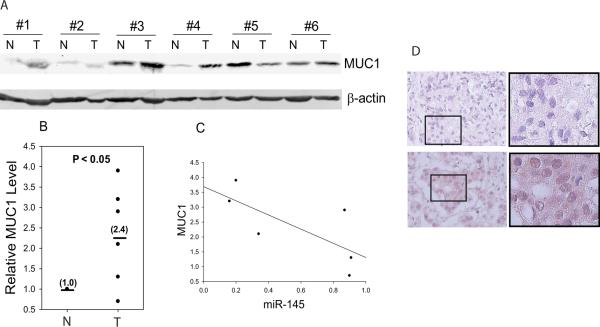
Finally, we examined MUC1 expression in matched breast tumor specimens to determine its clinical relevance. As shown in Fig. 6A, MUC1 level was visibly elevated in 4 of 6 cases of advanced breast cancer with average value of 2.4 compared to the matched normal tissue as 1.0 (Fig. 6B), which inversely correlated with miR-145 expression (Fig. 6C). We then determined whether metastasis status affects MUC1 expression by immunohistochemical staining breast cancer tissue microarrays and found that, in general, MUC1 level was higher in metastatic tumors than in non-metastatic tumors (Fig. 6D and S-Table 1), which is consistent with the previous reports that MUC1 correlates with poor survival in colorectal cancer (30) and breast cancer (31). Downregulation of miR-145 could derepress MUC1 as tumor cells progress and metastasize and thus, modulation of miR-145 may provide a therapeutic strategy for metastatic breast cancer.
Fig. 6. Expression of MUC1 in breast tumor specimens.
A and B, Western blot reveals upregulation of MUC1 in breast tumors (T) compared to matched normal breast tissue (N). C, A negative correlation between MUC1 and miR-145 in breast tumor specimens after normalization with normal tissue. D, Expression of MUC1 in breast tumor specimens with different metastasis status by immunohistochemistry, a: infiltrating ductal carcinoma without metastasis, and b: infiltrating ductal carcinoma with metastasis.
Discussion
We have previously shown that miR-145 is able to suppress tumor cell growth in both in vitro and in vivo in part through targeting the oncogene c-Myc (12). Although c-Myc has been implicated in indirect regulation of invasion and metastasis, our study suggests that factors other than c-Myc may play a more important role in miR-145-mediated suppression of cell invasion. The present study demonstrates that MUC1 is a direct target for miR-145. Ectopic expression of MUC1 and siRNA knockdown confirmed its invasion promoting activity. Moreover, miR-145-mediated suppression of MUC1 is dependent on the 3'-UTR. Finally, MUC1-induced cell invasion can be reversed by miR-145. Therefore, these results highlight the significance of miR-145 as a tumor suppressor in cell invasion and metastasis by targeting MUC1.
Increasing evidence suggests that MUC1 plays a role in invasion and metastasis (30). For example, MUC1 promotes invasion in breast cancer by interacting with β-catenin (29); moreover, MUC1 and β-catenin are coexpressed at the invasion front of colorectal carcinomas which correlates with poor prognosis in colorectal cancer (30). However, the precise role of MUC1 in invasion and metastasis as well as its regulation is not well understood. MUC1 is a member of a large mucin family, which are characterized by a variable number of tandem repeats (VNTR) responsible for its glycosylation. MUC1 belongs to type I membrane glycoprotein subfamily and possess a single membrane-spanning domain and a short cytoplasmic tail in addition to the extensive extracellular domain (22). MUC1 has 7 variants which differ mainly in the VNTR region. Since all variants share the same 3'-UTR, miR-145 is expected to be able to suppress all of the variants. Our western data supports this notion (Fig. 2B)
The role of MUC1 in invasion and metastasis has been demonstrated in different models. For example, the cytoplasmic tail of MUC1 was reported to enhance the invasion of MDA-MB-468 breast cancer cells expressing wild type GSK-3β and β-catenin (32), suggesting possible interactions between these proteins. Moreover, MUC1 expression is associated with increased steady-state levels of β-catenin in the cytoplasm and nucleus of breast carcinoma cells by blocking the GSK-3β-mediated phosphorylation of β-catenin, thus preventing proteosomal degradation (29). It is possible that the cytoplasmic tail of MUC1 serves as a scaffold protein, enabling interaction between different regulators or alternatively might compete for or sequester β-catenin. In some cell types, the MUC1 cytoplasmic tail is also involved in the transcriptional activation of β-catenin-TCF-binding sites and transcriptional activation of cyclin D1 (25). Consistent with this finding, we detected decreased levels of β-catenin and cyclin D1 in the miR-145 cells (Fig. 4A). Moreover, MUC1 may play an antiapoptotic role in response to cellular stresses by stimulating Akt and the antiapoptotic protein Bcl-X to attenuate genotoxin-induced apoptosis (33). A recent report suggests that this MUC1-mediated tumorigenesis is likely through the TGF-α signaling pathway (34).
Although MUC1 is often deregulated in tumors, information about its regulation is limited. In one report, MUC1 was shown to be induced by hypoxia in a lung adenocarcinoma cell line (35); a recent report indicates that MUC1 and galectin-3 oncoproteins function in a miRNA-dependent regulatory loop (36). N-glycosylated MUC1 C-terminal subunit increases galectin-3 mRNA levels by suppressing expression of miR-322 and thereby stabilizing galectin-3. However, it is not clear whether MUC1 itself is subject to miRNA regulation. Our study establishes the posttranscriptional regulation of MUC1 by miR-145.
Clinically, MUC1 overexpression occurs frequently in many types of cancer, including breast, ovarian, lung, colon, and pancreatic carcinomas (22, 23); its expression tends to increase as tumors progress (37, 38). Consistent with these reports, we also observed an increased level of MUC1 in more metastatic breast tumors compared to less metastatic breast tumors. However, the precise mechanism of MUC1-mediated metastasis remains to be elucidated. Mucin plays an important role in protecting normal cells from pathogen but tumors cells may use this protective shield to evade from immune system (39). In addition, MUC1 may enhance metastasis in part through interaction with adhesion molecules such as the endothelial protein ICAM-1 (40) or by serving as a ligand for galectin-3 (41).
Our study suggests that MUC1-mediated invasion may involve the oncogenic cadherin 11 because miR-145 also causes downregulation of cadherin 11 in MDA-MB-231 cells. It has been previously shown that MDA-MB-231 has lost the epithelial marker type I cadherins but it expresses an oncogenic cadherin 11 instead (27). Further evidence suggests that cadherin 11 is expressed in several types of cancers including breast cancer; moreover, cadherin 11 expression promotes the metastasis of prostate cancer cells to bone (42). Although the underlying mechanism remains to be determined, this downregulation of cadherin 11 by miR-145 is possibly through MUC1 and β-catenin. Given that MUC1 interacts with β-catenin and enhances the β- catenin level (29), it would be reasonable to speculate that this MUC1-mediated β-catenin expression also affects cadherin 11 and thus, a likely consequence of ectopic expression of miR-145 is a reduction of cadherin 11 through suppression of MUC1. In support of this, we found that miR-145 suppresses MUC1, β-catenin and cadherin 11 (Fig. 4A and B). Moreover, suppression of MUC1 by RNAi also downregulates β-catenin and cadherin 11 (Fig. 5A). Therefore, further characterization of this pathway will provide new insight into miR-145-mediated suppression of invasion and metastasis.
Supplementary Material
Acknowledgment
We are grateful to Dr. Joan Massagué for providing LM2-4142 cells. This study is supported by grants CA102630 from NCI.
Reference
- 1.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 2.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–61. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zamore P, DHaley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–24. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–71. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 9.Sempere LF, Christensen M, Silahtaroglu A, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 10.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 11.Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and Prognostic MicroRNAs in Stage II Colon Cancer. Cancer Res. 2008;68:6416–24. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 12.Sachdeva M, Zhu S, Wu F, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106:3207–12. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi B, Sepp-Lorenzino L, Prisco M, et al. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J Biol Chem. 2007;282:32582–90. doi: 10.1074/jbc.M702806200. [DOI] [PubMed] [Google Scholar]
- 14.Yu T, Wang XY, Gong RG, et al. The expression profile of microRNAs in a model of 7,12-dimethyl-benz[a]anthrance-induced oral carcinogenesis in Syrian hamster. J Exp Clin Cancer Res. 2009;28:64. doi: 10.1186/1756-9966-28-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordes KR, Sheehy NT, White MP, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–10. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minn AJ, Kang Y, Serganova I, et al. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest. 2005;115:44–55. doi: 10.1172/JCI22320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–36. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 19.Wu F, Chiocca S, Beck WT, Mo YY. Gam1-associated alterations of drug responsiveness through activation of apoptosis. Mol Cancer Ther. 2007;6:1823–30. doi: 10.1158/1535-7163.MCT-06-0771. [DOI] [PubMed] [Google Scholar]
- 20.Zhang XY, DeSalle LM, Patel JH, et al. Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein. Proc Natl Acad Sci U S A. 2005;102:13968–73. doi: 10.1073/pnas.0502330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thornton DJ, Rousseau K, McGuckin MA. Structure and function of the polymeric mucins in airways mucus. Annu Rev Physiol. 2008;70:459–86. doi: 10.1146/annurev.physiol.70.113006.100702. [DOI] [PubMed] [Google Scholar]
- 23.Ren J, Agata N, Chen D, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell. 2004;5:163–75. doi: 10.1016/s1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahanta S, Fessler SP, Park J, Bamdad C. A minimal fragment of MUC1 mediates growth of cancer cells. PLoS ONE. 2008;3:e2054. doi: 10.1371/journal.pone.0002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang L, Chen D, Liu D, et al. MUC1 oncoprotein blocks glycogen synthase kinase 3beta-mediated phosphorylation and degradation of beta-catenin. Cancer Res. 2005;65:10413–22. doi: 10.1158/0008-5472.CAN-05-2474. [DOI] [PubMed] [Google Scholar]
- 26.Huang L, Ren J, Chen D, et al. MUC1 cytoplasmic domain coactivates Wnt target gene transcription and confers transformation. Cancer Biol Ther. 2003;2:702–6. [PubMed] [Google Scholar]
- 27.Pishvaian MJ, Feltes CM, Thompson P, et al. Cadherin-11 is expressed in invasive breast cancer cell lines. Cancer Res. 1999;59:947–52. [PubMed] [Google Scholar]
- 28.Shibata T, Ochiai A, Gotoh M, Machinami R, Hirohashi S. Simultaneous expression of cadherin-11 in signet-ring cell carcinoma and stromal cells of diffuse-type gastric cancer. Cancer Lett. 1996;99:147–53. doi: 10.1016/0304-3835(95)04047-1. [DOI] [PubMed] [Google Scholar]
- 29.Schroeder JA, Adriance MC, Thompson MC, Camenisch TD, Gendler SJ. MUC1 alters beta-catenin-dependent tumor formation and promotes cellular invasion. Oncogene. 2003;22:1324–32. doi: 10.1038/sj.onc.1206291. [DOI] [PubMed] [Google Scholar]
- 30.Baldus SE, Monig SP, Huxel S, et al. MUC1 and nuclear beta-catenin are coexpressed at the invasion front of colorectal carcinomas and are both correlated with tumor prognosis. Clin Cancer Res. 2004;10:2790–6. doi: 10.1158/1078-0432.ccr-03-0163. [DOI] [PubMed] [Google Scholar]
- 31.Baldus SE, Engelmann K, Hanisch FG. MUC1 and the MUCs: a family of human mucins with impact in cancer biology. Crit Rev Clin Lab Sci. 2004;41:189–231. doi: 10.1080/10408360490452040. [DOI] [PubMed] [Google Scholar]
- 32.Lillehoj EP, Han F, Kim KC. Mutagenesis of a Gly-Ser cleavage site in MUC1 inhibits ectodomain shedding. Biochem Biophys Res Commun. 2003;307:743–9. doi: 10.1016/s0006-291x(03)01260-9. [DOI] [PubMed] [Google Scholar]
- 33.Raina D, Kharbanda S, Kufe D. The MUC1 oncoprotein activates the anti-apoptotic phosphoinositide 3-kinase/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J Biol Chem. 2004;279:20607–12. doi: 10.1074/jbc.M310538200. [DOI] [PubMed] [Google Scholar]
- 34.Pochampalli MR, Bitler BG, Schroeder JA. Transforming growth factor alpha dependent cancer progression is modulated by Muc1. Cancer Res. 2007;67:6591–8. doi: 10.1158/0008-5472.CAN-06-4518. [DOI] [PubMed] [Google Scholar]
- 35.Mikami Y, Hisatsune A, Tashiro T, Isohama Y, Katsuki H. Hypoxia enhances MUC1 expression in a lung adenocarcinoma cell line. Biochem Biophys Res Commun. 2009;379:1060–5. doi: 10.1016/j.bbrc.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Ramasamy S, Duraisamy S, Barbashov S, et al. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell. 2007;27:992–1004. doi: 10.1016/j.molcel.2007.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohlgraf KG, Gawron AJ, Higashi M, et al. Contribution of the MUC1 tandem repeat and cytoplasmic tail to invasive and metastatic properties of a pancreatic cancer cell line. Cancer Res. 2003;63:5011–20. [PubMed] [Google Scholar]
- 38.Nakamori S, Ota DM, Cleary KR, Shirotani K, Irimura T. MUC1 mucin expression as a marker of progression and metastasis of human colorectal carcinoma. Gastroenterology. 1994;106:353–61. doi: 10.1016/0016-5085(94)90592-4. [DOI] [PubMed] [Google Scholar]
- 39.Carraway KL, Ramsauer VP, Carraway CA. Glycoprotein contributions to mammary gland and mammary tumor structure and function: roles of adherens junctions, ErbBs and membrane MUCs. J Cell Biochem. 2005;96:914–26. doi: 10.1002/jcb.20612. [DOI] [PubMed] [Google Scholar]
- 40.Rahn JJ, Chow JW, Horne GJ, et al. MUC1 mediates transendothelial migration in vitro by ligating endothelial cell ICAM-1. Clin Exp Metastasis. 2005;22:475–83. doi: 10.1007/s10585-005-3098-x. [DOI] [PubMed] [Google Scholar]
- 41.Yu LG, Andrews N, Zhao Q, et al. Galectin-3 interaction with Thomsen-Friedenreich disaccharide on cancer-associated MUC1 causes increased cancer cell endothelial adhesion. J Biol Chem. 2007;282:773–81. doi: 10.1074/jbc.M606862200. [DOI] [PubMed] [Google Scholar]
- 42.Chu K, Cheng CJ, Ye X, et al. Cadherin-11 promotes the metastasis of prostate cancer cells to bone. Mol Cancer Res. 2008;6:1259–67. doi: 10.1158/1541-7786.MCR-08-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.