Abstract
Murine gammaherpesvirus-68 (MHV68) is genetically related to human Epstein-Barr virus and Kaposi’s sarcoma-associated herpesvirus and provides a tractable model to study gammaherpesvirus-host interactions in vivo and in vitro. The MHV68-encoded v-RCA product inhibits murine complement activation and shares sequence homology with other virus and host regulators of complement activation. Here we show that v-RCA is required for efficient MHV68 replication in primary murine macrophages, but not in murine embryonic fibroblasts. v-RCA deficient MHV68 mutant viruses display defects in viral DNA synthesis in infected macrophages. Importantly, attenuated growth of v-RCA mutant viruses is not rescued in macrophages lacking critical components of the complement system including C3, indicating that the macrophage-specific role of v-RCA in MHV68 replication is complement-independent. This contrasts with the situation in vivo in which attenuated neurovirulence of v-RCA mutant viruses is rescued in C3-deficient mice. This study shows a novel, complement independent cell-type-specific function of a gammaherpesvirus RCA protein.
Keywords: gammaherpesvirus, complement, MHV68, viral RCA
INTRODUCTION
Murine gammaherpesvirus-68 (MHV68, γHV68) is a rodent gammaherpesvirus genetically and biologically related to human gammaherpesviruses Epstein-Barr virus (EBV), Kaposi’s sarcoma-associated herpesvirus (KSHV), and simian herpesvirus saimirii (HVS) (Efstathiou, Ho et al., 1990a;Virgin, Latreille et al., 1997). In vivo, MHV68 establishes latency in a variety of cell types, including macrophages, B cells, and dendritic cells (Flano, Husain et al., 2000;Sunil-Chandra, Efstathiou et al., 1992;Weck, Kim et al., 1999a;Weck, Kim et al., 1999b;Willer & Speck, 2003). There are several lines of evidence indicating that MHV68-host interactions are determined by cell-type specific functions of viral genes. During the establishment of latency, expression of viral genes is dependent on cell type and stage of cell differentiation (Marques, Efstathiou et al., 2003). Further, MHV68-encoded proteins, such as M2 and v-cyclin, display cell-type specific effects in vitro and during latency in vivo (Herskowitz, Jacoby et al., 2005;Van Dyk, Virgin et al., 2003;Siegel, Herskowitz et al., 2008;Herskowitz, Siegel et al., 2008). The MHV68-encoded protein kinase orf36 is required for efficient MHV68 replication in primary bone marrow derived macrophages, but not mouse embryonic fibroblasts (MEFs) in vitro (Tarakanova, Leung-Pineda et al., 2007). From the host perspective, several cellular signaling pathways, such as NF-kB and interferon gamma signaling, function in a cell-type specific manner during MHV68 latency (Krug, Moser et al., 2007;Steed, Buch et al., 2007). Therefore, determining the cell type-specific activities of MHV68 genes in relevant primary cells such as macrophages is an important goal for linking studies of viral gene function to studies of the pathogenesis of infection (Virgin, 2007).
The MHV68 v-RCA protein is homologous to host and virus RCA proteins encoded by other herpes as well as poxviruses (Cummings, Waggoner et al., 2007;Kapadia, Molina et al., 1999;Kapadia, Levine et al., 2002). Both host and viral homologues of the MHV68 v-RCA protein inhibit the classical and alternative pathways of complement activation by multiple mechanisms, including accelerated decay of C3 convertase, degradation of activated complement factors, and prevention of complement deposition (Cummings, Waggoner, Tacke, & Hahn, 2007). Importantly, not all activities of this class of viral molecules are complement dependent. Vaccinia-encoded RCA protein (B5R), in addition to its complement regulatory functions, plays a role in virion morphogenesis, cell-to-cell spread, and induction of thick actin bundles in vitro (Herrera, del Mar et al., 1998;Mathew, Sanderson et al., 2001;Rodger & Smith, 2002). Vaccinia B5R, KSHV KCP, as well as cellular C4b binding protein and factor H also bind heparan sulfate proteoglycans, suggesting a potential role for viral RCA proteins in viral binding and entry (Mark, Lee et al., 2006;Mullick, Singh et al., 2005;Murthy, Smith et al., 2001;Blackmore, Hellwage et al., 1998;Prodinger, Hellwage et al., 1998;Blom, Kask et al., 2001). Together these data indicate that viral RCA proteins can have complement independent roles in infection.
MHV68 v-RCA is expressed with late kinetics and is not required for MHV68 growth in a mouse fibroblast cell line (Kapadia, Molina, van, V, Speck, & Virgin, 1999). Recombinant v-RCA and supernatants from MHV68 infected cells are sufficient to block complement deposition on zymosan beads (Kapadia, Molina, van, V, Speck, & Virgin, 1999). In vivo, absence of MHV68 v-RCA leads to a modest attenuation of acute viral replication in adult BL6 mice, However, v-RCA has an important role in neurovirulence of MHV68 in weanling CD-1 mice (Kapadia, Levine, Speck, & Virgin, 2002). CD-1 mice infected with a MHV68 mutant lacking v-RCA show decreased viral titers in numerous organs and increased survival as compared to wild type MHV68 infected mice (Kapadia, Levine, Speck, & Virgin, 2002). The virulence of v-RCA mutant virus in weanling mice is rescued in mice deficient in C3, a critical component of all pathways of complement activation (Kapadia, Levine, Speck, & Virgin, 2002). These data show that a major aspect of v-RCA function relates to inhibition of C3-dependent actions of complement in vivo. However, there are other in vivo phenotypes of v-RCA mutant virus where the role of complement has not been determined. For example, the lethality of v-RCA mutant virus is attenuated in interferon γ receptor deficient mice (Kapadia, Levine, Speck, & Virgin, 2002). It is not clear whether complement plays a role in this attenuation. While complement controls reactivation of wild type MHV68 from latency, v-RCA also seems to be dispensable for MHV68 reactivation from latency in BL6 and C3 -/- mice (Kapadia, Levine, Speck, & Virgin, 2002). These considerations led us to test the hypothesis that MHV68 v-RCA has complement independent effects relevant to viral replication. We focused on the possible role of v-RCA in primary macrophages as these cells harbor latent MHV68.
RESULTS AND DISCUSSION
MHV68 v-RCA is required for efficient viral replication in macrophages
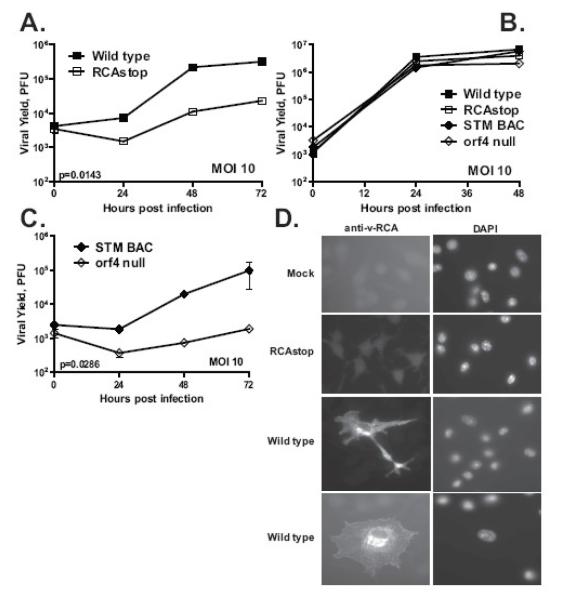
We previously found a MHV68 gene (orf36) required for viral replication in cultured primary macrophages but not embryonic fibroblasts (Tarakanova, Leung-Pineda, Hwang, Yang, Matatall, Basson, Sun, Piwnica-Worms, Sleckman, & Virgin, 2007). This suggested that assays performed in fibroblasts may not provide information on the relevant functions of MHV68 genes, a finding confirmed by recent studies demonstrating a role for the MHV68 v-cyclin in primary endothelial cells (Suarez & Van Dyk, 2008). To identify other MHV68 genes with cell-type specific requirements during viral replication, a panel of MHV68 mutants was examined for growth defects in primary bone marrow derived macrophages. Attenuated replication was observed, at both high and low multiplicities of infection, for a mutant virus containing a translational stop in v-RCA [RCAstop (Kapadia, Levine, Speck, & Virgin, 2002)] (Fig. 1A and data not shown). We confirmed that mutation of v-RCA had no effect on replication in mouse embryonic fibroblasts (Fig. 1B). The RCAstop MHV68 mutant was derived from wild type virus by insertion of two stop codons and a frame shift in the v-RCA open reading frame, and eliminates expression of v-RCA in a mouse fibroblast cell line (Kapadia, Levine, Speck, & Virgin, 2002).
Figure 1. MHV68 v-RCA is required for efficient viral replication in primary bone marrow derived macrophages.
Primary mouse macrophages (A, C) or mouse embryonic fibroblasts (B) were infected with 10PFU/cell of v-RCA deficient mutants or control wild type viruses in triplicate, samples were harvested at indicated time points and titered on BALB-3T12. D. Primary macrophages were mock-infected or infected with 5PFU/cell of indicated viruses and stained at 48 hours post infection with anti-v-RCA and DAPI to visualize nuclei. Shown is a representative result of at least three independent experiments.
To confirm that the defect in growth in macrophages observed in RCAstop is due to the altered open reading frame and not a secondary effect of the particular mutation in v-RCA, we tested a second independently constructed v-RCA mutant generated by transposon insertion at a site distinct from the translational stop present in the RCAstop MHV68 mutant (Song, Hwang et al., 2005). Compared to the control virus (STM BAC), growth of orf4 null was attenuated in macrophages (Fig. 1C). The consistency of phenotype between two v-RCA mutant viruses constructed independently strongly suggests that the phenotype of decreased growth in primary macrophages is due to mutation of orf4 encoding v-RCA.
In MHV68-infected 3T12 fibroblast cells, v-RCA protein is associated with the surface of infected cells and the virion envelope (Kapadia, Molina, van, V, Speck, & Virgin, 1999). As v-RCA is not essential for efficient growth of the virus in those cells, we considered the possibility that the sub-cellular distribution of v-RCA might differ between infected fibroblasts and macrophages. V-RCA staining in MHV68-infected macrophages was consistent with the endoplasmic reticulum and plasma membrane localization, suggesting that the subcellular distribution of v-RCA was similar in primary macrophages and fibroblasts [Fig. 1D, (Kapadia, Molina, van, V, Speck, & Virgin, 1999)]. As expected, no v-RCA specific staining was detected in mock-infected or RCAstop-infected primary macrophages (Fig. 1D).
V-RCA is expressed with late kinetics in primary MHV68-infected macrophages
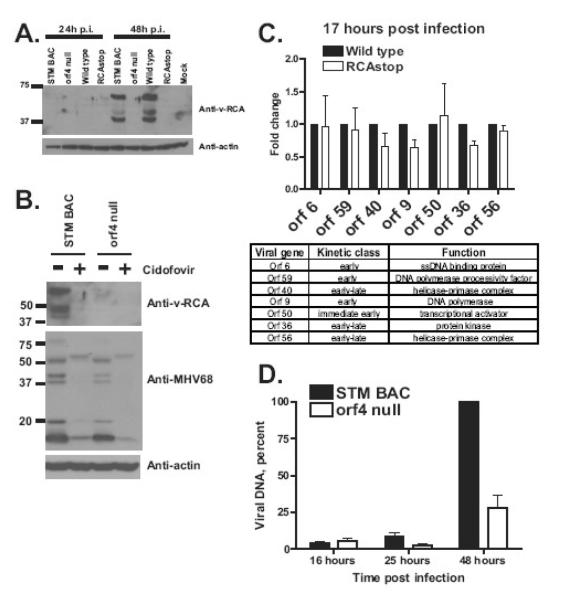
V-RCA is classified as a late viral protein based on its expression in 3T12 fibroblasts (Kapadia, Molina, van, V, Speck, & Virgin, 1999). To determine if this is correct in primary macrophages we performed western analysis to define the kinetics and transcriptional class of v-RCA expression in macrophages. v-RCA was undetectable in MHV-68 infected primary macrophages at 24 hours post infection, but accumulated by 48 hours post infection (Fig. 2A). As expected, mutation of orf4 eliminated v-RCA expression (Fig. 2A). When primary MHV-68 infected macrophages were treated with cidofovir, v-RCA expression was no longer detected at 48 hours post infection as compared to untreated controls (Fig. 2B, compare first two lanes). In addition to its effect on v-RCA expression, cidofovir treatment led to a reduction in expression levels of several MHV68 proteins detected by the polyclonal anti-MHV68 antibody [Fig. 2B, (Dal Canto, Virgin et al., 2000)]. Thus, similar to fibroblasts, v-RCA was expressed with late kinetics in MHV68-infected primary macrophages.
Figure 2. MHV68 v-RCA is expressed with late kinetics and is required for viral DNA synthesis in infected macrophages.
A, B. Primary macrophages were infected at 10 PFU/cell with indicated viruses, cellular lysates collected at indicated times post infection and analyzed by Western. Some of infected cells in B were treated with 1 microgram/ml of cidofovir for the duration of infection. C. Total RNA was harvested from primary macrophages infected at 5 PFU/cell for 17h and treated with DNase. Levels of indicated viral messages and host GAPDH were measured by qRT-PCR. Viral messages were normalized to corresponding GAPDH levels using delta-delta Ct method. Data was pooled from 2-4 independent experiments for each viral message. The table indicates kinetic class of expression and known functions of proteins encoded by measured viral transcripts. D. Primary macrophages were infected at 5 PFU/cell and total DNA harvested from identical number of infected cells at indicated time points. Levels of MHV68 DNA (EcoRI H fragment) were measured by Southern using viral genome as a probe. Data was pooled from at least three independent experiments.
MHV68 v-RCA is required for viral DNA synthesis in infected macrophages
Host and viral RCA proteins interact with heparan sulfate proteoglycans with a proposed effect on virus binding (Mark, Lee, Spiller, Villoutreix, & Blom, 2006;Mullick, Singh, Panse, Yadav, Bernet, & Sahu, 2005;Murthy, Smith, Ganesh, Judge, Mullin, Barlow, Ogata, & Kotwal, 2001;Blackmore, Hellwage, Sadlon, Higgs, Zipfel, Ward, & Gordon, 1998;Prodinger, Hellwage, Spruth, Dierich, & Zipfel, 1998;Blom, Kask, & Dahlback, 2001). Furthermore, human herpesvirus-6 and EBV utilize host RCA proteins or complement receptors for cell entry (Santoro, Kennedy et al., 1999;Fingeroth, Clabby et al., 1988). MHV68 v-RCA protein is an envelope component of MHV68 virions (Kapadia, Molina, van, V, Speck, & Virgin, 1999), raising the possibility that attenuation of v-RCA MHV68 mutants in primary macrophages (Fig. 1 A, C) could be due to defects early in infection. To address whether v-RCA is required early in MHV68 infection, levels of several viral transcripts were measured in primary macrophages infected with v-RCA deficient or wild type MHV68. Measured viral transcripts encode proteins critical for MHV68 DNA synthesis, lytic replication, or cell type-specific replication in macrophages (table, Fig. 2C). The kinetic class of examined MHV-68 transcripts had been defined (Martinez-Guzman, Rickabaugh et al., 2003).
Total RNA isolated from primary macrophages infected with wild type MHV68 or RCAstop mutant for 17 hours was subjected to qRT-PCR analysis. All transcripts examined, including transcripts from the immediate early and early transcriptional classes, were expressed comparably between RCAstop- and wild type-infected cells (Fig. 2C). Similar results were observed with the orf4 null mutant (data not shown). Comparable levels of immediate early and early viral transcripts in cells infected with control and v-RCA mutant viruses indicates that v-RCA does not play a significant role in cell entry, transport of the viral genome to the nucleus, or the initial stages of MHV68 gene expression.
We next considered the hypothesis that v-RCA is important for viral DNA synthesis. Total DNA was harvested from primary macrophages infected with STM BAC or orf4 null MHV68 mutant at several times post infection and subjected to Southern analysis using viral genome as a probe. Signal from the EcoRI H fragment (Efstathiou, Ho et al., 1990b) was measured over multiple experiments and the results pooled for analysis (Fig. 2D). Levels of MHV68 DNA were similar at 16 and 25 hours post infection in macrophages infected with STM BAC or orf4 null viruses (Fig. 2D, p>0.05). These results are consistent with v-RCA-independent expression of early viral genes (17 hours post infection, Fig. 2C), suggesting that v-RCA is dispensable for early events in MHV68 infection of primary macrophages. However, MHV68 DNA accumulation was significantly reduced at 48 hours post infection in macrophages infected with orf4 null MHV68 mutant as compared to the control STM BAC virus (Fig. 2D, p<0.01), indicating that vRCA is required for efficient MHV68 DNA synthesis in primary macrophages.
Complement component expression is not required for v-RCA-dependent MHV68 replication in primary macrophages
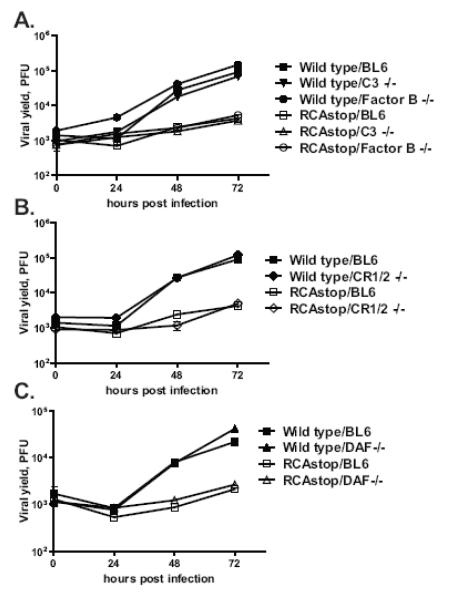
MHV68 v-RCA protein inhibits complement activation at the level or upstream of C3 in the complement cascade (Kapadia, Molina, van, V, Speck, & Virgin, 1999). Further, macrophages can express most if not all of the proteins in the complement system, including C3, in addition to multiple complement regulatory proteins (Morgan & Gasque, 1997). Therefore, we tested the hypothesis that v-RCA is required to counter an antiviral effect of C3 expression, or complement activation, in primary macrophage cultures. Macrophages were derived from mice deficient in C3 or in Factor B, a critical component of an alternative complement pathway, and infected with wild type MHV68 or the RCAstop virus mutant (Matsumoto, Fukuda et al., 1997;Circolo, Garnier et al., 1999). We examined factor B as a likely candidate for C3 activation in primary macrophage cultures since these cultures were not expected to contain antibodies capable of activation of the classical pathway of complement activation. Deletion of either Factor B or C3 failed to rescue the attenuated phenotype of MHV68 v-RCA mutant viruses (Fig. 3A). Identical results were obtained with the orf4 null mutant (data not shown). These results indicate that v-RCA has a C3 and Factor B independent function in MHV68 replication in primary macrophages.
Figure 3. Complement system is dispensable for gene 4-dependent MHV68 replication in primary macrophages.
Primary macrophages were isolated from mice with genetic deficiencies in components of complement pathway or RCA proteins. Macrophages were infected with 5PFU/cell of wild type MHC68 or RCAstop mutant and virus yield was determined at indicated times post infection.
Viral RCA, including MHV68 orf4, share homology with host RCA proteins, such as decay accelerating factor (DAF), complement receptors 1 and 2 (CR1, CR2), and membrane cofactor protein (MCP) (Kapadia, Molina, van, V, Speck, & Virgin, 1999). Each of the host RCA has a distinct function in the control of complement activity [reviewed in (Kim & Song, 2006)]. DAF prevents formation and accelerates decay of preexisting C3 and C5 convertases. MCP controls C3 activation by facilitating C3b cleavage. CR1 controls complement activity by combining functions of DAF and MCP. There are notable differences in the expression and genetic organization of RCA in human and mouse. The DAF gene is duplicated in mice (daf-1, daf-2) with ubiquitous expression of daf-1 and testis-specific expression of daf-2 (Kim & Song, 2006). CR1 and 2 are expressed from a single mouse gene through alternative splicing. MCP is not expressed by somatic cells of the mouse, and complement activation is instead controlled by a functional homologue Complement-receptor 1-related gene/protein Y (Crry) (Foley, Li et al., 1993). Unlike DAF and CR1/2, Crry is critical for embryonic survival (Xu, Mao et al., 2000).
We considered the hypothesis that the host RCA proteins might be involved in control of the replication of v-RCA mutant viruses. To examine whether host RCA proteins play a role in the attenuation of v-RCA deficient MHV68 mutants, macrophages were isolated from mice deficient in CR1/2 or DAF (Molina, Holers et al., 1996;Sun, Funk et al., 1999). The absence of host CR1/2 or DAF failed to rescue the defect in replication of RCAstop MHV68 mutant (Fig. 3B, C), suggesting that host RCA are dispensable for v-RCA function in MHV68 replication. Therefore, v-RCA requirement for efficient MHV68 replication in primary macrophages is independent of CR1/CR2 and or host RCA proteins.
This study identifies a macrophage-specific role for a gammaherpesvirus RCA protein in viral replication. Importantly, the connection between MHV68 complement regulatory protein and MHV68 replication was established in a primary, physiologically relevant cell type. While the absence of v-RCA did not affect early phases of MHV68 infection and did not selectively alter viral release, accumulation of MHV68 DNA was significantly impaired in cells infected with v-RCA deficient MHV68 mutants. Importantly, the requirement for v-RCA was independent of C3 and factor B and independent of the known murine RCA proteins DAF and CR1/2. Together these data suggest that v-RCA is multifunctional, playing a role in countering the activities of the classical and alternative C3 convertases under some circumstances, while playing a role in viral DNA synthesis in macrophages. In this respect, it appears that MHV68 has evolved to combine complement-dependent and complement-independent roles for proteins containing SCR domains in a single gene product. MHV68 vRCA is homologous to RCA encoded by herpesviruses and poxviruses. In the future studies it will be important to identify the mechanism of complement-independent function of MHV68 v-RCA, as it may be shared with other v-RCA proteins.
MATERIALS AND METHODS
Animals
C57BL/6J (BL6) mice were obtained from Jackson Laboratories (Bar Harbor, Maine) and bred at Washington University, St. Louis, MO. C3, Factor B, and CR1/2 deficient mice were provided by Dr. Diamond. DAF deficient mice were provided by Dr. Atkinson. All mice were housed in a specific-pathogen-free barrier facility at Washington University in accordance with federal and institutional guidelines. Bone marrow was harvested from mice between 7 and 10 weeks of age.
Virus infections and primary bone marrow macrophage derivation
Primary bone marrow macrophages were generated and infected as previously described (Tarakanova, Leung-Pineda, Hwang, Yang, Matatall, Basson, Sun, Piwnica-Worms, Sleckman, & Virgin, 2007). All samples were titered on NIH 3T12 fibroblasts.
Western blot analysis
Macrophages or fibroblasts were collected into laemmli buffer and analyzed as previously described (Lenschow, Giannakopoulos et al., 2005). Antibodies used were anti-gene 4 (Kapadia, Molina, van, V, Speck, & Virgin, 1999) (1:1000 dilution), anti-β actin (1:3000 dilution, clone AC-74 Sigma, Saint Louis, MO), and a secondary goat anti-mouse or anti-rabbit HRP-conjugated secondary antibody (Jackson Immunoresearch).
Immunofluorescence assay
Immunoflourescence assay was performed as previously described (Tarakanova, Leung-Pineda, Hwang, Yang, Matatall, Basson, Sun, Piwnica-Worms, Sleckman, & Virgin, 2007). Anti-gene 4 antibody was used at a 1:200 dilution.
Southern analysis
Total DNA was harvested from infected macrophages and 20 micrograms digested overnight with EcoRI. Digested DNA was electrophoresed through a 0.8% agarose gel in the presence of the Tris-Acetate-EDTA (TAE) buffer and denatured as follows: 30 minutes rinse in 0.2N hydrochloric acid, followed by two 30 min rinses in a Base solution (1.5M sodium chloride, 0.5M sodium hydroxide). DNA was transferred to a Nylon membrane (Hybond-N+, GE Healthcare, Piscataway, NJ) overnight using a standard capillary transfer approach in the presence of a Transfer buffer (3M sodium chloride, 8mM sodium hydroxide). Membrane with DNA was crosslinked by UV irradiation and blocked in the hybridization buffer (5X SSC, 1X Denhardt’s solution, 10% dextran sulfate, 1% SDS, 50% formamide, 0.1mg/ml sheared salmon sperm DNA) for 1-2 hours at 42 C. Megaprime DNA labeling system (GE Healthcare, Piscataway, NJ) was used to prepare a 32P-labeled probe with BAC MHV68 DNA as a template. Probe in fresh hybridization buffer was added to the blocked membrane and hybridization was conducted at 42C overnight. After a quick rinse in the wash buffer (0.1% SDS, 2X SSC) to remove most of radioactivity, membrane was washed at 65C twice, 15 min each wash. Signal was detected and quantified using Storm phosphorimager (GE Healthcare, Piscataway, NJ).
qRT-PCR quantitation of viral messages
Total RNA was harvested from infected cells using Trizol according to the manufacturers instructions (Invitrogen, Carlsbad, CA). Three micrograms of RNA were treated with RNase free DNAse (Ambion, Austin, TX) and split into two reactions. First half was subjected to reverse transcription using SSII Superscript (Invitrogen, Carlsbad, CA) and oligo-dT primers. Second half of DNase treated RNA was subjected to mock reverse transcription in the absence of the enzyme (-RT control). cDNA was serially diluted (8-fold), including –RT controls and measured, in triplicate for each dilution, by real time PCR using iCycler (Bio-Rad, Hercules, CA). Gene specific cDNA was amplified using probes in Table 1. Delta-delta Ct method was used to quantify relative abundance of each cDNA. Levels of viral messages were normalized to the corresponding GAPDH levels. Ct values of -RT controls did not exceed background levels
Table 1.
qRT-PCR primers.
| Viral gene |
Forward primer | Reverse primer |
|---|---|---|
| Gene 6 | GTTGCCAGATATCCCTAGGATGA | ACCTGGCTGGGTCAAGAGACT |
| Orf 59 | TGACTGGCAGGTTTTTGTATGC | GATGATCGTGAGGCCAATGG |
| Orf 40 | CTGCATTTTATCTCCACACAAAGC | TCAGTTGTAGTATGTCACACAGTTCCA |
| Orf 9 | TGCATGCAAGTTTGTCCAGTCT | CTTCCCCCAGTTACTCATTGTTTG |
| Orf 50 | AGAAACCCACAGCTCGCACTT | CAATATGCTGGACAGGCGTATC |
| Orf 36 | AGCAGCAACATCCACATTCC | AACTGCTGCCGTACCAAACA |
| Orf 56 | TGTCTGACTCATCAACACAGAACAA | GCATTGACTCATAAGTGTTGCAGTT |
| GAPDH | TGCCCCCATGTTTGTGATG | TGTGGTCATGAGCCCTTCC |
Acknowledgement
We would like to thank Darren Kreamalmayer for his outstanding expertise in animal breeding and members of the Virgin laboratory for helpful discussions. We are grateful to James Brien, Michael Diamond, Xiaobo Wu, and John Atkinson for help with the complement deficient mice. H.W.V. was supported by National Institutes of Health (NIH). V.L.T. is a Leukemia and Lymphoma society fellow. M.G. is supported by a National Science Foundation Graduate Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Blackmore TK, Hellwage J, Sadlon TA, Higgs N, Zipfel PF, Ward HM, Gordon DL. Identification of the second heparin-binding domain in human complement factor H. Journal of Immunology. 1998;160:3342–3348. [PubMed] [Google Scholar]
- Blom AM, Kask L, Dahlback B. Structural requirements for the complement regulatory activities of C4BP. Journal of Biological Chemistry. 2001;276:27136–27144. doi: 10.1074/jbc.M102445200. [DOI] [PubMed] [Google Scholar]
- Circolo A, Garnier G, Fukuda W, Wang XF, Hidvegi T, Szalai AJ, Briles DE, Volanakis JE, Wetsel RA, Colten HR. Genetic disruption of the murine complement C3 promoter region generates deficient mice with extrahepatic expression of C3 mRNA. Immunopharmacology. 1999;42:135–149. doi: 10.1016/s0162-3109(99)00021-1. [DOI] [PubMed] [Google Scholar]
- Cummings KL, Waggoner SN, Tacke R, Hahn YS. Role of complement in immune regulation and its exploitation by virus. Viral Immunology. 2007;20:505–524. doi: 10.1089/vim.2007.0061. [DOI] [PubMed] [Google Scholar]
- Dal Canto AJ, Virgin HW, Speck SH. Ongoing viral replication is required for gammaherpesvirus 68-induced vascular damage. Journal of Virology. 2000;74:11304–11310. doi: 10.1128/jvi.74.23.11304-11310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou S, Ho YM, Hall S, Styles CJ, Scott SD, Gompels UA. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. Journal of General Virology. 1990a;71:1365–1372. doi: 10.1099/0022-1317-71-6-1365. [DOI] [PubMed] [Google Scholar]
- Efstathiou S, Ho YM, Minson AC. Cloning and molecular characterization of the murine herpesvirus 68 genome. Journal of General Virology. 1990b;71:1355–1364. doi: 10.1099/0022-1317-71-6-1355. [DOI] [PubMed] [Google Scholar]
- Fingeroth JD, Clabby ML, Strominger JD. Characterization of A Lymphocyte-T Epstein-Barr Virus C3D Receptor (Cd21) Journal of Virology. 1988;62:1442–1447. doi: 10.1128/jvi.62.4.1442-1447.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flano E, Husain SM, Sample JT, Woodland DL, Blackman MA. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. Journal of Immunology. 2000;165:1074–1081. doi: 10.4049/jimmunol.165.2.1074. [DOI] [PubMed] [Google Scholar]
- Foley S, Li B, Dehoff M, Molina H, Holers VM. Mouse Crry/p65 is a regulator of the alternative pathway of complement activation. Eur.J.Immunol. 1993;23:1381–1384. doi: 10.1002/eji.1830230630. [DOI] [PubMed] [Google Scholar]
- Herrera E, del Mar LM, Blasco R, Isaacs SN. Functional analysis of vaccinia virus B5R protein: essential role in virus envelopment is independent of a large portion of the extracellular domain. Journal of Virology. 1998;72:294–302. doi: 10.1128/jvi.72.1.294-302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz J, Jacoby MA, Speck SH. The murine gammaherpesvirus 68 M2 gene is required for efficient reactivation from latently infected B cells. J Virol. 2005;79:2261–2273. doi: 10.1128/JVI.79.4.2261-2273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz JH, Siegel AM, Jacoby MA, Speck SH. Systematic mutagenesis of the murine gammaherpesvirus 68 M2 protein identifies domains important for chronic infection. Journal of Virology. 2008;82:3295–3310. doi: 10.1128/JVI.02234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia SB, Levine B, Speck SH, Virgin HW. Critical role of complement and viral evasion of complement in acute, persistent, and latent gamma-herpesvirus infection. Immunity. 2002;17:143–155. doi: 10.1016/s1074-7613(02)00369-2. [DOI] [PubMed] [Google Scholar]
- Kapadia SB, Molina H, van B, V, Speck SH, Virgin HW. Murine gammaherpesvirus 68 encodes a functional regulator of complement activation. J.Virol. 1999;73:7658–7670. doi: 10.1128/jvi.73.9.7658-7670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DD, Song WC. Membrane complement regulatory proteins. Clinical Immunology. 2006;118:127–136. doi: 10.1016/j.clim.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Krug LT, Moser JM, Dickerson SM, Speck SH. Inhibition of NF-kB Activation In Vivo Impairs Establishment of Gammaherpesvirus Latency. PLoS Pathog. 2007;3:97–118. doi: 10.1371/journal.ppat.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow DJ, Giannakopoulos NV, Gunn LJ, Johnston C, O’Guin AK, Schmidt RE, Levine B, Virgin HW. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J Virol. 2005;79:13974–13983. doi: 10.1128/JVI.79.22.13974-13983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark L, Lee WH, Spiller OB, Villoutreix BO, Blom AM. The Kaposi’s sarcoma-associated herpesvirus complement control protein (KCP) binds to heparin and cell surfaces via positively charged amino acids in CCP1-2. Molecular Immunology. 2006;43:1665–1675. doi: 10.1016/j.molimm.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Marques S, Efstathiou S, Smith KG, Haury M, Simas JP. Selective gene expression of latent murine gammaherpesvirus 68 in B lymphocytes. Journal of Virology. 2003;77:7308–7318. doi: 10.1128/JVI.77.13.7308-7318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Guzman D, Rickabaugh T, Wu TT, Brown H, Cole S, Song MJ, Tong L, Sun R. Transcription program of murine gammaherpesvirus 68. Journal of Virology. 2003;77:10488–10503. doi: 10.1128/JVI.77.19.10488-10503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew EC, Sanderson CM, Hollinshead R, Smith GL. A mutational analysis of the vaccinia virus B5R protein. Journal of General Virology. 2001;82:1199–1213. doi: 10.1099/0022-1317-82-5-1199. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Fukuda W, Circolo A, Goellner J, Strauss-Schoenberger J, Wang X, Fujita S, Hidvegi T, Chaplin DD, Colten HR. Abrogation of the alternative complement pathway by targeted deletion of murine factor B. Proceeding of the National Academy of Science. 1997;94:8720–8725. doi: 10.1073/pnas.94.16.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina H, Holers VM, Li B, Fung Y, Mariathasan S, Goellner J, Strauss-Schoenberger J, Karr RW, Chaplin DD. Markedly impaired humoral immune response in mice deficient in complement receptors 1 and 2. Proceeding of the National Academy of Science. 1996;93:3357–3361. doi: 10.1073/pnas.93.8.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BP, Gasque P. Extrahepatic complement biosynthesis: Where, when and why? Clinical and Experimental Immunology. 1997;107:1–7. doi: 10.1046/j.1365-2249.1997.d01-890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullick J, Singh AK, Panse Y, Yadav V, Bernet J, Sahu A. Identification of functional domains in Kaposica, the complement control protein homolog of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) Journal of Virology. 2005;79:5850–5856. doi: 10.1128/JVI.79.9.5850-5856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy KHM, Smith SA, Ganesh VK, Judge KW, Mullin N, Barlow PN, Ogata CM, Kotwal GJ. Crystal structure of a complement control protein that regulates both pathways of complement activation and binds heparan sulfate proteoglycans. Cell. 2001;104:301–311. doi: 10.1016/s0092-8674(01)00214-8. [DOI] [PubMed] [Google Scholar]
- Prodinger WM, Hellwage J, Spruth M, Dierich MP, Zipfel PF. The C-terminus of factor H: monoclonal antibodies inhibit heparin binding and identify epitopes common to factor H and factor H-related proteins. Biochemical Journal. 1998;331:41–47. doi: 10.1042/bj3310041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodger G, Smith GL. Replacing the SCR domains of vaccinia virus protein B5R with EGFP causes a reduction in plaque size and actin tail formation but enveloped virions are still transported to the cell surface. Journal of General Virology. 2002;83:323–332. doi: 10.1099/0022-1317-83-2-323. [DOI] [PubMed] [Google Scholar]
- Santoro F, Kennedy PE, Locatelli G, Malnati MS, Berger EA, Lusso P. CD46 is a cellular receptor for human herpesvirus 6. Cell. 1999;99:817–827. doi: 10.1016/s0092-8674(00)81678-5. [DOI] [PubMed] [Google Scholar]
- Siegel AM, Herskowitz JH, Speck SH. The MHV68 M2 protein drives IL-10 dependent B cell proliferation and differentiation. Plos Pathogens. 2008;4 doi: 10.1371/journal.ppat.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MJ, Hwang SM, Wong WH, Wu TT, Lee SM, Liao HI, Sun R. Identification of viral genes essential for replication of murine gamma-herpesvirus 68 using signature-tagged mutagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3805–3810. doi: 10.1073/pnas.0404521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed A, Buch T, Waisman A, Virgin HW. Interferon gamma blocks {gamma}-herpesvirus reactivation from latency in a cell type specific manner. Journal of Virology. 2007;81:6134–6140. doi: 10.1128/JVI.00108-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez AL, Van Dyk LF. Endothelial cells support persistent gammaherpesvirus 68 infection. Plos Pathogens. 2008;4 doi: 10.1371/journal.ppat.1000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Funk CD, Deng C, Sahu A, Lambris JD, Song WC. Role of decay-accelerating factor in regulating complement activation on the erythrocyte surface as revealed by gene targeting. Proceeding of the National Academy of Science. 1999;96:628–633. doi: 10.1073/pnas.96.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunil- Chandra NP, Efstathiou S, Nash AA. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. Journal of General Virology. 1992;73:3275–3279. doi: 10.1099/0022-1317-73-12-3275. [DOI] [PubMed] [Google Scholar]
- Tarakanova VL, Leung-Pineda V, Hwang S, Yang C-W, Matatall K, Basson M, Sun R, Piwnica-Worms H, Sleckman BP, Virgin HW. Gamma-Herpesvirus Kinase Actively Initiates a DNA Damage Response by Inducing Phosphorylation of H2AX to Foster Viral Replication. Cell Host and Microbe. 2007;1:275–286. doi: 10.1016/j.chom.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyk LF, Virgin HW, Speck SH. Maintenance of gammaherpesvirus latency requires viral cyclin in the absence of B lymphocytes. Journal of Virology. 2003;77:5118–5126. doi: 10.1128/JVI.77.9.5118-5126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW. In vivo veritas: pathogenesis of infection as it actually happens. Nat.Immunol. 2007;8:1143–1147. doi: 10.1038/ni1529. [DOI] [PubMed] [Google Scholar]
- Virgin HW, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. Complete sequence and genomic analysis of murine gammaherpesvirus 68. Journal of Virology. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weck KE, Kim SS, Virgin HW, Speck SH. B cells regulate murine gammaherpesvirus 68 latency. Journal of Virology. 1999a;73:4651–4661. doi: 10.1128/jvi.73.6.4651-4661.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weck KE, Kim SS, Virgin HW, Speck SH. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. Journal of Virology. 1999b;73:3273–3283. doi: 10.1128/jvi.73.4.3273-3283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer DO, Speck SH. Long-term latent murine Gammaherpesvirus 68 infection is preferentially found within the surface immunoglobulin D-negative subset of splenic B cells in vivo. Journal of Virology. 2003;77:8310–8321. doi: 10.1128/JVI.77.15.8310-8321.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Mao D, Holers VM, Palanca B, Cheng AM, Molina H. A critical role for murine complement regulator crry in fetomaternal tolerance [In Process Citation] Science. 2000;287:498–501. doi: 10.1126/science.287.5452.498. [DOI] [PubMed] [Google Scholar]