Abstract
Studies have shown the fundamental contribution of the yeast vacuole as a site for storage and detoxification of metals. Whereas the transmembrane proteins responsible for iron transport into and out of the vacuole have been identified in Saccharomyces cerevisiae, less information is available concerning the mobilization of vacuolar iron stores in Schizosaccharomyces pombe. In this study, we report the identification of a gene designated abc3+ that encodes a protein which exhibits sequence homology with the ABCC subfamily of ATP-binding cassette transporters. The transcription of abc3+ is induced by low concentrations of iron but repressed by high levels of iron. The iron-mediated repression of abc3+ required a functional fep1+ gene. Chromatin immunoprecipitation assays showed that Fep1 associates with the abc3+ promoter in vivo, in an iron-dependent manner. Microscopic analyses revealed that a functional Abc3-green fluorescent protein localizes to the membrane vacuole when iron levels were low. Abc3 was required for growth in low-iron medium in the absence of the transport system mediated by Fio1 and Fip1. abc3Δ cells exhibited increased levels of expression of the frp1+-encoded ferric reductase, suggesting a loss of Fep1 repression and, consequently, the activation of Fep1-regulated genes. When abc3+ was expressed using the nmt1+ promoter system, its induction led to a reduced transcriptional activity of the frp1+ gene. Because S. pombe does not possess vacuolar membrane-localized orthologs to S. cerevisiae Fth1, Fet5, and Smf3, our findings suggested that Abc3 may be responsible for mobilizing stored iron from the vacuole to the cytosol in response to iron deficiency.
All eukaryotes require iron for survival. The ability of this transition metal to exist in two different redox states makes it an essential component of the active centers of many enzymes and electron transporters (23). For instance, DNA synthesis, cell cycle progression, and energy-generating respiratory chain require iron. Paradoxically, the properties that make iron essential in these reactions could also make it toxic under certain conditions. Excess iron has the ability to unleash toxic oxygen radicals that can damage cellular components (18). Consequently, organisms must tightly regulate their internal iron load and must be able to respond to changes in iron levels by appropriately controlling iron acquisition, utilization, and compartmentalization in order to maintain homeostasis.
Studies in Saccharomyces cerevisiae have revealed that it possesses two genetically distinct systems for iron uptake (43). One of these systems requires that ferric iron [Fe3+] chelates be reduced to ferrous iron [Fe2+]. This task is performed by the cell surface reductases Fre1 and Fre2 that reduce extracellular Fe3+ chelates (8, 14). The Fe2+ generated by this action is, in turn, captured by an oxidase-permease complex formed by Fet3 and Ftr1 (4, 55, 57). Fet3 acts as a multicopper oxidase, converting Fe2+ to Fe3+, which is transported across the plasma membrane by the permease Ftr1. A clear interdependence between Fet3 and Ftr1 has been established since the trafficking of either protein to the cell surface requires the concomitant trafficking of the other. A Fet3 oxidase homolog called Fet5 and a Ftr1 permease homolog termed Fth1 are known to be present in S. cerevisiae (56, 60). These two proteins physically interact and form an oxidase-permease complex in the vacuole membrane (60). The permease Fth1 is dependent on the presence of Fet5 for exiting the secretory pathway, suggesting that both proteins are required during biosynthesis for their correct targeting to the vacuole membrane (60). The Fet5-Fth1 complex most likely mobilizes stored iron from the vacuole to the cytosol when cells undergo a transition from iron excess to iron-limiting conditions (60). S. cerevisiae possesses a second system for iron uptake in which siderophore-iron chelates are taken up by the ARN1-ARN4 transporters (42).
In Schizosaccharomyces pombe, studies have shown that Fe3+ is reduced to Fe2+ by the cell surface reductase Frp1 (49). Once reduced, Fe2+ is taken up by an oxidase-permease complex consisting of Fio1 and Fip1, which are the orthologs of Fet3 and Ftr1, respectively (5). Despite the fact that Fio1 and Fip1 share significant similarities to Fet3 and Ftr1, only the simultaneous expression of both S. pombe genes (fio1+ and fip1+) in S. cerevisiae fet3Δ cells is able to reconstitute high-affinity iron transport (5). The expression of S. pombe fio1+ alone in an S. cerevisiae fet3Δ disrupted strain does not result in complementation of the fet3Δ iron starvation defects (5). This observation suggests that, although Fio1 and Fet3 are homologous in function, Fio1 cannot assemble with endogenous S. cerevisiae Ftr1. This result further suggests that molecular differences may exist between the oxidase-permease complexes in these two fungal species. Orthologs of Fet5 and Fth1 have not yet been identified in S. pombe. BLAST searches for both Fet5- and Fth1-like proteins in the S. pombe genome database have revealed no S. pombe proteins with significant homologies. Although S. pombe vacuoles may serve as an important site for intracellular iron stores, transport of iron into and out of the vacuole has not been investigated in detail in fission yeast. The second pathway of iron uptake in S. pombe relies on the transport of iron-siderophore chelates, a process that primarily involves the hydroxamate-type siderophore ferrichrome (39, 52).
A critical issue for cells is the absolute requirement of being able to control iron concentrations in order to be able to rapidly respond to changes in extracellular iron levels. Exposure of S. pombe to elevated concentrations of iron is sensed by the GATA-type transcriptional repressor Fep1 (26, 38). An important response to Fep1 activation is the downregulation of the genes encoding the components of the high-affinity iron transport machinery, including frp1+, fio1+, and fip1+ (38, 50). Based on genomic profiling studies, it has been proposed that Fep1 and its orthologs are also required for the iron-regulated expression of the genes encoding the proteins involved in iron-sulfur cluster formation, in compartmentalization, and in the utilization of inorganic iron (22, 50). Once activated, Fep1 binds to the DNA sequences known as GATA elements [5′-(A/T)GATA(A/T)-3′], which are found in the promoters of its target genes. Conversely, when the iron concentration is limited inside the cell, Fep1 dissociates from the chromatin, thereby allowing transcription of its target genes to take place (21, 40).
Analysis of iron-regulated gene expression in S. pombe using DNA microarrays has identified several uncharacterized genes that are transcriptionally activated in response to iron deprivation (34). Although the roles of these genes remain unclear, their iron starvation-dependent induction suggests that they possess iron-related functions. One example of these genes is SPBC359.05, which is also called abc3+ (20). This gene encodes a putative transmembrane protein that exhibits sequence homology with the ABCC subfamily of ATP-binding cassette (ABC) transporters (9, 10, 15, 37). Members of the ABCC subfamily have a typical ABC “core” region, consisting of two homologous halves. Each half contains a membrane spanning domain (MSD) that includes six transmembrane spans and a nucleotide binding domain (NBD). The NBD harbors several conserved motifs designated Walker A, Walker B, and the signature motifs (15). The two halves of the core region are joined by a linker region (L1). A hallmark of the ABCC transporters is the presence of an additional N-terminal extension (NTE) that contains five putative transmembrane spans (MSD0) that are connected to the ABC core domain by a hydrophilic region (L0). A previous study has shown that S. pombe cells lacking a functional abc3+ gene are sensitive to cerulenin, an antibiotic that inhibits the biosynthesis of fatty acids (20). Using direct fluorescence microscopy, the expression of a green fluorescent protein (GFP)-tagged form of Abc3 transformed in wild-type S. pombe cells suggests that Abc3 may localize to the vacuole membrane, although neither its cellular localization nor its role within the cell has been firmly characterized (20). S. pombe is one of the yeast species that does not have any homologs of S. cerevisiae proteins Fet5, Fth1, and Smf3 (16). However, as shown for the maintenance of the concentration of intracellular copper (6), the fission yeast vacuole may contribute to the overall iron metabolism of the cell.
In the present study, we determined that abc3+ is regulated at the level of gene transcription, and its iron-dependent regulated expression requires a proximal GATA-type cis-acting element and a functional fep1+ gene. Using a chromatin immunoprecipitation (ChIP) approach, we show that Fep1 occupies the abc3+ promoter in the presence of high levels of iron, whereas iron deficiency results in a loss of Fep1 occupancy at the abc3+ promoter. Using an abc3+-GFP allele that retained wild-type function, we found that Abc3-GFP is localized to the vacuole membrane when iron levels are low but become undetectable upon exposure to iron. Cell fractionation experiments revealed that Abc3 is an integral membrane protein. The loss of Abc3 resulted in an elevated transcriptional activity of the frp1+ gene. In contrast, permanent expression of abc3+ lowered the steady-state levels of the frp1+ transcript. Taken together, these results strongly suggest that under iron-limiting conditions, Abc3 could serve to transport iron from the vacuole to the cytoplasm.
MATERIALS AND METHODS
Strains and media.
The S. pombe strains used in the present study were the wild-type FY435 (h+ his7-366 leu1-32 ura4-Δ18 ade6-M210) and eight isogenic mutant strains: abc3Δ (h+ his7-366 leu1-32 ura4-Δ18 ade6-M210 abc3Δ::KANr), pcl1Δ (h+ his7-366 leu1-32 ura4-Δ18 ade6-M210 pcl1Δ::KANr), fep1Δ (38), ctr6Δ (6), fep1Δ php4Δ (21), abc3Δ ctr6Δ (h+ his7-366 leu1-32 ura4-Δ18 ade6-M210 abc3Δ::KANr ctr6Δ::ura4+), fio1Δ fip1Δ (h+ his7-366 leu1-32 ura4-Δ18 ade6-M210 fio1-fip1Δ::KANr), and abc3Δ fio1Δ fip1Δ (h+ his7-366 leu1-32 ura4-Δ18 ade6-M210 abc3Δ::loxP fio1-fip1Δ::KANr). All S. pombe cells were cultured in either yeast extract plus supplements (YES) or selective Edinburgh minimal medium (EMM) lacking the specific amino acids required for integrative or nonintegrative plasmid selection (1). Unsupplemented EMM contained 74 nM iron, unless otherwise stated. Liquid cultures were seeded to an A600 of 0.5, allowed to grow to exponential phase (A600 of ∼1.0), and then treated with 250 μM 2,2′-dipyridyl (Dip) or 100 μM FeCl3 for 90 min, unless otherwise stated. Yeast transformations and manipulations were carried out by using standard techniques (1).
Plasmids.
A BamHI-EcoRI PCR-amplified fragment derived from the abc3+ promoter containing 584 bp of the 5′-noncoding region and the first 10 codons of the abc3+ gene was introduced into the BamHI-EcoRI-digested YEp357R vector (35). The abc3+ promoter region was isolated from YEp357Rabc3+-584lacZ via digestion with BamHI and Bsu36I. It was then swapped for the equivalent DNA restriction fragment in pSP1fio1+-1155lacZ (38) in order to generate pSP1abc3+-584lacZ. Two plasmids (pSP1abc3+-239lacZ and pSP1abc3+-114lacZ) harboring sequential deletions from the 5′ end of the abc3+ promoter were created by PCR from plasmid pSP1abc3+-584lacZ. Each PCR product obtained was purified, digested with BamHI and Bsu36I, and then used to replace the equivalent DNA restriction fragment in pSP1abc3+-584lacZ. Plasmids pSP1abc3+-584lacZ and pSP1abc3+-239lacZ were used to introduce mutations to either or both of the GATA boxes (positions −115 to −120 and positions −197 to −202 with respect to the A of the ATG codon of abc3+) [CCTGTC instead of TGATA(A/T)] via the overlap-extension method (19). The DNA sequence of each mutant was confirmed by dideoxy sequencing. In order to create both the wild-type and the mutant pCF83abc3+-239/-56lacZ fusion plasmids, the abc3+ promoter region (positions −239 to −56) was PCR amplified from both the wild-type and the mutant pSP1abc3+-584lacZ constructs. These PCR products were purified and inserted in their natural orientations into the XmaI and XhoI sites of the CYC1-lacZ fusion plasmid pCF83 (33).
PCR amplification of the abc3+ gene was carried out with primers designed to generate SpeI and XhoI restriction sites at the upstream and downstream termini of the ORF, respectively. The full-length gene was isolated from S. pombe strain FY435 genomic DNA. The PCR product was digested with SpeI and XhoI and cloned into the corresponding sites of the pBluescript SK vector (Stratagene, La Jolla, CA), creating plasmid pSKabc3+. Subsequently, the S. pombe abc3+ promoter region from position −808 upstream of the start codon of the abc3+ gene was isolated by PCR amplification and was then inserted into pSKabc3+ at the SstI and SpeI sites. This pSKabc3+ derivative was named pSKprom-abc3+. The SstI-XhoI DNA fragment was isolated from pSKprom-abc3+ and then inserted into SstI-XhoI-digested pSP1 or pJK148 plasmid, creating plasmids pSP1abc3+ and pJK148abc3+, respectively. The GFP coding sequence derived from pSF-GP1 (24) was isolated by PCR, using primers designed to generate XhoI and ApaI sites at the 5′ and 3′ termini, respectively, of the GFP gene. The resulting DNA fragment was used to clone the GFP gene into the pSP1abc3+ plasmid to which XhoI and ApaI restriction sites had previously been introduced by PCR and were placed immediately before the abc3+ stop codon. For this particular construct, named pSP1abc3+-GFP, the XhoI-ApaI GFP-encoded fragment was placed in frame with the C-terminal region of Abc3. The nmt1+ 41X promoter (13) up to position −1178 from the initiator codon of the nmt1+ gene was isolated by PCR and then swapped to replace the SstI-SpeI abc3+ promoter fragment in either pSP1abc3+ or pSP1abc3+-GFP. The resulting plasmids were named pSP-1178nmt41X-abc3+ and pSP-1178nmt41X-abc3+-GFP, respectively.
RNA analysis.
Total RNA was isolated by using the hot phenol method (7). RNA samples were quantified spectrophotometrically, and 15 μg of RNA per sample was used for the RNase protection protocol (34). DNA templates for antisense riboprobes (Table 1) were cloned into the BamHI and EcoRI sites of pBluescript SK. The resultant constructs were linearized with BamHI for subsequent antisense RNA labeling with [α-32P]UTP and the T7 RNA polymerase. 32P-labeled antisense lacZ RNA was generated from the HindIII-linearized plasmid pKSlacZ (38). The riboprobe derived from pSKact1+ (41) was used to probe act1+ mRNA as an internal control for the normalization purposes during quantification of the RNase protection products.
TABLE 1.
Riboprobes used to detect steady-state levels of transcripts
| Gene ID | Gene | Riboprobe length (bp) | Positions from the initiator codon | Source or reference |
|---|---|---|---|---|
| SPAC9E9.12c | abc1+ | 174 | +205 to +379 | This study |
| SPAC3F10.11c | abc2+ | 184 | +301 to +485 | This study |
| SPBC359.05 | abc3+ | 179 | +351 to +530 | This study |
| SPAC30.04c | abc4+ | 189 | +112 to +301 | This study |
| SPBC32H8.12c | act1+ | 151 | +334 to +485 | 41 |
| SPAC1F7.08 | fio1+ | 183 | +16 to +199 | This study |
| SPBC1683.09c | frp1+ | 190 | +117 to +307 | This study |
| SPCC737.09c | hmt1+ | 193 | +584 to +777 | This study |
| lacZ | 233 | +1188 to +1421 | 38 |
ChIP.
Cell growth conditions and the preparation of chromatin were carried out as described previously (21). Immunoprecipitation of TAP-tagged Fep1 with immunoglobulin G (IgG)-Sepharose beads and the subsequent elution of the immunocomplexes were performed as described previously (21). To reverse the formaldehyde cross-links, both the eluted DNA and the DNA of the input control were first incubated at 65°C for 18 h, followed by 2 h at 37°C in the presence of 50 μg of proteinase K. Free DNA was then purified by using phenol-chloroform-isoamyl alcohol (25:24:1) extraction in the presence of 0.4 M LiCl. After centrifugation and precipitation of the DNA by the addition of glycogen and ethanol, the precipitated DNA was resuspended in 100 μl of Tris-EDTA prior to PCR analysis.
PCR amplifications were performed essentially as described by Komarnitsky et al. (25), except that PCR program consisted of 2 min at 94°C, followed by 25 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C, and a final 4-min step at 72°C. Radiolabeled PCR products were purified by using Quick Spin columns (Roche Diagnostics, Indianapolis, IN) and were resolved in 6% polyacrylamide-1× Tris-borate-EDTA gels. PCR signals were quantitated by PhosphorImager scanning and then normalized with respect to both the input DNA reaction and the internal intergenic control primer pair (in order to correct for PCR efficiency and background signals). All experiments were conducted at least three times, and each experiment yielded similar results. Primers that span the abc3+ promoter region that included a functional GATA box were used for PCR analysis. The primers were designated by the name of the gene promoter, followed by the position of their 5′ ends relative to the translation initiation codon: abc3-a−181, 5′-CATAATGAGATTCAGCGCAAACGTTATTAGTC-3′; abc3-b−16, 5′-CCAAATACTTTATTAAAAGCGATGCTTAAGTC-3′; Intergenic-cII3860000-a, 5′-CGGTGCGTTTTTCTACGCGCATCTTC-3′; and Intergenic-cII3860000-b, 5′-GCCCAAGGCCCATCAACAATCTAACATG-3′.
Cerulenin sensitivity assay, organelle staining, and fluorescence microscopy.
Cells were grown to an A600 of 1.0 (∼2 × 107 cells) at 30°C. Each cell culture was diluted (∼2 × 104 cells) and incubated to a final volume of 4 ml in EMM containing cerulenin (1 μg/ml; Sigma-Aldrich). At the designated time intervals, total growth was determined spectrophotometrically at A600. Vacuole membrane staining using FM4-64 (Sigma-Aldrich) was performed as described previously (20), except that the cells were not resuspended in distilled water prior to microscopic analysis. Briefly, the cells were harvested and resuspended in EMM containing 16 μM FM4-64 for 30 min at 30°C. The cells were pelleted, resuspended in fresh EMM, and incubated at 30°C for an additional 90 min. They were then placed on microscope slides and viewed on a Nikon Eclipse E800 epifluorescence microscope (Nikon, Melville, NY) equipped with a Hamamatsu ORCA-ER cooled charge-coupled device camera (Hamamatsu, Bridgewater, NJ). The samples were analyzed using ×1,000 magnification with the following filters: 465 to 495 nm (GFP) and 510 to 560 nm (FM4-64). The cell fields shown in the present study are representative of a minimum of five independent experiments.
Preparation of S. pombe extracts, Western blot analysis, and spectrophotometric method using BPS-citric acid.
Protein extracts were prepared from logarithmic-phase cells that were grown in standard EMM or taken after their incubation in the presence of either 50 μM Dip or 50 μM FeCl3. The harvested cells were washed in HEGN100 buffer (20 mM HEPES [pH 7.9], 1 mM EDTA, 10% glycerol, 100 mM NaCl) and lysed with glass beads using the same buffer supplemented with 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and a complete protease inhibitor mixture (P8340; Sigma-Aldrich, St. Louis, MO). The cells were broken by using a FastPrep FP120 instrument (Bio 101; Thermo Electron Corp., Milford, MA; twice for 45 s at 4°C, with cooling intervals of 1 min in an ice bath). The resulting lysates were centrifuged at 100,000 × g for 30 min at 4°C. The supernatant was kept at 4°C, whereas the pellet fraction was resuspended in 0.2 ml of buffer A (phosphate-buffered saline [pH 7.4], 1 mM EDTA, 150 mM NaCl, 1% Triton X-100, 1 mM dithiothreitol, and the above-mentioned protease inhibitors) and incubated on ice for 30 min. Alternatively, the pellet fraction was resuspended and left untreated or was adjusted to 0.1 M Na2CO3 (to replace 1% Triton X-100). After incubation on ice, the pellet fraction was resuspended and centrifuged at 100,000 × g for 30 min at 4°C. Both supernatant and pellet fraction were resuspended in 2X sodium dodecyl sulfate (SDS) loading buffer (50 mM Tris-HCl [pH 6.8], 0.1 mM EDTA, 15% SDS, 0.01% bromophenol BLUE, and 150 mM dithiothreitol) containing 8.0 M urea and 4% β-mercaptoethanol, unless otherwise indicated. After a 30-min incubation at 37°C, the samples were resolved by SDS-polyacrylamide gel electrophoresis and visualized by Coomassie brilliant blue staining, or they were used for Western blot analysis. Monoclonal anti-GFP (clone B-2; Santa Cruz Biotechnology, Santa Cruz, CA) and monoclonal anti-HA (clone F-7; Santa Cruz Biotechnology) antibodies were used for protein expression analysis of the Abc3-GFP and Ctr6-HA4 fusion proteins, respectively. A monoclonal anti-PCNA antibody (clone PC10; Sigma-Aldrich) was used to detect PCNA as an internal control for soluble proteins. For measuring intracellular iron, the BPS-based spectrophotometric assay was carried out as described previously (45).
RESULTS
Molecular architecture of S. pombe Abc3.
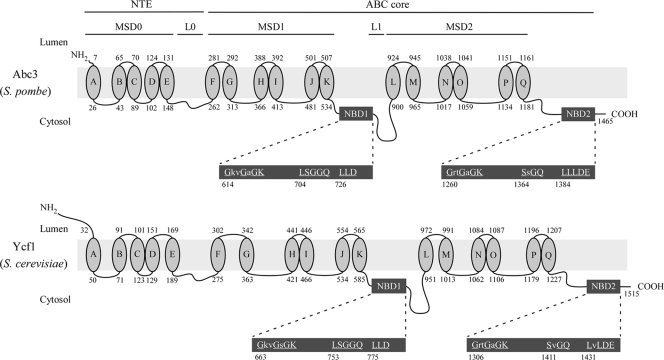
There are 11 genes that encode putative proteins related to the ABC family of transporters in S. pombe (20). Analysis of iron-regulated gene expression using DNA microarrays has identified abc3+ as one of the affected genes (34, 50). Because the transcript levels of abc3+ (SPBC359.05) showed increases of expression in response to iron deficiency (34), we hypothesized that the gene product (Abc3) could be part of a mechanism that was activated under conditions of iron deficiency. The hypothesis was tested by isolating the gene for further analysis. abc3+ encodes a polypeptide composed of 1,465 amino acid residues with a predicted molecular mass of 166.5 kDa. Abc3 is 42% sequence identical and 56% sequence similar to S. cerevisiae Ycf1, a prototypical member of the ABCC subfamily of ABC transporters (58). Similarly to Ycf1 and the other members of the ABCC subfamily, Abc3 possesses an ABC core domain that consists of two homologous halves, each half containing a membrane-spanning domain (MSD) with six putative transmembrane spans, and an NBD (Fig. 1). The two halves of Abc3 are connected by a putative cytosolic loop (L1). The Abc3 N-terminal portion strongly resembles the unique NTE that is a hallmark of proteins belonging to the subgroup ABCC (31). The Abc3 NTE is very hydrophobic, harboring five predicted transmembrane helices, in addition to a cytoplasmic linker region (L0). Three conserved motifs (614GkvGaGK620 [Walker A], 704LSGGQ708 [signature], and 726LLD728 [Walker B] for NBD1; 1260GrtGaGK1266 [Walker A], 1364SsGQ1367 [signature], and 1384LLLDE1388 [Walker B] for NBD2) are present within the NBD domains. These motifs may participate in the binding and hydrolysis of nucleotides such as ATP to energize the transport process. Sequence alignment analysis of the amino acid residues that compose the NBD1 and NBD2 of Abc3 and Ycf1 indicated that the spacing of the conserved motifs (Walker A, signature, and Walker B) in Abc3 was highly similar to that of Ycf1 (data not shown). Furthermore, Abc3 residues 614 to 620 (Walker A), 704 to 708 (signature), and 726 to 728 (Walker B) for NBD1, as well as residues 1260 to 1266 (Walker A), 1364 to 1367 (signature), and 1384 to 1388 (Walker B) for NBD2, were 90% identical to the Walker A, signature, and Walker B motifs found in Ycf1. Collectively, these observations strongly suggested that abc3+ encodes an ABC protein of the ABCC subfamily similar to Ycf1.
FIG. 1.
Structural features of Abc3 and its close homologue from S. cerevisiae Ycf1. Topological models of the S. pombe Abc3 (top) and S. cerevisiae Ycf1 (bottom) proteins are shown. The N-terminal extension (NTE) includes five membrane spans (denoted as MSD0; ovals A to E) and a cytosolic loop (L0). The ABC core domain consists of two homologous halves, each containing a transmembrane domain (denoted as MSD1 or MSD2) composed of six membrane spans (ovals F to K or ovals L to Q), and a nucleotide binding domain (denoted as NBD1 or NBD2). The halves are joined by a linker region (L1). NBD1 and NBD2 (enlarged dark gray boxes) contain residues that are found in other ABC transporters. Highly conserved residues are both underlined and in capital letters. The amino acid sequence numbers refer to the position relative to the first amino acid of each protein.
abc3+ transcript was induced under conditions of iron deficiency and negatively regulated by iron through Fep1.
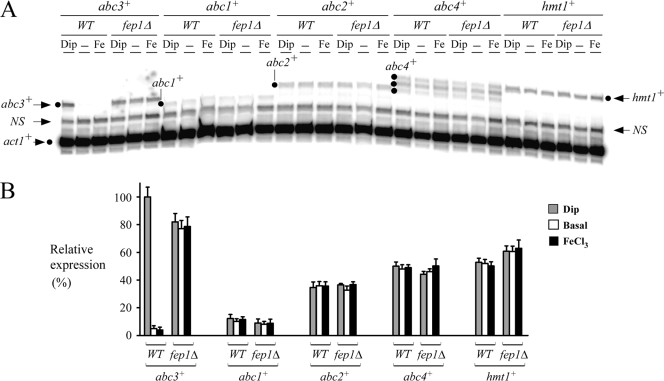
Our gene expression profiling data suggested that abc3+ gene expression was downregulated in the presence of iron (34). To independently verify the microarray data (34), S. pombe wild-type strain was left untreated or treated with Dip (250 μM) or FeCl3 (100 μM). In the presence of the iron chelator Dip, abc3+ mRNA levels were increased ∼17- to 20-fold compared to the low basal levels observed in the untreated cells (Fig. 2). In contrast, under iron-replete conditions, the transcript levels of abc3+ were repressed and remained approximately equal to those observed in the untreated cells (Fig. 2). The abc3+ increases in transcript levels observed under conditions of iron deficiency paralleled those observed by DNA microarray analysis (16.9-fold) (34). The results of the dependency of abc3+ gene expression on iron concentrations suggested that the iron-regulatory transcriptional repressor Fep1 could play a role in abc3+ gene regulation. This possibility was investigated by using an isogenic fep1Δ deletion strain of S. pombe. The results showed that the levels of abc3+ mRNA were constitutive and unresponsive to cellular iron status in the yeast mutant (Fig. 2), suggesting that the iron-mediated repression of abc3+ occurred through the activity of Fep1. Furthermore, these results revealed that wild-type abc3+ gene was regulated by iron in a manner identical to that of iron uptake genes such as frp1+, fio1+, and fip1+ (5, 27, 38).
FIG. 2.
abc3+ transcript levels are downregulated by iron in a Fep1-dependent manner. (A) Logarithmic-phase cultures of the isogenic strains FY435 (wild type [WT]) and BPY10 (fep1Δ) were left untreated (−) or were treated with Dip (250 μM) or FeCl3 (Fe) (100 μM) for 90 min. Total RNA was then prepared from each sample and analyzed by RNase protection assays. Steady-state levels of abc3+, abc1+, abc2+, abc4+, hmt1+, and act1+ mRNAs (indicated with dots and arrows) were analyzed in both the wild-type strain and a strain lacking the fep1+ allele. NS, nonspecific signal. (B) Graphic representation of the quantification of the results of three independent RNase protection assays, including the experiment shown in panel A. The values shown are the means of three replicates ± the standard deviations.
According to the ABC transporter classification system (10), S. pombe Abc3 is a member of the ABCC subfamily. In fission yeast, three other genes, namely, abc1+, abc2+, and abc4+, encode putative ABC transporters related to the ABCC subfamily (20). To examine whether they were differentially regulated as a function of iron availability, we investigated the profiles of expression of abc1+, abc2+, and abc4+ by RNase protection assays (Fig. 2). The hmt1+ gene (36) that encodes a member of the ABCB subfamily was also analyzed in RNase protection experiments. The results showed that the steady-state mRNA levels of abc1+, abc2+, abc4+, and hmt1+ in the wild-type strain were not regulated by iron depletion (Dip, 250 μM) or iron abundance (FeCl3, 100 μM) (Fig. 2). There were no significant changes in the levels of abc1+, abc2+, abc4+, and hmt1+ transcription in treated cells compared to the basal levels in untreated cells. To further examine whether abc1+, abc2+, abc4+, and hmt1+ transcription was controlled by Fep1, a fep1Δ null strain was grown in either the absence or the presence of either 250 μM Dip or 100 μM FeCl3. As shown in Fig. 2, the fep1Δ mutant had no significant effect on the expression of the above-mentioned genes. The bulk of these data led us to conclude that, among the five different ABC-encoded transporter genes tested, only abc3+ was regulated by iron through the activity of the transcription factor Fep1.
Analysis of abc3+ promoter sequences required to repress gene expression under basal and iron-replete conditions.
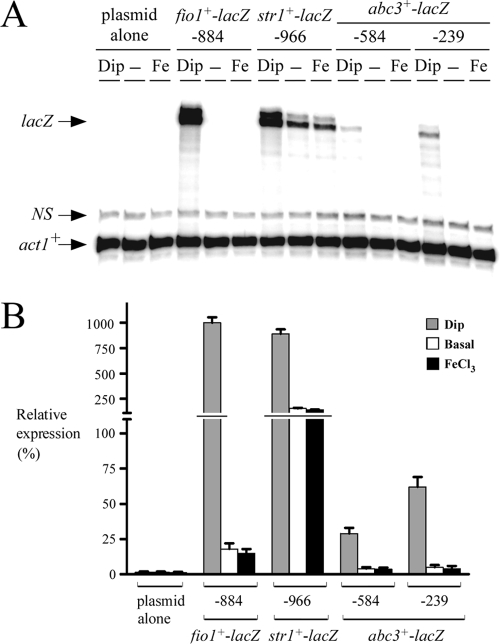
The fact that Fep1 was necessary for the repression of abc3+ transcription under basal and iron-replete conditions led us to investigate whether the abc3+ promoter region up to −584 from the start codon of the abc3+ ORF harbored GATA elements. Six copies of the repeated sequence 5′-(A/T)GATA(A/T)-3′ were identified at positions −115 to −120, −197 to −202, −391 to −396, −405 to −410, −418 to −423, and −511 to −516. To determine whether the GATA sequences played a role in the abc3+ regulation by iron, we further examined two regions of the abc3+ promoter that were fused upstream of and in-frame to the lacZ gene in pSP1-lacZ. The first region contained the promoter up to −584 from the initiator codon of the abc3+ gene, whereas the second region harbored a shorter promoter segment up to −239. lacZ mRNA expression from these two plasmids was analyzed by RNase protection assays. The results showed that both fusion promoters were able to repress lacZ mRNA expression in the presence of iron (Fig. 3). Plasmids pSP1abc3+-584lacZ and pSP1abc3+-239lacZ were repressed ∼7- and ∼12-fold, respectively, compared to their levels of expression in iron-starved cells. Unexpectedly, the overall magnitude of iron-regulated expression of the lacZ mRNA was slightly higher when the abc3+ promoter was further deleted to position −239 (Fig. 3). As expected, fio1+-884lacZ (38) and str1+-966lacZ (39) (assayed as controls) were derepressed (∼56- and ∼6-fold, respectively) after treatment with Dip (250 μM) (Fig. 3). In contrast, their expression was downregulated under basal or iron-replete conditions. Our data did not permit explanation of why the overall magnitude of the iron limitation-dependent activation of both fio1+-884lacZ and str1+-966lacZ gene expression was ∼1.5 orders of magnitude greater than that for the abc3+-lacZ fusion derivatives. However, the abc3+-lacZ fusion plasmids were undoubtedly regulated in response to iron levels (Fig. 3).
FIG. 3.
Iron responsiveness of the abc3+ promoter. (A) lacZ fusion genes harboring wild-type DNA fragments derived from the fio1+ (38), str1+ (39), and abc3+ promoters were analyzed by RNase protection assays. Total RNA was isolated from both control (untreated) cells (−) and cells treated with Dip (250 μM) or FeCl3 (100 μM). Plasmids pSP1fio1+-884lacZ and pSP1str1+-966lacZ were used (38, 39) as controls for iron-regulated gene expression. The lacZ and act1+ mRNA steady-state levels are indicated by arrows. NS, nonspecific signal. (B) Graphic representation of the quantification of the results of three independent RNase protection assays, including the experiment shown in panel A. Reporter gene activity values shown are the averages of triplicate determinations ± the standard deviations.
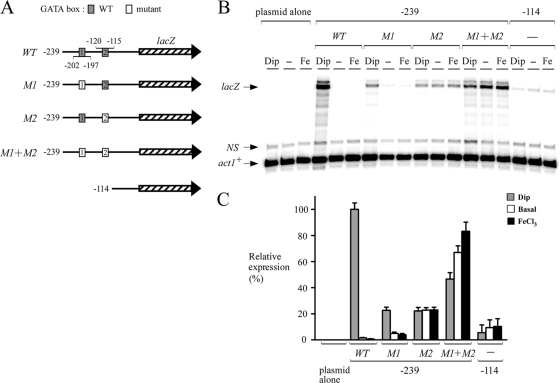
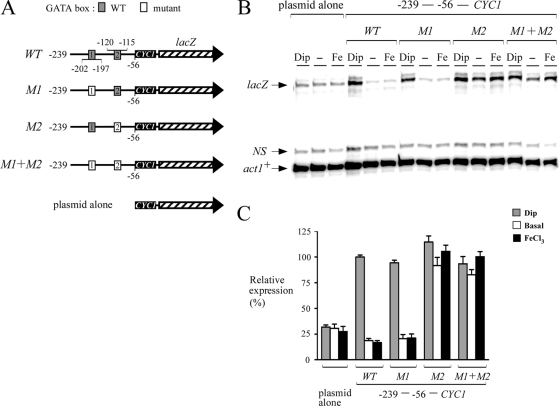
To gain further insight into the mechanism by which abc3+ expression was regulated by iron, and because of the observation that the promoter region between positions −239 and −1 was sufficient to drive the iron-dependent repression of the abc3+-lacZ gene, we examined whether the two putative GATA sequences present in this region (positions −115 to −120 and −197 to −202) could mediate gene expression as a function of iron availability. Multiple point mutations that mimic the changes known to abolish the binding of Fep1 to GATA boxes (38) were inserted in each one or both elements. Mutation of the base pairs within the−202TTATCA−197 element (GACGGC instead of TTATCA; denoted M1) had no effect on the iron-regulatable expression of the abc3+-239lacZ fusion (Fig. 4). However, the overall magnitude of the response decreased by ∼77% compared to cells containing the wild-type plasmid. When the second GATA element, −120TGATAT−115, was mutated (CCTGTC instead of TGATAT; denoted M2), a complete lack of iron responsiveness of the reporter gene was observed (Fig. 4). Surprisingly, when both (M1 + M2) GATA elements were mutated, the overall magnitude of the lacZ response was higher (Fig. 4). Although the results indicated that the steady-state levels of lacZ mRNA under both basal and iron-replete conditions were slightly increased by 1.4- and 1.8-fold, respectively, above the levels observed in cells treated with Dip, the reason for such expression profile is unknown (Fig. 4). When the abc3+ promoter was further deleted to position −114, lacZ transcript levels were very low, with no dramatic change as a function of iron availability (Fig. 4).
FIG. 4.
Analysis of the proximal abc3+ promoter sequences required to repress gene expression under both basal and iron-replete conditions. (A) Diagram representation of a 239-bp abc3+ promoter DNA fragment and its mutant derivatives assayed using the RNase protection protocol. The gray boxes indicate the wild-type GATA elements [5′-(T/A)GATA(T/A)-3′], whereas the white ones represent the mutant versions (5′-GCCGTC-3′). The hatched arrow represents the lacZ reporter gene. The nucleotide numbers refer to the positions of the GATA boxes relative to that of the abc3+ initiator codon. (B) Steady-state levels of lacZ mRNA from both the wild type (WT) and mutant GATA fusions (M1, M2, and M1+M2) were analyzed in the absence (−) or the presence of Dip (250 μM) or FeCl3 (Fe, 100 μM) for 90 min. The lacZ and act1+ (as controls) mRNA levels are indicated with arrows. NS, nonspecific signal. (C) Quantitation of the lacZ levels after the treatments shown in panel B. The values shown are the means of three replicates ± the standard deviations.
Based on the findings that the integrity of the second GATA element located between positions −120 and −115 was essential in triggering the iron repression of the abc3+-lacZ fusion, we examined whether this sequence could regulate a heterologous reporter gene in an iron-dependent manner. A short DNA segment derived from the abc3+ promoter (positions −239 to −56) was inserted in its natural orientation upstream of the minimal promoter of the CYC1 gene fused to lacZ in pCF83. The fact that the upstream region of lacZ in pCF83 contains the CYC1 minimal promoter may explain why low levels of lacZ transcript were detected from cells transformed with the plasmid alone. This promoter fusion was able to downregulate lacZ mRNA expression under both standard (untreated) and iron-replete conditions. In contrast, under conditions of iron deprivation, lacZ mRNA expression was induced (∼5- to 6-fold) compared to the transcript levels detected with either control (untreated) or iron-exposed cells (Fig. 5). When the first GATA element (positions −202 to −197) was mutated and the second one (positions −120 to −115) was left unchanged (wild-type), the steady-state levels of lacZ mRNA were decreased by ∼4- to 5-fold under basal and iron-replete conditions compared to the levels observed in iron-starved cells (Fig. 5). When the first GATA element was unaltered and the second one was mutated, iron-dependent downregulation of lacZ mRNA was compromised in a manner similar to that observed for the abc3-239M2-lacZ mutant (Fig. 4 and 5). When both GATA elements were mutated, a sustained and constitutive level of lacZ mRNA was observed regardless of the iron status (Fig. 5). Taken together, these results were consistent with the interpretation that the proximal promoter region of abc3+ contained a single functional GATA-type element, −120TGATAT−115, which was required for the transcriptional repression of abc3+ in response to iron.
FIG. 5.
A single GATA element in the abc3+ promoter is sufficient to regulate a heterologous reporter gene in an iron-dependent manner. (A) Schematic representation of a 183-bp abc3+ promoter DNA fragment and its mutant derivatives that were inserted into the minimal promoter of the CYC1 gene fused to lacZ (hatched arrow). The gray boxes shown in the abc3+ promoter region indicate the wild-type elements, while the white boxes represent the mutant versions. The nucleotide numbers refer to the position relative to that of the A of the ATG initiation codon of the abc3+ gene. (B) Total RNA was isolated from cells harboring the indicated abc3+-CYC1-lacZ promoter derivatives and the steady-state mRNA levels of both lacZ and act1+ (indicated with arrows) were analyzed by RNase protection experiments. Where indicated, the cells were left untreated (−) or were treated with Dip (250 μM) or FeCl3 (Fe, 100 μM). NS, nonspecific signal. The data illustrated represent the results of three independent experiments. (C) Normalized expression levels of abc3+-CYC1-lacZ mRNA. The values shown represent the averages of three independent determinations ± the standard deviations.
Iron triggered the association of Fep1 with the abc3+ promoter in vivo.
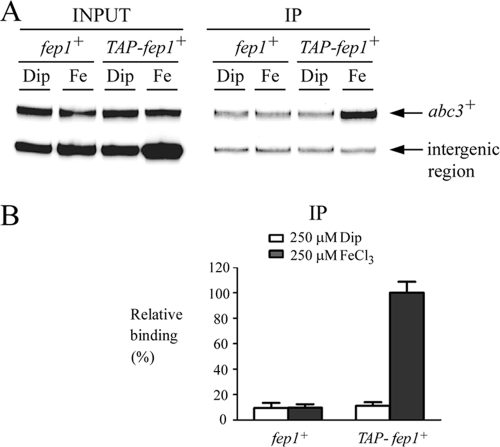
In previous studies, we reported that the fusion of TAP to the N terminus of Fep1 does not interfere with the latter's function (21). Furthermore, we created a fep1Δ php4Δ double-mutant strain in which the expression of TAP-fep1+ is disengaged from its transcriptional regulation by Php4, thereby ensuring its constitutive expression irrespective of the iron status (21). Taking advantage of this mutant, we used a ChIP method (21) to test whether the TAP-Fep1 fusion protein associates with the abc3+ promoter in vivo. fep1Δ php4Δ mutant cells expressing either untagged or TAP-tagged fep1+ alleles were precultivated in the presence of the iron chelator Dip (100 μM) to prevent any iron-dependent activation of Fep1 and, consequently, downregulation of target gene expression. Logarithmic-phase cells were harvested, washed, and resuspended in the same medium containing Dip (250 μM) or FeCl3 (250 μM) for 90 min. The cells were then fixed by formaldehyde treatment and chromatin was prepared to an average size of 500 bp. DNA fragments cross-linked to TAP-Fep1 were isolated by immunoprecipitation with an anti-mouse IgG that bound to the TAP tag. To determine the DNA sequences bound to TAP-Fep1, the cross-links were reversed, and DNA was analyzed by quantitative PCR using primer sets specific for the abc3+ promoter region encompassing the cis-acting element−120TGATAT−115 that confers iron responsiveness. An intergenic region on chromosome II (contig location 3860292 to 3860402) devoid of open reading frame (ORFs) and GATA-type cis-acting sequences was used as a control for unregulated and nontranscribed DNA. The results from ChIP analysis showed that TAP-Fep1 occupied the abc3+ promoter at high levels when cells had been incubated in the presence of iron (Fig. 6). The binding was iron dependent, exhibiting ∼9-fold-higher levels of abc3+ promoter DNA immunoprecipitated when the chromatin was prepared from cells grown in the presence of iron than from cells cultured in the presence of Dip (Fig. 6). As a control, untagged Fep1 immunoprecipitated the background levels of the abc3+ promoter region. These data were consistent with results showing repression of the abc3+ mRNA levels in response to iron. Furthermore, the data suggested that, under conditions of iron repletion, Fep1 was strongly associated with the abc3+ promoter in vivo and dissociated from this promoter in response to iron deprivation.
FIG. 6.
Fep1 binds to the abc3+ promoter in vivo in an iron-dependent manner. (A) ChIP analysis of the abc3+ promoter in fep1Δ php4Δ cells harboring an integrated untagged or TAP-tagged fep1+ allele. The cells were precultured in the presence of 100 μM Dip, allowed to grow to an A600 of ∼1.0, washed and then incubated (90 min) in the presence of 250 μM Dip or 250 μM FeCl3 (Fe). Chromatin was immunoprecipitated with anti-mouse IgG antibodies, and a specific region of the abc3+ promoter was analyzed by PCR to determine Fep1 occupancy. The top band represents the abc3+-specific signal, whereas the lower band is an internal background control derived from a nontranscribed region (intergenic region). (B) Quantitation of the PCR products obtained from anti-IgG immunoprecipitated (IP) chromatin. The results are representative of three independent experiments. The signals are expressed as the relative binding (%) and were calculated as percentages of the largest amount of chromatin measured. Input, input chromatin; IP, immunoprecipitated chromatin.
Vacuolar membrane protein Abc3 reduced cerulenin cytotoxicity and contributed to iron metabolism.
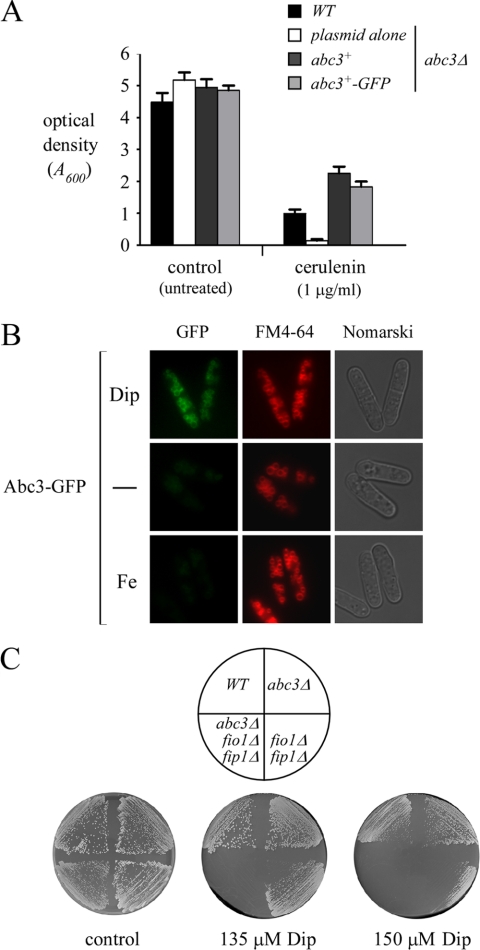
It has been shown that inactivation of Abc3 increased the sensitivity of S. pombe cells to cerulenin (20). Although the molecular basis of this growth inhibition remains unclear, we took advantage of this phenotype to determine whether insertion of GFP interfered with Abc3 function. As shown in Fig. 7A, the abc3Δ mutant strain exhibited an increased sensitivity to cerulenin compared to the wild-type strain. Whereas the wild-type strain showed a 77% inhibition of growth in medium containing 1 μg of cerulenin/ml, the abc3Δ mutant was ∼7-fold more sensitive to cerulenin. When the abc3Δ disruptant was transformed with the wild-type or GFP epitope-tagged abc3+ allele, cell resistance was restored to ∼184 to 226% of the wild-type starting strain (Fig. 7A). The Abc3-GFP fusion protein possessed Abc3 activity that was comparable to that of the wild-type (untagged) Abc3 protein (Fig. 7A). We next sought to determine the subcellular location of active Abc3-GFP in response to changing environmental iron concentrations. As shown in Fig. 7B, Abc3-GFP fluorescence was detected in the vacuole membranes of cells expressing the fusion allele under iron-limiting conditions. Abc3-GFP fluorescence colocalized with the vacuole-staining dye FM4-64, which was used as a marker to stain the vacuolar membrane. Consistent with the iron-dependent downregulation of the abc3+ gene expression, Abc3-GFP fluorescence levels were strongly reduced in cells grown under basal or elevated (100 μM) iron concentrations. Whereas there was an absence of fluorescence in cells that had been transformed with an empty vector, the abc3Δ deletion strain expressing GFP alone displayed a pattern of fluorescence that was distributed throughout the cytoplasm and nuclei of the cells (data not shown). These results led us to conclude that Abc3 functions at the vacuole membrane under conditions of iron deficiency.
FIG. 7.
Vacuolar localization of a functional Abc3-GFP fusion protein and the contribution of Abc3 to cell growth under low-iron conditions. (A) abc3Δ mutant cells were transformed with the indicated plasmids and their growth was measured in unmodified (control) Edinburgh minimal medium or that supplemented with cerulenin (1 μg/ml). Total growth relative to that determined in the absence of cerulenin (percent control growth) was evaluated by turbidimetry at A600. Each point represents the average of triplicates ± the standard deviation. (B) Expression of abc3+ in transformed abc3Δ cells. Exponentially growing cells (2 × 106 cells) were incubated in the absence (−) or the presence of Dip (50 μM) or FeCl3 (Fe, 50 μM) for 16 h at 30°C. Cells were analyzed by direct fluorescence microscopy for GFP. FM4-64 staining visualized the vacuolar membranes, and Nomarski optics were used to examine cell morphology. (C) The indicated S. pombe strains were streaked on EMM iron-poor media containing either 135 or 150 μM Dip and then incubated for 9 days at 30°C in order to test for cell viability. The strains were also streaked on EMM medium lacking Dip (control) and were incubated at 30°C for 4 days. WT, isogenic wild-type strain.
The observation that abc3+ transcription responded to low iron concentrations suggested that the Abc3 protein was involved in iron metabolism and that its expression was required under iron-limiting conditions. To begin to address this question, we created a set of isogenic strains that contained disruptions in genes known to play a role in iron transport. Gene deletions were introduced in the parental strain to disrupt the fio1+ and fip1+ genes that encode the cell surface oxidase-permease-based iron transport system in S. pombe (5). Subsequently, we inactivated the abc3+ locus in the context of a fio1Δ fip1Δ double mutant background. We tested whether these mutations affected the ability of cells to grow on media containing the iron chelator Dip. Although the abc3Δ single mutant cells exhibited no obvious defects, the deletion of abc3+ in the fio1Δ fip1Δ background resulted in a more severe growth defect than was observed with the fio1Δ fip1Δ double mutant strain (Fig. 7C). Thus, a role for Abc3 in mobilizing iron became apparent only when the Fio1-Fip1 iron transport system was missing. As a control, the wild-type parental strain was able to grow on medium containing either 135 or 150 μM Dip.
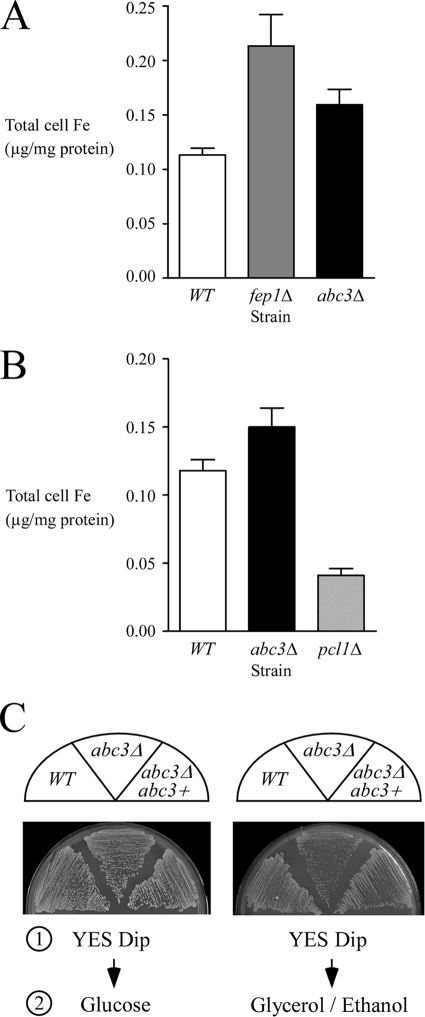
If Abc3 functions by exporting Fe, the rationale is that total cell Fe should increase in cells with abc3 deleted. To determine total cell Fe concentrations in wild-type versus abc3Δ null cells, we used a BPS-based spectrophotometric assay for quantitative measurement of iron (45). The wild-type cells exhibited a total Fe concentration of 0.11 μg/mg of protein. Interestingly, the abc3Δ and fep1Δ mutant strains displayed a total cell Fe concentration of 0.16 and 0.21 μg/mg of protein, respectively, which was 1.5 and 1.9 times higher than the wild-type strain (Fig. 8A). The total cell Fe content results revealed that Abc3 plays a role in Fe efflux. Deletion of abc3+ blocked Fe efflux, triggering Fe accumulation within the cells. For the fep1Δ mutant strain, the quantitative data revealed that fep1Δ cells accumulate Fe in excess of the physiological requirement (Fig. 8A). These results were fully consistent with the fact that in the absence of Fep1, there is lack of transcriptional repression of genes encoding components of the high-affinity Fe uptake machinery (38). We also measured total cell Fe content in an S. pombe pcl1Δ disruption strain (33). As observed for the vacuolar Fe importer Ccc1 in S. cerevisiae (28), deletion of the pcl1+ gene in S. pombe rendered cells sensitive to Fe compared to the wild-type strain (33; data not shown). Consistent with this, we observed that S. pombe cells lacking the putative vacuolar Fe importer Pcl1 contained 0.04 μg/mg of protein, which is three times weaker than the wild-type strain (Fig. 8B).
FIG. 8.
Iron accumulates in abc3Δ mutant cells. (A) The wild-type (WT), fep1Δ, and abc3Δ strains were grown in YES medium to the exponential phase (A600 of ∼1.0). The cells were harvested, and the iron content was determined by the BPS-based spectrophotometric method (45). The values indicated are the averages of triplicate measurements ± the standard deviations. (B) Logarithmic-phase cultures of the isogenic wild-type (WT), abc3Δ, and pcl1Δ strains were grown in EMM containing 0.74 μg of iron. The cell lysates were prepared from each culture and analyzed using the BPS-based spectrophotometric assay for quantitative measurement of iron. The values of total iron concentration shown are the means of three replicates ± the standard deviations. (C) An S. pombe strain bearing a disrupted abc3Δ allele was transformed with pJK148 (plasmid alone, abc3Δ) or pJK148abc3+ (abc3+). For step 1, cultures were grown in YES medium containing glucose and Dip (150 μM). For step 2, cells were washed in water, and equivalent amount of each culture was streaked onto fermentable (glucose) and nonfermentable (ethanol/glycerol) agar media, and incubated at 30°C for 4 and 7 days, respectively. WT, wild-type strain.
Previous studies in S. cerevisiae have shown that, during the transition from growth on glucose to growth on a nonfermentable carbon source, the vacuolar iron stores are redistributed within the cell and contribute to iron-requiring processes such as mitochondriogenesis (46). To test whether abc3Δ cells displayed a lower efficiency in making the switch from nonrespiratory to respiratory metabolism, cells were first grown on glucose under low-iron conditions. After a 4-day incubation on solid medium containing Dip (150 μM), the cells were transferred onto solid medium containing ethanol-glycerol, or onto medium containing glucose for control of viability. abc3Δ mutant cells were found to lag well behind their wild-type counterparts in converting to respiratory metabolism (Fig. 8C). In contrast, the parental wild-type cells were capable of quickly converting from nonrespiratory to respiratory growth when switched from a glucose- to an ethanol-glycerol-containing medium (Fig. 8C). Taken together with the microscopic data, the results were consistent with a role for Abc3 in mobilizing iron within the cell, supporting a possible function for this protein in the transmission of an intracellular pool of iron to extravacuolar metalloenzymes.
Expression and membrane association of Abc3.
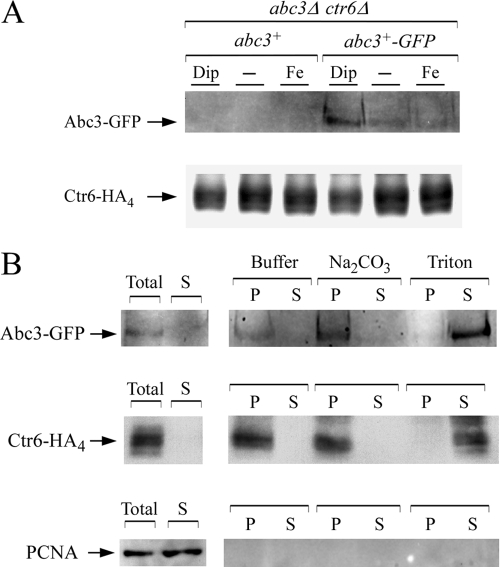
The iron-dependent regulated expression of abc3+, and its vacuolar detection under iron-limiting conditions, prompted us to examine the Abc3-GFP protein levels in both untreated cells and cells incubated under conditions of low and high levels of iron. abc3+-GFP and ctr6+-HA4 (6) fusion genes expressed under the control of their own promoters were cotransformed to an abc3Δ ctr6Δ double mutant disruption strain. The abc3Δ ctr6Δ mutant strain coexpressing the untagged abc3+ and ctr6+-HA4 alleles was used in parallel experiments. Cells coexpressing either the abc3+-GFP and ctr6+-HA4 alleles or the abc3+ and ctr6+-HA4 alleles were grown in the presence of the iron chelator Dip. Membrane fractions collected after ultracentrifugation were treated with Triton X-100, and the supernatants were fractionated by electrophoresis. The results of immunoblotting with an antibody directed against GFP showed that Abc3-GFP was detected after 16 h of treatment (Fig. 9A and data not shown). Consistent with the regulation of abc3+ mRNA levels, the Abc3-GFP protein levels were markedly reduced in cells grown for 16 h under basal and high-iron conditions. Immunoblot analyses of the Triton X-100-solubilized cell membranes were also carried out with an anti-HA antibody. In this case, results showed that Ctr6-HA4 was detected after 16 h in cultures containing Dip or iron, as well as under untreated (basal) conditions (Fig. 9A).
FIG. 9.
Abc3 is an integral membrane protein biosynthetically regulated by cellular iron levels. (A) S. pombe cells harboring an abc3Δ ctr6Δ double deletion were cotransformed with abc3+ and ctr6+-HA4, or with abc3+-GFP and ctr6+-HA4. The cotransformed cells were grown to early logarithmic phase and then treated with either Dip (50 μM) or FeCl3 (50 μM), or were left untreated for 16 h. Triton X-100-solubilized extracts were prepared and analyzed by immunoblotting with either anti-GFP or anti-HA antibody. The positions of the Abc3-GFP and Ctr6-HA4 proteins are indicated by the arrows. (B) abc3Δ ctr6Δ cells expressing Abc3-GFP and Ctr6-HA4 proteins were grown in the presence of 50 μM Dip. Total-extract preparation (Total) was subjected to ultracentrifugation at 100,000 × g. The membrane-containing pellet fraction was resuspended and either left untreated (buffer) or adjusted to either 0.1 M Na2CO3 or 1% Triton X-100 and then ultracentrifuged (100,000 × g). The supernatant (S) and pellet (P) fractions were separated on an SDS-polyacrylamide gel and then analyzed by Western blotting with an anti-GFP, an anti-HA4, or an anti-PCNA antibody.
The primary amino acid sequence of Abc3 suggests that it is integrated into cellular membranes. This possibility was investigated in the following manner. Cell membranes were obtained by ultracentrifugation of whole-cell extracts of cells grown under iron-limiting conditions. Soluble and detached peripheral membrane proteins present in the supernatants were precipitated, resuspended, and left untreated before analysis by immunoblot assays. The pellet fraction was resuspended and left untreated or was adjusted to 0.1 M Na2CO3 or 1% Triton X-100, and then refractionated at 100,000 × g. The results showed that in the absence of any treatment, Abc3-GFP and Ctr6-HA4 proteins were not detected in the supernatant fractions but only in the pellet fractions (Fig. 9B). An identical protein pattern was observed when the procedure had been carried out in the presence of 0.1 M Na2CO3, which is known to linearize membrane structures, releasing nonintegral membrane proteins into the soluble fraction. In the presence of Triton X-100, a nonionic detergent that solubilizes membranes, Abc3-GFP and Ctr6-HA4 were released from the membrane and were detected in the supernatant fractions, indicating that Abc3-GFP was an integral membrane protein as previously reported in the case of Ctr6-HA4 (6). On the other hand, the soluble PCNA protein was only found in the supernatant fraction.
Effects of deletion and expression of abc3+ on the transcriptional regulation of the frp1+ ferrireductase gene.
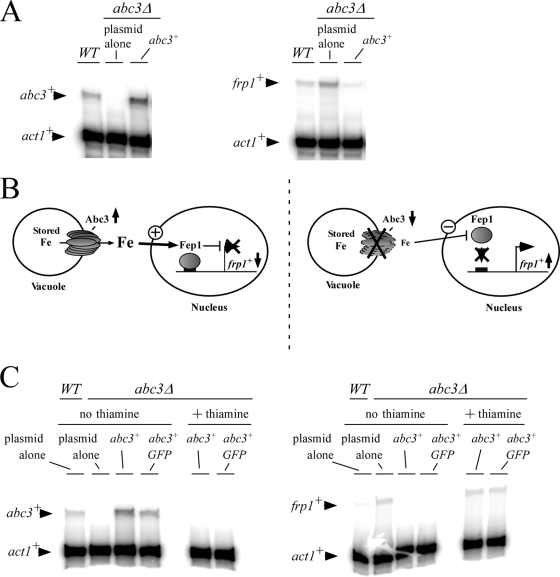
The observation that abc3+ was part of the transcriptional program that cells use to respond to low iron levels suggested that this protein was required during iron deficiency. Based on the fact of the vacuolar localization of Abc3, we reasoned that Abc3 could be important for providing iron to the cell from intracellular stores and that its activity could lead to the activation of Fep1. Under conditions of activation, Fep1 negatively regulates several genes, including those encoding components of the high-affinity iron transport machinery. To test how Abc3 expression influences cellular iron-dependent regulation, we investigated whether the levels of frp1+ transcript were increased in cells harboring an inactivated abc3+ gene. Cells were grown to early logarithmic phase in Edinburgh minimal basal medium. The data showed that the expression of frp1+ was increased ∼2.8-fold compared to the basal level of frp1+ transcripts detected in wild-type cells (abc3+) (Fig. 10A). When the abc3Δ mutant was transformed with the wild-type abc3+ allele, frp1+ mRNA levels decreased ∼1.8-fold under the basal levels observed in the wild-type starting strain (Fig. 10A). We concluded that deletion of the abc3+ gene (abc3Δ) increased the steady-state levels of frp1+ mRNA, suggesting a function for Abc3 in providing iron to the cell from intracellular stores. Its absence would lead to activation of the expression of the plasma membrane uptake machinery through the downregulation of the iron-sensing transcription factor Fep1.
FIG. 10.
Disruption of the abc3+ gene increases frp1+ mRNA levels, whereas its transcriptional activation decreases frp1+ expression. (A) Cells were grown to early logarithmic phase in Edinburgh minimal basal medium. abc3+, frp1+, and act1+ mRNA steady-state levels (indicated by the arrows) were determined in a wild-type strain (WT) and an abc3Δ disruption strain in which either an empty plasmid or a wild-type copy of the abc3+ gene was returned by transformation. The results are representative of three independent experiments. (B) The left side shows a proposed model for the iron-dependent repression of frp1+ transcription when Abc3 is active, while the right side shows how the inactivation of Abc3 function leads to frp1+ transcriptional induction. (C) Cells harboring an abc3Δ deletion were transformed with either pSP-1178nmt41X-abc3+ or pSP-1178nmt41X-abc3+-GFP and precultured in the presence of both thiamine (15 μM) and FeCl3 (10 μM). Early logarithmic cultures were transferred to thiamine-free minimal medium containing 74 nM iron for 16 h. The cells (A600 of ∼1.0) were then kept in thiamine-depleted medium or were transferred to thiamine-replete medium and grown for a further 60 min. After incubation, total RNA was prepared and analyzed by RNase protection assays. Steady-state levels of the abc3+, frp1+, and act1+ mRNAs (indicated with arrows) were probed. As controls, abc3+and frp1+ mRNA steady-state levels were probed in a wild-type strain harboring an empty plasmid.
To further investigate the ability of the cell to downregulate the transcription of frp1+ as an indicator of Abc3 activity, we utilized the nmt1+ inducible/repressible promoter system (32). The expression of abc3+ or abc3+-GFP under the control of the nmt1+ 41X promoter (13) permitted the repression of the synthesis of Abc3 in the presence of iron (10 μM), thereby ensuring intracellular accumulation of iron. Subsequently, the cells were harvested, washed, and resuspended in the same medium in the absence of iron. After induction for 60 min, the ability of Abc3 or Abc3-GFP to generate an iron-mediated signal that fostered repression of frp1+ transcription was analyzed. As shown in Fig. 10C, cells expressing abc3+ or abc3+-GFP triggered downregulation of the frp1+ mRNA levels (∼3- to 4-fold). In contrast, treatment of the cells with thiamine to repress Abc3 synthesis induced the upregulation of frp1+ mRNA which exhibited steady-state levels similar to that observed in abc3Δ null cells. Taken together, the results strongly suggested that iron levels were compromised sufficiently in abc3Δ mutant cells to induce the activation of the frp1+ gene. In contrast, when abc3+ was induced under the control of the nmt1+ promoter, a larger pool of labile iron may become available and activate Fep1, which in turn downregulates the frp1+ transcription levels.
DISCUSSION
Members of the ABC protein superfamily include transporters that are involved in the translocation of a wide variety of substrates across membranes (15). ABC transporters are classified into seven subfamilies (ABCA to ABCG) based on conserved sequences within their amino acid sequences (10). S. pombe has 11 putative ABC transporters, four of which (Abc1, Abc2, Abc3, and Abc4) are members of the ABCC subfamily (20). A previous report has localized a GFP-tagged form of Abc1 to the endoplasmic reticulum and the Abc2-GFP, Abc3-GFP, and Abc4-GFP fusion proteins to the vacuolar membrane (20).
The amino acid identities between Abc3 and Abc1, Abc2, and Abc4 are 23.6, 62.8, and 22.8%, respectively, while the amino acid similarities are 39.7, 74.7, and 38.3%, respectively. Although Abc2 and Abc3 exhibit the highest percentage of amino acid identity and similarity, it has been shown that these two proteins are functionally distinct with respect to vacuolar accumulation of glutathione-conjugated compounds, including both the adenine biosynthetic intermediate phosphoribosylaminoimidazole and monochlorobimane (20). In addition, although Abc2 and Abc4 possess much less homology (23.7% identity; 40% similarity), the two proteins share similar functions in vivo, and differ considerably from Abc3 and Abc1 (20). Given the unrelated localizations of Abc3 and Abc1 and the lack of common phenotypes associated with their respective gene deletions (20), the probability that Abc3 and Abc1 share a biological role is low.
As would be expected for genes regulated by Fep1, putative Fep1 consensus binding sites were found in the abc3+ promoter. The removal of four GATA boxes from the 5′ end of the abc3+ promoter had no apparent effect on either the iron- or the Dip-regulated expression of the abc3+-lacZ fusion gene. The overall magnitude of the response was found to be even higher when the DNA between positions −584 and −239 was deleted. From there, we used two independent reporter gene assays, one using the endogenous abc3+ minimal promoter and the second using a heterologous CYC1 minimal promoter. Both approaches revealed that the integrity of the GATA sequence located between positions −120 to −115 was essential to the iron-dependent repression of abc3+. Thus, abc3+ is the second Fep1-regulated gene to be the subject of negative transcriptional regulation via the presence of a single GATA element, the first reported example being str1+ (39). When iron is in excess in Ustilago maydis, siderophore transporter gene expression is negatively regulated at the transcriptional level by Urbs1 (2, 3). Similarly to Fep1, Urbs1 has two Cys2/Cys2-type zinc fingers located within its DNA-binding domain. Analogous to the situation for Fep1, in vitro DNA-binding assays have shown that Urbs1 can specifically interact with a single GATA element (2). However, as opposed to Fep1, Urbs1 requires the presence of two GATA boxes for its in vivo function (2). These observations may indicate differences in the use of amino acids that serve to interact with DNA between Fep1 and Urbs1. Alternatively, identification and characterization of additional target genes in U. maydis may reveal that some of them are negatively regulated through binding of Urbs1 to a single GATA element. A sequence comparison between many functional GATA boxes found that a variation occurred within the consensus cis-acting 5′-(A/T)GATA(A/T)-3′ element of target gene promoters that respond to Fep1. We have observed that when the GATA element contained the following sequence, 5′-ATC(A/T)GATA(A/T)-3′, the iron-regulatory response was more consistent and almost invariably operative. Examples included the strongest upstream 5′-−803ATCTGATAA−795-3′ element of fio1+, the 5′-−873ATCAGATAA−865-3′ element of str1+, both the 5′-−191ATCAGATAT−183-3′ and the 5′-−168ATCTGATAA−160-3′ elements of php4+, and the 5′-−123ATCTGATAT−115-3′ element of abc3+. Furthermore, other groups have also observed that the extended 5′-ATC(A/T)GATA(A/T)-3′ motif was significantly over-represented in target gene promoters that responded to U. maydis Ursb1 and A. fumigatus SreA, two fungal iron-responsive GATA factors (12, 51).
Yeast studies have shown that the vacuole is an important storage compartment for metals, either as a means of detoxifying the cell or as a reservoir of metal that enable the cell to grow under metal-deficient conditions (6, 47, 48, 53, 54). In the case of S. cerevisiae, when the cells dispose of ample iron, Ccc1 mediates the import of iron into the vacuole (28). In the case of S. pombe, pcl1+ encodes a putative ortholog of S. cerevisiae Ccc1. As is observed for Ccc1, the deletion of the pcl1+ gene renders the cells sensitive to iron compared to the wild-type strain (33; data not shown). Two distinct mechanisms are used for vacuolar iron mobilization when S. cerevisiae cells respond to a shift from sufficient to low iron concentrations. One mechanism involves the NRAMP homologue Smf3, which localizes to the vacuolar membrane and helps to mobilize vacuolar stores of iron (44). The other mechanism involves the vacuolar membrane-resident Fth1/Fet5 complex, which transports stored iron out of the vacuole, resulting in a subsequent redistribution within the cell (60). In S. pombe, vacuolar iron mobilization in response to iron deficiency has not been established and may function differently because of a lack of homologs to the S. cerevisiae Smf3, Fth1, and Fet5. In fission yeast, Pdt1 is the only protein homologous to the NRAMP family of metal transporters (59). Analysis of protein localization has shown that Pdt1 is detected in periphery of the nucleus and the cell perimeter, suggesting a biological role in the endoplasmic reticulum and, perhaps, at the cell surface (59). Furthermore, genetic and functional studies have shown that Pdt1 and Pmr1 cooperatively regulate cell morphogenesis through an as-yet-uncharacterized manganese-dependent homeostatic mechanism (30).
In the present study, our data do not allow us to establish a relationship between cerulenin and Fe homeostasis. However, it has been previously shown that cerulenin is an effective inhibitor of sterol synthesis in yeast (61). Interestingly, like abc3+, the mRNA levels of genes involved in sterol synthesis increase upon Fe depletion (34). One can envision that in the absence of Abc3, less Fe is available for Fe-requiring proteins involved in sterol synthesis, making cerulenin more effective to inhibit sterol biosynthesis and then cell growth.
Given the fact that genetic studies have implicated the vacuole as a player in iron storage (28, 54), and assuming that vacuolar iron is present in a usable form, we suggest that S. pombe Abc3 is an intracellular transporter that mobilizes stores of iron from the organelle for redistribution throughout the cell. This model is supported by six experimental results. First, a functional Abc3-GFP fusion protein was localized to the membrane vacuole under iron-limiting conditions. Second, deletion of the abc3+ gene led to elevated transcriptional activity of frp1+ to presumably compensate for iron-poor conditions. Third, when abc3+ was induced from the nmt1+ promoter, there was inhibition of frp1+ gene expression. Fourth, abc3Δ cells contained more total cell iron than wild-type cells. Fifth, in S. pombe cells lacking the high-affinity plasma membrane transporting complex Fio1 and Fip1, Abc3 was required for growth in low-iron medium. Sixth, the fact that abc3+ was transcriptionally regulated by iron in the same direction as the genes encoding components of the high-affinity iron uptake machinery (e.g., frp1+, fip1+, and fio1+) suggests a function for Abc3 in iron utilization as opposed to iron detoxification.
Fungi such as Aspergillus nidulans, A. fumigatus, and U. maydis do not possess Fth1 orthologs (16). As in the case of S. pombe Abc3, A. nidulans AtrH, A. fumigatus Afu3g03430 and Afu3g03670, and U. maydis Fer6 are all ABC-like proteins that are regulated at the level of gene transcription (12, 16, 51). Their loci are induced in iron-depleted cells and repressed in iron-replete cells. Although the protein localization of these putative ABC-type transporters has not yet been ascertained, the fission yeast S. pombe may represent an attractive model system for understanding the involvement of these proteins in A. nidulans, A. fumigatus, and U. maydis with respect to iron intracellular transport and homeostasis.
One feature of the members of the ABCC subfamily of transporters that distinguishes them from other ABC transporters is their ability to transport substrates in the form of glutathione conjugates or complexes (17, 29). In the presence of excess iron, the vacuole is postulated to function as a storage compartment, preventing detrimental levels of iron accumulation in the cytosol (28). Intravacuolar iron may be bound in a bio-unavailable form such as Fe3+ to polyphosphates or other molecules. In response to iron deficiency, Abc3 may mobilize stored iron either in an inorganic form or in the form of iron conjugates. This possibility remains speculative since a putative interaction between Abc3 and inorganic iron or organic iron conjugates is unknown.
It is interesting that our data concerning Abc3 are reminiscent of those observed in the case of the IDI7 protein isolated from barley root cells, which is a member of the ABC superfamily of transporters (62). IDI7 localizes to the vacuolar surface in plant cells that is known as the tonoplast. As observed in the case of Abc3, IDI7 is expressed only in cells grown under conditions of iron deprivation. More recently, a third ABC transporter, named NtPDR3, has been found to be induced in iron-starved tobacco cells (11). Considering this information, one can envision the possibility that a new group of ABC transporters, including Abc3, IDI7, and NtPDR3, may be responsible for mobilizing intravacuolar stores of iron when cells face iron deprivation.
Acknowledgments
We are grateful to Gilles Dupuis and William Home for critically reading the manuscript. We thank Jude Beaudoin for stimulating discussion and suggestions.
B.P. and M.J. are the recipients of studentships from the Foundation of Stars for Children's Health Research and the Faculty of Medicine and Health Sciences of the Université de Sherbrooke, respectively. A.M. is the recipient of a studentship from the Fonds de la Recherche en Santé du Québec (FRSQ). This study was supported by Natural Sciences and Engineering Research Council of Canada grant MOP-238238-01 to S.L. S.L. is the recipient of a Senior Investigator Scholarship from FRSQ.
Footnotes
Published ahead of print on 13 November 2009.
REFERENCES
- 1.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 2.An, Z., B. Mei, W. M. Yuan, and S. A. Leong. 1997. The distal GATA sequences of the sid1 promoter of Ustilago maydis mediate iron repression of siderophore production and interact directly with Urbs1, a GATA family transcription factor. EMBO J. 16:1742-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, Z., Q. Zhao, J. McEvoy, W. M. Yuan, J. L. Markley, and S. A. Leong. 1997. The second finger of Urbs1 is required for iron-mediated repression of sid1 in Ustilago maydis. Proc. Natl. Acad. Sci. U. S. A. 94:5882-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Askwith, C., D. Eide, A. Van Ho, P. S. Bernard, L. Li, S. Davis-Kaplan, D. M. Sipe, and J. Kaplan. 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Askwith, C., and J. Kaplan. 1997. An oxidase-permease-based iron transport system in Schizosaccharomyces pombe and its expression in Saccharomyces cerevisiae. J. Biol. Chem. 272:401-405. [DOI] [PubMed] [Google Scholar]
- 6.Bellemare, D. R., L. Shaner, K. A. Morano, J. Beaudoin, R. Langlois, and S. Labbé. 2002. Ctr6, a vacuolar membrane copper transporter in Schizosaccharomyces pombe. J. Biol. Chem. 277:46676-46686. [DOI] [PubMed] [Google Scholar]
- 7.Chen, D., W. M. Toone, J. Mata, R. Lyne, G. Burns, K. Kivinen, A. Brazma, N. Jones, and J. Bähler. 2003. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14:214-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dancis, A., D. G. Roman, G. J. Anderson, A. G. Hinnebusch, and R. D. Klausner. 1992. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc. Natl. Acad. Sci. U. S. A. 89:3869-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean, M. 2005. The genetics of ATP-binding cassette transporters. Methods Enzymol. 400:409-429. [DOI] [PubMed] [Google Scholar]
- 10.Decottignies, A., and A. Goffeau. 1997. Complete inventory of the yeast ABC proteins. Nat. Genet. 15:137-145. [DOI] [PubMed] [Google Scholar]
- 11.Ducos, E., S. Fraysse, and M. Boutry. 2005. NtPDR3, an iron-deficiency inducible ABC transporter in Nicotiana tabacum. FEBS Lett. 579:6791-6795. [DOI] [PubMed] [Google Scholar]
- 12.Eichhorn, H., F. Lessing, B. Winterberg, J. Schirawski, J. Kamper, P. Muller, and R. Kahmann. 2006. A ferroxidation/permeation iron uptake system is required for virulence in Ustilago maydis. Plant Cell 18:3332-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsburg, S. L. 1993. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 21:2955-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgatsou, E., and D. Alexandraki. 1994. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottesman, M. M., and S. V. Ambudkar. 2001. Overview: ABC transporters and human disease. J. Bioenerg. Biomembr. 33:453-458. [DOI] [PubMed] [Google Scholar]
- 16.Haas, H., M. Eisendle, and B. G. Turgeon. 2008. Siderophores in fungal physiology and virulence. Annu. Rev. Phytopathol. 46:149-187. [DOI] [PubMed] [Google Scholar]
- 17.Haimeur, A., G. Conseil, R. G. Deeley, and S. P. Cole. 2004. The MRP-related and BCRP/ABCG2 multidrug resistance proteins: biology, substrate specificity and regulation. Curr. Drug Metab. 5:21-53. [DOI] [PubMed] [Google Scholar]
- 18.Halliwell, B., and J. M. Gutteridge. 1992. Biologically relevant metal ion-dependent hydroxyl radical generation. FEBS Lett. 307:108-112. [DOI] [PubMed] [Google Scholar]
- 19.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 20.Iwaki, T., Y. Giga-Hama, and K. Takegawa. 2006. A survey of all 11 ABC transporters in fission yeast: two novel ABC transporters are required for red pigment accumulation in a Schizosaccharomyces pombe adenine biosynthetic mutant. Microbiology 152:2309-2321. [DOI] [PubMed] [Google Scholar]
- 21.Jbel, M., A. Mercier, B. Pelletier, J. Beaudoin, and S. Labbé. 2009. Iron activates in vivo DNA binding of Schizosaccharomyces pombe transcription factor Fep1 through its amino-terminal region. Eukaryot. Cell 8:649-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung, W. H., A. Sham, R. White, and J. W. Kronstad. 2006. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 4:e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan, C. D., and J. Kaplan. 25 August 2009. Iron acquisition and transcriptional regulation. Chem. Rev. [Epub ahead of print.]. [DOI] [PubMed]
- 24.Kim, J., and J. P. Hirsch. 1998. A nucleolar protein that affects mating efficiency in Saccharomyces cerevisiae by altering the morphological response to pheromone. Genetics 149:795-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labbé, S., B. Pelletier, and A. Mercier. 2007. Iron homeostasis in the fission yeast Schizosaccharomyces pombe. Biometals 20:523-537. [DOI] [PubMed] [Google Scholar]
- 27.Labbé, S., M. M. O. Peña, A. R. Fernandes, and D. J. Thiele. 1999. A copper-sensing transcription factor regulates iron uptake genes in Schizosaccharomyces pombe. J. Biol. Chem. 274:36252-36260. [DOI] [PubMed] [Google Scholar]
- 28.Li, L., O. S. Chen, D. McVey Ward, and J. Kaplan. 2001. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 276:29515-29519. [DOI] [PubMed] [Google Scholar]
- 29.Li, Z. S., Y. P. Lu, R. G. Zhen, M. Szczypka, D. J. Thiele, and P. A. Rea. 1997. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc. Natl. Acad. Sci. U. S. A. 94:42-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda, T., R. Sugiura, A. Kita, M. Saito, L. Deng, Y. He, L. Yabin, Y. Fujita, K. Takegawa, H. Shuntoh, and T. Kuno. 2004. Pmr1, a P-type ATPase, and Pdt1, an Nramp homologue, cooperatively regulate cell morphogenesis in fission yeast: the importance of Mn2+ homeostasis. Genes Cells 9:71-82. [DOI] [PubMed] [Google Scholar]
- 31.Mason, D. L., and S. Michaelis. 2002. Requirement of the N-terminal extension for vacuolar trafficking and transport activity of yeast Ycf1p, an ATP-binding cassette transporter. Mol. Biol. Cell 13:4443-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 33.Mercier, A., B. Pelletier, and S. Labbé. 2006. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 5:1866-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mercier, A., S. Watt, J. Bähler, and S. Labbé. 2008. Key function for the CCAAT-binding factor Php4 to regulate gene expression in response to iron deficiency in fission yeast. Eukaryot. Cell 7:493-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myers, A. M., A. Tzagoloff, D. M. Kinney, and C. J. Lusty. 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299-310. [DOI] [PubMed] [Google Scholar]
- 36.Ortiz, D. F., L. Kreppel, D. M. Speiser, G. Scheel, G. McDonald, and D. W. Ow. 1992. Heavy metal tolerance in the fission yeast requires an ATP-binding cassette-type vacuolar membrane transporter. EMBO J. 11:3491-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paumi, C. M., M. Chuk, I. Chevelev, I. Stagljar, and S. Michaelis. 2008. Negative regulation of the yeast ABC transporter Ycf1p by phosphorylation within its N-terminal extension. J. Biol. Chem. 283:27079-27088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelletier, B., J. Beaudoin, Y. Mukai, and S. Labbé. 2002. Fep1, an iron sensor regulating iron transporter gene expression in Schizosaccharomyces pombe. J. Biol. Chem. 277:22950-22958. [DOI] [PubMed] [Google Scholar]
- 39.Pelletier, B., J. Beaudoin, C. C. Philpott, and S. Labbé. 2003. Fep1 represses expression of the fission yeast Schizosaccharomyces pombe siderophore-iron transport system. Nucleic Acids Res. 31:4332-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelletier, B., A. Mercier, M. Durand, C. Peter, M. Jbel, J. Beaudoin, and S. Labbé. 2007. Expression of Candida albicans Sfu1 in fission yeast complements the loss of the iron-regulatory transcription factor Fep1 and requires Tup co-repressors. Yeast 24:883-900. [DOI] [PubMed] [Google Scholar]
- 41.Peter, C., J. Laliberté, J. Beaudoin, and S. Labbé. 2008. Copper distributed by Atx1 is available to copper amine oxidase 1 in Schizosaccharomyces pombe. Eukaryot. Cell 7:1781-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philpott, C. C. 2006. Iron uptake in fungi: a system for every source. Biochim. Biophys. Acta 1763:636-645. [DOI] [PubMed] [Google Scholar]
- 43.Philpott, C. C., and O. Protchenko. 2008. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 7:20-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Portnoy, M. E., X. F. Liu, and V. C. Culotta. 2000. Saccharomyces cerevisiae expresses three functionally distinct homologues of the Nramp family of metal transporters. Mol. Cell. Biol. 20:7893-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rad, A. M., B. Janic, A. S. M. Iskander, H. Soltanian-Zadeh, and A. S. Arbab. 2007. Measurement of quantity of iron in magnetically labeled cells: comparison among different UV/VIS spectrometric methods. Biotechniques 43:627-636. [DOI] [PubMed] [Google Scholar]
- 46.Raguzzi, F., E. Lesuisse, and R. R. Crichton. 1988. Iron storage in Saccharomyces cerevisiae. FEBS Lett. 231:253-258. [DOI] [PubMed] [Google Scholar]
- 47.Ramsay, L. M., and G. M. Gadd. 1997. Mutants of Saccharomyces cerevisiae defective in vacuolar function confirm a role for the vacuole in toxic metal ion detoxification. FEMS Microbiol. Lett. 152:293-298. [DOI] [PubMed] [Google Scholar]
- 48.Rees, E. M., J. Lee, and D. J. Thiele. 2004. Mobilization of intracellular copper stores by the Ctr2 vacuolar copper transporter. J. Biol. Chem. 279:54221-54229. [DOI] [PubMed] [Google Scholar]
- 49.Roman, D. G., A. Dancis, G. J. Anderson, and R. D. Klausner. 1993. The fission yeast ferric reductase gene frp1+ is required for ferric iron uptake and encodes a protein that is homologous to the gp91-phox subunit of the human NADPH phagocyte oxidoreductase. Mol. Cell. Biol. 13:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rustici, G., H. van Bakel, D. H. Lackner, F. C. Holstege, C. Wijmenga, J. Bähler, and A. Brazma. 2007. Global transcriptional responses of fission and budding yeast to changes in copper and iron levels: a comparative study. Genome Biol. 8:R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrettl, M., H. S. Kim, M. Eisendle, C. Kragl, W. C. Nierman, T. Heinekamp, E. R. Werner, I. Jacobsen, P. Illmer, H. Yi, A. A. Brakhage, and H. Haas. 2008. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 70:27-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schrettl, M., G. Winkelmann, and H. Haas. 2004. Ferrichrome in Schizosaccharomyces pombe, an iron transport and iron storage compound. Biometals 17:647-654. [DOI] [PubMed] [Google Scholar]
- 53.Simm, C., B. Lahner, D. Salt, A. LeFurgey, P. Ingram, B. Yandell, and D. J. Eide. 2007. Saccharomyces cerevisiae vacuole in zinc storage and intracellular zinc distribution. Eukaryot. Cell 6:1166-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh, A., N. Kaur, and D. J. Kosman. 2007. The metalloreductase Fre6p in Fe-efflux from the yeast vacuole. J. Biol. Chem. 282:28619-28626. [DOI] [PubMed] [Google Scholar]
- 55.Singh, A., S. Severance, N. Kaur, W. Wiltsie, and D. J. Kosman. 2006. Assembly, activation, and trafficking of the Fet3p.Ftr1p high affinity iron permease complex in Saccharomyces cerevisiae. J. Biol. Chem. 281:13355-13364. [DOI] [PubMed] [Google Scholar]
- 56.Spizzo, T., C. Byersdorfer, S. Duesterhoeft, and D. Eide. 1997. The yeast FET5 gene encodes a FET3-related multicopper oxidase implicated in iron transport. Mol. Gen. Genet. 256:547-556. [DOI] [PubMed] [Google Scholar]
- 57.Stearman, R., D. S. Yuan, Y. Yamaguchi-Iwai, R. D. Klausner, and A. Dancis. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552-1557. [DOI] [PubMed] [Google Scholar]
- 58.Szczypka, M. S., J. A. Wemmie, W. S. Moye-Rowley, and D. J. Thiele. 1994. A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J. Biol. Chem. 269:22853-22857. [PubMed] [Google Scholar]
- 59.Tabuchi, M., T. Yoshida, K. Takegawa, and F. Kishi. 1999. Functional analysis of the human NRAMP family expressed in fission yeast. Biochem. J. 344(Pt. 1):211-219. [PMC free article] [PubMed] [Google Scholar]
- 60.Urbanowski, J. L., and R. C. Piper. 1999. The iron transporter Fth1p forms a complex with the Fet5 iron oxidase and resides on the vacuolar membrane. J. Biol. Chem. 274:38061-38070. [DOI] [PubMed] [Google Scholar]
- 61.Waring, A. J., and G. G. Laties. 1977. Inhibition of the development of induced respiration and cyanide-insensitive respiration in potato tuber slices by cerulenin and dimethylaminoethanol. Plant Physiol. 60:11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamaguchi, H., N. K. Nishizawa, H. Nakanishi, and S. Mori. 2002. IDI7, a new iron-regulated ABC transporter from barley roots, localizes to the tonoplast. J. Exp. Bot. 53:727-735. [DOI] [PubMed] [Google Scholar]