Abstract
The pathogen Campylobacter fetus comprises two subspecies, C. fetus subsp. fetus and C. fetus subsp. venerealis. Although these taxa are highly related on the genome level, they are adapted to distinct hosts and tissues. C. fetus subsp. fetus infects a diversity of hosts, including humans, and colonizes the gastrointestinal tract. In contrast, C. fetus subsp. venerealis is largely restricted to the bovine genital tract, causing epidemic abortion in these animals. In light of their close genetic relatedness, the specific niche preferences make the C. fetus subspecies an ideal model system to investigate the molecular basis of host adaptation. In this study, a subtractive-hybridization approach was applied to the genomes of the subspecies to identify different genes potentially underlying this specificity. The comparison revealed a genomic island uniquely present in C. fetus subsp. venerealis that harbors several genes indicative of horizontal transfer and that encodes the core components necessary for bacterial type IV secretion. Macromolecular transporters of this type deliver effector molecules to host cells, thereby contributing to virulence in various pathogens. Mutational inactivation of the putative secretion system confirmed its involvement in the pathogenicity of C. fetus subsp. venerealis.
Campylobacter species are Gram-negative epsilonproteobacteria highly adapted to mucosal surfaces. The majority are human and/or animal pathogens (19, 61). The 18 species comprising the genus Campylobacter display a high degree of host and tissue specificity, which makes them excellent models to study host-pathogen relationships (25). The most prominent member, Campylobacter jejuni, is a commensal of the chicken intestine and the major cause of human bacterial diarrhea (74). Comparative analysis of Campylobacter genomes has revealed a process of genome decay—supported by a small genome size (about 1.5 Mb) and the loss of metabolic genes—consistent with successful adaptation to a specific niche (41). Campylobacter genomes are among the densest bacterial genomes known, with about 95% coding sequence. Despite this evidence of reduction, plasticity in genetic composition remains evident, as strain-specific genes comprise a substantial proportion of the entire repertoire of 1,500 to 1,800 genes (16, 23, 25, 56).
This study focuses on the species Campylobacter fetus, which is represented by the two subspecies C. fetus subsp. fetus and C. fetus subsp. venerealis. Although the two taxa are genetically closely related, they exhibit striking tissue and host specificity. C. fetus subsp. fetus is a human, as well as animal, pathogen. Human infection results in serious systemic disease, especially in immunocompromised people. C. fetus subsp. fetus is the Campylobacter species most often isolated from human blood (75), and it is considered an emerging pathogen (9). The infection mode shares similarities with that of Salmonella enterica serovar Typhi. Orally acquired C. fetus subsp. fetus penetrates the intestinal mucosa, leading to bacteremia, and subsequent excretion via the biliary tract leads to secondary colonization of the intestine (9). Colonization of reproductive organs induces abortion in sheep and to a lesser extent in cattle, and very rarely in humans (11). C. fetus subsp. fetus can also be isolated from the intestinal tracts of birds and reptiles (78, 80). In contrast, C. fetus subsp. venerealis is host restricted. It is isolated primarily from the bovine genital tract and causes the epidemic disease bovine venereal campylobacteriosis (BVC). The reservoir of C. fetus subsp. venerealis is the penile prepuce of the bull. Transmission to cows occurs at coitus or during artificial insemination, and infection leads to endometritis, abortion, and infertility (28). Since BVC is a worldwide problem with substantial economic consequences, diagnosed cases must be registered (75) and import and export of bovine semen and embryos for cattle breeding requires statutory preclusion of C. fetus infection (2). Despite the distinct niche preferences of the C. fetus subspecies, they show high genetic relatedness, complicating the task of correct subspecies identification (46, 62, 81). Their population structure is clonal, and C. fetus subsp. venerealis is thought to represent a bovine clone of C. fetus (81).
In this study, we employed the C. fetus subspecies to investigate the genetic basis for their host and tissue specificities. A genomic subtractive-hybridization approach was taken to identify subspecies-specific genomic fragments. This led to the discovery of a genomic island exclusively present on the chromosome of the host-adapted subspecies C. fetus subsp. venerealis. This island harbors a type IV secretion system (T4SS), as well as mobility genes (insertion sequence [IS] transposases and phage integrases) and shares substantial homology and similar structure with resistance plasmids found in other Campylobacter species. These features are indicative of a horizontally acquired genetic element. Finally, mutational analysis of genes within the island substantiates its involvement in C. fetus subsp. venerealis virulence.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and identification of bacterial isolates.
Campylobacter and Escherichia coli strains were grown as previously described (48). The 112 Campylobacter strains used in this study are listed in Table 1 and Table S1 in the supplemental material. Subspecies were identified biochemically according to growth in the presence of 1% (wt/vol) glycine and the reduction of 0.1% sodium selenite in liquid culture (82). Additionally, a subspecies-specific PCR assay was applied to all isolates (46). Equivocal results were clarified using amplified fragment length polymorphism fingerprinting (83) and pulsed-field gel electrophoresis (PFGE) (30, 62).
TABLE 1.
Bacterial strains used in this studya
| Strainb | Descriptionc | Source or reference |
|---|---|---|
| C. fetus subsp. venerealis ATCC 19438 (VR) | Type strain; vaginal mucus of heifer | ATCC |
| C. fetus subsp. venerealis 3-18 | Kmr, Cfv ATCC 19438, virB9::PCc-aphA-3 | This study |
| C. fetus subsp. venerealis 1a (V1) | Bovine | 31 |
| C. fetus subsp. venerealis 3 (V3) | Bovine | 31 |
| C. fetus subsp. venerealis D96 (V4) | Bovine | 31 |
| C. fetus subsp. venerealis G91 (V5) | Bovine | 31 |
| C. fetus subsp. venerealis TH15 (V6) | Bovine | 31 |
| C. fetus subsp. venerealis TH24 (V7) | Bovine | 31 |
| C. fetus subsp. venerealis 80/4172 (V8) | Bovine | 46 |
| C. fetus subsp. venerealis 108/4111 (V9) | Bovine | 46 |
| C. fetus subsp. venerealis 121/4401 (V10) | Aborted bovine fetus | 46 |
| C. fetus subsp. venerealis 84-112 (V81) | Bovine isolate; genital secretion | 64 |
| C. fetus subsp. venerealis V81_SK1 | Kmr; Cfv 84-112; virD4::PrpsJ-aphA-3 | This study |
| C. fetus subsp. venerealis V81_JL1 | Kmr; Cfv 84-112; cdtB::PgatC-aphA-3 | This study |
| C. fetus subsp. fetus ATCC 27374 (FR) | Type strain; brain of sheep fetus | ATCC |
| C. fetus subsp. fetus D (F1) | Bovine | 31 |
| C. fetus subsp. fetus B398/2 SK (F5) | Bovine | 31 |
| C. fetus subsp. fetus B88 (F6) | Bovine | 31 |
| C. fetus subsp. fetus H88 (F7) | Bovine | 31 |
| C. fetus subsp. fetus S88 (F8) | Bovine | 31 |
| C. fetus subsp. fetus 94/4256 (F9) | Aborted bovine fetus | 46 |
| C. fetus subsp. fetus 107/4172 (F10) | Aborted bovine placenta | 46 |
| C. fetus subsp. fetus 133/4369 (F11) | Aborted bovine fetus | 46 |
| C. fetus subsp. fetus 12 (F12) | Human blood | 51 |
| E. coli DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 86 |
| E. coli S17-1 λpir | Tpr Smr; recA thi pro hsdR−M− RP4::2-Tc::Mu::Km Tn7 λpir | 21 |
Additional strains used in analysis of gene distribution are listed in Table S1 in the supplemental material.
Strain abbreviations used in the figures are shown in parentheses.
Kmr, kanamycin resistance phenotype; Smr, streptomycin resistance phenotype; Tpr, trimethoprim resistance phenotype; Cfv, C. fetus subsp. venerealis.
DNA manipulations.
Plasmids were purified from E. coli by alkaline lysis or with the QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany). Chromosomal Campylobacter DNA was isolated either using a lysozyme lysis method (29) or with cetyl trimethyl ammonium bromide (CTAB) (4).
RDA.
The oligonucleotides used are described in Table 2. PCR-based genomic subtractive analysis was done according to the method of Lisitsyn et al. (54) with modifications as described by Hubank and Schatz (45). C. fetus reference strains (C. fetus subsp. venerealis ATCC 19438 and C. fetus subsp. fetus ATCC 27374) were each used once as driver and once as tester DNA, respectively, in independent experiments. Ten μg genomic DNA was treated with 12 U of Sau3AI at 37°C for 5 h. The partial digest was purified (QIAquick PCR Purification Kit; Qiagen, Hilden, Germany) and used as driver DNA. To prepare tester DNA, 0.5 μg of digested (driver) DNA and 4 μl of a mixture containing primers R Bgl24 and R Bgl12 (10 μM each) were heated to 56°C in a total volume of 16 μl and gradually cooled to 12°C. Overnight ligation at 16°C with 800 U of T4 ligase (New England Biolabs, Beverly, MA) in a final volume of 20 μl followed. The products were diluted with 80 μl double-distilled H2O (ddH2O), and 1 μl was amplified by PCR in 25 μl containing 0.5 μl Advantage Polymerase (Clontech, Mountain View, CA), 10 μM primer R Bgl24, and 0.2 mM deoxynucleotide triphosphates (dNTPs). The 3′ ends were filled for 5 min at 75°C, followed by 20 cycles of 94°C for 30 s and 72°C for 2 min and a final extension for 10 min at 72°C. The products were column purified (QIAquick) and designated tester DNA. Ten ng of tester DNA from one subspecies and 1 μg of driver DNA from the other were heated to 98°C for 1.5 min and then hybridized overnight at 63°C in a final volume of 16 μl containing 50 mM HEPES·HCl, pH 8.0, 0.5 M NaCl, 0.2 mM EDTA. The hybridization products were diluted to 100 μl with ddH2O and incubated at 63°C for 10 min. Selective PCR amplification of 1 μl of the hybridization products was performed under the conditions described above. The purified products (QIAquick) were incubated with 20 U of mung bean nuclease (New England Biolabs) in the buffer provided for 30 min at 30°C. One μl served as a template for subsequent PCR amplification under the same conditions. To perform a second round of RDA, the PCR products were digested with 12 U of Sau3AI to remove adaptors and purified (QIAquick). The J Bgl24 adaptors were ligated to these DNA fragments using the oligonucleotide J Bgl12 as a linker. Hybridization, amplification using primer J Bgl24, and mung bean nuclease digestion were performed as described above. To perform a third round of RDA, the PCR products were again digested with Sau3AI and purified. Adaptor N Bgl24 was ligated to these DNA fragments using N Bgl12 as a linker. Hybridization, amplification with primer N Bgl24, and nuclease digestion were repeated. The PCR products resulting from the final selective amplification step with primer N Bgl24 represent the enriched tester-specific fragments, which were separated on 1% agarose gels. The resulting bands, which ranged in size from 300 to 650 bp, were excised, eluted from the gel slices, and amplified by PCR using primer N Bgl24. The adaptors were removed with Sau3AI, and the DNA fragments were ligated to pBluescript II KS(−) (Stratagene, La Jolla, CA). For each ligation reaction, 16 to 36 independent transformants were analyzed further. About 500 ng of plasmid DNA from the transformants was transferred to nylon membranes with a vacuum manifold (48). Two identical blots were produced for each set of clones. Genomic DNA (15 ng) of each subspecies was digested with Sau3AI and radioactively labeled with 2.5 U of Klenow polymerase and 20 μCi of [α-32P]dCTP by using the Prime-a-Gene labeling kit according to the manufacturer's specifications (Promega, Mannheim, Germany). Each pair of blots was hybridized to gene probes as described previously (48).
TABLE 2.
Oligonucleotide primers
| Oligonucleotide | Sequence (5′-3′) | Reference and/or applicationa |
|---|---|---|
| R Bgl24 | AGCACTCTCCAGCCTCTCACCGCA | RDA (45) |
| R Bgl12 | GATCTGCGGTGA | RDA (45) |
| J Bgl24 | ACCGACGTCGACTATCCATGAACA | RDA (45) |
| J Bgl12 | GATCTGTTCATG | RDA (45) |
| N Bgl24 | AGGCAACTGTGCTATCCGAGGGAA | RDA (45) |
| N Bgl12 | GATCTTCCCTCG | RDA (45) |
| Int 11 | GCTTTAACACGTCCGCCC | nt 6453 to 6470 (virB3/4) |
| VirB4-6 | CTTGTCTTTTGGTGTGCTATCTGC | nt 24353 to 24330 (virB3/4) |
| VirB8-1 | ACCTAGTTCGCTTACGACAG | nt 12063 to 12082 (virB8) |
| VirB8-3 | CTGTTACAAACTCAGGCACAAAT | nt 12378 to 12356 (orf13) |
| VirB8-4 | CCTCTGTCGTAAGCGAACT | nt 12085 to 12067 (virB8) |
| VirB8-14 | CCAGCGTAAATGAAGCCG | nt 11699 to 11716 (virB8) |
| VirB9-PstI-fwd | GCCTGCAGAATACAAAGCGATAAGC | nt 12825 to 12843 |
| VirB9-HindIII-rev | CTAAGCTTGCTCGTAGGCAATACG | nt 14005 to 13988 |
| VirB9-1 | CTTTGGGATATTTATTGCGTGC | nt 13074 to 13053 (virB9) |
| VirB9-2 | CTGGGAGGTTATGGATAAGG | nt 13221 to 13240 (virB9) |
| VirB9_KmR_fwd | GCAAAGAAGGTTTTGTAAGC | nt 13142 to 13161 |
| KmR_rev | GACGCATTTATCGTCTTGG | aphA-3 (nt 220 to 202); M26832b |
| VirB10-1 | CAGCAAAGATGGCGTAACTC | nt 14617 to 14636 (virB10) |
| VirB10-2 | CTTCAAGTGCAGTGTTTGCC | nt 14969 to 14950 (virB10) |
| VirB11-1 | ATGAGATAGCTTATAATGGTGG | nt 15173 to 15194 (virB11) |
| VirB11-2 | TTGGTTTCATTCTTAGGCAGC | nt 16806 to 16786 (virB11) |
| UP1 | ACGTATTGCCTACGAGCAAC | nt 13987 to 14006 (virB10) |
| UP3 | ATTTGAAGAAAGCGGATCTGC | nt 14299 to 14279 (virB10) |
| UP4 | CCAACTAATTTATCGCCACTTC | nt 13835 to 13814 (virB9) |
| 3Dv 3' | CGGTTGCGATGCTAATACACTA | nt 19253 to 19274 (orf 21) |
| 3DV 5' | TTTATCGCTTGGTAGCGTATTG | nt 19514 to 19493 (orf 21) |
| Trans 4 | ATCACTCAGCAGCAAGAGCG | nt 22739 to 22758 (orf 26) |
| Top/traE1 | TTGCACGTGCGAGTTCAGGC | nt 23294 to 23275 (orf 27) |
| Top/traE4 | ATTGCAAAAGAGTGTGAAGATA | nt 24008 to 24029 (orf 27) |
| TraE7 | GATCTAGTGGCAGGTGTTCC | nt 24867 to 24848 (orf 27) |
| TaxB2 | CATACAATGTTCTAGCCGAGC | nt 16951 to 16971 (virD4) |
| TaxB3 | CCATCAGGCTGATTTGCTTC | nt 17424 to 17409 (virD4) |
| TaxB6 | GCCATTTTGTTTGATTTTGATACG | nt 14696 to 14673 (virD4) |
| Inter_1 | CATAAATGCCGTAGTTCCGG | nt 45575 to 45594 (orf 57) |
| Inter_2 | CGAACCTAATTTATCTATAATAGG | nt 3152 to 3175 (orf 3) |
| 0033_NotI_f | TTGCGGCCGCGATATAATTGACTTATAACATTTAT | Cff 82-40, upstream of rpsJ (nt 33094 to 33118); NC_00859b |
| 0033_PstI_r | ATTCTGCAGTGTTTTTCCTTTAAAAATAACTTGTCA | Cff 82-40, upstream of rpsJ (nt 33548 to 33522); NC_00859b |
| SK_virD4_PstI_f | GTACTGCAGAAGAGAGCCTAAGGAACAAGT | nt 16058 to 16078 |
| SK_virD4_XmaI_r | TAGCCCGGGTCATCGTCTAGCTCATCATAAT | nt 18265 to 18244 |
| VirD4_EcoRI_f | TGAAATTCTGATAATGCAGATGGACAGC | nt 16667 to 16686 |
| VirD4_XhoI _r | ACTCGAG TTTGCTTCAAGTTGCGATCC | nt 17416 to 17397 |
| PstI_cdt_KO-fwd | ATCTGCAGAACGACAAATGTAAGCACTCA | Cff 82-40 (nt 28892 to 28827); NC_00859b |
| NotI_cdt_KO-rev | ATGCGGCCGCCGTTTGGATTTTCAAATGTTCC | Cff 82-40 (nt 26916 to 26937); NC_00859b |
| 1146_NotI_f | TTGCGGCCGCAATAGTATCCTTAACATAAAATTTT | Cff 82-40, upstream of gatC (nt 113712 to 113745); NC_00859b |
| 1146_PstI_r | ATTCTGCAGAGAATACAACTCCATTTTTGATT | Cff 82-40, upstream of gatC (nt 1136947 to 1136925); NC_00859b |
| P2_NotI_f | TTGCGGCCGCTTTGACTTCGCAGGTCAGCA | nt 5760 to 5779 |
| P2-1_PstI_r | ATTCTGCAGTATTAGGGTAGAAAGTTTTGTCATC | nt 6081 to 6057 |
Nucleotide positions correspond to GenBank accession no. EU443150 if not otherwise noted; Cff, C. fetus subsp. fetus.
Nucleotide position relative to the indicated GenBank accession number.
Library construction and DNA sequencing.
A genomic DNA library of C. fetus subsp. venerealis ATCC 19438 was constructed according to standard procedures (4). Briefly, bacterial DNA was isolated following lysozyme lysis, partially digested with Sau3AI, and subsequently cloned into pBluescript II KS(−). The insert size range was 3 to 5 kb. To analyze the DNA sequence flanking the 3Dv clone (see below), a chromosome-walking strategy was applied. Transformants of E. coli were transferred to nitrocellulose membranes by colony lifting (69). In each screening cycle, the 5′ and the 3′ termini of the cloned inserts provided the next generation of 32P-radiolabeled DNA probes. Nucleotide sequence data were generated by cycle sequencing with the BigDye Termination Cycle Sequencing Ready Reaction Kit v.3.1 (Applied Biosystems, Foster City, CA). The products were resolved on an Applied Biosystems ABI 310 genetic analyzer. Sequence gaps were closed with PCR using C. fetus subsp. venerealis ATCC 19438 DNA as a template. Contig assembly was performed with the SeqMan module from Lasergene (DNAStar Inc., Madison, WI).
Gene annotation.
Open reading frames (ORFs) were predicted using ARTEMIS (68) with a cutoff value of 50 bp. Additional annotation criteria were the presence of a probable initiation codon, allowing for alternative bacterial initiation codons, as well as appropriate spacing to a ribosome-binding site (88). Annotation of the identified ORFs was accomplished on the basis of similarity searches using BLASTX (http://www.ncbi.nlm.nih.gov/BLAST/) against the nonredundant database on the protein level, as well as by using the NCBI conserved-domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Sequence comparisons were made using MegAlign from Lasergene (DNAStar). Analysis of secondary structural properties and detection of specific domains or motifs in the predicted proteins was carried out with SMART (http://smart.embl-heidelberg.de), SCANPROSITE (http://www.expasy.org/tools/scanprosite/), and MOTIF (http://motif.genome.ad.jp). The tandem repeats finder program (http://tandem.bu.edu/) was applied, as well as ARAGORN (http://130.235.46.10./ARAGORN/), to search the sequence for tRNA genes.
Survey of genomic-island gene prevalence.
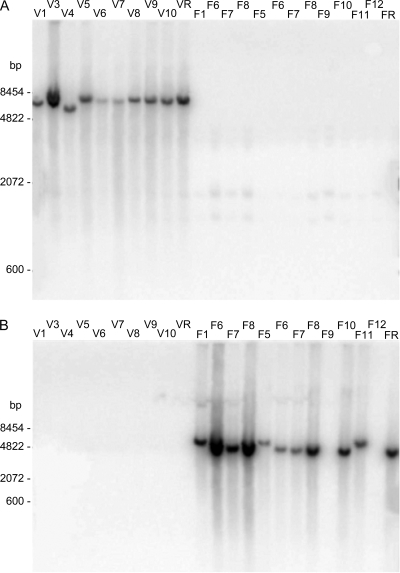
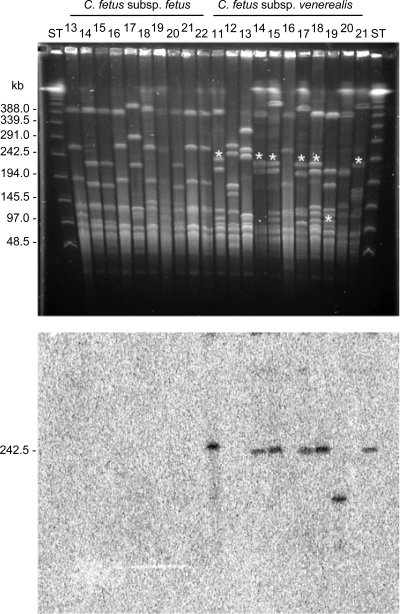
Southern hybridization and/or PCR analyses were performed with an extended C. fetus strain collection (see Table S1 in the supplemental material). For Southern analysis, DNA from a random selection of 22 C. fetus subsp. fetus strains and 38 C. fetus subsp. venerealis strains was subjected to Southern analyses (see Fig. 2; see Table S1 in the supplemental material). Probes specific to the genomic island were constructed by PCR amplification with the oligonucleotide primers 3Dv5′ and 3Dv3′ (orf21); Top/traE4 and TraE7 (orf27); VirB8-1 and VirB9-1, spanning from virB8 to virB9; VirB10-1 and VirB10-2 (virB10); and TaxB2 and TaxB3 (virB4) (Table 2). The DNA fragments were radiolabeled internally and hybridized as described previously (48). Additionally, PFGE was used to resolve SmaI-macrorestricted DNA originating from 10 C. fetus subsp. fetus and 11 C. fetus subsp. venerealis strains according to a recently described protocol (30) (see Fig. 4). Southern hybridizations were performed with the orf21 probe.
FIG. 2.
Distribution of hybridization signals indicating the subspecies specificity of genomic fragments 3Dv and 4Ch. Genomic DNA (4 μg) from a selection of reference and field strains of C. fetus was digested with 20 U HaeIII, resolved on 1% agarose Tris-acetate-EDTA (TAE) gels, and transferred to nitrocellulose for Southern analysis. The identification numbers above the lanes designate isolates of the strain collection, as listed in Table 1. Samples F6 to F8 were loaded twice. The first set (left) contained 5-fold more genomic DNA. (A) A 32P-labeled probe derived from clone 3Dv hybridized specifically to complementary sequences exclusively present in C. fetus subsp. venerealis strains (lanes V1 to VR). (B) A 32P-labeled-probe derived from clone 4Ch hybridized to genomic DNA from the majority of C. fetus subsp. fetus strains tested (lanes F1 to FR), but not to DNA from C. fetus subsp. venerealis strains (lanes V1 to VR).
FIG. 4.
The C. fetus subsp. venerealis genomic island exhibits a conserved position of chromosome insertion. PFGE of SmaI-digested C. fetus DNA (top), followed by Southern hybridization with a radioactively labeled orf21 probe (bottom), reveals a typical resolution of the unique gene region on a 240-kb fragment. The asterisks (top) identify hybridized fragments. The identification numbers above the lanes designate isolates of the strain collection, as listed parenthetically in Table S1 in the supplemental material. A PFGE-DNA ladder (New England Biolabs) served as a standard (ST).
All strains were also tested with PCR for the presence of at least two different island-specific regions. In addition to the primer combinations mentioned above, the screen applied Trans4 and Top/traE1, spanning from orf26 to orf27; Int 11 and VirB4-6 (virB3 and virB4); VirB8-1 and VirB8-3 (virB8 and virB9); VirB9-2 and UP4 (virB9); UP1 and VirB10-2 (virB10); VirB11-1 and VirB11-2 (virB11); and TaxB2 and TaxB6 (virD4).
Mutational analysis.
A virB9 mutant of C. fetus subsp. venerealis ATCC 19438 was constructed by insertion mutagenesis. A 1.2-kb fragment carrying the entire gene was amplified with VirB9-PstI-fwd and VirB9-HindIII-rev. The products were digested, purified, and cloned into PstI/HindIII-cut pBluescript II KS(−), generating pBlueKS−/VirB9 (Table 3). A 1.4-kb SmaI fragment of pILL600 (52) containing the aphA-3 gene was inserted into the EcoRI site of pBlueKS−/VirB9, yielding pGG5. The suicide vector pGG5 was brought into C. fetus subsp. venerealis ATCC 19438 via an electrotransformation protocol originally developed for C. jejuni (35, 84) and adapted for C. fetus (48). Generally, for each mutagenesis, kanamycin-resistant (Kmr) mutants were analyzed via Southern hybridization with linearized fragments of vector, aphA-3, and the targeted gene to confirm the site specificity and copy number of the integration. The orientation of the inserted Kmr cassette was determined by either restriction mapping or PCR and DNA sequencing. Growth on solid media and in liquid culture was indistinguishable for the wild-type and mutant strains. Identical morphology and motility were observed with phase-contrast microscopy.
TABLE 3.
Plasmids used in this study
| Plasmid | Relevant features | Reference or source |
|---|---|---|
| pBluescript II KS(−) | bla lacZ | Stratagene |
| pBlueKS−/VirB9 | virB9 cloned in the MCSa of pBluescript II KS(−) | This study |
| pGG5 | aphA-3 from pILL600 inserted in virB9 of pBlueKS−/VirB9 | This study |
| pILL600 | Source of aphA-3 | 52 |
| pRYVL1 | Φ (PsapA-aphA-3) in pRYGG1, BamHI, and PstI linker between PsapA and aphA-3 | 48 |
| pRYBM3 | Φ (PrpsJ-aphA-3); PsapA of pRYVL1 replaced by PrpsJ | This study |
| pRYBM5 | Φ (PgatC-aphA-3); PsapA of pRYVL1 replaced by PgatC | This study |
| pBlue-oriT | mobIncP of pILL570 in the pBluescript II KS(−) MCS | 48 |
| pSK5 | virD4 cloned into pBlue-oriT | This study |
| pSK6 | virD4 of pSK5 interrupted with PrpsJ-aphA-3 of pRYBM3 | This study |
| pBlue-oriT-cdt | cdtB cloned in pBlue-oriT | This study |
| pJL1 | cdtB of pBlue-oriT-cdt interrupted with Φ(PgatC-aphA-3) | This study |
| pRYSK8 | Φ (PT4SS-1.2-aphA-3); PsapA of pRYVL1 replaced by PT4SS-1.2 | This study |
| pRYSS1 | Φ (PsapA-aphA-3) in pRYGG1 | 48 |
| pRYVL2 | Φ (PsapA-virD4-aphA-3) in pRYGG1 | 48 |
MCS, multiple-cloning site.
virD4 of C. fetus subsp. venerealis 84-112 (V81) was disrupted. The sap promoter in pRYVL1 (48) was replaced with the rpsJ promoter amplified with 0033_NotI_f and 0033_PstI_r from C. fetus subsp. fetus 82-40. This fragment was joined with NotI and PstI sites upstream of aphA-3 to create the cloning intermediate pRYBM3. virD4 from C. fetus subsp. venerealis ATCC 19438 was amplified with SK_virD4_PstI_f and SK_virD4_XmaI_r. The PCR product and vector pBlue-oriT (48) were cut with PstI and XmaI and ligated to generate pSK5. The Kmr cassette, containing the rpsJ promoter-aphA-3 fusion, was excised from pRYBM3 with NotI and EcoRI, blunted, and ligated with the blunted HindIII fragment of pSK5. To create virD4 mutants, the resulting suicide vector, pSK6, was transferred from E. coli S17-1 λpir via conjugation to C. fetus subsp. venerealis 84-112 (V81) as described previously (48).
Construction of a cdtB mutant required replacing the sapA promoter of pRYVL1 with a 220-bp gatC promoter, which was amplified from C. fetus subsp. venerealis ATCC 19438 with 1146_NotI_f and 1146_PstI_r. The product and pRYVL1 were ligated via NotI and PstI ends to create pRYBM5. A 1.9-kb fragment carrying cdtB was amplified from C. fetus subsp. venerealis ATCC 19438 with PstI_cdt_KO-fwd and NotI_cdt_KO-rev. The products and vector pBlue-oriT were ligated via NotI and PstI ends to generate the cloning intermediate pBlue-oriT-cdt. The gatC promoter-aphA-3 fusion was excised via NotI/EcoRI from pRYBM5, blunted, and joined to the blunted XbaI sites of pBlue-oriT-cdt. To create mutant strains, the resulting suicide vector, pJL1, was conjugatively mobilized from E. coli S17-1 λpir to C. fetus subsp. venerealis 84-112 (V81) as described previously (48).
Construction of the virD4 expression plasmid pRYVL2 and the empty vector control pRYSS1 was published previously (48). For complementation experiments, plasmids were mobilized from E. coli S17-1 λpir to the mutant or wild-type strain.
Expression analysis of vir genes.
Wild-type and Cfv 3-18 mutant strains were grown on Columbia blood agar (CBA) plates for 72 h in a microaerobic atmosphere (48). Total RNA was isolated using RNAeasy (Qiagen). One μg of total RNA was treated with 2 U of RQ1-DNase (Promega) according to the manufacturer's specifications. For first-strand cDNA synthesis, 200 ng template was mixed with 2 pmol of antisense oligonucleotide and 0.1 mM dNTP and incubated at 65°C for 5 min. After being chilled on ice, 4 μl of 5-fold first-strand buffer and 2 μl of 0.1 M dithiothreitol (DTT) were added, and the mixture was warmed to 37°C prior to the addition of 200 U of SuperScript II reverse transcriptase (Invitrogen, Paisley, United Kingdom). The 20-μl mixture was incubated at 37°C for 50 min and then inactivated at 70°C for 5 min. For second-strand synthesis, 1 μl of the first-strand product was incubated with 2.5 μl of 5-fold buffer, 0.5 μl of a 10 mM dNTP mixture, 10 pmol of each primer, and 0.5 μl of Advantage DNA polymerase (Clontech) in a final volume of 25 μl.
Cell culture and virulence assays.
HeLa cells were routinely grown in Dulbecco's modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal calf serum (FCS). Caco-2 cells were grown in Eagle's minimum essential medium (EMEM) supplemented with 10% FCS and 1% nonessential amino acids (Invitrogen). All cells were cultured without antibiotics at 37°C and 7% CO2.
A continuous coculture assay was used to assess bacterial cytotoxicity. HeLa cells were seeded in 6-well plates (5 × 105 cells per well in 3 ml). At 24 h, the cell counts of 3 wells were determined. In the remaining wells, the medium was replaced with standard medium plus 10% brucella broth (1) and then inoculated with bacteria at a multiplicity of infection (MOI) of 100. A triplicate set of cultures was harvested every 24 h over at least 4 days. Media were collected and diluted, and the bacterial cell counts were determined on CBA plates. HeLa cells were washed twice with 2 ml of phosphate-buffered saline (PBS), and the monolayers were trypsinized. The cells were enumerated with the CASY cell counter system (Innovatis AG, Reutlingen, Germany). For each experiment, cell counts were compared with those of 2 independent mock-infected controls.
Gentamicin protection assays were performed with cultured Caco-2 cells; 5 × 104 cells were seeded per well in 24-well tissue culture plates and incubated for 2 days to reach 80% confluence. Each well was washed once with medium before inoculation with a bacterial suspension (MOI = 1,000). The number of living bacteria was verified at infection by growth on CBA plates. Infection progressed for up to 5 h before the medium containing the bacteria was removed. The wells were washed once with 500 μl medium containing 400 μg/ml gentamicin and then incubated with 1 ml medium plus 400 μg/ml gentamicin for 3 h to kill extracellular bacteria. The medium was removed, pooled, and centrifuged at 5,000 rpm for 15 min. The pelleted material was washed with PBS and then spread on CBA plates to detect surviving extracellular bacteria. To release intracellular bacteria, eukaryotic cells were washed with 500 μl medium without drug and then incubated with 500 μl PBS containing 0.1% Triton X-100 for 20 min. Fresh medium was added. Suspensions were diluted, spread on CBA plates, and incubated for 4 days. For each measurement, cell counts were determined in triplicate, and assays were repeated on 3 different days. The values shown represent a summary of all counts of the 3 independent experiments. Statistical analysis used SigmaStat software (version 2.03; SPSS Inc., Chicago, IL). The (paired) t test or the Mann-Whitney rank sum test was used to assess the significance of variation exhibited in cell killing or invasion levels.
Nucleotide sequence accession number.
The sequence of the C. fetus subsp. venerealis ATCC 19438 genomic island was deposited in GenBank under the accession number EU443150.
RESULTS
Identification of subspecies-specific gene fragments.
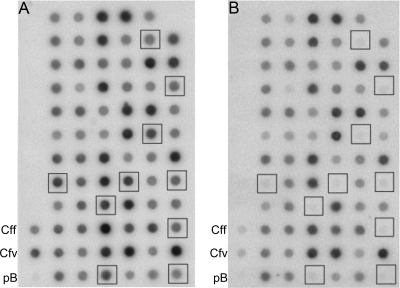
RDA was applied to the genomes of the C. fetus subspecies. Two independent RDA reactions, each comprising three rounds of selective enrichment, were performed. Ultimately, the recombinant DNA of 800 clones was isolated and analyzed by hybridization. Pairs of duplicate dot blots representing all clones were differentially probed with either radioactively labeled C. fetus subsp. venerealis ATCC 19438 DNA or C. fetus subsp. fetus ATCC 27374 DNA (Fig. 1). A total of 46 clones that were apparently unique to a subspecies were identified (34 specific for C. fetus subsp. venerealis and 12 specific for C. fetus subsp. fetus). Clones showing weak differences in hybridization signals between the two reference strains were not further analyzed. Those appearing to be different for one of the subspecies were sequenced. The 34 clones specific for C. fetus subsp. venerealis ATCC 19438 corresponded to six independent DNA fragments, whereas the 12 clones that were different for C. fetus subsp. fetus ATCC 27374 represented a single gene region (Table 4). To survey the prevalence of these genomic fragments within different C. fetus strains, each isolated fragment was radioactively labeled and hybridized to blotted genomic DNA from selected C. fetus reference and field strains (Table 4 and Fig. 2).
FIG. 1.
RDA with C. fetus subsp. venerealis DNA as tester DNA reveals candidate clones for subspecies-specific genomic fragments. Typical results for the analysis are illustrated with duplicate dot blots of 71 clones. Blot A was hybridized with Sau3A1-digested and 32P-labeled genomic C. fetus subsp. venerealis ATCC 19438 DNA. Blot B was hybridized with an equivalently prepared C. fetus subsp. fetus ATCC 27374 genomic DNA probe. The squares indicate clones specific for the C. fetus subsp. venerealis reference strain (no hybridization with the C. fetus subsp. fetus probe). DNA (500 ng) from reference strains were applied as positive controls (Cff and Cfv) as marked, and vector DNA (300 ng) was used to control for plasmid hybridization (pB).
TABLE 4.
Gene fragments identified during RDA and their annotation
| Clone | Best match (accession no.) | Length (nt) | % Identitya | % Similaritya | E value | No. of clones obtained |
Subspecies distribution (no./total) in Southern analysisb |
||
|---|---|---|---|---|---|---|---|---|---|
| 1st RDA | 2nd RDA | Cfv | Cff | ||||||
| 4Ch | GDP-mannose 4,6-dehydratase; Thiomicrospira crunogena XCL-2 (ABB42272.1) | 419 | 65 | 80 | 2 × 10−47 | 5 | 0 | 0/10 | 8/10 |
| 3Dv | Hypothetical protein pTet_29; Campylobacter jejuni 81-176 (YP_247557.1) | 354 | 42 | 56 | 9 × 10−10 | 0 | 1 | 10/10 | 0/10 |
| 3Cp | Flagellin B; Campylobacter fetus subsp. fetus 82-40 (ZP_01073199.1) | 366 | 70 | 86 | 1 × 10−23 | 4 | 6 | 9/10 | 2/10 |
| 1Au | TrbE; Agrobacterium tumefaciens (NP_059757.1) | 571 | 40 | 60 | 1 × 10−27 | 3 | 0 | 5/10 | 1/10 |
| 1Du | TrbJ; Rhizobium sp. NGR234 (NP_443810.1) | 544 | 21 | 51 | 1 × 10−06 | 14 | 0 | 5/10 | 1/10 |
| 1Bu | DNA gyrase subunit A; Campylobacter jejuni (AAS88433.1) | 348 | 99 | 99 | 1 × 10−59 | 1 | 0 | 4/10 | 0/10 |
| 1Ac | Conserved hypothetical protein; Campylobacter coli RM2228 (EAL57391.1) | 404 | 58 | 78 | 0.025 | 7 | 5 | 2/5 | 0/7 |
As revealed by BlastX analysis.
Cfv, C. fetus subsp. venerealis; Cff, C. fetus subsp. fetus.
The fragment in clone 4Ch was detected in 8 of 10 C. fetus subsp. fetus strains and absent in all tested C. fetus subsp. venerealis strains (Fig. 2B). Sequence homology implied that 4Ch encodes GDP-mannose 4,6-dehydratase, an important enzyme for lipopolysaccharide (LPS) biosynthesis. Conversely, the other six fragments identified in the initial screen were confirmed by Southern hybridization to harbor gene regions predominantly or exclusively detectable among C. fetus subsp. venerealis strains (Table 4). For example, all of the C. fetus subsp. venerealis strains tested (n = 10) and none of the 10 C. fetus subsp. fetus strains harbored gene fragment 3Dv (Fig. 2A). This apparently unique locus was chosen for detailed analysis.
Characteristics of the C. fetus subsp. venerealis-specific genomic island. (i) General features.
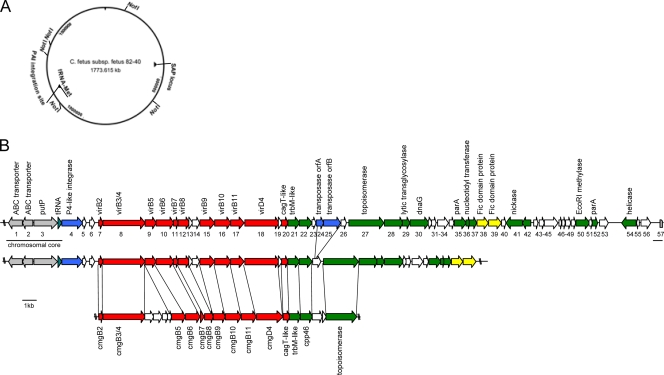
DNA sequence flanking the subspecies-specific fragment 3Dv was obtained by chromosome walking within a C. fetus subsp. venerealis ATCC 19438 genomic library. A total of 45,627 bp of continuous sequence with an average 5.75-fold coverage was produced. The overall G+C content of the sequence was 32.8%, and it contained 57 ORFs, all but six of them oriented for transcription in the clockwise direction on the DNA plus strand. A linear representation and an annotation of the ORFs are presented in Fig. 3 and Table 5.
FIG. 3.
Schematic representation of the C. fetus subsp. venerealis-specific genomic island. (A) The site of integration of the genomic island (PAI) is projected onto a map of the sequenced genome of C. fetus subsp. fetus 82-40 in the vicinity of a methionyl-tRNA gene. The SAP locus denotes the localization of the surface layer protein locus. (B) Annotated genomic island of C. fetus subsp. venerealis ATCC 19438 (top) and data from the partially completed C. fetus subsp. venerealis 84-112 (V81) genome (middle) compared to the highly syntenic resistance plasmid pCC31 of C. coli (bottom). The site of island insertion into the chromosomal core (gray, underlined) follows a tRNA gene. Gene designations are given as numbers under the gene map, and putative functional assignments are shown above. The corresponding functional groups are highlighted with color, including mobility genes, such as IS transposases and phage integrases (blue), a type IV secretion system (red), putative effector molecules (yellow), genes of apparent plasmid origin (green), and genes of unknown function (white). The scale bar represents 1 kb.
TABLE 5.
Annotation of genes found on the C. fetus subsp. venerealis ATCC 19438 chromosome
| ORF | Gene or product | Codon region (bp) | Length (aa)a | No. of identities/total (%)b | No. of positives/total (%)b | E value | Best match/accession no./note |
|---|---|---|---|---|---|---|---|
| 1 | Amino acid ABC transporter, ATP-binding protein | 885-157 | 242 | 242/242 (100) | 242/242 (100) | 1 × 10−133 | Amino acid ABC transporter, ATP-binding protein (C. fetus subsp. fetus 82-40)/YP_892388 |
| 2 | Amino acid ABC tansporter, permease protein | 1600-878 | 240 | 239/239 (100) | 239/239 (100) | 6 × 10−130 | Amino acid ABC transporter, permease protein, His-Glu-Gln-Arg-opine family (C. fetus subsp. fetus 82-40)/YP_892387.1 |
| 3 | putP | 1706-3221 | 504 | 503/504 (99) | 503/504 (99) | 0 | Sodium/proline symporter (C. fetus subsp. fetus 82-40)/YP_892386.1/slipped-strand mispairing, truncated due to premature stop codon |
| tRNA (CAT)d | 3382-3458 | CFF8240_1228; tRNA-Met | |||||
| 4 | Bacteriophage P4-like integrase | 3653-4906 | 417 | 125/408 (30) | 201/408 (49) | 1 × 10−42 | Site-specific recombinase, phage integrase family (Campylobacter curvus 525.92)/EAU00342.1 |
| 5 | Unknown | 5019-5210 | 63 | NAc | NA | NA | No database match; putative pseudogene |
| 6 | Unknown | 5522-5764 | 80 | 24/32 (75) | 25/32 (78) | 2 × 10−5 | Hypothetical protein CFF8240_0645 (C. fetus subsp. fetus 82-40)/YP_891820.1/only N-terminal 33-aa match |
| 7 | virB2 | 6194-6415 | 73 | 31/73 (42) | 49/73 (67) | 1 × 10−8 | TraC/VirB2, pCU110 (C. upsaliensis RM3195)/ZP_00371896/TTG initiation codon |
| 8 | virB3/virB4 | 6426-9215 | 929 | 438/930 (47) | 626/930 (67) | 0 | Hypothetical protein CJJHB9313, pTet (C. jejuni HB93-13)/ZP_01072315.1 |
| 9 | virB5 | 9225-10016 | 263 | 74/249 (29) | 125/249 (50) | 9 × 10−18 | Hypothetical protein, pVT745_p33 (Actinobacillus actinomycetemcomitans)/NP_067580.1 |
| 10 | virB6 | 10026-11201 | 391 | 113/351 (32) | 177/351 (50) | 1 × 10−32 | Hypothetical protein CHAB381_0178 (C. hominis ATCC BAA-381)/YP_063431.1 |
| 11 | virB7 | 11203-11451 | 82 | 12/25 (48) | 18/25 (72) | 1.3 | Hypothetical protein NCU09307 (Neurospora crassa OR74A)/XP_963949.1/putative lipoprotein; contains a transmembrane domain |
| 12 | virB8 | 11452-12156 | 234 | 140/225 (62) | 175/225 (77) | 3 × 10−75 | CmgB8 (C. hominis ATCC BAA-381)/YP_001405784.1 |
| 13 | Unknown | 12159-12434 | 91 | 21/56 (37) | 30/56 (53) | 4.7 | Conserved hypothetical protein (Talaromyces stipitatus ATCC 10500)/EED17168.1 |
| 14 | Unknown | 12438-12926 | 162 | 22/58 (37) | 37/58 (63) | 2 | Putative stage III sporulation protein AH (Clostridium difficile 630)/YP_001087692.1 |
| 15 | virB9 | 12988-13929 | 313 | 187/306 (61) | 248/306 (81) | 1 × 10−105 | P-type conjugative-transfer protein VirB9 (C. hominis ATCC BAA-381)/YP_001405783.1 |
| 16 | virB10 | 13922-15088 | 388 | 77/346 (51) | 234/346 (67) | 3 × 10−83 | CmgB10 (C. hominis ATCC BAA-381)/YP_001405782.1 |
| 17 | virB11 | 15085-16080 | 331 | 205/306 (66) | 245/306 (80) | 3 × 10−117 | P-type DNA transfer ATPase VirB11 (C. hominis ATCC BAA-381)/YP_001405780.1 |
| 18 | virD4 | 16080-18287 | 735 | 365/654 (55) | 478/654 (73) | 0 | CmgD4 (C. hominis ATCC BAA-381)/YP_001405778.1 |
| 19 | Unknown | 18291-18512 | 73 | 22/61 (36) | 32/61 (52) | 0.01 | Hypothetical protein, excisionase family (C. jejuni RM1221)/YP_178561.1/contains DNA binding domain |
| 20 | virB7/cagT homolog | 18530-18997 | 155 | 65/135 (48) | 89/135 (65) | 2 × 10−28 | Cpp44 (C. hominis ATCC BAA-381)/YP_001405777.1 |
| 21 | trbM homolog | 18951-19658 | 235 | 89/247 (36) | 127/247 (51) | 3 × 10−32 | YggA-like protein, pCC178 (C. coli RM2228)/ZP_00370966.1 |
| 22 | Unknown | 19660-20637 | 325 | 70/263 (26) | 111/263 (42) | 3 × 10−4 | Hypothetical protein CHAB381_0167 (C. hominis ATCC BAA-381)/YP_001405775.1/contains AAA-ATPase motif and tansmembrane domain |
| 23 | Unknown | 20669-20953 | 94 | 32/82 (39) | 51/82 (62) | 2 × 10−8 | Hypothetical protein CHAB381_0166 (C. hominis ATCC BAA-381)/putative pseudogene |
| 24 | Transposase OrfA | 21045-21500 | 151 | 121/145 (83) | 133/145 (91) | 1 × 10−68 | ISHp608 transposase OrfA (Helicobacter pylori)/AAL06574.1 |
| 25 | Transposase OrfB | 21478-22578 | 366 | 203/382 (53) | 264/382 (69) | 4 × 10−100 | ISHp608 transposase OrfB (Helicobacter pylori)/AAL06585.1 |
| 26 | Unknown | 22604-23104 | 166 | 52/138 (37) | 81/138 (58) | 3 × 10−15 | Hypothetical protein CHAB381_0166 (C. hominis ATCC BAA-381)/YP_001405774.1/putative pseudogene |
| 27 | DNA topoisomerase IA | 23249-25498 | 749 | 386/740 (52) | 529/740 (71) | 0 | Cpp49, pCC31 (C. coli)/YP_063443.1 |
| 28 | Unknown | 25508-26776 | 422 | 60/234 (25) | 102/234 (43) | 7 × 10−10 | Hypothetical protein CHAB381_0162 (C. hominis ATCC BAA-381)/YP_001405770.1/ putative membrane protein |
| 29 | Lytic transglycosylase | 26773-27270 | 165 | 62/113 (54) | 81/113 (71) | 4 × 10−28 | Lytic transglycosylase (Helicobacter pullorum MIT 98-5489)/ZP_03660875.1 |
| 30 | dnaG | 27267-28487 | 406 | 114/247 (46) | 162/247 (65) | 6 × 10−57 | Hypothetical protein CHAB381_0154 (C. hominis ATCC BAA-381)/YP_001405763.1 |
| 31 | Unknown | 28484-28738 | 84 | 22/47 (46) | 31/47 (65) | 0.88 | Hypothetical protein CHAB381_0151 (C. hominis ATCC BAA-381)/YP_001405760.1 |
| 32 | Unknown | 28735-28950 | 71 | 21/56 (37) | 30/56 (53) | 2.1 | Helicase domain protein (Magnetococcus sp. MC-1) (DnaJ domain-containing protein)/YP_865923.1 |
| 33 | Unknown | 29153-29863 | 236 | 30/120 (25) | 57/120 (47) | 0.075 | Transcriptional regulator (Enterococcus faecalis V583)/NP_815906.1 |
| 34 | Unknown | 29931-30248 | 105 | 25/68 (36) | 37/68 (54) | 0.15 | Cpp23 (Helicobacter pullorum MIT 98-5489)/ZP_03661237.1/contains a signal peptide motif |
| 35 | parA | 30273-30935 | 220 | 117/222 (52) | 170/222 (76) | 3 × 10−66 | Hypothetical protein HcinC1_10026 (Helicobacter cinaedi CCUG 18818)/ZP_03659243.1 |
| 36 | Nucleotidyltransferase | 31003-31314 | 103 | 59/102 (57) | 79/102 (77) | 9 × 10−26 | Nucleotidyltransferase family protein (Helicobacter cinaedi CCUG 18818)/ZP_03659230.1 |
| 37 | Unknown | 31307-31738 | 143 | 42/113 (37) | 77/113 (68) | 7 × 10−16 | Hypothetical protein CHAB381_1342 (C. hominis ATCC BAA-381)/YP_001406891.1/DUF86 protein superfamily |
| 38 | Fic protein | 31742-32578 | 278 | 134/220 (60) | 171/220 (77) | 1 × 10−75 | Hypothetical protein HcinC1_00045 (Helicobacter cinaedi CCUG 18818)/ZP_03657318.1/ |
| 39 | Fic protein | 32578-33498 | 306 | 102/243 (41) | 146/243 (60) | 2 × 10−46 | Hypothetical protein CLOSS21_03156 (Clostridium sp. SS2/1)/ZP_02440650.1 |
| 40 | Unknown | 33498-33767 | 89 | 26/75 (34) | 36/75 (48) | 1 | DEAD box ATP-dependent DNA helicase (Clostridium acetobutylicum ATCC 824)/NP_349354.1 |
| 41 | Unknown | 35113-33776 | 445 | 152/387 (39) | 228/387 (58) | 4 × 10−60 | Cpp17 (C. hominis ATCC BAA-381)/YP_001405759.1/putative nickase |
| 42 | Unknown | 35691-35110 | 193 | 26/92 (28) | 56/92 (60) | 3 × 10−5 | Cpp18, pCC31 (C. coli)/YP_063413.1 |
| 43 | Unknown | 35948-36217 | 89 | 18/56 (32) | 32/56 (57) | 0.99 | ATPase domain-containing protein (Desulfococcus oleovorans Hxd3)/YP_001527917.1 |
| 44 | Unknown | 36359-36751 | 130 | 28/95 (29) | 51/95 (53) | 0.59 | Hypothetical protein CHAB381_0165 (Campylobacter hominis ATCC BAA-381)/YP_001405773.1/cpp50 homolog |
| 45 | Unknown | 36742-37797 | 351 | NA | NA | NA | No database match |
| 46 | Unknown | 37875-38042 | 55 | NA | NA | NA | No database match |
| 47 | Unknown | 38039-38299 | 86 | 24/57 (42) | 34/57 (59) | 2.5 | LOC495429 protein (Xenopus laevis)/AAH84946.1 |
| 48 | Unknown | 38313-38705 | 130 | 32/80 (40) | 39/80 (48) | 8 × 10−12 | Hypothetical protein PPSIR1_31413 (Plesiocystis pacifica SIR-1)/ZP_01910965.1 |
| 49 | Unknown | 38695-39078 | 127 | 22/79 (27) | 40/79 (50) | 1.2 | Hypothetical protein Tlet_0960 (Thermotoga lettingae TMO)/YP_001470590.1 |
| 50 | EcoRI methylase/methyltransferase | 39108-40106 | 332 | 246/323 (76) | 290/323 (89) | 3 × 10−148 | EcoRI methylase/methyltransferase (Campylobacter rectus RM3267)/ZP_03609883.1 |
| 51 | Unknown | 40153-40431 | 92 | 9/53 (35) | 28/53 (52) | 5.3 | Conserved hypothetical protein (C. rectus RM3267)/ZP_03608654.1 |
| 52 | Unknown | 40400-40774 | 124 | 71/89 (79) | 83/89 (93) | 3 × 10−35 | ParA protein (C. rectus RM3267)/ZP_03609888.1/plasmid-partitioning protein |
| 53 | Unknown | 40951-41577 | 208 | 108/189 (57) | 147/189 (77) | 5 × 10−55 | Hypothetical protein CAMRE0001_3153 (C. rectus RM3267)/ZP_03609899.1/homolog on pVir |
| 54 | Unknown | 42682-43710 | 342 | 69/206 (33) | 118/206 (57) | 4 × 10−23 | DnaB-like helicase-like (Sulfurimonas denitrificans DSM 1251)/YP_393316.1/possibly phage derived |
| 55 | Unknown | 43745-43981 | 79 | 0.61 | Hypothetical protein CFF8240_1710 (C. fetus subsp. fetus 82-40) | ||
| 56 | Unknown | 44112-44801 | 229 | 59/230 (25) | 106/230 (46) | 6 × 10−7 | Hypothetical protein HH0748 (Helicobacter hepaticus ATCC 51449)/NP_860279.1/putative transcriptional regulator, XRE family; helix-turn-helix protein |
| 57 | Methyl-accepting chemotaxis protein | 45493-45626 | NA | 134/134 (100) | 134/134 (100) | 0 | CFF8240_1227 (C. fetus subsp. fetus 82-40)/ABK81741.1/partial sequence |
aa, amino acids.
As predicted by Blast analyses.
NA, not applicable.
CAT, tRNA with anticodon CAT.
(ii) The chromosomal core and site of insertion.
The first three ORFs of the analyzed chromosomal locus are similarly arranged and nearly identical to homologs from C. fetus subsp. fetus 82-40 (GenBank accession number NC_008599). orf1 and orf2 are presumably part of an ABC transporter dedicated to histidine and glutamine uptake (43), and orf3 is an apparent homolog of a sodium/proline symporter. orf3 may be a pseudogene, as an additional guanosine inserted within a poly(G) tract (nucleotide [nt] 2141) creates a premature stop codon. Nucleotides 3381 to 3459 encode a methionyl-tRNA, which is also evident in C. fetus subsp. fetus 82-40.
Pathogenicity islands (PAIs) are typically inserted into the backbone genome at specific sites that are frequently tRNA genes followed by direct repeats. The latter serve as recognition sites for the integration of bacteriophages (36, 70). Our sequence showed a 2.4-fold tandem repeat (nt 3471 to 3517) shortly after the methionyl-tRNA gene, followed by orf4, which is homologous to a bacteriophage P4-like integrase gene. Integrase genes of this type are also found in other campylobacters and on different integrative plasmids and pathogenicity islands. Two additional mobility genes are present about 15 kb downstream. orf24 and orf25 are highly homologous to the Helicobacter pylori transposable element ISHp608 (47). This type of IS is always comprised of the consecutive elements orfA and orfB. A well-conserved DDEK motif, which has been suggested to be the catalytically active site, is evident in orf25 (47) and is characteristic of the IS3 family of IS elements, typically found in epsilonproteobacterial species and in retroviral integrases (55). Interestingly, orf24 and orf25 are absent in the partially analyzed strain C. fetus subsp. venerealis 84-112 (Fig. 3). The 3′ boundary of the island shows a 19-bp remnant of a tRNA gene (nt 45138 to 45156). The chromosomal core starts with orf57, which encodes a methyl-accepting chemotaxis protein with an identical sequence in C. fetus subsp. fetus 82-40.
(iii) Type IV secretion system.
Sequences distal to orf6 exhibit strong homology to genes commonly required for assembly of a T4SS. This class of macromolecular transfer systems functions to translocate proteins and nucleoprotein complexes from donor to recipient cells in processes related to bacterial conjugation. In pathogenic bacteria, T4SSs transmit effector molecules with virulence functions to eukaryotic hosts. At least 10 protein constituents typically mediate the transmission of substrates. Homologs of the VirB2-11 and VirD4 proteins of the agrobacterial paradigm T4SS were recognized, and the organization of the vir gene homologs in C. fetus subsp. venerealis is also conserved (Fig. 3). A significant proportion of the orf7 product matches TraC/VirB2 of pCU110 of Campylobacter upsaliensis. The downstream orf8 product comprises a fused VirB3/VirB4 homolog. The orf9 and orf10 products represent VirB5 and VirB6 homologs, respectively. orf11 is homologous to the lipoprotein motif-containing gene cmgb7 (VirB7) of the Campylobacter coli plasmid pCC31 (8). orf12 and orf15 to orf18 encode the T4SS proteins VirB8, VirB9, VirB10, VirB11, and VirD4. orf20 is highly homologous to cpp44, a VirB7 homolog of pTet from C. jejuni, and also to the cagT gene of H. pylori. Lytic transglycosylases are common to bacterial type III and type IV secretion systems (89). They degrade the peptidoglycan within the periplasm, which facilitates the assembly of the envelope-spanning secretion apparatus (50). Consistent with the observation that VirB1-like transglycosylases are not universally required for transporter assembly, no VirB1 homolog was recognized on the C. fetus subsp. venerealis island. Alternatively, the distally located orf29, encoding a putative lytic transglycosylase, may provide that function. The T4SS exhibits the most apparent synteny and homology with the resistance (R) plasmids pCC31 of C. coli and pTet of C. jejuni (8, 26), as well as an uncharacterized locus within the Campylobacter hominis genome (GenBank accession no. NC_009714.1).
Importantly, the C. fetus subsp. venerealis locus contains two putative effector molecules, possibly transported by the T4SS. Both orf38 and orf39 encode a Fic (filamentation induced by cyclic AMP [cAMP]) domain (HPFXXGNG) (87). Bacterial Fic domain proteins disrupt host cell processes after their transmission into eukaryotic cells. In Legionella pneumophila, Coxiella burnetii, and Bartonella henselae, Fic domain-containing proteins are translocated via a T4SS (67, 72). Indeed, fusion of the putative effector proteins to Cre recombinase for analysis in CRAfT (Cre recombinase reporter assay for translocation) (72) provides evidence that recognition and secretion of the product of orf39, at least, belong to the functional repertoire of this T4SS (S. Kienesberger, G. Gorkiewicz, and E. L. Zechner, unpublished data).
(iv) Plasmid-borne genes and genes of unknown function.
Besides the T4SS noted above, several ORFs show homology to components typically carried by plasmids. Among them are homologs to cpp genes found on the C. coli plasmid pCC31 (cpp45 [orf21], cpp46 [orf22], cpp49 [orf27], cpp17 [orf41], cpp18 [orf42], and cpp50 [orf44]). orf28 shows significant homology to a gene encoding a hypothetical protein of the Campylobacter lari plasmid pCL46, and orf37 matches a gene encoding a hypothetical protein found on pCU110 from C. upsaliensis. Moreover, orf33 matches an Enterococcus faecalis transcriptional regulator of the two-component response regulator class. The amino acid sequence contains a short helix-turn-helix motif also found in DeoR-like transcriptional regulators, which are known to regulate genes localized on pathogenicity islands (37). orf36 encodes a putative nucleotidyltransferase, which is a plasmid-encoded enzyme responsible for resistance to aminoglycosides also found in Campylobacter (59), and orf50 encodes an EcoRI methylase. A detailed annotation of the genes found on the genomic locus is given in Table 5.
In conclusion, the characteristics of this subspecies-specific sequence, as well as the organizations and putative functions of the genes carried, imply that the chromosome of C. fetus subsp. venerealis bears a genomic island. The 5′ and 3′ borders flanking the island are identical to those of C. fetus subsp. fetus 82-40. The genomic island harbors a conjugation-related T4SS and bears homology to several plasmid-borne genes, and it displays high homology and synteny to the R plasmids pCC31 (Fig. 3) and pTet found in C. coli and C. jejuni, respectively.
The genomic island is uniquely present in C. fetus subsp. venerealis.
The distribution of the genomic island was surveyed in a collection of 112 C. fetus isolates (see Table S1 in the supplemental material). To this end, Southern hybridization analysis was performed with chromosomal DNA from a random selection of C. fetus strains. The gene-specific probes corresponded to orf21, orf27, virB10, and virD4, as well as to a region spanning from virB8 to virB9. To expand the analysis, genomic DNA from all strains of the collection was subjected to PCR analyses with a minimum of two different primer combinations targeting different loci from the island and adding information about the presence of virB4 and virB11. In every case where the hybridization and PCR approaches were applied to a particular gene in the same strain, consistent results were obtained with both tests. All results are summarized in Table S1 in the supplemental material. None of the test genes were detected in any of the 45 C. fetus subsp. fetus strains investigated. In contrast, of the 67 C. fetus subsp. venerealis strains, 51 (76%) harbored all assessed genes, and 10 scored positively for at least one of the analyzed genes. Hybridization and/or PCR were negative for all genes tested in only 6 C. fetus subsp. venerealis strains (9%). To assess whether these strains harbor a remnant of the island, PCR primers were designed to hybridize upstream (orf3) and downstream (orf57) of the insertion site (Inter_1 and Inter_2 [Table 2]). Amplification of C. fetus subsp. fetus 82-40 DNA, which harbors no genomic island, and five of the six “negative” C. fetus subsp. venerealis strains gave the same 721-bp product, indicating that no additional DNA was inserted at the site (not shown). In contrast, DNA from one strain (V13) did not support amplification across this position (not shown). The quality of the V13 template DNA was verified by a control amplification of a gene outside of the genomic island. This finding implies that a remnant of the island may exist in this strain, but the remaining region lacks the genes investigated thus far. Alternatively, DNA of a different origin may have been integrated at this site.
To gain complementary data about whether the site of island integration is conserved in “positive” C. fetus subsp. venerealis strains, 21 C. fetus strains were examined by SmaI macrorestriction and PFGE, followed by hybridization with the island-specific orf21 probe (Fig. 4). Hybridization was observed for 7 of the 11 C. fetus subsp. venerealis strains, as expected, and was again completely absent in all C. fetus subsp. fetus strains. Six of the seven positive samples exhibited a band about 240 kb in size, while one isolate displayed a hybridization signal of about 140 kb. This finding implies that the genomic island is located at similar chromosomal positions in the majority of C. fetus subsp. venerealis strains. Moreover, comparison of the sequence of the C. fetus subsp. venerealis ATCC 19438 island with a 33.7-kb sequence newly generated from the C. fetus subsp. venerealis 84-112 (V81) genome (32) revealed that the site of insertion is conserved. Indeed, the loci were nearly identical. Although the C. fetus subsp. venerealis 84-112 genome lacks the IS transposase (orf24 and orf25) (Fig. 3), only 3 base changes were detected within the remaining shared sequence (data not shown).
Taken together, our findings indicate a complete (100%) absence of homologous sequences from the C. fetus subsp. fetus strains surveyed and a high frequency of detection for at least some genes in C. fetus subsp. venerealis isolates (91%). We conclude that within the species C. fetus this locus is apparently subspecies specific. Evidence for genetic insertion at a conserved position is absent in a minority of strains, however, suggesting that the presence of these genes is not universal for C. fetus subsp. venerealis. These data also suggest that the genomic island may maintain the capacity for lateral transmission.
vir genes are expressed in C. fetus subsp. venerealis.
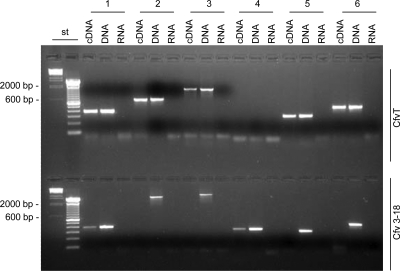
In the agrobacterial paradigm T4SS, and in all similar systems studied thus far, the conserved block of virB-virD4 genes is organized as a transcriptional unit (15, 17). We identified a region upstream of the vir genes with an increased A+T value and similarity to other Campylobacter promoters (49, 88). Functional promoter activity was confirmed when this putative regulatory region 5′ to orf7 (nt 5760 to 6081) replaced the existing promoter regulating aphA-3 in vector pRYVL1 (48). The test construction, pRYSK8, was introduced into C. fetus subsp. venerealis ATCC 19438 via conjugation. Derivatives of this vector backbone, which do not provide a C. fetus-specific promoter upstream of aphA-3, do not support an antibiotic-resistant phenotype (48). Kanamycin-resistant transconjugants were obtained at high frequencies after transconjugation of pRYSK8, confirming promoter activity for the putative vir regulatory sequence under laboratory conditions (data not shown). Moreover, application of reverse transcriptase-PCR generated cDNA of virB8, virB9, virB10, and virD4 from the mRNA pool of C. fetus subsp. venerealis ATCC 19438 (Fig. 5). Transcripts were isolated following growth on CBA plates, again demonstrating expression under laboratory conditions. Notably, the insertion of a Kmr cassette in virB9 (mutant Cfv 3-18) had polar effects on the transcription of downstream genes, consistent with the hypothesized operon organization (Fig. 5).
FIG. 5.
Comparative analysis of C. fetus subsp. venerealis ATCC 19438 (CfvT) and the virB9 mutant (Cfv 3-18) vir gene expression. Shown are reverse transcriptase-PCR products generated from cDNA obtained from the strains indicated (right) for the targeted genes shown (top). Control reactions included genomic DNA (DNA) and DNase-treated RNA without reverse transcription (RNA), as indicated above the lanes. Products (10 μl) were separated on a 2% agarose-TAE gel with 500 ng of λ HindIII marker and 1 μg of a 100-bp DNA ladder (st). Primers (Table 2) hybridized as follows: (1) VirB8-4 and VirB8-14 within virB8; (2) UP4 and VirB9-2 in virB9 or (3) VirB9_HindIII_rev and VirB9_PstI_fwd within virB9 and flanking the insertion site of aphA-3; (4) KmR_rev and VirB9_KmR_fwd, spanning from virB9 to aphA-3; (5) UP3 and UP1 in virB10 and (6) TaxB3 and TaxB2 in virD4.
Mutation of virB9 reduced bacterial killing of eukaryotic cells.
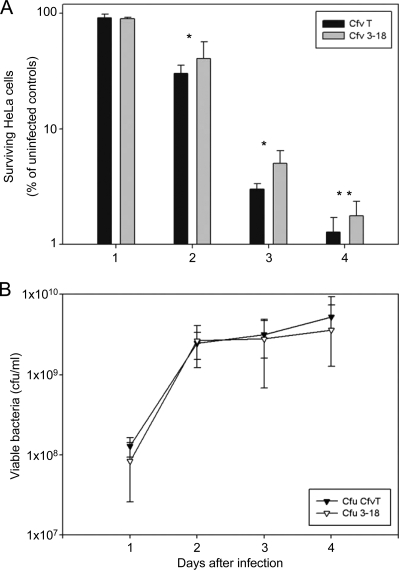
To test for the putative virulence functions conferred by the island, we used a basic coculture experiment that measured the toxic effects of pathogenic bacteria on cultured human cell lines (1). HeLa cells were infected with the wild-type strain (C. fetus subsp. venerealis ATCC 19438) or the isogenic virB9 mutant (Cfv 3-18). Cocultivation continued for 4 days, and eukaryotic cells and bacteria were enumerated daily. Compared to the uninfected controls, viable HeLa cells diminished drastically when cocultured with bacteria, and only between 1 and 2% of the cells survived until day 4. By comparison, infection with the virB9 mutant Cfv 3-18 killed fewer HeLa cells than the wild-type strain (Fig. 6). A significant difference between the cytotoxic effects of the two strains was observed at day 2 and continued to the end of the experiment. At day 2 of cocultivation with the wild-type strain, only 30.3% ± 5.26% of the HeLa cells remained viable compared to the uninfected control. In contrast, cocultivation with the virB9 mutant resulted in 40.7% ± 15.91% of viable HeLa cells (wild type versus Cfv 3-18; P = 0.034). Living cells decreased further until day 4 of the experiment, and only 1.3% ± 0.42% of wild-type-infected HeLa cells remained viable, whereas 1.8% ± 0.6% of Cfv 3-18-infected cells remained viable compared to the uninfected control. Thus, the virB9 mutant killed about 30% fewer cells than the wild-type strain at day 4 of cocultivation, and this result was reproducible in three independent experiments with statistical significance (wild type versus Cfv 3-18; P = 0.001; paired t test) (Fig. 6).
FIG. 6.
The virB9 mutant Cfv 3-18 shows an impaired cytotoxic phenotype in continuous coculture with HeLa cells. HeLa cells were grown to confluence and then infected in triplicate at an MOI of 100 with either C. fetus subsp. venerealis ATCC 19438 (CfvT) or Cfv 3-18. (A) The number of surviving HeLa cells relative to uninfected control cultures is indicated. (B) The number of viable bacteria in the cell culture medium was determined daily. No statistical difference between CfvT and Cfv 3-18 was observed. The values shown are averages of 3 independent experiments. Statistical significance is indicated: *, P < 0.05; **, P < 0.005 (CfvT versus Cfv 3-18). The error bars indicate standard deviations.
Disruption of virD4, but not cdtB, attenuates bacterial invasion of eukaryotic cells.
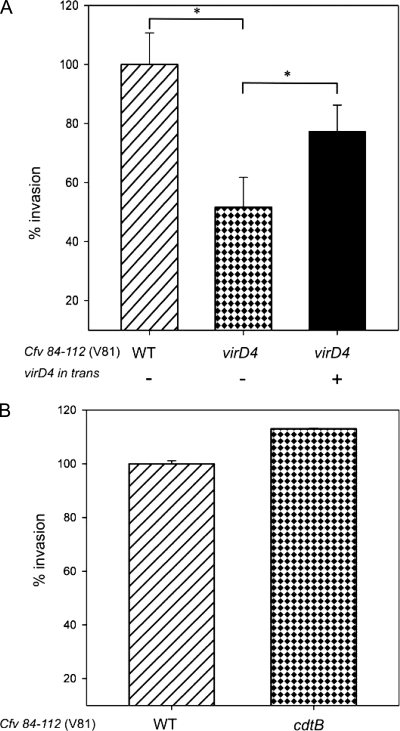
The initial indication gained by the coculture experiments that mutational inactivation of vir genes attenuates the virulence of C. fetus subsp. venerealis was analyzed further. Invasion of eukaryotic cells is a hallmark property of bacterial pathogens. The reference strain, C. fetus subsp. venerealis ATCC 19438, tested as described above for cytotoxic effects, is not invasive when used in eukaryotic cell culture experiments (data not shown). For this analysis, therefore, we used the bovine isolate C. fetus subsp. venerealis 84-112 (V81), which exhibits invasion levels comparable to those of C. jejuni (44). A standard gentamicin protection assay was applied to Caco-2 cells, as this in vitro invasion model is established for Campylobacter (44). We chose to disrupt virD4, since the gene is important to the virulence of various pathogens harboring pathogenicity-associated T4SSs (24, 71), and as it is the last gene in the predicted transcriptional unit, complementation of the mutation would not need to include downstream genes. Caco-2 cells were infected with the wild-type strain (V81) and an isogenic virD4 mutant (V81_SK1). Five hours postinfection, the culture was treated with gentamicin to kill extracellular bacteria. Intracellular bacteria were protected from the antibiotic and were enumerated following eukaryotic cell lysis. Wild-type V81 cells that survived gentamicin treatment were recovered at a ratio of 0.38 per Caco-2 cell, meaning that a maximum of 38% of these cells harbored intracellular bacteria. A similar invasion frequency was achieved over a range of 2 to 7 h of cocultivation before the addition of antibiotic (not shown), in good agreement with prior results for Campylobacter (44). Carriage of the vector control did not alter the invasion proficiency of V81. Accordingly, this wild-type value was set to 100% for comparison to the mutant strains (Fig. 7A). The isogenic virD4 mutant harboring the vector control lost 42% of the wild-type invasiveness, as bacteria were recovered at the significantly reduced ratio of 0.22 per Caco-2 cell (wild type versus virD4 mutant; P = 0.012). The extent of attenuation due to the virD4 disruption is in line with invasion phenotypes observed in a mutational analysis of the pVir-encoded T4SS components of C. jejuni (7). The presence of the virD4 expression construction pRYVL2 in the mutant background enhanced invasion levels significantly compared to the mutant carrying the vector control, reaching 79% of the wild-type level (mutant complemented with virD4 versus virD4 mutant; P = 0.018) (Fig. 7A).
FIG. 7.
virD4 mutation attenuates bacterial invasion. (A) Caco-2 cells were infected with C. fetus subsp. venerealis 84-112 harboring the vector control (left) (V81[pRYSS1]), the isogenic virD4 mutant harboring the vector control (middle) (V81_SK1[pRYSS1]), or the mutant strain harboring virD4 in trans (right) (V81_SK1[pRYVL2]). Extracellular bacteria were killed with gentamicin after 5 h, and the relative number of intracellular bacteria per Caco-2 cell was compared to that of the wild type (WT) (set to 100%). (B) For a negative control of the invasion phenotype, Caco-2 cells were infected with wild-type C. fetus subsp. venerealis 84-112 (V81) or the isogenic mutant in cdtB (V81_JL1) for 3 h. The values shown are the means of three independent experiments plus standard deviations; *, P < 0.02.
To confirm the specific involvement of VirD4 in this model of host cell invasion, we performed a similar mutational analysis of an independent virulence gene not located in the C. fetus genomic island. C. fetus, like other campylobacters, produces a cytolethal distending toxin (CDT), which is thought to be a major virulence factor but does not influence invasion ability (3, 38, 53, 65, 66). The exotoxin is composed of three subunits, including the active component CdtB. The cdtB gene of V81 was inactivated by insertion mutagenesis, and the capacity of the resulting mutant (V81_JL1) to invade Caco-2 cells was compared to that of the parent strain, V81. Intracellular bacteria were recovered from 23% and 26% of Caco-2 cells following infection with the cdtB mutant or the isogenic parent strain, respectively. This difference was not statistically significant (P = 0.362) (Fig. 7B). It is important to note that the invasiveness of the wild-type strain, V81, was not limited to the established human colonic epithelial cell line Caco-2 but was similarly exhibited by the cultured human trophoblast cell line ACH-3P (39 and data not shown).
In summary these experiments identified two virulence properties of C. fetus subsp. venerealis that are manifest in vitro: cytotoxicity against HeLa cells and the ability to invade human epithelial and placenta cells. These virulence attributes were functionally linked to expression of the putative T4SS components VirB9 and VirD4. In contrast, mutational inactivation of CdtB had no effect on the invasiveness of strain V81, validating the specificity of our mutant phenotypes. In conclusion, we propose that vir components of the T4SS encoded by the C. fetus subsp. venerealis genomic island contribute to the virulence properties of the pathogen.
DISCUSSION
A genomic subtractive-hybridization approach was used to investigate the genetic basis potentially underlying the distinct niche preferences displayed by C. fetus subspecies. In this report, we focused on the characteristics of a genomic island that is uniquely present in C. fetus subsp. venerealis and the potential virulence functions contributed by the island to the subspecies. An extended survey of our collection of diverse C. fetus strains from all over the world using gene hybridization and PCR revealed that none of the C. fetus subsp. fetus isolates harbored the island, whereas the set of genes we tested was conserved in the majority of the C. fetus subsp. venerealis isolates. The island seems to represent a major fraction of the genomic difference between the two subspecies as revealed by a recently sequenced genome of C. fetus subsp. venerealis 84-112 (32). Moreover, a recent paper reporting the genome analysis of C. fetus subsp. venerealis AZUL-94 also found that a major difference between the two subspecies was genes encoding type IV secretion components (57).
In general, genomic islands increase the fitness of their hosts by providing new functions. PAIs, a subset of genomic islands, have recently been recognized as a repository of virulence genes in many organisms. Acquisition of PAIs by horizontal gene transfer is clearly responsible for the conversion of many weakly pathogenic or nonpathogenic organisms into major pathogens. Consistent with this model, the island specific to C. fetus subsp. venerealis carries homologs of genes known to be involved in virulence. Notably, the island carries conserved sequence and structural homology to the virB2-virB11 and virD4 loci, named for the prototypical T-DNA transfer system of Agrobacterium tumefaciens, which are typically required for bacterial type IV secretion (15). Secretion systems of this type are complex machineries evolutionarily related to bacterial conjugation systems (20, 90). Many operate in pathogenic bacteria, such as A. tumefaciens, Helicobacter pylori, Bartonella spp., Brucella spp., Bordetella pertussis, and Legionella pneumophila, to translocate effector molecules across the Gram-negative envelope and into bacterial, plant, or mammalian cells (15, 17). The translocated effectors generally have a central function in host-pathogen interactions, e.g., they induce inflammatory responses and cytoskeletal alterations or facilitate the intracellular lifestyle of the bacterial pathogen (6). Experimental evidence has unequivocally established the importance of the T4SS as a pathogenicity factor in each of the systems mentioned. In contrast, identifying the protein substrates of the systems in question has been difficult in most organisms. Accordingly, it has been surprising to identify some systems, such as the Bartonella T4SS, in which genes for the protein effectors are tandemly arrayed immediately downstream of the virB-virD4 operon (72). In the C. fetus genomic island, two putative effector molecules downstream of the virB-virD4 operon were identified based on the presence of a Fic domain. Fic domain-containing proteins translocated by T4S or T3S systems have been identified in several pathogens, including Legionella pneumophila, Coxiella burnetii, and Vibrio parahaemolyticus (67). Moreover, this domain is conserved in the translocated effectors BepA-BepC of Bartonella (72). Fic domain proteins disrupt host cell processes following delivery into eukaryotic host cells. Preliminary data suggest that the protein encoded by orf39 is recognized as a factor for T4 secretion (Kienesberger et al., unpublished). Its potential contribution to virulence is the focus of current investigation.
The species C. fetus is currently divided into two subspecies based on their habitats and biological and phenotypic properties (e.g., glycine tolerance), as well as the diseases they cause (81). C. fetus subsp. fetus strains may be either serotype A or serotype B, whereas C. fetus subsp. venerealis strains are restricted to serotype A (58, 64). These serotypes are associated with differences in LPS structures and also with the type of the surface layer protein (SLP) (76). The C. fetus SLPs form a proteinaceous capsule on the cell surface, which mediates resistance to complement C3b binding. SLPs further undergo antigenic variation to protect against antibody-mediated opsonization, thus contributing to both persistence and systemic infection (12-14, 33, 34, 63, 75, 79). The SLP is a constituent of both C. fetus subspecies, and it represents an ancient, highly conserved constituent of the C. fetus genome, not a pathogenicity island (76). Recent multilocus sequence typing (MLST) data have indicated that C. fetus is genetically homogeneous, in contrast to other Campylobacter species. It displays a highly clonal population structure, wherein C. fetus subsp. venerealis represents a bovine clone (81). The clonality within C. fetus suggests that genetic change accumulates slowly by vertical transmission of point mutations. Additionally, genetic diversity may be limited by the lack of natural competence in C. fetus (77). Other campylobacters, like C. jejuni or H. pylori, show extensive genetic variation, vertically as well as horizontally, and these continuous genetic alterations are important traits of the pathogens for colonizing and persisting in their habitats (10, 18, 85).
Given the phylogenetic perspective that C. fetus subsp. venerealis represents a bovine clone of C. fetus subsp. fetus with limited genetic variation, it is intriguing to speculate that the island discovered in this study contributes to the host tropism and therefore the unique pathogenicity of C. fetus subsp. venerealis. The stringent subspecies prevalence of this island is consistent with this hypothesis. Interestingly, in Bartonella, it was shown that a T4SS originating from a conjugation system was acquired by the pathogen, ultimately serving as a virulence trait (73). The absence of the island in some C. fetus subsp. venerealis isolates underscores its probable origin as a mobile genetic element. Several well-characterized T4SSs have been shown to exhibit dual functionality by maintaining the capacity to mobilize plasmids conjugatively to other bacterial cells (6, 15, 22). Mutagenic analysis of C. fetus subsp. venerealis isolate 84-112 (V81) verified that this system has also retained conjugative functions (Kienesberger et al., unpublished). Consistent with that observation, the structure of the C. fetus genomic island is highly syntenic to resistance plasmids found in other Campylobacter species, pCC31 and pTet (8, 26). Some T4SS components also mediate DNA uptake, thus conferring a natural competence on organisms such as C. jejuni or H. pylori (7, 42). The C. fetus subspecies are refractory to DNA uptake by transformation, and therefore, competence is not a function linked to the identified locus (48, 77). High-level homology to an uncharacterized genomic island of C. hominis also exists. The question of whether this genome region represents a symbiosis island conferring increased fitness on this nonpathogenic species remains open (36, 37). Other genomic islands with similarity to plasmids are also present in certain campylobacters, for example, the 71-kb Helicobacter hepaticus ATCC 51449 genomic island HHGI1 shares high homology with the C. coli plasmid pCC178, and the Wolinella succinogenes DSM 1740 genomic island, WsuGI, is syntenic to the C. jejuni plasmid pVir (5, 23, 25). Mobility genes, like IS elements or prophages, present on such islands facilitate gene shuffling between plasmids and the core genome and contribute to genome diversity (40). Laterally acquired DNA accounts for up to 5 to 6% of the genomes of certain campylobacters (23). Interestingly, the T4SS located on the C. fetus genomic island and its syntenic counterparts on pCC31 and pTet show a significant level of homology to T4SSs found in the mucosal pathogens Actinobacillus actinomycetemcomitans and Brucella spp., suggesting a possible common ancestry (8, 27, 60).
In conclusion, we propose that in the species C. fetus, the presence of the virB-virD4 locus is apparently unique to the subspecies C. fetus subsp. venerealis. All indications suggest that the locus is part of a mobile genetic element. Consistent with this expected mobility, the genomic island is not universally found in the C. fetus subsp. venerealis isolates we tested. Six strains currently lack an insertion at this site on the genome. Lastly, the question arises whether the ancestral locus or independently evolved homologs have also spread among other campylobacters. Emerging evidence suggests that this is the case (57). As the genomic database expands, a wealth of information concerning the mobile gene pool is certain to emerge. The challenge will be to understand adaptations in distinct organisms that create functional diversity using shared and unique genes. In this study, mutational inactivation of virB9 and virD4 unambiguously verified a contribution of the genomic island to the virulence properties of C. fetus subsp. venerealis. Given the genetic homogeneity of C. fetus subspecies, the discovery of the C. fetus subsp. venerealis genomic island warrants further analysis of the properties it provides to this host-adapted mucosal pathogen.
Supplementary Material
Acknowledgments
We thank J. Wagenaar, M. van Bergen, A. Burnens, and S. Hum for providing C. fetus strains. We are grateful to S. Häusler for excellent technical assistance and to J. Liebergesell, M. M. Joainig, B. Munk and M. Monschein for their contribution to this work.
This study was supported by the Austrian FWF P18607-B12 (E.L.Z.) and FWF P20479-B05 (G.G.) and the Austrian Academy of Science's DOC scholarship, awarded to S.K.
Footnotes
Published ahead of print on 6 November 2009.
Supplemental material for this article may be found at http://jb.asm.org./.
REFERENCES
- 1.Amieva, M. R., R. Vogelmann, A. Covacci, L. S. Tompkins, W. J. Nelson, and S. Falkow. 2003. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300:1430-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 1996. OIE manual of standards for diagnostic tests and vaccines, 3rd ed., p. 256-266. Office International des Epizooties, Paris, France.
- 3.Asakura, M., W. Samosornsuk, M. Taguchi, K. Kobayashi, N. Misawa, M. Kusumoto, K. Nishimura, A. Matsuhisa, and S. Yamasaki. 2007. Comparative analysis of cytolethal distending toxin (cdt) genes among Campylobacter jejuni, C. coli and C. fetus strains. Microb. Pathog. 42:174-183. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 5.Baar, C., M. Eppinger, G. Raddatz, J. Simon, C. Lanz, O. Klimmek, R. Nandakumar, R. Gross, A. Rosinus, H. Keller, P. Jagtap, B. Linke, F. Meyer, H. Lederer, and S. C. Schuster. 2003. Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl. Acad. Sci. U. S. A. 100:11690-11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Backert, S., and T. F. Meyer. 2006. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9:207-217. [DOI] [PubMed] [Google Scholar]
- 7.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in the virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batchelor, R. A., B. M. Pearson, L. M. Friis, P. Guerry, and J. M. Wells. 2004. Nucleotide sequences and comparison of two large conjugative plasmids from different Campylobacter species. Microbiology 150:3507-3517. [DOI] [PubMed] [Google Scholar]
- 9.Blaser, M. J. 1998. Campylobacter fetus—emerging infection and model system for bacterial pathogenesis at mucosal surfaces. Clin. Infect. Dis. 27:256-258. [DOI] [PubMed] [Google Scholar]
- 10.Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: biology and disease. J. Clin. Invest. 113:321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaser, M. J., D. N. Newell, S. A. Thompson, and E. L. Zechner. 2008. Pathogenesis of Campylobacter fetus, p. 401-428. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 12.Blaser, M. J., and Z. Pei. 1993. Pathogenesis of Campylobacter fetus infections: critical role of the high molecular weight S-layer proteins in virulence. J. Infect. Dis. 167:696-706. [DOI] [PubMed] [Google Scholar]
- 13.Blaser, M. J., P. F. Smith, J. A. Hopkins, I. Heinzer, J. H. Bryner, and W. L. Wang. 1987. Pathogenesis of Campylobacter fetus infections: serum resistance associated with high-molecular-weight surface proteins. J. Infect. Dis. 155:696-706. [DOI] [PubMed] [Google Scholar]
- 14.Blaser, M. J., P. F. Smith, J. E. Repine, and K. A. Joiner. 1988. Pathogenesis of Campylobacter fetus infections: failure of encapsulated Campylobacter fetus to bind C3b explains serum and phagocytosis resistance. J. Clin. Invest. 81:1434-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Champion, O. L., S. Al-Jaberi, R. A. Stabler, and B. W. Wren. 2008. Comparative genomics of Campylobacter jejuni, p. 63-71. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 17.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. van Putten, L. M. Schouls, and B. Duim. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 19.Debruyne, L., D. Gevers, and P. Vandamme. 2008. Taxonomy of the family Campylobacteraceae, p. 3-25. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 20.de la Cruz, F., L. S. Frost, R. J. Meyer, and E. L. Zechner. 2009. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Micriobiol. Rev. [Epub ahead of print.] doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed]
- 21.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 22.Ding, Z., K. Atmakuri, and P. J. Christie. 2003. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 11:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eppinger, M., C. Baar, G. Raddatz, D. H. Huson, and S. C. Schuster. 2004. Comparative analysis of four Campylobacterales. Nat. Rev. Microbiol. 2:872-885. [DOI] [PubMed] [Google Scholar]
- 24.Fischer, W., J. Püls, R. Buhrdorf, B. Gebert, S. Odenbreit, and R. Haas. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42:1337-1348. [DOI] [PubMed] [Google Scholar]
- 25.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friis, L. M., C. Pin, D. E. Taylor, B. M. Pearson, and J. M. Wells. 2007. A role for the tet(O) plasmid in maintaining Campylobacter plasticity. Plasmid 57:18-28. [DOI] [PubMed] [Google Scholar]
- 27.Galli, D. M., J. Chen, K. F. Novak, and D. J. Leblanc. 2001. Nucleotide sequence and analysis of conjugative plasmid pVT745. J. Bacteriol. 183:1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia, M. M., M. D. Eaglesome, and C. Rigby. 1983. Campylobacters important in veterinary medicine. Vet. Bull. 53:793-818. [Google Scholar]
- 29.Giesendorf, B. A., H. Goossens, H. G. Niesters, A. Van Belkum, A. Koeken, H. P. Endtz, H. Stegeman, and W. G. Quint. 1994. Polymerase chain reaction-mediated DNA fingerprinting for epidemiological studies on Campylobacter spp. J. Med. Microbiol. 40:141-147. [DOI] [PubMed] [Google Scholar]
- 30.Gorkiewicz, G., G. Feierl, R. Zechner, and E. L. Zechner. 2002. Transmission of Campylobacter hyointestinalis from a pig to a human. J. Clin. Microbiol. 40:2601-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorkiewicz, G., G. Feierl, C. Schober, F. Dieber, J. Kofer, R. Zechner, and E. L. Zechner. 2003. Species-specific identification of campylobacters by partial 16S rRNA gene sequencing. J. Clin. Microbiol. 41:2537-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorkiewicz, G., S. Kienesberger, C. Gülly, and E. L. Zechner. 2009. Comparative genome analysis of Campylobacter fetus subspecies reveals loci important for virulence and host adaption, abstr. P-99. Abstr. 15th Int. Workshop Campylobacter, Helicobacter Rel. Organisms (CHRO), Niigata, Japan.
- 33.Grogono-Thomas, R., J. Dworkin, M. J. Blaser, and D. G. Newell. 2000. Roles of the surface layer proteins of Campylobacter fetus subsp. fetus in ovine abortion. Infect. Immun. 68:1687-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grogono-Thomas, R., M. J. Blaser, M. Ahmadi, and D. G. Newell. 2003. Role of S-layer protein antigenic diversity in the immune responses of sheep experimentally challenged with Campylobacter fetus subsp. fetus. Infect. Immun. 71:147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guerry, P., R. Yao, R. A. Alm, D. H. Burr, and T. J. Trust. 1994. Systems of experimental genetics for Campylobacter species. Methods Enzymol. 235:474-481. [DOI] [PubMed] [Google Scholar]
- 36.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 37.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 38.Hickey, T. E., A. L. McVeigh, D. A. Scott, R. E. Michielutti, A. Bixby, S. A. Carroll, A. L. Bourgeois, and P. Guerry. 2000. Campylobacter jejuni cytolethal distending toxin mediates release of interleukin-8 from intestinal epithelial cells. Infect. Immun. 68:6535-6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiden, U., C. Wadsack, N. Prutsch, M. Gauster, U. Weiss, H. G. Frank, U. Schmitz, C. Fast-Hirsch, M. Hengstschläger, A. Pötgens, A. Rüben, M. Knöfler, P. Haslinger, B. Huppertz, M. Bilban, P. Kaufmann, and G. Desoye. 2007. The first trimester human trophoblast cell line ACH-3P: a novel tool to study autocrine/paracrine regulatory loops of human trophoblast subpopulations—TNF-alpha stimulates MMP15 expression. BMC Dev. Biol. 7:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofreuter, D., and R. Haas. 2002. Characterization of two cryptic Helicobacter pylori plasmids: a putative source for horizontal gene transfer and gene shuffling. J. Bacteriol. 184:2755-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofreuter, D., V. Novik, and J. E. Galán. 2008. Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 4:425-433. [DOI] [PubMed] [Google Scholar]
- 42.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 43.Hosie, A. H., and P. S. Poole. 2001. Bacterial ABC transporters of amino acids. Res. Microbiol. 152:259-270. [DOI] [PubMed] [Google Scholar]
- 44.Hu, L., and D. J. Kopecko. 2000. Interactions of Campylobacter with eukaryotic cells: gut luminal colonization and mucosal invasion mechanisms, p. 191-215. In I. Nachamkin, and M. J. Blaser (ed.), Campylobacter, 2nd ed., ASM Press, Washington, DC.
- 45.Hubank, M., and D. G. Schatz. 1994. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 22:5640-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hum, S., K. Quinn, J. Brunner, and S. L. On. 1997. Evaluation of a PCR assay for identification and differentiation of Campylobacter fetus subspecies. Aust. Vet. J. 75:827-831. [DOI] [PubMed] [Google Scholar]
- 47.Kersulyte, D., B. Velapatino, G. Dailide, A. K. Mukhopadhyay, Y. Ito, L. Cahuayme, A. J. Parkinson, R. H. Gilman, and D. E. Berg. 2002. Transposable element ISHp608 of Helicobacter pylori: nonrandom geographic distribution, functional organization, and insertion specificity. J. Bacteriol. 184:992-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kienesberger, S., G. Gorkiewicz, M. M. Joainig, S. R. Scheicher, E. Leitner, and E. L. Zechner. 2007. Development of experimental genetic tools for Campylobacter fetus. Appl. Environ. Microbiol. 73:4619-4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kienesberger, S., M. M. Joainig, G. Gorkiewicz, and E. L. Zechner. 2007. Promoter analysis and extension of experimental genetics tools for manipulation of Campylobacter fetus. Zoonoses Public Health 54(Suppl. 1):S60. [Google Scholar]
- 50.Koraimann, G. 2003. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol. Life Sci. 60:2371-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krause, R., S. Ramschak-Schwarzer, G. Gorkiewicz, W. J. Schnedl, G. Feierl, C. Wenisch, and E. C. Reisinger. 2002. Recurrent septicemia due to Campylobacter fetus and Campylobacter lari in an immunocompetent patient. Infection 30:171-174. [DOI] [PubMed] [Google Scholar]
- 52.Labigne-Roussel, A., P. Courcoux, and L. Tompkins. 1988. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J. Bacteriol. 170:1704-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lara-Tejero, M., and J. E. Galan. 2000. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 290:354-357. [DOI] [PubMed] [Google Scholar]
- 54.Lisitsyn, N., N. Lisitsyn, and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 55.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller, W. G. 2008. Comparative genomics of Campylobacter species other than Campylobacter jejuni, p. 73-95. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 57.Moolhuijzen, P. M., A. E. Lew-Tabor, B. M. Wlodek, F. G. Agüero, D. J. Comerci, R. A. Ugalde, D. O. Sanchez, R. Appels, and M. Bellgard. 2009. Genomic analysis of Campylobacter fetus subspecies: identification of candidate virulence determinants and diagnostic assay targets. BMC Microbiol. 9:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moran, A. P., D. T. O'Malley, T. U. Kosunen, and I. M. Helander. 1994. Biochemical characterization of Campylobacter fetus lipopolysaccharides. Infect. Immun. 62:3922-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nirdnoy, W., C. J. Mason, and P. Guerry. 2005. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob. Agents Chemother. 49:2454-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Novak, K. F., B. Dougherty, and M. Pelaez. 2001. Actinobacillus actinomycetemcomitans harbours type IV secretion system genes on a plasmid and in the chromosome. Microbiology 147:3027-3035. [DOI] [PubMed] [Google Scholar]
- 61.On, S. L. 1996. Identification methods for campylobacters, helicobacters, and related organisms. Clin. Microbiol. Rev. 9:405-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.On, S. L., and C. S. Harrington. 2001. Evaluation of numerical analysis of PFGE-DNA profiles for differentiating Campylobacter fetus subspecies by comparison with phenotypic, PCR and 16S rDNA sequencing methods. J. Appl. Microbiol. 90:285-289. [DOI] [PubMed] [Google Scholar]
- 63.Pei, Z., and M. J. Blaser. 1990. Pathogenesis of Campylobacter fetus infections: role of surface array proteins in virulence in a mouse model. J. Clin. Invest. 85:1036-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pérez-Pérez, G. I., M. J. Blaser, and J. H. Bryner. 1986. Lipopolysaccharide structures of Campylobacter fetus are related to heat-stable serogroups. Infect. Immun. 51:209-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pickett, C. L., E. C. Pesci, D. L. Cottle, G. Russell, A. N. Erdem, and H. Zeytin. 1996. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB gene. Infect. Immun. 64:2070-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pickett, C. L. 2000. Campylobacter toxins and their role in pathogenesis, p. 179-190. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 67.Roy, C. G., and S. Mukherjee. 2009. Bacterial FIC proteins AMP up infection. Sci. Signal. 2:pe14. [DOI] [PubMed] [Google Scholar]
- 68.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 69.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 70.Schmidt, H., and M. Hensel. 2004. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17:14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schulein, R., and C. Dehio. 2002. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol. Microbiol. 46:1053-1067. [DOI] [PubMed] [Google Scholar]
- 72.Schulein, R., P. Guye, T. A. Rhomberg, M. C. Schmid, G. Schröder, A. C. Vergunst, I. Carena, and C. Dehio. 2005. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc. Natl. Acad. Sci. U. S. A. 102:856-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seubert, A., R. Hiestand, F. de la Cruz, and C. Dehio. 2003. A bacterial conjugation machinery recruited for pathogenesis. Mol. Microbiol. 49:1253-1266. [DOI] [PubMed] [Google Scholar]
- 74.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infection, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 75.Thompson, S. A., and M. J. Blaser. 2000. Pathogenesis of Campylobacter fetus infections, p. 321-347. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 76.Tu, Z. C., F. E. Dewhirst, and M. J. Blaser. 2001. Evidence that the Campylobacter fetus sap locus is an ancient genomic constituent with origins before mammals and reptiles diverged. Infect. Immun. 69:2237-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tu, Z. C., T. M. Wassenaar, S. A. Thompson, and M. J. Blaser. 2003. Structure and genotypic plasticity of the Campylobacter fetus sap locus. Mol. Microbiol. 48:685-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tu, Z. C., G. Zeitlin, J. P. Gagner, T. Keo, B. A. Hanna, and M. J. Blaser. 2004. Campylobacter fetus of reptile origin as a human pathogen. J. Clin. Microbiol. 42:4405-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tu, Z. C., C. Gaudreau, and M. J. Blaser. 2005. Mechanisms underlying Campylobacter fetus pathogenesis in humans: surface-layer protein variation in relapsing infections. J. Infect. Dis. 191:2082-2089. [DOI] [PubMed] [Google Scholar]
- 80.Tu, Z. C., W. Eisner, B. N. Kreiswirth, and M. J. Blaser. 2005. Genetic divergence of Campylobacter fetus strains of mammal and reptile origins. J. Clin. Microbiol. 43:3334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Bergen, M. A., K. E. Dingle, M. C. Maiden, D. G. Newell, L. van der Graaf-Van Bloois, J. P. van Putten, and J. A. Wagenaar. 2005. Clonal nature of Campylobacter fetus as defined by multilocus sequence typing. J. Clin. Microbiol. 43:5888-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Véron, M., and R. Chatelain. 1973. Taxonomic study of the genus Campylobacter Sebald and Véron and designation of the neotype strain for the type species, Campylobacter fetus (Smith and Taylor) Sebald and Véron. Int. J. Syst. Bacteriol. 23:122-134. [Google Scholar]
- 83.Wagenaar, J. A., M. A. van Bergen, D. G. Newell, R. Grogono-Thomas, and B. Duim. 2001. Comparative study using amplified fragment length polymorphism fingerprinting, PCR genotyping, and phenotyping to differentiate Campylobacter fetus strains isolated from animals. J. Clin. Microbiol. 39:2283-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wassenaar, T. M., B. N. Fry, and B. A. van der Zeijst. 1993. Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene 132:131-135. [DOI] [PubMed] [Google Scholar]
- 85.Wiesner, R. S., D. R. Hendrixson, and V. J. DiRita. 2003. Natural transformation of Campylobacter jejuni requires components of a type II secretion system. J. Bacteriol. 185:5408-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Worby, C. A., S. Mattoo, R. P. Kruger, L. B. Corbeil, A. Koller, J. C. Mendez, B. Zekarias, C. Lazar, and J. E. Dixon. 2009. The fic domain: regulation of cell signaling by adenylylation. Mol. Cell 34:93-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wosten, M. M., M. Boeve, M. G. Koot, A. C. van Nuene, and B. A. van der Zeijst. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zahrl, D., M. Wagner, K. Bischof, M. Bayer, B. Zavecz, A. Beranek, C. Ruckenstuhl, G. E. Zarfel, and G. Koraimann. 2005. Peptidoglycan degradation by specialized lytic transglycosylases associated with type III and type IV secretion systems. Microbiology 151:3455-3467. [DOI] [PubMed] [Google Scholar]
- 90.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 2000. Conjugative-DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool: bacterial plasmids and gene spread. Harwood Academic Publishers, Amsterdam, The Netherlands.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.