Tyr131 plays an important role in determining the substrate specificity and thermal stability of azoreductase 1 from P. aeruginosa (pAzoR1). The structure shows that the substrate methyl red binds deeper in the active site of the mutant, which is reflected in the change in enzymic properties.
Keywords: paAzoR1, azoreductases
Abstract
Azoreductase 1 from Pseudomonas aeruginosa strain PAO1 (paAzoR1) catalyses the activation of the prodrug balsalazide and reduces the azo dye methyl red using reduced nicotinamide adenine dinucleotide cofactor as an electron donor. To investigate the mechanism of the enzyme, a Y131F mutation was introduced and the enzymic properties of the mutant were compared with those of the wild-type enzyme. The crystallographic structure of the mutant with methyl red bound was solved at 2.1 Å resolution and compared with the wild-type structure. Tyr131 is important in the architecture of the active site but is not essential for enzymic activity.
1. Introduction
Azoreductases catalyse the reductive cleavage of azo bonds at the expense of NAD(P)H. Azoreductases in intestinal microflora are essential for activation of azo prodrugs in the treatment of inflammatory bowel disease. The anti-inflammatory agent 5-aminosalicylic acid can either be administered directly or is given in a prodrug formulation via an azo cross-link to an inert carrier molecule, with the prodrug being activated in the colon by azoreductases expressed by gastrointestinal microflora (Peppercorn & Goldman, 1972 ▶; Dissanayake & Truelove, 1973 ▶; Hanauer, 1996 ▶).
We have described an oxygen-insensitive azoreductase from Pseudomonas aeruginosa (paAzoR1; PDB code 2v9c), which activates the azo prodrug balsalazide with relatively high specificity compared with common reference azo dyes (Wang et al., 2007 ▶). The three-dimensional structure of paAzoR1 was the first azoreductase structure to be reported with a flavin mononucleotide (FMN) prosthetic group and an azo substrate (methyl red) bound. From the structure, Tyr131 was identified as the potential key electron-transferring residue as it is suitably located to transfer protons during the reduction reaction. Secondly, when an equivalent mutation (Y196F) was made to old yellow enzyme 1, an azoreductase from Sacharomyces carlsbergensis, the oxidative half-reaction was slowed dramatically by nearly six orders of magnitude (Kohli & Massey, 1998 ▶). A recombinant mutant paAzoR1 in which Tyr131 is replaced by phenylalanine has been made and the enzymatic characterization and structure of the mutant with substrate bound are described here.
2. Materials and methods
2.1. Site-directed mutagenesis and transformation
The pET28b(+) plasmid vector containing the sequence of paazor1 was isolated from a 6 ml culture of Escherichia coli BL21 (DE3) using a QIAprep Spin Miniprep Kit (Qiagen). Site-directed mutagenesis was achieved using a QuikChange II kit (Stratagene). The reaction mixture (50 µl), which contained 5 µl 10× reaction buffer, 50 ng pET28b(+) plasmid as template, 125 ng of both Y131F forward (5′-CGTCGCCCAGTTCCGGCCGCTGC-3′) and Y131F reverse (5′-GCAGCGGCCGGAACTGGGCGACG-3′) primers and 1 µl dNTP mix, was subjected to thermo-cycling: 30 s at 368 K and 12 cycles of 30 s at 368 K, 1 min at 328 K and 6 min at 341 K, followed by 2 min on ice. The reaction mixture was digested (10 U DpnI, 1 h, 310 K) to cleave all template DNA before being purified by gel electrophoresis in ethidium-bromide-stained agarose [1.0%(w/v)]. The sequence of the insert was confirmed and the pET28b(+)-Y131F plasmid vector was transformed into E. coli BL21 (DE3) pLysS as described previously (Wang et al., 2007 ▶).
2.2. Protein production, purification and characterization
Like wild-type paAzoR1, the Y131F mutant was expressed with a hexahistidine tag; the protein was then purified via affinity purification as described previously (Wang et al., 2007 ▶). Spectra were recorded from 600 to 300 nm and the enzymic activities were determined as described previously (Wang et al., 2007 ▶). Thermostability was determined by incubating the enzyme (10 min) from 277 to 363 K followed by 1 min on ice before adding methyl red and NADPH and measuring the loss of absorbance as described in Wang et al. (2007 ▶). Initial velocities were also measured at different concentrations of substrate and cofactor [from 5 to 40 µM methyl red and from 0.1 to 2.0 mM NAD(P)H], while the concentration of the other substrate was kept constant (Wang et al., 2007 ▶). In each case, linearity was established. Apparent K m and V max values were obtained from Lineweaver–Burk plots using the initial linear rates expressed in µM s−1.
2.3. Protein crystallization and structural determination
Crystals of the mutant enzyme in the presence of methyl red (Y131F_MRE) were grown by sitting-drop vapour diffusion. Protein drops were prepared by mixing equal volumes (1 µl) of protein solution (23 mg ml−1 in sterile water with 2 mM methyl red) with mother liquor (Molecular Dimensions JCSG-plus screen condition E8; 0.1 M sodium acetate pH 4.5, 1.0 M diammonium hydrogen phosphate) in a 24-well CrystalPlate (Axygen Bioscience). Crystals were briefly transferred to a cryoprotectant solution consisting of 1:3 glycerol:mother liquor prior to freezing in liquid nitrogen. Data were collected on beamline ID14-EH4 at the European Synchrotron Radiation Facility (ESRF, Grenoble) using a Quantum 315 ADSC CCD detector. Data were integrated with iMOSFLM v.0.5.2 (Leslie, 1992 ▶) and scaled and merged with SCALA (Collaborative Computational Project, Number 4, 1994 ▶). The structure was solved using the molecular-replacement program Phaser (Read, 2001 ▶), with the paAzoR1 structure (PDB code 2v9c) as a model. The atomic model of Y131F_MRE was rebuilt and refined with Coot (Emsley & Cowtan, 2004 ▶), REFMAC 5.4 (Murshudov et al., 1997 ▶) and PHENIX (Adams et al., 2002 ▶). Initial translation, liberation and screw (TLS) parameters were determined with the program TLSMD (Painter & Merritt, 2006 ▶) and TLS refinement was performed with PHENIX (Adams et al., 2002 ▶). Noncrystallographic symmetry (NCS) restraints (tight main chain and moderate side chain) were applied to the two protein chains in the asymmetric unit. Water molecules were added with PHENIX (Adams et al., 2002 ▶), Coot (Emsley & Cowtan, 2004 ▶) and ARP/wARP (Morris et al., 2003 ▶). Model validation was performed with MolProbity (Davis et al., 2007 ▶) and multimer analysis was performed using PISA (Krissinel & Henrick, 2005 ▶). Data-collection and refinement statistics are shown in Table 1 ▶.
Table 1. Data-collection and refinement statistics for Y131F_MRE.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P3121 |
| Unit-cell parameters (Å, °) | a = b = 82.81, c = 109.05, α = β = 90, γ = 120 |
| Wavelength (Å) | 0.97650 |
| Resolution range (Å) | 43.35–2.10 (2.20–2.10) |
| Unique reflections | 25785 (3689) |
| Rmerge† | 0.066 (0.474) |
| 〈I/σ(I)〉 | 20.7 (4.0) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 7.0 (7.2) |
| Wilson B value (Å2) | 34 |
| Refinement and model statistics | |
| Resolution (Å) | 43.4–2.10 (2.18–2.10) |
| Rwork‡ | 0.182 (0.210) |
| Rfree‡ | 0.217 (0.281) |
| No. of residues (chain A/chain B) | 192/192 |
| No. of water molecules | 269 |
| Additional molecules | 1 glycerol |
| Total No. of atoms | 3495 |
| R.m.s. deviation from ideal bond lengths§ (Å) | 0.008 |
| R.m.s. deviation from ideal bond angles§ (°) | 0.831 |
| Mean B factor (Å2) | 35 |
| Ramachandran statistics¶ (%) | |
| Preferred region | 96.8 |
| Allowed region | 2.7 |
| Outliers | 0.5 |
R
merge = 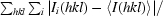
 , where I
i(hkl) is the intensity of the ith observation of unique reflection hkl.
, where I
i(hkl) is the intensity of the ith observation of unique reflection hkl.
R
work and R
free = 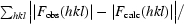
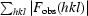 for the working set and test set (5%) of reflections, respectively, where |F
obs(hkl)| and |F
calc(hkl)| are the observed and calculated structure-factor amplitudes for reflection hkl.
for the working set and test set (5%) of reflections, respectively, where |F
obs(hkl)| and |F
calc(hkl)| are the observed and calculated structure-factor amplitudes for reflection hkl.
The idealized protein-geometry libraries used were those of Engh & Huber (1991 ▶).
The Ramachandran plot was calculated in MolProbity (Davis et al., 2007 ▶).
3. Results and discussion
3.1. Production of purified recombinant Y131F
Y131F was successfully overexpressed in E. coli following induction with 0.25 mM IPTG. The recombinant protein was affinity purified on Ni–NTA, eluting in the 250 mM imidazole wash. Approximately 200 mg of pure Y131F paAzoR1 was produced per litre of culture (a similar yield to that of wild-type paAzoR1; Wang et al., 2007 ▶). Like the native protein, Y131F paAzoR1 is soluble at over 40 mg ml−1 in 20 mM Tris–HCl buffer pH 8. The theoretical molecular mass of Y131F paAzoR1 with a hexahistidine tag is 25 024.1 Da. The migration of Y131F on SDS–PAGE corresponds to approximately 31 kDa (data not shown), a phenomenon that was also observed for paAzoR1 (Wang et al., 2007 ▶).
3.2. Characterization of Y131F
The UV–Vis absorbance spectrum of Y131F paAzoR1 exhibits a typical flavoprotein signature and is indistinguishable from that of wild-type paAzor1 (Wang et al., 2007 ▶). The spectral properties indicate that the protein is folded because the λmax is shifted by approximately 10 nm, with an absorbance shoulder at around 490 nm, compared with the spectrum of FMN (Zenno et al., 1996 ▶; Duurkens et al., 2007 ▶). When Y131F paAzoR1 is denatured with SDS, the shoulder at 490 nm is lost and the spectrum resembles that of FMN alone, suggesting that FMN is noncovalently bound to the protein as is the case for paAzoR1 (Wang et al., 2007 ▶). Spectral studies also show that the flavin cofactor of Y131F is reduced by either NADH or NADPH and also by dithionite. The milder reducing agent dithiothreitol does not reduce FMN in Y131F paAzoR1 (data not shown).
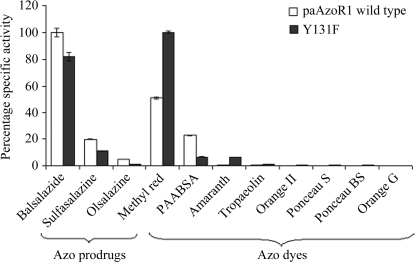
Expression of the mutated enzyme results in active protein, which suggests but does not prove that Y131F paAzoR1 is correctly folded (Fig. 1 ▶). The substrate specificity is similar but not identical to that of paAzoR1 (Fig. 1 ▶). The most notable changes are in the activity of Y131F paAzoR1 towards methyl red and balsalazide (Fig. 1 ▶). There is an increase in the V max for methyl red, whilst the V max for balsalazide is decreased (Table 2 ▶). The K m for balsalazide is very similar for the mutant and paAzoR1, while the K m for methyl red is decreased (Table 2 ▶). As a result, there is a threefold increase in the specificity constant (k cat/K m) for methyl red of Y131F paAzoR1 (429 mM −1 s−1 compared with 137 mM −1 s−1 for the wild type), while the constant for balsalazide decreases by ∼40% (234 mM −1 s−1 compared with 373 mM −1 s−1 for the wild type).
Figure 1.
Comparison of the substrate profiles of purified recombinant wild-type and Y131F paAzoR1. The percentage specific activity towards the best substrate for each enzyme was taken as 100%. Results are expressed as mean ± standard deviation from triplicate determinations. The enzymic reaction mixture (200 µl) contained 10 µg purified enzyme and 50 µM azo compound in 20 mM Tris–HCl pH 8.0, 0.3 M NaCl. The reaction was initiated by adding 0.5 mM NADPH for paAzoR1 and the loss of colour was assessed as described in §2.
Table 2. Comparison of paAzoR1 and Y131F.
Apparent values of the Michaelis–Menten constants (K m), maximum velocities (V max) and turnover numbers (k cat) of purified recombinant paAzoR1 with different substrates are reported. The term apparent indicates that the kinetic constants were determined by varying the concentration of one substrate while keeping the concentration of the other constant. The values for methyl red and balsalazide were determined with NADPH (at 500 µM) as the electron donor and the values for NADH and NADPH were determined with methyl red at 50 µM as the azo substrate (Wang et al., 2007 ▶).
| Apparent kinetic constants | MRE | Balsalazide | NADH | NADPH | |
|---|---|---|---|---|---|
| PaAzoR1 | Vmax (µM s−1) | 0.28 ± 0.01 | 0.81 ± 0.02 | 0.22 ± 0.01 | 0.74 ± 0.03 |
| Km (µM) | 92.7 ± 7.6 | 98.6 ± 4.2 | 538 ± 21 | 1197 ± 33 | |
| kcat (s−1) | 12.7 ± 0.5 | 36.8 ± 1.1 | 10.0 ± 0.5 | 33.6 ± 2.0 | |
| Y131F | Vmax (µM s−1) | 0.42 ± 0.02 | 0.53 ± 0.02 | 0.55 ± 0.02 | 0.78 ± 0.04 |
| Km (µM) | 44.5 ± 2.2 | 102.9 ± 3.2 | 965 ± 25 | 535 ± 25 | |
| kcat (s−1) | 19.1 ± 0.9 | 24.1 ± 0.3 | 25.2 ± 0.9 | 35.5 ± 1.0 |
There is also a change in the kinetic parameters for reduced nicotinamide adenine dinucleotide cofactor such that the catalytic constant of Y131F paAzoR1 is 2.5-fold greater than that of wild-type paAzoR1 for NADH, whilst the catalytic constant for NADPH remains relatively unchanged. Interestingly, in the NADH-dependent azoreductases from E. coli (Nakanishi et al., 2001 ▶), Salmonella typhimurium (PDB code 1t5b; R. Zhang, R. Wu, F. Collart & A. Joachimiak, unpublished work), Enterococcus faecalis (Chen et al., 2004 ▶) and the NADH-preferring paAzoR2 and paAzoR3 the residue corresponding to Tyr131 is a conserved proline, which has a similar hydrophobicity index to phenylalanine. These data strongly suggest that Tyr131 is not the source of the proton that triggers the reaction sequence.
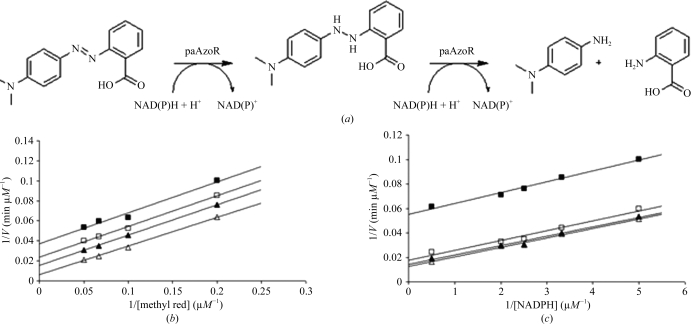
Many flavin-containing oxidoreductases (Lambeth et al., 1976 ▶; Lambeth & Kamin, 1977 ▶; Li et al., 1995 ▶; St Maurice et al., 2007 ▶), including the related E. coli azoreductase (Nakanishi et al., 2001 ▶), have been shown to act via a ping-pong bi-bi mechanism. Owing to its structural/functional similarity to the E. coli azoreductase, it was assumed that paAzoR1 follows the same mechanism (Wang et al., 2007 ▶). This was proven to be correct by analysis of the enzyme kinetics exhibited by wild-type paAzoR1 (Fig. 2 ▶). This mechanism requires two molecules of NAD(P)H to sequentially reduce FMN to FMNH2, which in turn reduces the azo substrate.
Figure 2.
Proposed catalytic reaction of paAzoRs. (a) Azoreductase reduces methyl red to N,N′-dimethyl-p-phenylenediamine and 2-aminobenzoic acid with the consumption of 2 mol NAD(P)H. FMN serves as the electron mediator between NAD(P)H and methyl red in a proposed double ping-pong bi-bi biomechanism. (b) Lineweaver–Burk plots showing the ping-pong bi-bi mechanism in the azoreduction of methyl red by paAzoR1 with NADPH as the electron donor. The plots show the effect of varying the methyl red concentration; the concentrations tested were 5, 10, 15 and 20 µM in 20 mM Tris–HCl buffer containing 0.3 M NaCl and 5 µg paAzoR1. The reactions were initiated with NADPH at concentrations of 0.2 mM (solid squares), 0.3 mM (open squares), 0.4 mM (solid triangles) and 0.5 mM (open triangles) in a total volume of 200 µl. (c) Lineweaver–Burk plots showing the ping-pong bi-bi mechanism in the azoreduction of methyl red by paAzoR1 with NADPH as the electron donor. The plots show the effect of varying the concentration of NADPH. The NADPH concentrations tested were 0.2, 0.3, 0.4, 0.5 and 2 mM in 20 mM Tris–HCl buffer, 0.3 M NaCl. The reactions were initiated by adding NADPH to a reaction mixture containing 5 µg paAzoR1 and methyl red at 5 µM (solid squares), 15 µM (open squares), 20 µM (solid triangles) or 25 µM (open triangles) in a total volume of 200 µl. In each case the rate is the initial rate of reduction of methyl red.
3.3. Effects of the mutation on folding
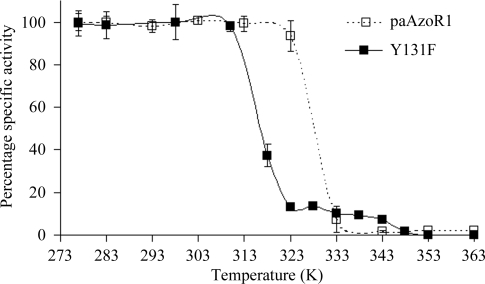
Whilst wild-type paAzoR1 loses 50% of its activity after incubation for 10 min at 328 K, Y131F paAzoR1 loses 50% of its activity after exposure to 315 K for 10 min (Fig. 3 ▶). As loss of activity has previously been shown in other enzymes to correlate with thermal unfolding (Kawamura et al., 2003 ▶), this indicates that the mutant is less thermostable than the wild-type enzyme. This is an unusual result, as the introduction of more hydrophobic residues normally leads to an increase in thermostability (Zhou et al., 2008 ▶).
Figure 3.
Comparison of the thermostability of wild-type and Y131F paAzoR1. The assay mixture (200 µl) contained 10 µg heat-treated Y131F (solid squares) or wild-type paAzoR1 (open squares) and 50 µM methyl red in 20 mM Tris–HCl pH 8.0 containing 0.3 M NaCl; the reaction was initiated by adding 0.5 mM NADPH. Results are expressed as mean ± standard deviation from three determinations.
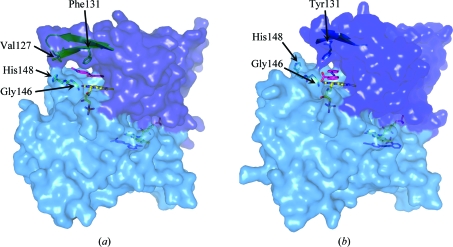
To ensure that the change in thermostability did not arise from the mutant being incorrectly folded, the crystal structure of the Y131F protein (PDB code 3keg) was determined following cocrystallization with methyl red (Fig. 4 ▶).
Figure 4.
Surface representations of (a) Y131F paAzoR1 and (b) wild-type paAzoR1 showing the active site. The surfaces were generated using PyMOL (DeLano, 2002 ▶). The β-strands on which residue 131 lies are shown under the surface along with the loop comprising Gly146, Gly147 and His148. FMN and methyl red are shown in stick view with yellow and pink C atoms, respectively. The side chains of residue 131, Val127 and His148 are shown as sticks.
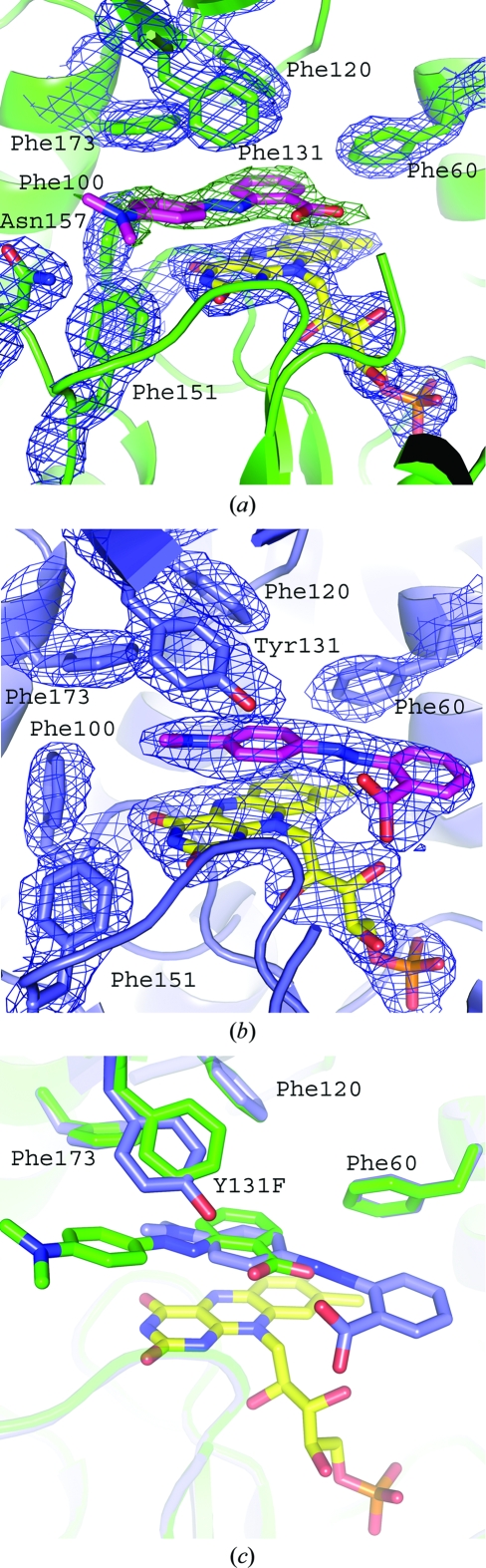
The data-collection statistics for Y131F_MRE are shown in Table 1 ▶. The overall fold is the same as that of wild-type paAzoR1 (Fig. 4 ▶), with an r.m.s.d. of 0.29 Å as calculated using secondary-structure matching in CCP4MG with 384 Cα atoms (Potterton et al., 2004 ▶). Although the overall fold is the same as that of wild-type paAzoR1, differences are seen within the active site. The orientations of methyl red within the active sites of Y131F and wild-type paAzoR1 are shown in Fig. 5 ▶. In Y131F_MRE the Phe131 ring is rotated by ∼15° (Fig. 5 ▶ c) relative to the orientation of the aromatic ring of Tyr131 in the wild-type structure. This movement, together with removal of the hydroxyl group of Tyr131, leads to an increase in the minimum distance between residue 131 and FMN of approximately 1.1 Å, increasing the distance from 5.5 to 6.6 Å. The result is that the substrate methyl red can fit into the increased gap. This allows methyl red to bind deeper in the active-site cavity of the mutant enzyme, resulting in a shorter distance between the methyl red azo bond and N5 of FMN (4.2 Å compared with 6.3 Å). This structural finding agrees with the conclusion that Tyr131 is not acting as a proton source. There are no hydrogen bonds evident between methyl red and either of the proteins. The rotation of methyl red within the active site in the structure of Y131F results in a significantly different binding pose for the substrate in the two structures (Fig. 5 ▶). The substrate is displaced further from the protein surface such that the aniline ring no longer forms stacking interactions with the central ring of the isoalloxazine group; instead, it binds in a pocket formed by the side chains of Phe100, Phe131, Gly147, Phe151, Asn157 and Phe173 (Fig. 5 ▶ a), all of which lie within 4 Å of the aniline ring. The aniline ring is replaced in its interactions with the central ring of FMN, Phe60, Phe120 and Phe173 by the benzoate ring, which binds in an overlapping position to that of the aniline ring of methyl red in the wild-type structure. The carboxylate group of methyl red is positioned within the solvent channel occupied by the azo bond in the wild-type enzyme structure. The azo bond forms interactions with the pyrimidine ring of FMN and is also in close contact with Phe173 in Y131F. The binding site of the azo bond and the aniline ring in Y131F is pushed deeper into the active-site cleft. The open solvent channel in which the benzoate ring and azo bond are found within the wild-type enzyme are occupied in Y131F by the carboxylate moiety only. The change in the mode of binding of methyl red to Y131F is compatible with the lower K m for methyl red as a substrate of the Y131F mutant (Table 2 ▶). The orientation of methyl red binding to paAzoR1 is not optimal for accepting a hydride ion from the N5 position of FMN in the wild-type enzyme. It is proposed that the Y131F_MRE structure in which the N5 bond of FMN is closer to the azo bond of the substrate methyl red could represent a closer approximation to an intermediate stage of methyl red in the enzyme–substrate complex during the azo-reduction reaction.
Figure 5.
Binding of methyl red to paAzoR1. (a) Binding of methyl red to Y131F-mutant paAzoR1. The blue electron density represents the refined 2F o − F c map contoured at 1σ. The green electron density represents the first F o − F c map calculated after initial phasing contoured at 2σ, showing the position of methyl red binding. (b) Binding of methyl red to wild-type paAzoR1. The structure is based upon PDB entry 2v9c. As in (a) blue electron density represents the refined 2F o − F c map. In both (a) and (b) methyl red is shown with pink C atoms, while FMN is shown with yellow C atoms. (c) Overlaid structures of methyl red binding to wild-type and Y131F-mutant paAzoR1. Side chains and methyl red molecules have the same colour C atoms. Blue C atoms are for wild-type paAzoR1. Green C atoms are for Y131F-mutant paAzoR1.
Whilst being compatible with a very similar overall fold, the subtle change in amino acid when Tyr131 is replaced by phenylalanine affects the thermostability of the enzyme.
Although Tyr131 is found in azoreductases which have sequences that are only 36–46% identical to paAzoRI (Supplementary Fig. 1 ▶ 1; Altschul et al., 1997 ▶; Hall, 1999 ▶; Thompson et al., 1994 ▶), Tyr131 is not universally conserved and is replaced by hydrophobic residues including phenylalanine. However, systematic activity information is not available for all of these azoreductases which would a priori discount Tyr131 having an essential role. Our studies show that the hydroxyl hydrogen from Tyr131 is not directly involved in the reduction of substrate but identify that Tyr131 is important in defining the architecture of the active site for both the azo substrate methyl red and the nicotinamide cofactor.
Supplementary Material
PDB reference: azoreductase 1, 3keg, r3kegsf
Supplementary material file. DOI: 10.1107/S1744309109044741/kw5016sup1.pdf
Footnotes
Supplementary material has been deposited in the IUCr electronic archive (Reference: KW5016).
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res.25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Chen, H., Wang, R. F. & Cerniglia, C. E. (2004). Protein Expr. Purif.34, 302–310. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B. III, Snoeyink, J., Richardson, J. S. & Richardson, D. C. (2007). Nucleic Acids Res.35, W375–W383. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). PyMOL Molecular Viewer. DeLano Scientific, Palo Alto, California, USA.
- Dissanayake, A. S. & Truelove, S. C. (1973). Gut, 14, 923–926. [DOI] [PMC free article] [PubMed]
- Duurkens, R. H., Tol, M. B., Geertsma, E. R., Permentier, H. P. & Slotboom, D. J. (2007). J. Biol. Chem.282, 10380–10386. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Engh, R. A. & Huber, R. (1991). Acta Cryst. A47, 392–400.
- Hall, T. A. (1999). Nucleic Acids Symp. Ser.41, 95–98.
- Hanauer, S. B. (1996). N. Engl. J. Med.334, 841–848. [DOI] [PubMed]
- Kawamura, A., Sandy, J., Upton, A., Noble, M. & Sim, E. (2003). Protein Expr. Purif.27, 75–84. [DOI] [PubMed]
- Kohli, R. M. & Massey, V. (1998). J. Biol. Chem.273, 32763–32770. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2005). CompLife 2005, edited by M. R. Berthold, R. Glen, K. Diederichs, O. Kohlbacher & I. Fischer, pp. 163–174. Berlin/Heidelberg: Springer-Verlag.
- Lambeth, J. D. & Kamin, H. (1977). J. Biol. Chem.252, 2908–2917. [PubMed]
- Lambeth, J. D., McCaslin, D. R. & Kamin, H. (1976). J. Biol. Chem.251, 7545–7550. [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Li, R., Bianchet, M. A., Talalay, P. & Amzel, L. M. (1995). Proc. Natl Acad. Sci. USA, 92, 8846–8850. [DOI] [PMC free article] [PubMed]
- Morris, R. J., Perrakis, A. & Lamzin, V. S. (2003). Methods Enzymol.374, 229–244. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Nakanishi, M., Yatome, C., Ishida, N. & Kitade, Y. (2001). J. Biol. Chem.276, 46394–46399. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2006). Acta Cryst. D62, 439–450. [DOI] [PubMed]
- Peppercorn, M. A. & Goldman, P. (1972). J. Pharmacol. Exp. Ther.181, 555–562. [PubMed]
- Potterton, L., McNicholas, S., Krissinel, E., Gruber, J., Cowtan, K., Emsley, P., Murshudov, G. N., Cohen, S., Perrakis, A. & Noble, M. (2004). Acta Cryst. D60, 2288–2294. [DOI] [PubMed]
- Read, R. J. (2001). Acta Cryst. D57, 1373–1382. [DOI] [PubMed]
- St Maurice, M., Cremades, N., Croxen, M. A., Sisson, G., Sancho, J. & Hoffman, P. S. (2007). J. Bacteriol.189, 4764–4773. [DOI] [PMC free article] [PubMed]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res.22, 4673–4680. [DOI] [PMC free article] [PubMed]
- Wang, C. J., Hagemeier, C., Rahman, N., Lowe, E., Noble, M., Coughtrie, M., Sim, E. & Westwood, I. (2007). J. Mol. Biol.373, 1213–1228. [DOI] [PubMed]
- Zenno, S., Koike, H., Kumar, A. N., Jayaraman, R., Tanokura, M. & Saigo, K. (1996). J. Bacteriol.178, 4508–4514. [DOI] [PMC free article] [PubMed]
- Zhou, X. X., Wang, Y. B., Pan, Y. J. & Li, W. F. (2008). Amino Acids, 34, 25–33. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: azoreductase 1, 3keg, r3kegsf
Supplementary material file. DOI: 10.1107/S1744309109044741/kw5016sup1.pdf
PDB reference: azoreductase 1, 3keg, r3kegsf
Supplementary material file. DOI: 10.1107/S1744309109044741/kw5016sup1.pdf