Abstract
To identify new components in the phytochrome (phy) signaling network in Arabidopsis, we used a sensitized genetic screen for deetiolation-defective seedlings. Two allelic mutants were isolated that exhibited reduced sensitivity to both continuous red and far-red light, suggesting involvement in both phyA and phyB signaling. The molecular lesions responsible for the phenotype were shown to be mutations in the Arabidopsis PSEUDO-RESPONSE REGULATOR7 (PRR7) gene. PRR7 is a member of a small gene family in Arabidopsis previously suggested to be involved in circadian rhythms. A PRR7–β-glucuronidase fusion protein localized to the nucleus, implying a possible function in the regulation of photoresponsive gene expression. Consistent with this suggestion, prr7 seedlings were partially defective in the regulation of the rapidly light-induced genes CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), observable as a premature increase in expression level during the second peak of the biphasic induction profile that is elicited upon initial exposure of dark-grown seedlings to light. A similar 3- to 6-h coordinated advance in peak free-running expression of CCA1, LHY, and TIMING-OF-CAB1, which are considered to encode the molecular components of the circadian oscillator in Arabidopsis, was observed in entrained fully green prr7 seedlings compared with wild-type seedlings. Collectively, these data suggest that PRR7 functions as a signaling intermediate in the phytochrome-regulated gene expression responsible for both seedling deetiolation and phasing of the circadian clock in response to light.
INTRODUCTION
Light has a profound impact on plant growth and development. It influences numerous developmental processes, such as seed germination, seedling deetiolation, neighbor sensing, and phototropism, enabling plants to grow in a way that optimizes their ability to capture light for photosynthesis. Plants possess a number of informational photoreceptors, including phytochromes, cryptochromes, and phototropins, which enable them to monitor specific regions of the light spectrum and to regulate the changes in gene expression that underlie their responses to the light environment (for a recent review, see Quail, 2002). Light also controls photoperiodic responses, such as flowering, through the circadian clock, thereby permitting the synchronization of reproduction with seasonal progression (Mouradov et al., 2002, and references therein). The circadian clock also allows the plant to anticipate and prepare for the regular daily changes in the light environment (Harmer et al., 2000). Both the phytochromes (red/far-red light receptors) and the cryptochromes (blue light receptors) mediate the entrainment of the circadian clock by light (Somers et al., 1998a). Because many light-regulated processes in plants, including elongation growth of stems and hypocotyls, leaf movements, stomatal opening, and transcription of CHLOROPHYLL A/B BINDING PROTEIN (CAB) genes, also are modulated by the circadian clock (Dowson-Day and Millar, 1999), it is not unlikely that one or more components of the central oscillator are positioned as integral intermediates at an early point in the light signaling pathway.
Genetic screens have generated mutants that have helped to define the photosensory and physiological roles of the various members of the phytochrome family (designated phyA to phyE in Arabidopsis). The evidence indicates that the different family members have differential, albeit partially overlapping, photosensory and/or physiological functions (Quail, 1998). Mutants also have provided insight into how the phytochrome signal is transmitted. The dual responses of light inhibition—cell elongation in seedling hypocotyls and the simultaneous promotion of cell expansion in cotyledons during seedling deetiolation—provide valuable phenotypic markers of light responsiveness. For example, a bona fide light signaling mutant may be distinguished from a mutant affected globally in cell elongation when both of these reciprocal responses are considered. Screens for mutants with reduced sensitivity to light or hypersensitivity to light have identified numerous loci putatively involved in phytochrome signaling (reviewed by Hudson, 2000), and this pathway seems to be far from saturated. Although some components appear to be required for deetiolation in response to either continuous far-red light (FRc) or continuous red light (Rc) exclusively, many appear to affect both pathways.
In addition, many Arabidopsis mutants that show defects in seedling photomorphogenesis during deetiolation also are affected in clock function in entrained deetiolated seedlings. These include early flowering3 (elf3), which has reduced sensitivity to both Rc and continuous blue light (Reed et al., 2000), gigantea (gi) and sensitivity to red light reduced, which are specifically insensitive to Rc (Huq et al., 2000; Staiger et al., 2003), zeitlupe, which is hypersensitive to Rc (Somers et al., 2000), and timing-of-cab1 (toc1), which has reduced sensitivity to both Rc and FRc (Mas et al., 2003). The apparent specificity of these defects for red and/or far-red light indicates that these mutations interfere with phytochrome-dependent seedling photomorphogenesis. Because almost all of these components are regulated by the circadian clock in entrained seedlings, they may be involved in the diurnal control of growth or function as feedback components that regulate light signaling to the clock and other plant light responses. This observation indicates that the circadian system shares many components with the phytochrome signaling pathway.
Considerable progress has been made in recent years in identifying the molecular components of the central oscillator in Arabidopsis (Harmer et al., 2001). There is now considerable evidence that TOC1, along with CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), are integral components of the circadian oscillator that function in a temporally offset feedback loop to generate mutually opposed oscillations (Yanovsky and Kay, 2001). CCA1 and LHY encode myb-related transcription factors that have been independently implicated in the maintenance of circadian rhythms in Arabidopsis. When the rhythmic expression of either of these genes is ablated, many circadian outputs display altered rhythmicity (Schaffer et al., 1998; Wang and Tobin, 1998; Matsushika et al., 2002b). Recent studies with plants reduced in both LHY and CCA1 activity show that these genes have partially redundant functions in the control of circadian rhythms (Alabadi et al., 2002; Mizoguchi et al., 2002). The TOC1 locus was identified genetically in a screen for mutants that misexpress a transgenic CAB-luciferase reporter gene (Millar et al., 1995). The mutant was further shown to have aberrant circadian rhythms in numerous outputs under all conditions; thus, it was believed to encode a component important for central oscillator function (Somers et al., 1998b). More recent studies with the toc1-2 null mutant have demonstrated that TOC1 is required for CCA1 and LHY transcriptional oscillations, providing compelling evidence for the feedback loop that suggests that these components likely constitute the circadian oscillator in Arabidopsis (Alabadi et al., 2001).
Transcription of both CCA1 and LHY is induced within 1 h of exposure of dark-grown seedlings to Rc or FRc in a phytochrome-dependent manner (Wang et al., 1997; Tepperman et al., 2001; J. Tepperman and P. Quail, unpublished data). Evidence that PHYTOCHROME-INTERACTING FACTOR3 (PIF3) binds to a fragment of the CCA1 promoter in vitro and that Rc-induced expression of CCA1 is attenuated in PIF3-deficient seedlings suggests a possible direct mechanism by which CCA1 transcription is controlled by light via phytochrome action (Martinez-Garcia et al., 2000). This transcriptional control may constitute the link between light signals perceived by phytochromes and the induction of the initial oscillations of the clock components that occurs during seedling deetiolation as well as the subsequent synchronization of these oscillations to daily cycles of light and darkness in fully green seedlings.
Recent evidence has shown that TOC1 is required for normal phytochrome-dependent seedling deetiolation in Rc and FRc and for the phytochrome-dependent induction of CCA1 by pulsed red light (Mas et al., 2003). TOC1 is a member of a small gene family in Arabidopsis (Imamura et al., 1998). These genes were independently named Arabidopsis PSEUDO-RESPONSE REGULATORS (APRRs, now synonymous with PRRs) because the proteins lack the conserved, phospho-accepting Asp of the bacterial response regulators (Makino et al., 2000). Close examination of the expression pattern of this gene family revealed that transcripts for each gene oscillate with a circadian expression pattern under free-running conditions, suggesting that these genes are regulated by the circadian clock (Makino et al., 2000; Matsushika et al., 2000). Plants that constitutively express PRR5 or PRR9 exhibit a hypersensitive seedling deetiolation phenotype in Rc and flower early, indicating that the aberrant expression of these genes interferes with normal phytochrome responses (Matsushika et al., 2002a; Sato et al., 2002). The precise molecular function of the PRRs is unknown, but TOC1, also known as PRR1, was shown to interact with PIF3 and a related basic helix-loop-helix transcription factor protein in vitro (Makino et al., 2002). This finding raises the possibility that TOC1 also may play a role in phytochrome-dependent transcriptional regulation.
To identify potential novel components of phytochrome signaling, we used a sensitized screen that exploits the synergistic effects of the phyA and phyB mutations on seedling deetiolation in Rc. phyA null mutants are almost completely insensitive to FRc but have a normal response to higher fluence rates of Rc (Parks and Quail, 1993). phyB null mutants have a more pleiotropic phenotype. Seedlings exhibit reduced sensitivity to Rc as well as reduced chlorophyll accumulation and early flowering, implicating phyB in both early light responses during deetiolation and the repression of flowering (Reed et al., 1993). phyA phyB double mutants have an even more severe defect in Rc perception at the seedling deetiolation stage, which indicates that phyA plays a role in Rc perception that is secondary to that of phyB (Reed et al., 1994; Smith et al., 1997). By screening for Rc-insensitive mutants in the phyA-101 null mutant (Quail et al., 1994) background, we hoped to enhance the phenotype of mutants in the phyB signaling pathway, with the goal of identifying components specific to the phyB response. This screen has allowed the identification of a new mutant defective in both the phytochrome-mediated response to Rc and FRc and the regulation of the circadian clock.
RESULTS
Isolation of a Light-Insensitive Mutant
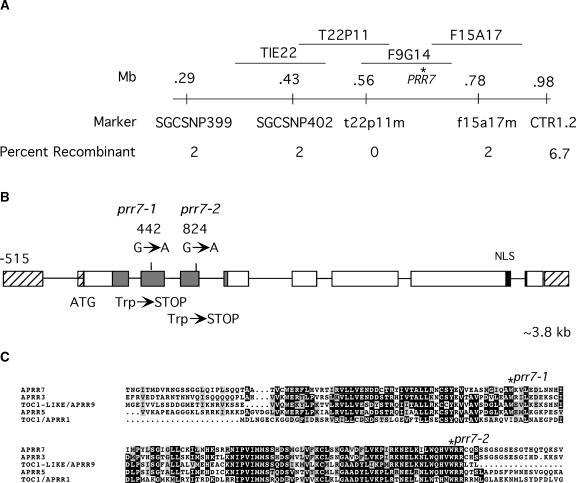
Many seedlings were selected in a screen for reduced sensitivity to Rc, but two independently isolated M3 lines, which were suspected to be novel mutants, are described here. Allelism tests performed with M4 plants from these lines showed that the long-hypocotyl phenotype was not complemented in the F1 generation, which demonstrated that these two lines constituted one complementation group (data not shown). Further tests showed that these mutant alleles complemented the phyA-101 phyB-1 double mutant and thus did not represent new alleles of phyB (data not shown). The long-hypocotyl phenotype of each line was mapped in the F2 generation from a cross of the mutant in the phyA-101 (RLD ecotype) background to phyA-211 (Columbia ecotype) (Quail et al., 1994) using PCR-based markers. The mutant locus was mapped to the top of chromosome V, distal to the marker CTR1.2, in a region not known to contain any other loci involved in photomorphogenesis. Recombinant F2 individuals on either side of the mutant locus defined a region spanned by four BACs (Figure 1A).
Figure 1.
Identification of the prr7 Locus.
(A) Map positions of the PRR7 locus and closely linked markers on chromosome 5.
(B) Structure of the PRR7 gene, showing the positions of the prr7-1 and prr7-2 mutations and the putative nuclear localization signal (NLS; black rectangle). The gray shaded area represents the conserved receiver domain shown in (C).
(C) Predicted protein sequence alignment of the conserved receiver domain of the TOC1/APRR1 family showing the positions of the stop codons introduced by prr7-1 and prr7-2.
Candidate gene sequencing in this region revealed lesions in Arabidopsis PRR7 (At5g02810) in both mutant lines (Figure 1B). This gene is a member of a small family that includes TOC1. This family has been studied with respect to gene expression and has been referred to as the TOC1/APRR1 family. Here, in keeping with community guidelines for mutant gene nomenclature, our mutants were given the three-letter abbreviation prr7. Both alleles contain G-to-A transversions that would result in early termination of the predicted protein in a highly conserved domain homologous with the receiver domain (Figures 1B and 1C).
PRR7 Is a Nuclear Protein
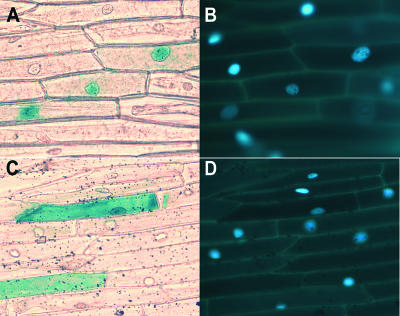
A PROSITE (Hofmann et al., 1999) scan revealed a potential bipartite nuclear localization signal near the C terminus of PRR7 (amino acids 680 to 696). A PRR7–β-glucuronidase (GUS) fusion protein was expressed transiently in leek epidermal cells after bombardment with a plasmid carrying a strong 35S promoter upstream of a fusion of the PRR7 coding region with the GUS marker gene. The GUS activity in dark-maintained leek epidermal peels transformed with the PRR7-GUS fusion protein was observed primarily in the nucleus (Figures 2A and 2B). In parallel experiments, GUS alone was detected throughout the cytoplasm (Figures 2C and 2D). These data provide evidence that PRR7 likely is a nuclear protein.
Figure 2.
PRR7 Is Localized to the Nucleus.
(A) Bright-field image showing that PRR7-GUS is localized to the nucleus in leek epidermal peels.
(B) 4′,6-Diamidino-2-phenylindole (DAPI) staining of the cells shown in (A).
(C) Bright-field image showing that the GUS control is distributed throughout the cell.
(D) DAPI staining of the cells shown in (C).
prr7 Seedlings Have Reduced Sensitivity to Rc and FRc, and the Rc Phenotype Is Enhanced by phyA-101
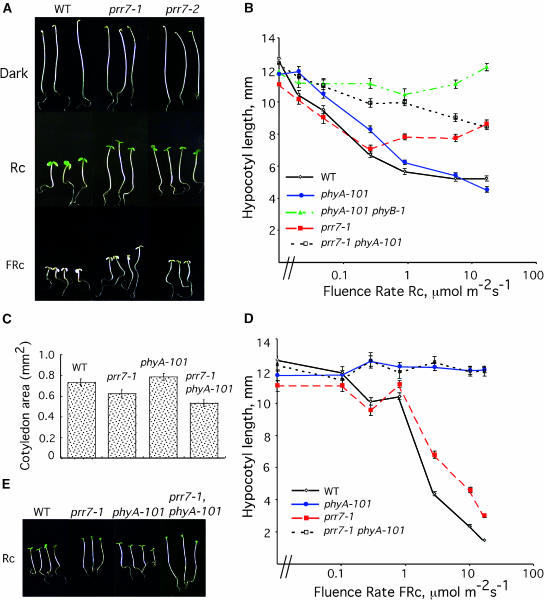
Fluence rate response curves in Rc and FRc were determined to quantitatively characterize the effect of the prr7 mutation on seedling sensitivity to these two diagnostic wavelengths. prr7 seedlings were 1 to 2 mm taller than wild-type seedlings at all fluence rates >0.1 μmol·m−2·s−1 Rc (Figures 3A and 3B), an effect that indicates reduced responsiveness to light of this wavelength. The reduced sensitivity of prr7 to Rc also was evident in the expansion of the cotyledons. Cotyledon expansion was reduced in prr7 with respect to the wild type (Figure 3C). Sensitivity to FRc also was reduced in prr7 with respect to the wild type (Figures 3A and 3D). Comparison of phyA prr7 double mutants with monogenic mutants showed that the prr7 mutant phenotype in Rc was more dramatic in the phyA-101 background, especially in the range from 0.1 to 10 μmol·m−2·s−1 (Figures 3B and 3D). However, in FRc (Figure 3D), phyA prr7 double mutant seedlings were indistinguishable from phyA seedlings, which suggests that phyA and prr7 do not have an additive effect under these light conditions. This is not surprising considering that phyA is the sole photoreceptor for far-red light and that phyA seedlings are essentially blind to FRc. prr7 seedlings did not have a long-hypocotyl phenotype when grown under continuous white light with a fluence rate of 150 μmol·m−2·s−1 (data not shown).
Figure 3.
prr7 Mutants Have Reduced Sensitivity to both Rc and FRc.
(A) Wild-type RLD (WT) and prr7 mutants grown at 25°C in darkness, Rc (17 μmol·m−2·s−1), or FRc (10.2 μmol·m−2·s−1).
(B) Rc fluence rate response curves for prr7.
(C) Cotyledon expansion in Rc is inhibited in prr7. Seedlings were grown in Rc (0.79 μmol·m−2·s−1).
(D) FRc fluence rate response curve for prr7.
(E) The prr7 mutant phenotype is enhanced by phyA-101. Seedlings were grown in Rc (0.89 μmol·m−2·s−1).
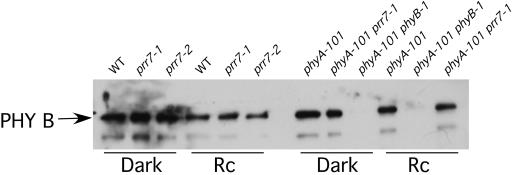
The Hyposensitive Phenotype of prr7 Mutants Is Not the Result of Reduced Levels of phyB
To test the possibility that PRR7 is involved in the control of PHYB expression, the levels of phyB protein in wild-type and prr7 seedlings were analyzed. Figure 4 shows that phyB levels were normal in the mutants both in the dark and under Rc. This finding suggests that the prr7 phenotype is not a consequence of altered phyB levels and that PRR7 likely functions downstream of the phyB photoreceptor in Rc signal transduction.
Figure 4.
phyB Levels Are Not Affected in prr7.
Immunoblot of total protein extracted from 3-day-old seedlings grown either in darkness or Rc and probed with monoclonal antibody to phyB. WT, wild-type ecotype RLD.
Light-Induced Gene Expression in prr7
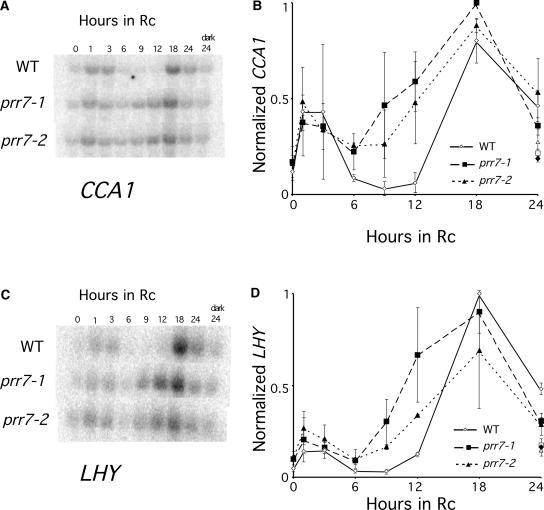
To test the involvement of PRR7 in the regulation of early-responding, light-induced genes (Tepperman et al., 2001), we compared the transcriptional induction of CCA1 and LHY in response to Rc in the mutant and the wild type. In the wild type, CCA1 and LHY both were induced rapidly and transiently in response to light, exhibiting biphasic patterns with an acute peak of expression at 1 h after transfer to light, followed by a trough from 6 to 12 h and a second peak at ∼18 h (Figures 5A and 5B), as observed previously (Wang et al., 1997; Schaffer et al., 1998; Wang and Tobin, 1998; Tepperman et al., 2001; J. Tepperman and P. Quail, unpublished data). No oscillation in CCA1 transcript level was detectable in dark-grown wild-type seedlings during the 24-h period preceding the transfer to light (data not shown), establishing that these oscillations occurred only in response to the light signal under these conditions. The prr7 mutants also showed an initial peak and partial immediate downregulation of CCA1 expression upon first exposure to light, but CCA1 transcript levels began to increase again prematurely at 6 h and peaked at 18 h, coincident with the peak in the wild type at this level of resolution (Figure 5A). Thus, the depth of the trough of CCA1 expression was attenuated in prr7.
Figure 5.
Rc-Induced Expression of CCA1 and LHY Is Altered in prr7.
Induction of CCA1 (A) and LHY (B) in wild-type RLD (WT), prr7-1, and prr7-2. Seedlings were grown for 96 h in darkness and transferred to Rc. Tissue was collected at the times indicated. Average values for two biological replicates are plotted, and error bars represent ranges. One representative RNA gel blot is shown for each transcript.
Figure 5B shows a similar but less severe defect in the expression of LHY in the prr7 mutants. These results suggest that PRR7 may be required for the negative regulation of CCA1 and LHY during the early trough in expression of these genes in Rc. A similar pattern of CCA1 expression was observed in FRc (data not shown). We observed a small difference in responsiveness between the prr7-1 and prr7-2 alleles both at 1 h after transfer to Rc and again at 18 to 24 h after transfer to Rc, but these differences are not likely to be significant in this experiment.
Circadian Clock–Regulated Gene Expression in prr7
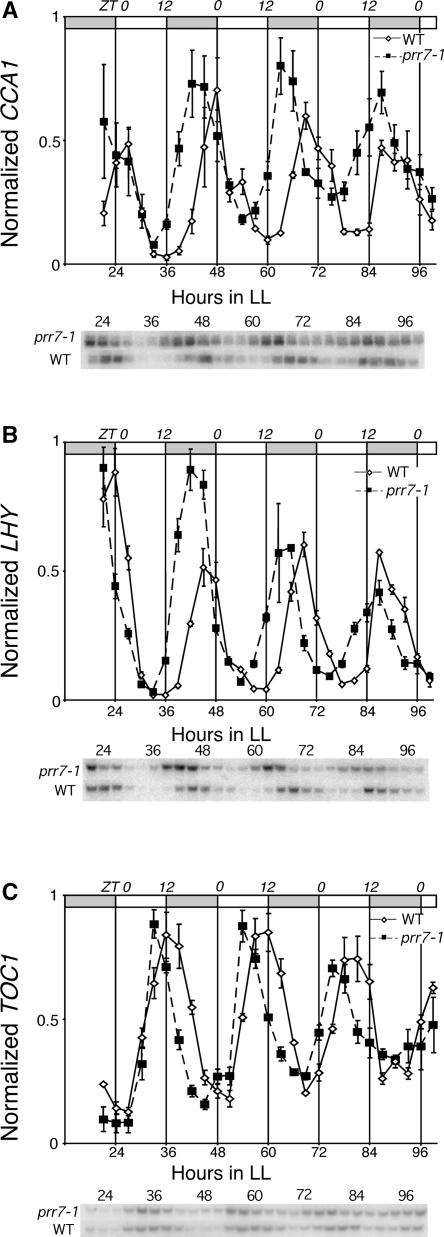
As a result of the proposed role of PRR7 in circadian clock function (Makino et al., 2001) and the initial misexpression of the clock genes CCA1 and LHY at early stages after transfer to red light observed here, we examined the circadian clock–regulated expression of these genes in prr7-1. Figure 6A shows that CCA1 mRNA oscillated with circadian rhythmicity in free-running continuous light conditions in fully green, wild-type seedlings that had been entrained to 12-h-light/12-h-dark cycles. In these seedlings, CCA1 expression began to increase in anticipation of subjective dawn to peak at zeitgeber time 0 (ZT0) to ZT3. prr7-1 also showed robust oscillations in CCA1 under these conditions (Figure 6A). However, the peaks of CCA1 expression in the mutant occurred before subjective dawn at ZT18 to ZT21 and in each cycle occurred 3 to 6 h before the wild-type peaks. CCA1 expression in the wild type increased most rapidly from ZT15 to ZT18, but in prr7-1, the trough was attenuated and CCA1 levels had already started to increase at ZT12. The mean period of expression for CCA1 was 20.7 ± 2.3 h in the wild type and 20.7 ± 2.8 h in prr7-1.
Figure 6.
Circadian Oscillations of Clock Transcripts Are Altered in prr7.
Expression in prr7-1 mutants and wild-type ecotype RLD (WT) under free-running continuous light conditions (LL) for CCA1 mRNA (A), LHY mRNA (B), and TOC1 mRNA (C). Shaded areas in the bars at top represent subjective night, and the upper axis indicates zeitgeber time. Expression level measurements were performed by RNA gel blot quantitation, and the values were corrected for gel loading relative to 18s rRNA hybridization. These values then were scaled such that the highest expression value for each experiment was 1, and the resulting values were averaged. The mean values from three independent biological replicates are plotted. Error bars represent standard errors of the mean. A representative gel blot image of one replicate is shown.
A similar phenomenon was observed for LHY, which also oscillated robustly in prr7-1 and the wild type, with peak expression occurring at ZT0 in the wild type and at ZT21 in the mutant. The mean free-running period of LHY expression was calculated to be 21 ± 2.6 h in the wild type and 22 ± 2.1 h in prr7-1. The average periods of expression of CCA1 and LHY did not differ significantly between the mutant and the wild type at the resolution level of this experiment. Amplitude values for both CCA1 and LHY were comparable between prr7 and the wild type, but with a tendency for expression to be higher in the troughs of the oscillations in prr7. The same is true for the TOC1 transcript (Figure 6C), for which peak expression in the wild type occurred at ZT9 to ZT12 but occurred earlier in prr7 at ZT6 to ZT9. Thus, the peak of expression for all three of the genes examined occurred earlier in prr7-1, without a detectable aberration in period length. These results indicate that the absence of PRR7 causes a significant, coordinated shift in the phasing of the oscillatory expression of the central components of the circadian clock.
DISCUSSION
In this study, a sensitized genetic screen was used to identify new loci that are required for the Rc-induced deetiolation response. Two mutant alleles of PRR7 were identified. Both contain a recessive mutation that results in early termination of the predicted protein in the conserved receiver domain, and neither is likely to make a functional protein. A reduction in the levels or activity of the phyB photoreceptor would have been sufficient to account for the insensitivity to Rc that we observed in prr7. However, no reduction in the levels of phyB were detected in the mutant by immunoblot analysis. Therefore, these data provide evidence that PRR7 functions downstream of phyB in the Rc-induced deetiolation response. The reduced response of prr7 to Rc in both the cotyledons and the hypocotyl was enhanced by the phyA-101 mutation. prr7 also had a defect in its responsiveness to FRc, apparent as reduced inhibition of hypocotyl elongation. We interpret these data to indicate that PRR7 functions in a positive manner downstream of the convergence of the phyA and phyB signaling pathways, which regulate seedling photomorphogenesis in Rc and FRc.
Evidence has emerged in recent years to indicate that phytochromes can enter the nucleus to directly regulate gene expression in response to light (Kircher et al., 1999; Huq et al., 2003). A number of proteins implicated in phytochrome signaling are known to be localized to the nucleus, including CCA1, PIF3, PIF4, FAR-RED-IMPAIRED RESPONSE1, SUPPRESSOR OF PHYA1, GI, ELF3, and TOC1 (Wang et al., 1997; Ni et al., 1998; Hoecker et al., 1999; Hudson et al., 1999; Huq et al., 2000; Makino et al., 2000; Strayer et al., 2000; Huq and Quail, 2002). The nuclear localization of PRR7 suggests a possible regulatory function in phytochrome-controlled gene expression.
Consistent with this possibility, phytochrome-dependent gene induction was affected in prr7. Here, we show that PRR7 was required for the negative regulation of CCA1 and LHY in seedlings in Rc during a defined temporal window in the early phases of deetiolation (Figure 5). A biphasic waveform pattern of expression for CCA1 and LHY was detectable in wild-type etiolated seedlings upon transfer to Rc. After the initial rapid induction of CCA1 and LHY, which peaked at ∼1 to 3 h after transfer from darkness to Rc, transcript levels declined sharply, remained low for 6 h, and then increased again at 12 h. By contrast, in both prr7-1 and prr7-2, CCA1 and LHY transcript levels increased prematurely between 6 and 12 h. The prr7 mutants had fourfold to sixfold higher levels of CCA1 and LHY transcript than the wild type during the 6- to 12-h period after transfer to Rc. This early and precise misexpression caused by the loss of PRR7 indicates that it functions as a negative modulator of the inductive phytochrome signaling pathway, suggesting that PRR7 plays a role in controlling the initial establishment of CCA1 and LHY oscillations.
prr7 also displayed a clear defect in the sustained circadian expression pattern of LHY and CCA1 (Figures 6A and 6B). In entrained, fully green prr7 seedlings under free-running conditions of continuous white light, the peak expression of both genes was shifted by 3 to 6 h earlier than in the wild type. The rising phase of the CCA1 and LHY transcriptional oscillations that occurred earlier in prr7 resembled the premature increase in transcript level observed after the initial transfer of dark-grown seedlings to Rc (Figure 5). Oscillation of TOC1 mRNA under free-running conditions was shifted similarly in prr7 (Figure 6C), demonstrating that mutation in PRR7 affected the clock genes expressed in both the morning (CCA1 and LHY) and evening (TOC1) phases of the diurnal cycle and that the interlocked, opposed oscillations of CCA1/LHY and TOC1 were maintained in register in this mutant. No differences in the mean free-running period length between the mutant and the wild type were detectable at the resolution of the assays used here.
The evidence that the expression of these genes is perturbed in the mutant after the initial transfer of seedlings to Rc (in the absence of an entrained circadian clock) suggests that the function of PRR7 may be to attenuate phytochrome signaling to the central oscillator during a specific temporal window. The loss of PRR7 appears to result in the early derepression of CCA1 and LHY expression in what would normally be the trough of the waveform. Therefore, we propose that the perturbation observed here in prr7 during the light-induced initiation of the oscillations in expression of clock-component genes, which occurred upon first exposure of dark-grown seedlings to light, is reiterated daily at dawn in the dark-to-light transition under diurnal cycles and is propagated under free-running conditions in entrained seedlings to generate a phase advance of the circadian oscillations in the clock-component transcripts. Consistent with this conclusion, phyB has been implicated in the control of circadian phase in white light (Salome et al., 2002).
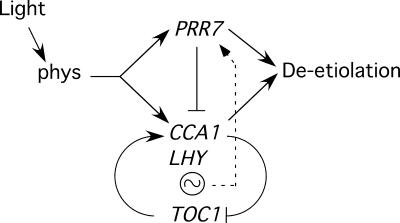
A simplified model of the proposed dual function of PRR7 in the control of seedling deetiolation and phasing of the circadian clock is presented in Figure 7. The observation that PRR7 appears to act positively in phytochrome-regulated deetiolation but negatively in controlling the clock suggests early branching in the phytochrome signaling pathway upstream of these two target processes. However, because CCA1, LHY, and TOC1 also have been shown to function in regulating deetiolation (Schaffer et al., 1998; Wang and Tobin, 1998; Mas et al., 2003), these two pathway branches presumably converge somewhere downstream (Figure 7). The initial light-induced transcriptional activation of CCA1 during deetiolation is postulated to be a direct target of the phytochrome photoreceptor via interaction with PIF3, with few if any other signaling intermediates intervening (Martinez-Garcia et al., 2000). PRR7 functions during the first 24 h of deetiolation to antagonize this initial light-induced expression in a temporally specific manner, creating a trough in the profile and determining the timing of the rise of the second peak of expression (Figure 5). It is proposed that under established circadian conditions, PRR7 continues to function in this manner, thereby controlling the phasing of the oscillator by determining the timing of release from repression of CCA1/LHY expression at each cycle.
Figure 7.
Model of the Proposed PRR7 Function in Phytochrome-Regulated Seedling Deetiolation and Phasing of the Circadian Oscillator.
Light perceived by phytochromes induces both seedling deetiolation and oscillations in expression of the central clock–component genes CCA1, LHY, and TOC1. PRR7 acts positively in the pathway that controls seedling photomorphogenesis but negatively in the circadian cycle by transiently repressing or delaying CCA1 and LHY expression during a temporally defined window and by antagonizing or repressing the promotive activity of TOC1. PRR7 expression also is controlled by the circadian clock.
The mechanism of repression is unknown. However, in principle, it appears likely that PRR7 antagonizes the promotive action of TOC1 on CCA1/LHY expression in some way (Figure 7). It is uncertain whether this antagonism by PRR7 is direct or indirect. The expression profiles of PRR7 and TOC1 appear to overlap sufficiently for PRR7 to act directly (Matsushika et al., 2000). On the other hand, because PRR7 is a member of the “PRR quintet” of genes that are expressed sequentially in the order PRR9, PRR7, PRR5, PRR3, PRR1/TOC1, it is possible that the effect is exerted indirectly through PRR5 and/or PRR3 in a cascade-like manner. Because the expression of this quintet, including PRR7, also is controlled by the clock in a feedback manner (Figure 7), it has been proposed that these five components may constitute a second interlocking loop involved in regulating circadian oscillations under diurnal conditions (Makino et al., 2001; Eriksson and Millar, 2003). Regardless, it seems reasonable to propose that this antagonism of TOC1 action is imposed at the post-transcriptional level, modulating only the onset of the period over which TOC1 can exert its promotive activity on CCA1/LHY transcription, without altering the onset of TOC1 expression. This configuration permits the phasing of the circadian oscillations to be varied without changing the peak-to-peak period. Possible mechanisms of such post-transcriptional regulation might include inhibitory protein–protein interactions, providing transient sequestration or inactivation of TOC1, and direct transcriptional repression, by competitive displacement of TOC1 from CCA1/LHY promoter target sites.
The model presented here exemplifies the concept that certain key early components of the phytochrome signaling network have been recruited to function also as integral components of the circadian oscillator, thereby imposing oscillatory behavior on a broad spectrum of downstream genes in the light-regulated transcriptional cascade (Harmer et al., 2000; Eriksson and Millar, 2003). Such a configuration provides an elegant mechanism by which the extensive and complex network of light-regulated processes on which the plant depends can be attuned to the diurnal light cycle. The strategy used here of directly monitoring the behavior of the molecular components of the clock during the initial induction of oscillations upon first transfer from darkness to light provides a powerful means of dissecting the primary sequence of events involved in inducing and establishing the necessary feedback loops that drive the oscillator under steady state conditions. We anticipate that the use of this strategy, in conjunction with mutants compromised in various aspects of light responsiveness, will provide additional insights into the hierarchy and interactions among the molecular components involved in the function and regulation of the circadian clock.
METHODS
Mutagenesis and Screening
A total of 40,000 Arabidopsis thaliana phyA-101 (ecotype RLD) seeds were treated with 0.3% ethyl methanesulfonate for 12 h, washed, and planted on soil. After stratification for 4 days at 4°C, flats were transferred to the greenhouse under continuous white light. M2 seeds were harvested from families of 1,000 M1 plants and desiccated for 14 days in Drierite (W.A. Hammond Drierite Company, Xenia, OH). M2 seeds were sown on growth media (Valvekens et al., 1988) without sucrose, stratified for 5 days at 4°C, given a synchronizing 3-h white light treatment and 21-h dark treatment, and then grown for 72 h in continuous red light (Rc; 2 to 4 μmol·m−2·s−1) on vertical plates at 21°C (a total of 96 h from germination). Light sources and fluence measurements have been described elsewhere (Huq et al., 2000). Seedlings with long hypocotyls were selected and allowed to self. Rescreening of the resulting M3 seeds identified a number of lines with a heritable long-hypocotyl phenotype. M3 seeds were plated on biliverdin-supplemented medium to eliminate chromophore biosynthetic mutants (Parks and Quail, 1991). M3 plants were crossed to the phyA-101 phyB-1 introgressed line (described by Smith et al., 1997) to identify new alleles of the phyB photoreceptor mutant. Candidate M3 lines were backcrossed to RLD and crossed to phyA-211 in the Columbia ecotype (Reed et al., 1994) for mapping.
Mapping and Sequencing
The mutant locus was mapped from the F2 population of the mutant by phyA-211 cross using simple sequence length polymorphism, cleaved amplified polymorphic sequence, and single nucleotide polymorphism markers polymorphic between RLD and Columbia. Plant DNA was prepared according to Edwards et al. (1991).
Nuclear Localization Experiments
The PRR7 open reading frame was amplified by PCR from cDNA using primers containing restriction sites for ClaI and XbaI and inserted into the modified pRTL2-GUS/NiaDBam vector described by Hoecker et al. (1999). Constructs were sequenced for accuracy. Leek (Allium porrum) epidermal peels were bombarded with this construct (35S:GUS-APRR7) and pRTL2-GUS and incubated in darkness for 24 h. Bombardment and GUS staining were performed as described by Ni et al. (1998).
Seedling Growth Conditions and Measurements
The F2 progeny of the first backcross to RLD were analyzed with derived-cleaved amplified polymorphic sequence markers (Neff et al., 1998) to select lines homozygous for prr7-1 and prr7-2. These plants also were tested for the phyA-101 mutation to obtain prr7-1 phyA-101 sibling lines for fluence response measurements. Seeds were sown, stratified, and synchronized as described above. After 72 h in Rc or continuous far-red light (96 h after germination) on horizontal plates containing growth media (Valvekens et al., 1988) without sucrose, seedlings were photographed with a digital camera and hypocotyl length was measured using NIH Image software (Bethesda, MD). For cotyledon area measurements, seedlings were sown as described above except that after 96 h of Rc (120 h after germination) at a fluence rate of 0.79 μmol·m−2·s−1, cotyledons were flattened to the agar surface and photographed with a digital camera and the area of individual cotyledons was measured using NIH Image software.
RNA Isolation
For red-light transcriptional induction experiments, seedlings were sown and stratified as described above but after synchronization were grown in darkness for 93 h before transfer to Rc (7.9 μmol·m−2·s−1) (time 0). Samples for RNA were taken at specified intervals. For circadian entrainment experiments, seeds were plated and stratified and immediately transferred to a 12-h-light/12-h-dark photoperiod for 6 days at a constant temperature of 21°C. On day 7, seedlings were transferred to continuous white light at a fluence rate of 150 μmol·m−2·s−1, and seedlings were collected at 3-h intervals. Seedling RNA was isolated and analyzed using the Qiagen Plant RNeasy kit (Valencia, CA) according to the manufacturer's instructions. Two to 10 μg of total RNA was separated on 1.2% formaldehyde gels and blotted to Ambion Brightstar membranes (Austin, TX). Hybridization and washes were performed at 65°C according to Church and Gilbert (1984). Probes were labeled by random priming using the Redi-Prime II kit (Amersham Pharmacia). Probes for CCA1 and LHY were described by Martinez-Garcia et al. (2000). The probe for TOC1 was amplified by PCR from genomic DNA using primers described by Makino et al. (2002).
Signal was quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA) and ImageQuant for Mac version 1.2 and corrected for loading using the 18s rRNA hybridization signal. These values were normalized so that the highest value for each transcript was equal to 1. For circadian experiments, the mean of three biological replicates was plotted, and error bars represent standard errors of the mean. For red light transcriptional induction experiments, the average normalized value (corrected for even loading with 18s rRNA signal, with the highest value set to 1) of two biological replicates was plotted, and the ends of the error bars represent the actual normalized values of each replicate. Period estimates for CCA1 and LHY oscillations were the average time between peaks of expression from three biological replicates over three cycles.
Protein Isolation and Immunoblot Analysis
Seedling protein isolation and immunoblotting and detection procedures were performed as described by Martinez-Garcia et al. (1999). Total protein was isolated from 3-day-old seedlings grown in darkness or in Rc and separated by SDS-PAGE (8%). Ten micrograms of total protein was loaded for dark-grown seedlings, and 30 μg was loaded for Rc-grown seedlings. To detect PHYB, membranes were probed with the monoclonal antibodies B1 and B7 (1:500 dilution each) described by Hirschfield et al. (1998).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Peter H. Quail, quail@nature.berkeley.edu.
Acknowledgments
We thank members of our laboratory for helpful discussion, M. Hudson and D. Somers for critical reading of the manuscript, A. Smith for technical assistance, and D. Hantz for excellent plant care. This work was supported by National Institutes of Health Grant GM47475, Department of Energy Grant DE-FG03-87ER13742, and U.S. Department of Agriculture–Agricultural Research Service Current Research Information Service number 5335-21000-017-00D.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015065.
References
- Alabadi, D., Oyama, T., Yanovsky, M.J., Harmon, F.G., Mas, P., and Kay, S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. [DOI] [PubMed] [Google Scholar]
- Alabadi, D., Yanovsky, M.J., Mas, P., Harmer, S.L., and Kay, S.A. (2002). Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 12, 757–761. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson-Day, M.J., and Millar, A.J. (1999). Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 17, 63–71. [DOI] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, M.E., and Millar, A.J. (2003). The circadian clock: A plant's best friend in a spinning world. Plant Physiol. 132, 732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L., Hogenesch, J.B., Straume, M., Chang, H.-S., Han, B., Zhu, T., Wang, X., Kreps, J.A., and Kay, S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Harmer, S.L., Panda, S., and Kay, S.A. (2001). Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 17, 215–253. [DOI] [PubMed] [Google Scholar]
- Hirschfield, M., Tepperman, J.M., Clack, T., Quail, P.H., and Sharrock, R.A. (1998). Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics 149, 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284, 496–499. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., Bucher, P., Falquest, L., and Bairoch, A. (1999). The PROSITE database, its status in 1999. Nucleic Acids Res. 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, M., Ringli, C., Boylan, M.T., and Quail, P.H. (1999). The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 13, 2017–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, M.E. (2000). The genetics of phytochrome signalling in Arabidopsis. Semin. Cell Dev. Biol. 11, 475–483. [DOI] [PubMed] [Google Scholar]
- Huq, E., Al-Sady, B., and Quail, P.H. (2003). Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 35, 660–665. [DOI] [PubMed] [Google Scholar]
- Huq, E., and Quail, P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21, 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq, E., Tepperman, J.M., and Quail, P.H. (2000). GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 97, 9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura, A., Hanaki, N., Umeda, H., Nakamura, A., Suzuki, T., Ueguchi, C., and Mizuno, T. (1998). Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 2691–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., and Nagy, F. (1999). Light quality–dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino, S., Kiba, T., Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., Taniguchi, M., Ueguchi, C., Sugiyama, T., and Mizuno, T. (2000). Genes encoding pseudo-response regulators: Insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 41, 791–803. [DOI] [PubMed] [Google Scholar]
- Makino, S., Matsushika, A., Kojima, M., Oda, Y., and Mizuno, T. (2001). Light response of the circadian waves of the APRR1/TOC1 quintet: When does the quintet start singing rhythmically in Arabidopsis? Plant Cell Physiol. 42, 334–339. [DOI] [PubMed] [Google Scholar]
- Makino, S., Matsushika, A., Kojima, M., Yamashino, T., and Mizuno, T. (2002). The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana. I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 43, 58–69. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Monte, E., and Quail, P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20, 251–257. [DOI] [PubMed] [Google Scholar]
- Mas, P., Alabadi, D., Yanovsky, M., Oyama, T., and Kay, S.A. (2003). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15, 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushika, A., Imamura, A., Yamashino, T., and Mizuno, T. (2002. a). Aberrant expression of the light-inducible and circadian-regulated APRR9 gene belonging to the circadian-associated APRR1/TOC1 quintet results in the phenotype of early flowering in Arabidopsis thaliana. Plant Cell Physiol. 43, 833–843. [DOI] [PubMed] [Google Scholar]
- Matsushika, A., Makino, S., Kojima, M., and Mizuno, T. (2000). Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 41, 1002–1012. [DOI] [PubMed] [Google Scholar]
- Matsushika, A., Makino, S., Kojima, M., Yamashino, T., and Mizuno, T. (2002. b). The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana. II. Characterization with CCA1-overexpressing plants. Plant Cell Physiol. 43, 118–122. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., Carre, I.A., Strayer, C.A., Chua, N.-H., and Kay, S.A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267, 1161–1163. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.-R., Carre, I.A., and Coupland, G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2, 629–641. [DOI] [PubMed] [Google Scholar]
- Mouradov, A., Cremer, F., and Coupland, G. (2002). Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14 (suppl.), S111.–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Neff, J.D., Chory, J., and Pepper, A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 14, 387–392. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Parks, B.M., and Quail, P.H. (1991). Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell 3, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, B.M., and Quail, P.H. (1993). hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell 5, 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H. (1998). The phytochrome family: Dissection of functional roles and signalling pathways among family members. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353, 1399–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Phytochrome photosensory signaling networks. Nat. Rev. Mol. Cell. Biol. 3, 85–93. [DOI] [PubMed] [Google Scholar]
- Quail, P.H., Briggs, W.R., Chory, J., Hangarter, R.P., Harberd, N.P., Kendrick, R.E., Koornneef, M., Parks, B., and Sharrock, R.A. (1994). Spotlight on phytochrome nomenclature. Plant Cell 6, 468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Bastow, R.M., Solomon, K.S., Dowson-Day, M.J., Elumalai, R.P., and Millar, A.J. (2000). Independent action of ELF3 and phyB to control hypocotyl elongation and flowering time. Plant Physiol. 122, 1149–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salome, P.A., Michael, T.P., Kearns, E.V., Fett-Neto, A.G., Sharrock, R.A., and McClung, C.R. (2002). The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol. 129, 1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, E., Nakamichi, N., Yamashino, T., and Mizuno, T. (2002). Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the TOC1/APRR1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol. 43, 1374–1385. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Smith, H., Xu, Y., and Quail, P.H. (1997). Antagonistic but complementary actions of phytochromes A and B allow optimum seedling de-etiolation. Plant Physiol. 114, 637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D.E., Devlin, P.F., and Kay, S.A. (1998. a). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282, 1488–1494. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Webb, A.A.R., Pearson, M., and Kay, S.A. (1998. b). The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125, 485–494. [DOI] [PubMed] [Google Scholar]
- Staiger, D., Allenbach, L., Salathia, N., Fiechter, V., Davis, S.J., Millar, A.J., Chory, J., and Fankhauser, C. (2003). The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev. 17, 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Mas, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. [DOI] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.-Y., Kenigsbuch, D., Sun, L., Harel, E., Ong, M.S., and Tobin, E.M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.-Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2001). Signaling networks in the plant circadian system. Curr. Opin. Plant Biol. 4, 429–435. [DOI] [PubMed] [Google Scholar]