Abstract
MicroRNAs (miRNAs) are ∼21-nucleotide noncoding RNAs that have been identified in both animals and plants. Although in animals there is direct evidence implicating particular miRNAs in the control of developmental timing, to date it is not known whether plant miRNAs also play a role in regulating temporal transitions. Through an activation-tagging approach, we demonstrate that miRNA 172 (miR172) causes early flowering and disrupts the specification of floral organ identity when overexpressed in Arabidopsis. miR172 normally is expressed in a temporal manner, consistent with its proposed role in flowering time control. The regulatory target of miR172 is a subfamily of APETALA2 (AP2) transcription factor genes. We present evidence that miR172 downregulates these target genes by a translational mechanism rather than by RNA cleavage. Gain-of-function and loss-of-function analyses indicate that two of the AP2-like target genes normally act as floral repressors, supporting the notion that miR172 regulates flowering time by downregulating AP2-like target genes.
INTRODUCTION
MicroRNAs (miRNAs) are noncoding RNAs of ∼21 nucleotides in length that have been identified in both animals and plants (Lagos-Quintana et al., 2001, 2002; Lau et al., 2001; Lee and Ambros, 2001; Llave et al., 2002a; Mourelatos et al., 2002; Park et al., 2002; Reinhart et al., 2002). They are processed from longer precursor transcripts that range in length from ∼70 to 200 nucleotides, and these precursor transcripts have the ability to form stable hairpin structures. In animals, the enzyme involved in processing miRNA precursors is called Dicer, an RNase III–like protein (Grishok et al., 2001; Hutvagner et al., 2001; Ketting et al., 2001). Plants also have a Dicer-like enzyme encoded by DICER-LIKE1 (Schauer et al., 2002), and recent evidence indicates that it, like Dicer from animals, is involved in processing the hairpin precursors to generate mature miRNAs (Park et al., 2002; Reinhart et al., 2002). Furthermore, it is becoming clear from recent work that at least some miRNA hairpin precursors originate as longer polyadenylated transcripts, and several different miRNAs and associated hairpins can be present in a single transcript (Lagos-Quintana et al., 2001; Lee et al., 2002).
In animals, there is direct evidence indicating a role for specific miRNAs in development. The lineage (lin)-4 and lethal (let)-7 miRNAs in the worm Caenorhabditis elegans have been found to control temporal development, based on the phenotypes generated when the genes producing the lin-4 and let-7 miRNAs are mutated (Lee et al., 1993; Reinhart et al., 2000). In addition, both miRNAs display a temporal expression pattern consistent with their roles in developmental timing. Other animal miRNAs display developmentally regulated patterns of expression, both temporal and tissue specific (Lagos-Quintana et al., 2001, 2002; Lau et al., 2001; Lee and Ambros, 2001), leading to the hypothesis that miRNAs, in many cases, may be involved in the regulation of important developmental processes. Recent studies in Drosophila and mammals have associated specific developmental phenotypes with reduced expression of specific miRNAs (Brennecke et al., 2003; Xu et al., 2003), providing support for the proposed developmental role of miRNAs. In plants, the differential expression patterns of many miRNAs also suggest a role in development (Llave et al., 2002a; Park et al., 2002; Reinhart et al., 2002). Not much is known, however, about the functioning of plant miRNAs in specific developmental processes, because phenotypes associated with the overexpression or underexpression of specific miRNAs have not been found.
miRNAs appear to regulate target genes by binding to complementary sequences located in the transcripts produced by these genes. In lin-4 and let-7, the target sites are located in the 3′ untranslated regions of the target mRNAs (Lee et al., 1993; Wightman et al., 1993; Reinhart et al., 2000; Slack et al., 2000). Binding of the lin-4 or let-7 miRNA appears to cause the downregulation of steady state levels of the protein encoded by the target mRNA without affecting the transcript itself (Olsen and Ambros, 1999). On the other hand, recent evidence suggests that in some cases miRNAs can cause specific RNA cleavage of the target transcript within the target site (Hutvagner and Zamore, 2002; Llave et al., 2002b). miRNAs entering the RNA cleavage pathway are analogous to the 21- to 25-nucleotide short interfering RNAs generated during RNA interference in animals and post-transcriptional gene silencing in plants (reviewed by Pickford and Cogoni, 2003) and likely are incorporated into an RNA-induced silencing complex that is similar or identical to that seen for RNA interference.
It has been suggested that the mode of regulation used by a particular miRNA (i.e., either translational regulation or RNA cleavage) may be determined by the extent of sequence complementarity between the miRNA and its target (Rhoades et al., 2002). miRNAs that have a significant number of mismatches and bulges when aligned to their targets are predicted to cause translational regulation, as in the cases of lin-4 and let-7. On the other hand, a miRNA with perfect or nearly perfect complementarity with its target is predicted to cause RNA cleavage, and recent studies in both plants and animals have provided support for this idea (Hutvagner and Zamore, 2002; Llave et al., 2002b; Tang et al., 2003). Plant miRNAs share perfect or nearly perfect complementarity with their targets (Rhoades et al., 2002), which implies that many of the plant miRNAs regulate their targets by an RNA cleavage mechanism.
One striking observation about the putative target transcripts of plant miRNAs is that most of them encode members of transcription factor families that have been implicated in plant developmental patterning or cell differentiation (Rhoades et al., 2002). Plant miRNAs thus are likely to control developmental decisions by downregulating important developmental transcription factors. To date, however, there are limited data directly demonstrating a role for miRNAs in plant development. Llave et al. (2002b) have shown that a transcript for a SCARECROW-like transcription factor is a target of the Arabidopsis miRNA miR171; however, these studies were performed in a heterologous species, and no plant phenotype associated with miR171 was reported. Another miRNA, miR165/166, is predicted to target the homeodomain/Leu zipper transcription factor genes PHABULOSA (PHB) and PHAVOLUTA (PHV) (Rhoades et al., 2002), and particular mutant alleles of these genes contain single-base mutations within the miR165/166 target site. In these alleles, the PHB and PHV gene products are expressed ectopically (McConnell et al., 2001), suggesting that miR165/166 normally limits the domain of PHB and PHV expression, and RNA cleavage studies have demonstrated that PHB and PHV are in fact targets of miR165/166 (Tang et al., 2003). Nonetheless, mutants of miR165/166 have not been reported; therefore, the true function of miR165/166 in plant development can only be inferred by the phb and phv mutant phenotypes.
Using a forward-genetics approach, we have identified a miRNA that confers a developmental phenotype in Arabidopsis. This miRNA, miR172 (Park et al., 2002), causes early flowering and defects in floral organ identity when overexpressed. The predicted target of miR172 is a small subfamily of APETALA2-like transcription factors (Okamuro et al., 1997), including APETALA2 (AP2) itself. Surprisingly, miR172 appears to downregulate the AP2-like target genes via a translational mechanism rather than by RNA cleavage, even though it shares nearly perfect complementarity with these targets. In a recent report, Chen (2003) used a reverse-genetics approach to characterize miR172. Whereas that study focused primarily on miR172 regulation of AP2, we have extended the analysis to other AP2-like targets of miR172. Specifically, we have determined that overexpression of one of the AP2-like target genes, TARGET OF EAT1 (TOE1), causes late flowering. This result, in conjunction with loss-of-function analyses of TOE1 and another target gene, TOE2, indicates that at least some of the AP2-like genes targeted by miR172 normally function as floral repressors. Finally, we present a model that postulates that downregulation of AP2-like target genes by miR172 during early wild-type seedling development relieves floral repression by these genes, resulting in the promotion of flowering.
RESULTS
Overexpression of EARLY ACTIVATION TAGGED Causes Early Flowering and an ap2 Phenotype
Flowering time in Arabidopsis is controlled by several environmental and endogenous pathways, and >80 genes participate in regulation of the floral transition in this species (Simpson and Dean, 2002). In an attempt to identify novel genes involved in flowering time control, we screened an activation-tagging population (Weigel et al., 2000) (see Methods) for mutants displaying altered flowering time. Several early- and late-flowering lines were identified, and here we describe two of these mutants, early activation tagged, dominant (eat-D) and toe1-1D (see below). eat-D flowers extremely early, as indicated by visual inspection (Figure 1B) and by measuring rosette leaf number (Table 1). In addition, EAT-D displays floral defects (Figure 1D) that are virtually identical to those observed for strong ap2 mutant alleles (Bowman et al., 1991), including the complete absence of petals and the transformation of sepals to carpels.
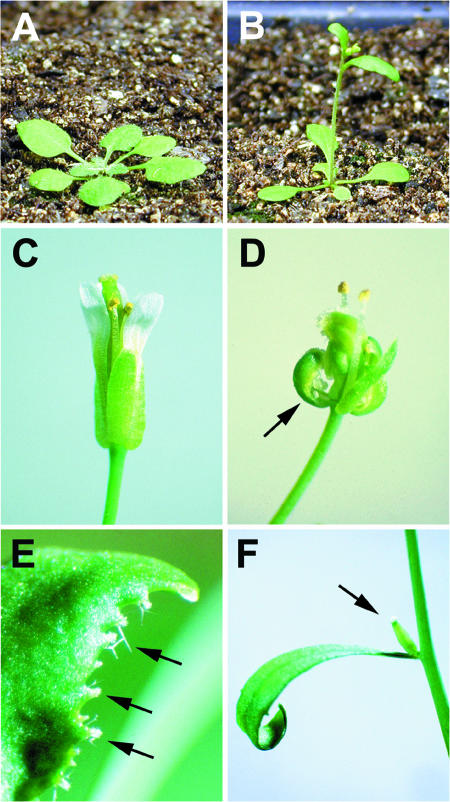
Figure 1.
EAT Overexpression Phenotype.
(A) Wild-type (Columbia [Col-0] ecotype) plant, 3.5 weeks old.
(B) eat-D plant, 3.5 weeks old.
(C) Wild-type flower.
(D) eat-D flower. Note the absence of second-whorl organs (petals). The arrow indicates a sepal with ovules along the margins and stigmatic papillae at the tip. The eat-D phenotype is virtually identical to that of strong ap2 mutant alleles.
(E) Cauline leaf margin from a 35S-EAT plant. Arrows indicate bundles of stigmatic papillae projecting from the margin.
(F) Solitary gynoecium (arrow) emerging from the axil of a cauline leaf of a 35S-EAT plant.
Table 1.
Flowering Times and Floral Phenotypes of Lines Used in This Study
| Genotype | Rosette Leaf No. (Average) a |
SD | Floral Phenotype |
|---|---|---|---|
| Wild type | 11.4 | 1.2 | Wild type |
| Wild type, short days | 36.7 | 5.2 | ap2 plus additional b |
| eat-D | 3.1 | 0.8 | ap2 |
| 35S-EAT | 2.0 | 0.2 | ap2 plus additional b |
| 35S-EAT, short days | 6.1 | 1.2 | Wild type |
| 35S-eatdel | 11.1c | 1.1 | Wild type |
| 35S-miR172a-1 | 2.1 | 0.3 | ap2 plus additional b |
| toe1-1D | 22.5 | 2.1 | Wild type |
| 35S-TOE1 | 28.6 | 3.6 | Wild type |
| toe1-2 | 8.7 | 0.6 | Wild type |
| toe2-1 | 10.2 c | 1.4 | Wild type |
| toe1-2 toe2-1 | 6.0 | 0.8 | Wild type |
Flowering time was determined by counting the number of rosette leaves, and floral phenotypes were observed visually. All plants were in the Col-0 genetic background and were grown in long-day conditions (16 h of light and 8 h of dark), except as indicated (short days [8 h of light and 16 h of dark]).
Average values from at least 10 plants per line.
See text and Figure 1 for details.
No statistically significant difference compared with the wild type. All other lines were significantly different from the wild type (Student's t test, P < 0.0001).
We mapped the activation-tagged T-DNA insert in eat-D to chromosome 5, between the annotated genes At5g04270 and At5g04280 (Figure 2A). We then used 5′ and 3′ rapid amplification of cDNA ends (RACE) PCR with primers located within this region to identify a 1.4-kb transcript, which we named EAT, that is upregulated in eat-D (Figure 2A; see also Figure 4A). When the 1.4-kb EAT cDNA was fused to the constitutive 35S promoter of Cauliflower mosaic virus and the resultant 35S-EAT construct was introduced into wild type Columbia (Col-0) plants by Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998), the 35S-EAT transformants displayed the identical early-flowering and ap2-like phenotypes seen in EAT-D (Table 1, Figures 1B and 1D). 35S-EAT plants had a normal response to photoperiod, flowering later under short days than under long days (Table 1). Many of the 35S-EAT transformants occasionally displayed floral defects in addition to those seen in eat-D, including stigmatic papillae on cauline leaf margins (Figure 1E) and the formation of a complete or partial flower rather than a secondary inflorescence in the axils of cauline leaves (Figure 1F). Therefore, ectopic expression of EAT in 35S-EAT plants affects both flowering time and the specification of floral organ identity.
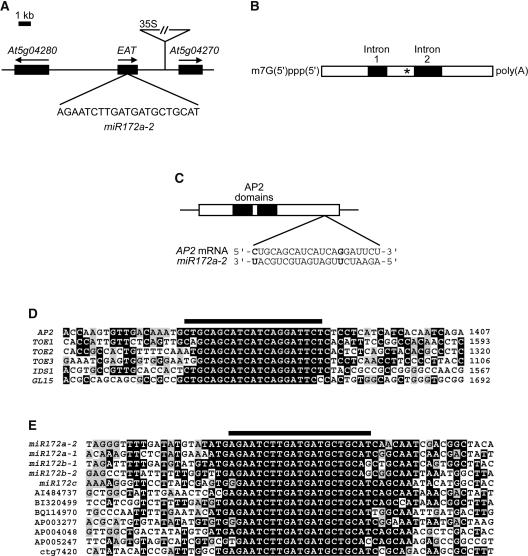
Figure 2.
EAT Contains a miRNA That Is Complementary to AP2.
(A) Location of EAT on chromosome 5. The T-DNA insertion and the orientation of the 35S enhancers are indicated. The 21-nucleotide sequence corresponding to miR172a-2 is shown below the EAT gene.
(B) Structure of the EAT precursor transcript. The polyadenylated and 5′ capped, unspliced transcript is shown, along with the positions of two introns. Two alternatively spliced transcripts, one with both introns removed and the other with intron 2 spliced out, also accumulate in wild-type plants. The location of the miR172a-2 miRNA is indicated with an asterisk.
(C) The putative 21-nucleotide miR172a-2/AP2 RNA duplex is shown below the gene structure of AP2. Two mismatches are indicated in boldface type. The boxed area delineates the coding region (ATG to stop) of the AP2 transcript, and the black boxes show the locations of the two AP2 domains.
(D) Alignment of the AP2 21-nucleotide target site (black bar at top) and surrounding sequence with three other Arabidopsis AP2 family members and two maize AP2 genes (IDS1 and GL15). Nucleotide positions of the sequences within each cDNA sequence are indicated at right. Black boxes indicate nucleotides that are highly similar to other family members, and gray boxes show nucleotides that are less well conserved.
(E) Alignment of miR172a-2 miRNA (black bar at top) and surrounding sequence with other miR172 sequences from Arabidopsis (miR172a-1, miR172b-1, miR172b-2, and miR172c), tomato (AI484737), soybean (BI320499), potato (BQ114970), and rice (AP003277, AP004048, AP005247, and ctg7420). Black and gray boxes are as described for (D).
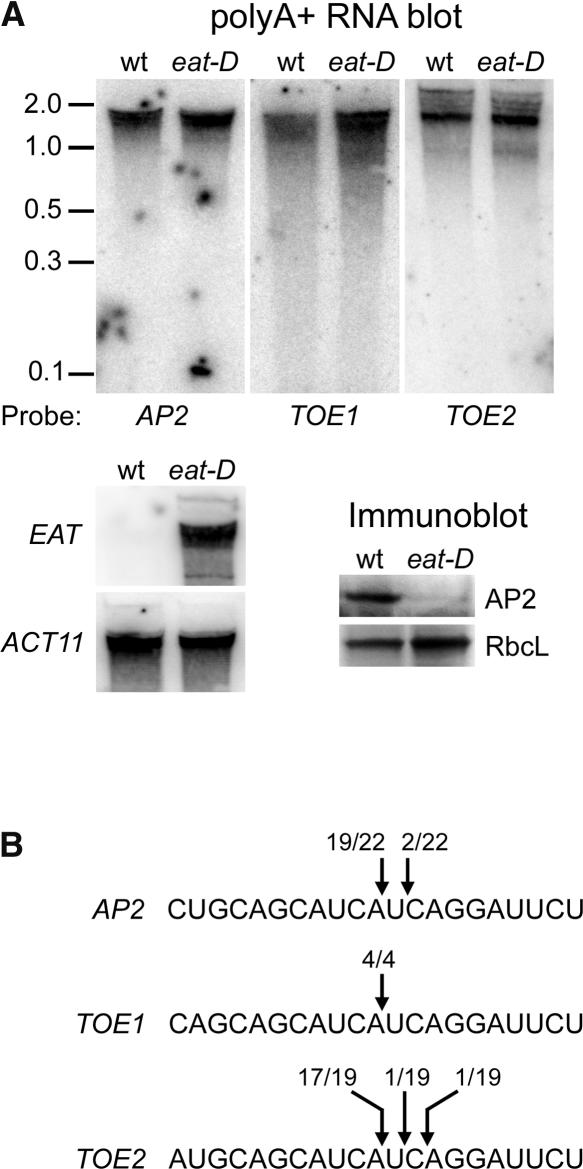
Figure 4.
miR172 Downregulates AP2.
(A) At top and bottom left are RNA gel blots of 1 μg of poly(A+) RNA from the wild type (wt) and eat-D. RNA from each genotype was isolated from floral buds. The blots were hybridized with probes specific for the AP2-like genes indicated below or at left. At bottom right are immunoblots of proteins from wild-type (wt) and eat-D floral buds probed with AP2 antibody. AP2 is severely reduced in eat-D. RbcL, large subunit of ribulose-1,5-bisphosphate carboxylase as a loading control.
(B) Target sites for miR172 in three AP2-like transcripts, with the 5′ ends of RNA cleavage products that were identified by 5′ RACE PCR indicated by arrows. The numbers of 5′ RACE clones sequenced that correspond to each cleavage product are indicated.
The EAT Transcript Is the Precursor RNA for a Member of the miR172 Family
EAT produces a 1417-nucleotide noncoding RNA that is predicted to be 5′ capped and polyadenylated (Figure 2B), based on our RACE-PCR methodology (see Methods). We also identified two splice variants of EAT: a short form in which two introns are removed, and an intermediate form in which only one intron (intron 2) is spliced out. All three transcripts are present at similar levels in wild-type plants (M.J. Aukerman, unpublished results). Basic Local Alignment Search Tool (BLASTN and BLASTX) searches of several databases with the 1.4-kb EAT cDNA revealed no extensive nucleotide or predicted amino acid sequence identity between EAT and any other gene. However, we did identify a 21-nucleotide stretch in the middle of the EAT transcript that is identical to miR172a-2 (Figure 2A), a recently identified miRNA (Park et al., 2002). Both splice variants of EAT also contain this 21-nucleotide sequence (Figure 2B, asterisk). To confirm the functional importance of the miR172a-2 sequence within the EAT cDNA, we generated a mutant form of EAT in which the miR172a-2 sequence was deleted and made a construct consisting of this mutant EAT cDNA, eatdel, driven by the 35S promoter. Transgenic plants carrying this 35S-eatdel construct flowered with the same number of leaves as the wild type and had normal flowers (Table 1), indicating that the miR172a-2 sequence is necessary to confer both the flowering-time and floral organ identity phenotypes seen in EAT-overexpressing lines.
As noted by Park et al. (2002), the 21-nucleotide miR172 miRNA has the potential to form an RNA duplex with a sequence near the 3′ end of the coding region of AP2 (Figure 2C). It is likely, then, that overexpression of miR172a-2 causes the observed ap2 phenotype by binding to this target site in AP2, leading to a reduction in AP2 expression (see below). Similar to other plant miRNAs, miR172 shares nearly perfect complementarity with its target (19 of 21 nucleotides). The target site for miR172 is found in a similar location in three other Arabidopsis AP2 family members that we have named TOE1, TOE2, and TOE3 (corresponding to At2g28550, At5g60120, and At5g67180, respectively) and in the maize AP2 genes INDETERMINATE SPIKELET1 (Chuck et al., 1998) and GLOSSY15 (Moose and Sisco, 1996) (Figure 2D). We also found the miR172 target site in AP2 family members from many other plant species, including soybean, rice, wheat, tomato, and pea (M.J. Aukerman, unpublished results). The conservation of this 21-nucleotide sequence in a subset of AP2 genes from both monocots and dicots suggests that this sequence is of functional significance and is likely a cis-acting factor that regulates the expression of these particular AP2 family members in response to miR172-like miRNAs.
There is an additional copy of the miR172a-2 miRNA in the Arabidopsis genome on chromosome 2 (miR172a-1), and miR172a-2 is highly similar to three other Arabidopsis loci (Figure 2E). Like the miR172a-2 miRNA, all four reiterations of the sequence are in intergenic regions (i.e., between the Arabidopsis genes currently annotated in GenBank). In addition, the sequence was found in ESTs from tomato, potato, and soybean, and four copies were found in the genomic sequence of rice (Figure 2E). To determine whether another member of the miR172 family, miR172a-1, would confer a phenotype similar to that of miR172a-2, we generated a construct containing the 35S promoter fused to the genomic region surrounding miR172a-1. Plants containing the 35S-miR172a-1 construct flowered early and displayed an ap2 phenotype (Table 1), indicating that miR172a-1 behaves in an identical manner to miR172a-2 when overexpressed.
Expression Analysis of miR172
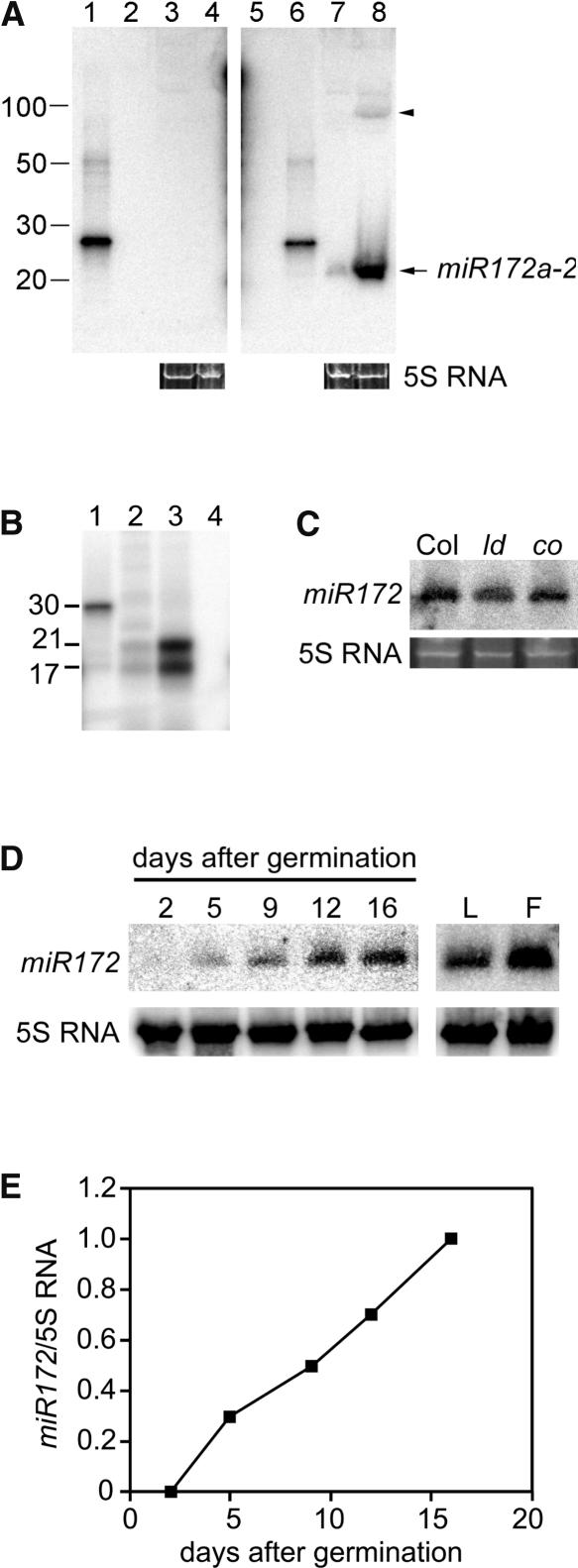
RNA gel blot analysis using a probe antisense to the miR172a-2 miRNA identified a small single-stranded RNA of 21 to 25 nucleotides accumulating to much higher levels in eat-D plants relative to the wild type (Figure 3A). The small amount of transcript seen in the wild type presumably represents endogenous levels of not only the miR172a-2 miRNA but also its family members, which are similar enough to cross-hybridize with the probe. The predicted miR172a-2 hairpin precursor is 95 nucleotides in length (Park et al., 2002), and we detected a transcript of approximately that size accumulating in eat-D that likely represents unprocessed miR172a-2 hairpin precursor (Figure 3A, arrowhead). S1 nuclease mapping of the miR172a-2 miRNA (Figure 3B) provided independent confirmation of the 5′ end of miR172 reported by Park et al. (2002).
Figure 3.
miR172 Expression.
(A) RNA gel blot of 50 μg of total RNA from wild-type Col-0 (lanes 3 and 7) and eat-D (lanes 4 and 8). RNA was isolated from the aboveground parts of plants that had already bolted. Blots were probed with sense (lanes 1 to 4) or antisense (lanes 5 to 8) oligonucleotides to miR172a-2 miRNA. One hundred picograms of sense oligonucleotide (lanes 2 and 6) or antisense oligonucleotide (lanes 1 and 5) was loaded as a hybridization control. Nucleotide length markers are indicated at left. The arrowhead shows the putative hairpin precursor of miR172a-2. Ethidium bromide–stained 5S RNA served as a loading control for lanes 3, 4, 7, and 8.
(B) S1 nuclease mapping of miR172a-2 miRNA. A 5′ end labeled probe was undigested (lane 1) or digested after hybridization to total RNA from wild-type (lane 2), eat-D (lane 3), or tRNA (lane 4) plants. RNA samples in lanes 2 and 3 were isolated from aboveground parts of plants that had already bolted. Nucleotide length markers are indicated at left.
(C) RNA gel blot of 30 μg of total RNA from wild-type Col-0 (Col), luminidependens (ld), or constans (co) probed with antisense oligonucleotide to miR172. Plants were harvested at 16 days after germination. 5S RNA, loading control.
(D) RNA gel blot analysis of 30 μg of total RNA from wild-type seedlings harvested at 2, 5, 9, 12, and 16 days after germination or from mature leaves (L) and floral buds (F). Probes for miR172 and 5S RNA are indicated at left.
(E) Quantification of miR172 transcript abundance (normalized to 5S RNA) with respect to developmental time (i.e., days after germination). The normalized miR172 RNA level at 16 days after germination was set arbitrarily to 1.
The expression of miR172 was regulated temporally, with no miR172 transcripts detected 2 days after germination, and progressively more steady state transcript accumulation was seen as the plant approached flowering (Figures 3D and 3E). The miR172 transcripts also accumulated after flowering had occurred in both leaves and floral buds. We assessed the levels of miR172 in two mutants, luminidependens and constans, and found that miR172 expression was not altered in either of these mutants (Figure 3C), indicating that miR172 is not found downstream of these genes. In addition, the levels of the miR172a-2 precursor transcript were identical in plants grown in long days versus those grown in short days (M.J. Aukerman, unpublished results).
miR172 Is a Translational Repressor of Its AP2-Like Target Genes
Immunoblot analysis with an antibody specific for AP2 (see Methods) indicated that the AP2 protein was reduced dramatically in the miR172a-2 overexpression (eat-D) line relative to the wild type, whereas the AP2 transcript accumulated to normal levels in eat-D (Figure 4A). Likewise, mRNA levels for the AP2-like target genes TOE1 and TOE2 were not reduced in the eat-D line. These data suggest that the miR172a-2 miRNA negatively regulates AP2 (and likely its other targets as well) at the level of translation and not by RNA cleavage. This result was unexpected in light of the report from Kasschau et al. (2003), in which cleavage products derived from the miR172 target sites in the AP2-like genes were identified. We used the same reverse transcriptase–mediated (RT) PCR procedure described by those authors and confirmed that cleavage products can be detected for the AP2-like target genes in both wild-type and eat-D plants (Figure 4B). Nevertheless, we detected no cleavage products of the appropriate size on RNA gel blots (Figure 4A), even when miR172 was overexpressed.
For each AP2-like gene, a gene-specific probe that recognizes sequences 3′ to the miR172 target site was used for hybridization; therefore, we expected to detect 3′ cleavage products in the region of the gel corresponding to fragment lengths of 200 to 500 nucleotides (e.g., the predicted AP2 cleavage product is ∼300 nucleotides in length). Cleavage products corresponding to the 5′ end of each transcript would not be detected, because the RNA samples were poly(A+) selected. Because we did not detect miR172-directed 3′ cleavage products by RNA gel blot analysis, the cleavage products detected by PCR likely represent a very small fraction of the total transcript population produced from the AP2-like genes. This notion is supported by the fact that overexpression of miR172 did not change the steady state levels of the AP2-like target genes (Figure 4A). We conclude that miR172 regulates its target genes by a translational mechanism and that the small amount of RNA cleavage observed likely is the result of overlap between the translational and RNA cleavage pathways (see Discussion for more details).
Two AP2-Like Targets of miR172 That Function as Floral Repressors
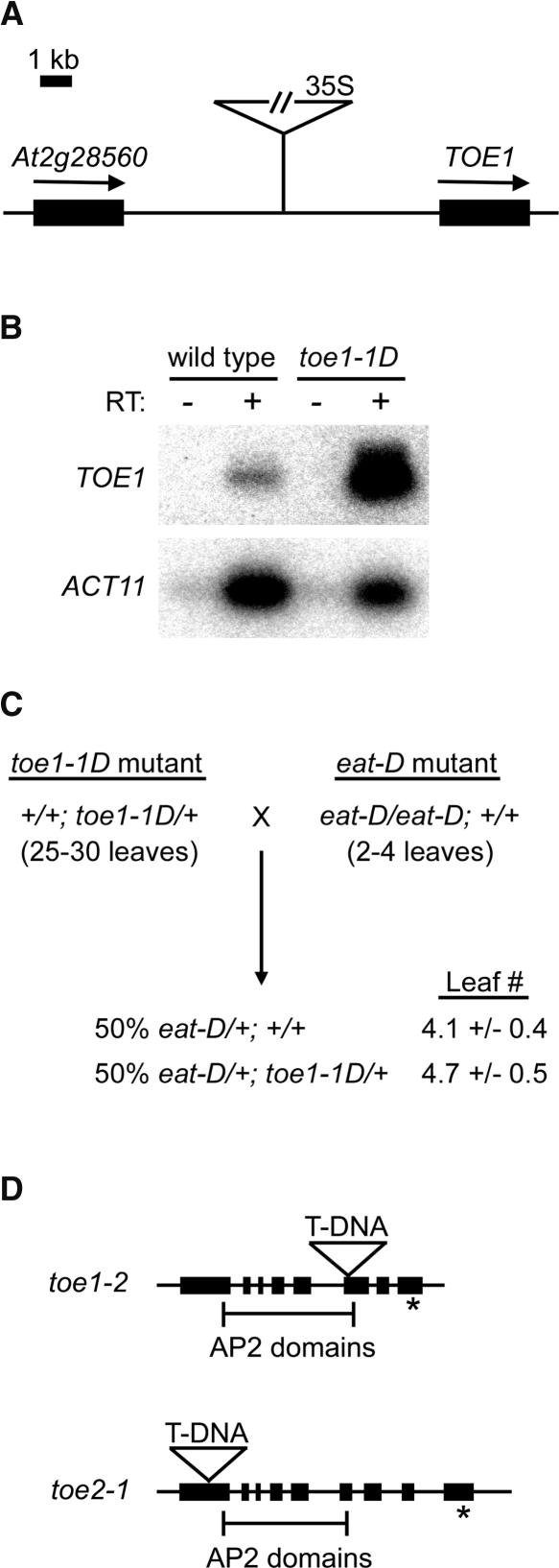
In the same genetic screen that identified the early-flowering eat-D mutant, we identified an activation-tagged late-flowering mutant called toe1-1D. toe1-1D displayed no additional phenotypes besides late flowering (Table 1), and the late-flowering phenotype cosegregated with a single T-DNA insertion (M.J. Aukerman, unpublished results). Sequence analysis of the T-DNA insert in toe1-1D indicated that the 4× 35S enhancer was located ∼5 kb upstream of TOE1 (Figure 5A), one of the AP2-like target genes that contains a target site for miR172a-2 (Figure 2D). Semiquantitative RT-PCR analysis using primers specific for TOE1 indicated that the transcript corresponding to this gene was expressed at 23-fold higher levels in toe1-1D relative to the wild type (Figure 5B). To confirm that overexpression of TOE1 causes late flowering, we fused a genomic region containing the entire TOE1 coding region to the 35S promoter and created transgenic plants containing this construct. Transgenic 35S-TOE1 plants flowered much later than wild-type plants and also later than toe1-1D (Table 1). This late-flowering phenotype was observed in multiple independent transformants (M.J. Aukerman, unpublished results).
Figure 5.
Activation-Tagged and Loss-of-Function Alleles of AP2-Like Target Genes.
(A) Location of the T-DNA insert in toe1-1D. The 35S enhancers are located ∼5 kb upstream of TOE1 (At2g28550).
(B) Semiquantitative RT-PCR analysis of TOE1 and ACT11 expression in the wild type versus toe1-1D. RNA from each genotype was isolated from floral buds. Inclusion of reverse transcriptase (RT) is indicated by + or − above the gels.
(C) eat-D suppresses the phenotype of toe1-1D. Flowering-time phenotypes (expressed as rosette leaf number) of F1 plants derived from the cross of a toe1-1D heterozygote to an eat-D homozygote. Approximately 50% of the F1 plants contained the toe1-1D allele, based on PCR genotyping; these plants were early flowering, indicating the suppression of toe1-1D (which overexpresses TOE1) by eat-D (which overexpresses miR172). In this experiment, wild-type control plants flowered with ∼11 leaves.
(D) Locations of the T-DNA inserts in toe1-2 and toe2-1. Exons are indicated by solid bars, and the target sequences for miR172 are indicated with asterisks. Brackets delineate the two AP2 domains contained in each gene.
To assess the ability of miR172a-2 to regulate TOE1, a plant homozygous for eat-D (and thus overexpressing miR172a-2) was crossed to a plant heterozygous for toe1-1D (and thus overexpressing TOE1). In this cross, all of the F1 progeny were expected to contain one copy of eat-D and 50% of the F1 progeny also should have had one copy of toe1-1D. F1 progeny were scored for the presence or absence of the toe1-1D allele by PCR and also were scored for flowering time. All of the F1 plants were early flowering regardless of whether or not they contained a copy of the toe1-1D allele (Figure 5C), indicating that overexpression of miR172a-2 can suppress the effects of TOE1 overexpression. This result is consistent with a model in which miR172a-2 directly downregulates TOE1 by binding to its cognate target site. We observed late-flowering toe1-1D segregants in the subsequent generation (M.J. Aukerman, unpublished results), which excludes the possibility that early flowering in the F1 generation was attributable to cosuppression occurring at the TOE1 locus.
To determine the effects of reducing TOE1 function, we identified plants containing a T-DNA insertion in TOE1 (Figure 5D). In addition, we identified a T-DNA mutant for TOE2, a closely related AP2-like gene that also contains the miR172a-2 target sequence. Given the locations of the inserts either upstream of (toe2-1) or within (toe1-2) the highly conserved AP2 domain, it is likely that both mutations result in the complete loss of function of the respective gene. We determined that plants homozygous for the toe1-2 T-DNA allele were slightly early flowering relative to the wild type, whereas toe2-1 plants were not significantly early flowering (Table 1). The two mutants were crossed, and the double mutant was identified by PCR genotyping. The toe1-2 toe2-1 double mutant was earlier flowering than either individual mutant (Table 1), suggesting that the genes have overlapping function. The early-flowering phenotype of toe1-2 toe2-1 is consistent with a model in which miR172 causes early flowering by downregulating several AP2-like floral repressors, including TOE1 and TOE2. Interestingly, toe1-2 toe2-1 is not as early flowering as miR172-overexpressing lines (cf. 35S-EAT; Table 1), which suggests that other AP2-like targets of miR172a-2 (e.g., AP2 itself or TOE3) also act as floral repressors. Because ap2 single mutants are not early flowering (Bowman et al., 1991), any potential negative regulation of flowering by AP2 must be normally masked by genetic redundancy.
DISCUSSION
Using an activation-tagging approach, we have identified a miRNA involved in the control of flowering time and floral organ identity. Overexpression of EAT, which produces a precursor transcript for miR172a-2, causes extremely early flowering and floral organ identity defects. The predicted target of miR172 is a small subfamily of AP2-like genes, including AP2 itself, and higher levels of miR172 are associated with the downregulation of AP2 gene expression at the translational level. In a recent report, miR172 was characterized by a reverse-genetics approach (Chen, 2003), and our findings support the general conclusions of that work. Whereas Chen focused primarily on miR172 regulation of AP2 and its effects on floral organ identity, we have found that two of the other AP2-like targets of miR172, TOE1 and TOE2, function in the control of flowering time as floral repressors. Because the target site sequences of these other AP2-like target genes are virtually identical to that of AP2, it is highly likely that they also are downregulated by miR172, which would explain the early-flowering phenotype associated with miR172 overexpression.
Based on the overexpression phenotypes of both miR172a-1 and miR172a-2 (Table 1), one would expect that loss-of-function alleles of these miRNAs would have the opposite phenotype. This prediction may be difficult to test, however, because of the high degree of sequence similarity among the predicted mature miR172 miRNAs (Figure 2E), which makes it likely that the members of the miR172 gene family are functionally interchangeable. Indeed, insertional mutations within either the miR172a-1 or miR172a-2 precursor gene do not cause any flowering-time or floral morphology defects (M.J. Aukerman, unpublished results), suggesting that there is genetic redundancy within the miR172 family. On the other hand, dicer-like1 mutants have lower overall levels of the entire miR172 family (Park et al., 2002; Kasschau et al., 2003) and display flowering-time and floral morphology defects that are opposite those of miR172 overexpression (Schauer et al., 2002). These observations provide independent evidence to support a role for miR172 in the regulation of flowering time and floral organ identity.
Although several different groups have identified plant miRNAs, the structure of any full-length plant miRNA precursor transcript was not known previously. We have found that EAT produces a 1.4-kb noncoding RNA that represents the full-length precursor of miR172a-2. The precursor transcript is polyadenylated and 5′ capped and appears to enter the splicing pathway, and these features strongly suggest that it is normally transcribed by RNA polymerase II (Pol II). It is unclear whether all or most of the other plant miRNA precursors also are Pol II transcripts. RNA polymerase III (Pol III) promoters have been used in mammalian systems to express hairpin precursors of miRNAs (McManus et al., 2002), and it is possible that some miRNA precursors are naturally transcribed by Pol III. Nevertheless, we have identified other plant miRNA sequences embedded within longer transcripts in EST collections (M.J. Aukerman, unpublished results), suggesting that many of the plant miRNA precursor transcripts are produced by Pol II rather than by Pol III.
RNA gel blot and immunoblot analyses (Figure 4A) indicated that miR172 downregulates its AP2-like target genes at the level of translation rather than by RNA cleavage. Although in animals there are several examples of translational regulation by miRNAs (Olsen and Ambros, 1999; Reinhart et al., 2000; Kawasaki and Taira, 2003), miR172 is the only plant miRNA that has been found to regulate its targets at the translational level. The identification of miR172-directed RNA cleavage products by PCR (Figure 4B) (Kasschau et al., 2003) appears to contradict this model of translational regulation; however, these cleavage products presumably represent a very small fraction of the total AP2 transcript population, because they are undetectable on RNA gel blots (Figure 4A). Furthermore, AP2 transcript abundance was unaffected by miR172a-2 overexpression, whereas AP2 was downregulated significantly (Figure 4A), indicating that regulation occurs at the level of translation. This unexpected result suggests that the regulation of target sequences by miRNAs may be more complex than was proposed previously, as we discuss further below.
It has been suggested that many plant miRNAs enter the RNA interference pathway exclusively as a result of their perfect or nearly perfect complementarity to putative targets (Rhoades et al., 2002; Tang et al., 2003). For example, miR171 has perfect complementarity to its SCARECROW-like target gene and has been shown to direct the cleavage of its target transcript (Llave et al., 2002b). A few mismatches between a miRNA and its target gene do not appear to abolish miRNA-directed RNA cleavage (Tang et al., 2003). In fact, most other plant miRNAs naturally have one, two, or three mismatches with their target genes; nonetheless, cleavage products from many of these target genes can be identified by PCR (Kasschau et al., 2003). Interestingly, miR172 itself contains only two mismatches with AP2 (Figure 2C), yet clearly it does not operate via an RNA cleavage mechanism. On the other hand, the fact that small amounts of RNA cleavage products do accumulate suggests overlap between the cleavage pathway and a second pathway operating at the translational level. Indeed, recent evidence suggests that miRNA-directed translational and RNA cleavage pathways may share some of the same molecular components (Hutvagner and Zamore, 2002; Mourelatos et al., 2002). It is possible that in the case of miR172 this overlap causes a small amount of target gene RNA cleavage as a side effect of translational regulation. In fact, it may turn out that other plant miRNAs normally regulate their targets at the translational level, even though cleavage products corresponding to their targets can be detected by PCR (Kasschau et al., 2003). Because these cleavage products are possible by-products of the proposed overlap between the two pathways and thus may not reflect a significant reduction of steady state target transcript levels, PCR-based detection of target RNA cleavage may give misleading results in these cases.
Although we have demonstrated that the regulation of miRNA target transcripts can occur at the level of translation in plants, the biochemical nature of the translational repression is not known. In C. elegans, lin-4 blocks the translation of its target transcript after the transcript has associated with ribosomes (Olsen and Ambros, 1999). Whether this postinitiation block also occurs in the case of miR172 and its AP2-like targets is unclear. Wheat germ extracts have been used to study miRNA-directed cleavage, and it may be possible to use such an in vitro system to characterize translational repression by miRNAs at the biochemical level. It also is unclear how miR172, which shares nearly perfect complementarity with its target, is directed away from the RNA cleavage pathway and into a translational repression pathway. We suggest a sequence determinant, within either miR172 or its target site, that functions as a recognition site for one or more of the components responsible for translational repression. This sequence determinant could be a single nucleotide, and in this regard it is interesting that miR172 is the only plant miRNA known to have an A residue at its 5′ end. As more plant miRNAs are discovered, it will be interesting to see if others begin with A and whether these also use a translational mode of regulation.
The target site for miR172 is found not only in AP2 but also in a few other AP2-like genes, including TOE1 and TOE2 (Figure 2D). Because ap2 mutants are not early flowering but miR172 overexpression lines are, we reasoned that miR172 controls flowering time by downregulating an AP2-like target other than AP2 itself. We have obtained data indicating that TOE1, and to a lesser extent TOE2, control flowering time and therefore are likely targets of miR172. Activation tagging of TOE1, followed by analysis of the toe1-2 loss-of-function mutant, indicated that TOE1 normally acts as a floral repressor (Table 1). These results are consistent with miR172 overexpression causing early flowering by downregulating TOE1. Interestingly, although toe2-1 was not significantly early flowering, the toe1-2 toe2-1 double loss-of-function mutant was earlier flowering than toe1-2, and this finding suggests that these two genes overlap in function. Further gene redundancy likely exists within this family of miR172 target genes, and we predict that triple and quadruple mutant combinations will more closely resemble the miR172 overexpression lines with regard to flowering time.
The loss-of-function phenotypes for TOE1 and TOE2 suggest that they are targets of miR172, and two additional pieces of evidence further solidify this connection. First, miR172 overexpression fully suppressed the late-flowering phenotype of TOE1 overexpression (Figure 5C), consistent with miR172 directly downregulating TOE1. Second, miR172-directed target site RNA cleavage products corresponding to TOE1 and TOE2 were recovered by PCR (Figure 4B) (Kasschau et al., 2003). Even though these cleavage products likely are side effects of translational control (see above), the fact that even a small amount of such cleavage products corresponding to these two target genes accumulate in wild-type plants strongly suggests that they normally are targeted by miR172. RNA cleavage products have been detected for all four predicted Arabidopsis miR172 targets (Kasschau et al., 2003), raising the possibility that any AP2-like gene containing the 21-nucleotide target site sequence, or a close variant, is a target of miR172. Indeed, the conservation of this sequence in a subset of AP2 genes from both monocots and dicots (Figure 2D) (M.J. Aukerman, unpublished results) strongly suggests that this sequence is a target site for miR172-like miRNAs in many plant species. This indicates that miR172 participates in flowering-time and floral organ identity control in plants other than Arabidopsis, which is a subject for future study.
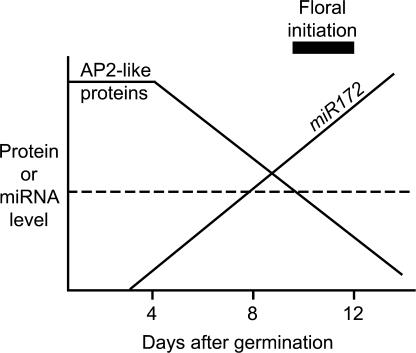
Consistent with its proposed role in the regulation of flowering time, miR172 expression was regulated temporally (Figures 3D and 3E). Therefore, we propose a model (Figure 6) that predicts that the temporal upregulation of miR172 family members during seedling development leads to the temporal downregulation of TOE1 and TOE2 (and perhaps AP2 and/or TOE3). This model further predicts that flowering is triggered once the levels of these floral repressors decrease below a critical threshold. However, we cannot confirm that TOE1 and TOE2 are downregulated during this same period, because antibodies for these proteins are unavailable. Certain aspects of miR172, in particular its temporal expression pattern and its regulation of target genes at the translational level, are reminiscent of lin-4 and let-7, two miRNAs that control developmental timing in C. elegans (Feinbaum and Ambros, 1999; Reinhart et al., 2000). The fact that miR172 also affects timing in plants (Figure 1B, Table 1) indicates that both plants and animals use miRNAs to control temporal events during development.
Figure 6.
Model Depicting the Temporal Regulation of Flowering by miR172.
In this model, the temporal expression of miR172 causes temporal downregulation of the AP2-like target genes at the level of translation, which triggers flowering once the target proteins decrease below a critical threshold (dotted line). The onset of floral initiation is indicated by the black bar.
Flowering time in Arabidopsis is controlled by endogenous and environmental mechanisms, and there are many genes implicated in the regulation of flowering time (reviewed by Simpson and Dean, 2002). These genes have been assigned to several pathways based on their response to photoperiod, vernalization, and hormones. Although it is clear that at least some of the regulation of these pathways is performed by transcription factors, most notably those of the MADS box family, no evidence has been presented to date of any involvement of AP2 family members in this process or of miRNAs. Here, we provide evidence for the involvement of both miRNAs and AP2-like proteins in flowering time. Although we have not yet assigned miR172 to a specific flowering-time pathway, we found that lines overexpressing miR172 have a normal response to photoperiod (Table 1). Furthermore, mutations that affect either the autonomous (luminidependens) or photoperiod (constans) pathways did not change the levels of miR172 (Figure 3C), suggesting that it is not a downstream component of these pathways. It is possible that miR172 and its targets function within a flowering-time pathway that is poorly characterized, such as the pathway that regulates flowering in response to intrinsic developmental age (Simpson and Dean, 2002) or the ambient-temperature-response pathway (Blazquez et al., 2003). Future physiological experiments with miR172 overexpression lines and the AP2-like loss-of-function mutants, coupled with transcript analysis of other flowering-time genes, should allow a more precise placement of miR172 within the flowering pathway network and help to identify downstream components. Interestingly, certain flowering-time genes, such as FLOWERING LOCUS T and SUPPRESSOR OF OVEREXPRESSION OF CO1, display a temporal expression pattern similar to that of miR172 and thus are potentially downstream of miR172 and the AP2-like floral repressors.
METHODS
Plant Material and Growth Conditions
All experiments were performed on the Columbia (Col-0) ecotype of Arabidopsis thaliana. Plants were grown in long days (16 h of light, 8 h of dark) under cool-white light at 22°C, except where noted.
Activation Tagging
Arabidopsis plants were transformed by a modified version of the floral-dip method (Clough and Bent, 1998) in which Agrobacterium tumefaciens cell suspension was applied to plants by direct watering from above. The T-DNA vector used, pHSbarENDs, contained four copies of the 35S enhancer of Cauliflower mosaic virus adjacent to the right border, an arrangement similar to that described by Weigel et al. (2000). Transformed plants were selected with glufosinate (Basta) and screened for flowering time, which resulted in the identification of the early-flowering mutant eat-D. A single T-DNA cosegregating with early flowering was identified in eat-D, and thermal asymmetric interlaced PCR was performed to amplify sequences adjacent to the left and right borders of the T-DNA. To identify transcripts upregulated in eat-D, we probed RNA gel blots containing RNA extracted from wild-type (Col-0) and eat-D plants. Probes for the genes At5g04270 and At5g04280 (Figure 2A) detected no difference between the wild type and eat-D, whereas a probe from the intergenic region identified an ∼1.4-kb transcript that was expressed at significantly higher levels in eat-D than in the wild type. 5′ and 3′ rapid amplification of cDNA ends PCR was performed to isolate the full-length EAT cDNA using the GeneRacer kit (Invitrogen, Carlsbad, CA) and RNA extracted from wild-type floral buds. The fact that the GeneRacer protocol allows selective amplification of 5′ capped messages as well as the use of oligo(dT) to prime first-strand cDNA synthesis indicated that the EAT transcript is normally 5′ capped and polyadenylated.
Transgenic Constructs
We amplified the entire 1.4-kb EAT transcription unit from Col-0 genomic DNA using primers located at the 5′ and 3′ ends of the gene that also contained XhoI sites (5′-GACTACTCGAGCACCTCTCACTCCCTTTCTCTAAC-3′ and 5′-GACTACTCGAGGTTCTCAAGTTGAGCACTTGAAAAC-3′). To generate the 35S-EAT construct, the XhoI-cut EAT was inserted into the XhoI site of binary vector pBE851 between the 35S promoter of Cauliflower mosaic virus and a β-phaseolin terminator. The 35S-eatdel construct was generated as follows. The 1.4-kb EAT cDNA (in a modified pBluescript SK+ vector [Stratagene] lacking BamHI and HindIII sites) was digested with BamHI and HindIII, releasing a 159-bp fragment containing the miR172a-2 sequence and hairpin precursor. The vector fragment containing the EAT cDNA with the microRNA (miRNA) and hairpin sequences deleted was gel-purified. Oligonucleotides that reconstructed the entire 159-bp fragment except for the 21-nucleotide miRNA sequence were annealed and ligated into the digested EAT cDNA, generating a modified EAT cDNA lacking the 21-nucleotide miRNA sequence. This eatdel cDNA was ligated into pBE851 as described above.
To generate 35S-miR172a-1, primers were designed to amplify a 1.9-kb genomic fragment surrounding miR172a-1 (5′-CCGCTCGAGATCTGTGTTCTAGATCTCACCAGGTC-3′ and 5′-CCGCTCGAGGAGTGAAGACATATTATATTAATTGAGGTAC-3′). This fragment encompasses a full-length cDNA sequence, RAFL21-01-E23, deposited in GenBank. The amplified fragment was digested with XhoI and inserted into pBE851 as described above. To generate the 35S-TOE1 construct, a 2-kb genomic fragment containing the entire TOE1 coding region was amplified with primers containing KpnI sites at the ends (5′-CCGGTACCATGTTGGATCTTAACCTCAACGCTGATTC-3′ and 5′-CCGGTACCTTAAGGGTGTGGATAAAAGTAACCACGTGTTG-3′). After KpnI digestion of the PCR product, the KpnI sites were converted to XhoI sites by ligation of an eight-nucleotide adapter oligonucleotide, and the fragment was ligated into XhoI-cut pBE851. All constructs were transformed into Agrobacterium, and Col-0 was transformed with the Agrobacterium strains by floral dip (Clough and Bent, 1998).
Analysis of miR172 and AP2 Expression
Total RNA was isolated from wild-type and eat-D plants at the stages and tissues indicated in the figure legends using TRIZOL reagent (Sigma). Fifty micrograms of each RNA was subjected to electrophoresis on a 15% TBE-Urea Criterion gel (Bio-Rad) and electroblotted onto Hybond-N+ filter paper (Amersham) using a TransBlot-SD apparatus (Bio-Rad). The filter then was hybridized at 37°C overnight in UltraHyb-Oligo buffer (Ambion, Austin, TX) with 32P-labeled oligonucleotides. The oligonucleotides were 30-mers that corresponded to either the sense or antisense strand of miR172a-2, with four or five nucleotides of flanking sequence on each side (5′-ATATGAGAATCTTGATGATGCTGCATCAAC-3′ and 5′-GTTGATGCAGCATCATCAAGATTCTCATAT-3′). The filters were washed twice at 37°C in buffer containing 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.5% SDS. For S1 analysis, probe was made by end-labeling an oligonucleotide (5′-ATGCAGCATCATCAAGATTCTCATATACAT-3′) with T4 polynucleotide kinase and 32P. Hybridization and processing of S1 reactions were performed using standard protocols (Ausubel et al., 1995).
RNA gel blot analysis to determine AP2 levels was performed on poly(A)-enriched RNA from floral buds. The RNA was subjected to electrophoresis on 5% TBE-Urea Criterion gels, blotted as described above, and incubated with 32P-labeled probes specific for AP2, TOE1, TOE2, EAT, or ACT11. The probes were amplified by PCR using Col-0 genomic DNA as template with the following primers: AP2 (5′-TTTCCGGGCAGCAGCAACATTGGTAG-3′ and 5′-GTTCGCCTAAGTTAACAAGAGGATTTAGG-3′); TOE1 (5′-TTTCCGGCCACAACCTCCCAATGACAATG-3′ and 5′-TTTTGAAAGGGGACTAGAGTGTTGAGAG-3′); TOE2 (5′-TACACGCCCTCCTTCATCCACAGCTATTC-3′ and 5′-GAAACTACAGCTTGTGTGCTGAGCCAAAC-3′); and ACT11 (5′-ATGGCAGATGGTGAAGACATTCAG-3′ and 5′-GAAGCACTTCCTGTGGACTATTGATG-3′). The EAT probe fragment was isolated by digesting the EAT cDNA in pBluescript SK+ (see above) with XhoI and gel-purifying the insert. Basic Local Alignment Search Tool (BLAST) searches with the sequences contained in each AP2-like gene probe indicated that each probe was gene specific. UltraHyb buffer was used for hybridization to the AP2-like genes (Ambion), and subsequent processing was as described in the UltraHyb protocol. Immunoblot analysis of AP2 was performed on proteins extracted from floral buds. After electrophoresis on a 10% SDS-PAGE gel, proteins were transferred to a Hybond-P membrane (Amersham) and incubated with an antibody specific for AP2 (aA-20; Santa Cruz Biotechnology, Santa Cruz, CA). The AP2 antibody specifically recognizes an ∼45-kD protein that corresponds to AP2, because in an ap2 null mutant this 45-kD protein is absent (Chen, 2003). The blot was processed using the ECL-Plus kit (Amersham).
Identification of toe1-1D, toe1-2, and toe2-1
toe1-1D was identified in the same manner as described for eat-D above, except that semiquantitative reverse transcriptase–mediated (RT) PCR was used to confirm the overexpression of TOE1 in toe1-1D (Figure 3B). Oligonucleotides used to detect TOE1 expression were 5′-CAAATGTGGTAGATGGGAAGCTAGGATG-3′ and 5′-CCAGTTACTCATCATCCCTTCAGCTTCAC-3′. RT-PCR of TOE1 was performed for 22 cycles to ensure logarithmic amplification. Control primers for ACT11 were 5′-ATGGCAGATGGTGAAGACATTCAG-3′ and 5′-GAAGCACTTCCTGTGGACTATTGATG-3′. RT-PCR products were subjected to electrophoresis on 2% agarose gels, blotted onto Hybond-N+, and hybridized with 32P-labeled probes for TOE1 and ACT11. toe1-2 (SALK_069677) and toe2-1 (SALK_065370) were identified in the collection of Salk mutants available from the Arabidopsis Stock Center. PCR amplification was used to confirm the presence of T-DNA inserts within both genes.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact M.J. Aukerman, milo.j.aukerman@usa.dupont.com.
Accession Number
The GenBank accession number for RAFL21-01-E23 is AK118705.
NOTE ADDED IN PROOF
Recently, a microRNA controlling leaf morphogenesis was described (Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. [2003]. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263).
Acknowledgments
We thank M. Mucha for technical assistance; W. Park, S. Tingey, S. Martino-Catt, and M. Albertsen for helpful discussions; and K. Glassman for critical reading of the manuscript. We also thank the anonymous reviewers for their comments.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.016238.
References
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1995). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
- Blazquez, M.A., Ahn, J.H., and Weigel, D. (2003). A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 33, 168–171. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Smyth, D.R., and Meyerowitz, E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. (2003). bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36. [DOI] [PubMed] [Google Scholar]
- Chen, X. (2003). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science, in press. [DOI] [PMC free article] [PubMed]
- Chuck, G., Meeley, R.B., and Hake, S. (1998). The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 12, 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Feinbaum, R., and Ambros, V. (1999). The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans. Dev. Biol. 210, 87–95. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., and Zamore, P.D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Kawasaki, H., and Taira, K. (2003). Hes1 is a target of microRNA-23 during retinoic-acid-induced differentiation of NT2 cells. Nature 423, 838–842. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735–739. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., and Ambros, V. (2001). An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.T., Kim, S., and Kim, V.N. (2002). MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21, 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. (2002. a). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002. b). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- McManus, M.T., Petersen, C.P., Haines, B.B., Chen, J., and Sharp, P.A. (2002). Gene silencing using micro-RNA designed hairpins. RNA 6, 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose, S.P., and Sisco, P.H. (1996). Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev. 10, 3018–3027. [DOI] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. (2002). miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro, J.K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, P.H., and Ambros, V. (1999). The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216, 671–680. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickford, A.S., and Cogoni, C. (2003). RNA-mediated gene silencing. Cell. Mol. Life Sci. 60, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Schauer, S.E., Jacobsen, S.E., Meinke, D.W., and Ray, A. (2002). DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 11, 487–491. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289. [DOI] [PubMed] [Google Scholar]
- Slack, F.J., Basson, M., Liu, Z., Ambros, V., Horvitz, H.R., and Ruvkun, G. (2000). The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5, 659–669. [DOI] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., and Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. [DOI] [PubMed] [Google Scholar]
- Xu, P., Vernooy, S.Y., Guo, M., and Hay, B.A. (2003). The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13, 790–795. [DOI] [PubMed] [Google Scholar]