Abstract
DNA damage by adduct formation is a critical step for the initiation of carcinogenesis. Aromatic amines are strong inducers of environmental carcinogenesis. Their DNA adducts are known to exist in an equilibrium between the major groove (B) and base-displaced stacked (S) conformations. However, the factors governing such heterogeneity remain unclear. Here we conducted extensive calorimetry/NMR/CD studies on the model DNA lesions caused by fluorinated 2-aminfluorene (FAF) and 4-aminobiphenyl (FABP) in order to gain thermodynamic and kinetic insights into the S/B-conformational equilibrium. We demonstrate that there are large differences in enthalpy-entropy compensations for FABP and FAF. The small and flexible FABP exclusively adopts the less perturbed B-conformer, thus resulting in small enthalpy (ΔΔH 2.7 kcal/mol)/entropy (ΔΔS 0.7 eu) change. In contrast, FAF stacks better and exists as a mixture of B and S conformers, thus contributing to large enthalpy (ΔΔH 13.4 kcal/mol)/entropy (ΔΔS 34.2 eu) compensation. van't Hoff analysis of dynamic 19F NMR data indicated ΔHB/S = 4.1 kcal/mol in favor of the B conformer and ΔSB/S = 15.6 cal mol−1 k−1 in favor of intercalated S conformer. These findings demonstrate that the favorable entropy of the S conformer over B conformer determines the S/B-population ratios at physiological temperatures.
The human genome is under constant assault from various sources of endogenous and exogenous chemical and radiation insult. Adduct formation is an important type of DNA damage that has been implicated in the etiology of various human cancers. Although occurring at a low frequency, DNA-adducts in certain functionally active regions of the genome can elicit devastating molecular and cellular consequences (1). For example, mutations on the p53 tumor suppressor gene that alters transcriptional activity are responsible for approximately 50% of all cancers (2, 3). Elucidating the mechanisms of adduct-induced mutagenesis is an essential first step to developing effective strategies for risk assessment and novel chemotherapy targets (4–6).
We are particularly interested in the structure-function relationship of arylamine-DNA adducts, as they display a unique sequence-dependent conformational heterogeneity (7, 8). Depending on the location of the hydrophobic carcinogen on the DNA, the prototype arylamine carcinogen 2-aminofluore (AF) adduct can exist as a mixture of two well-defined conformations: base-displaced stacked (S) or major groove B-type (B) (Figure 1)(9, 10). The S/B population ratio is dependent upon the 3'-flanking sequence, that is, 3'-purine promotes S-conformation (11, 12). The intrinsic fluorescence of 2-aminopurine positioned on the adducted strand flanking the AF on its 5'-side has been utilized to emphasize the important role of base stacking and hydrophobic interactions for sequence-dependent S/B equilibrium (13). Quantum mechanics calculations have further indicated the involvement of electrostatic interaction (12). In addition, S/B equilibrium is dependent on the size, co-planarity, and topology of the arylamine carcinogen. For example, B conformation is predominant for the flexible one-ring aniline (14) and two-ring aminobiphenyl (ABP) adducts (15), whereas the larger amine adducts such as 1-aminopyrene (AP)(16) and isoquinoline (IQ) exist predominantly as S and/or wedge type (W) conformers (17–19), the latter resulting from the carcinogen being pressed further into the narrow minor groove. However, the food-borne heterocyclic amine imidazopyridine (PhIP) is similar in size to AF but differs in topology so that the freely-rotatable phenyl at C6 can be accommodated by the minor groove (20). As a result, a significant shift in the equilibrium towards the S conformer occurs in a temperature-dependent manner (20).
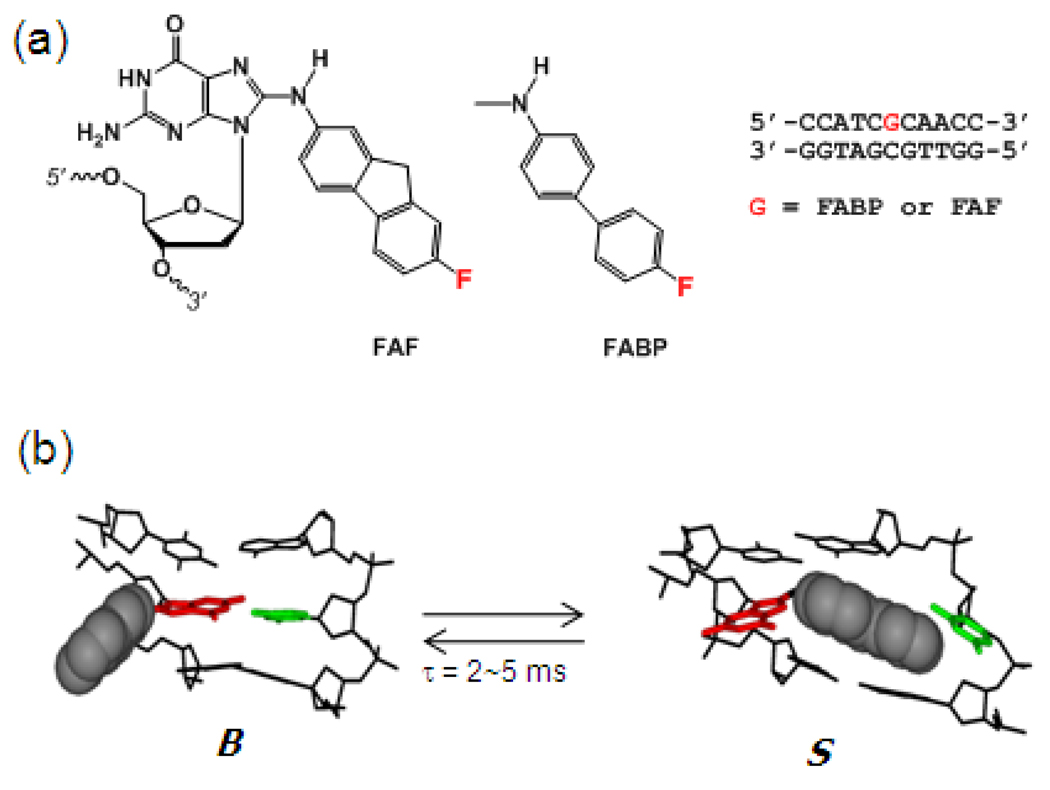
Fig. 1.
(a) Chemical structures of FABP and FAF-modified DNA. (b) AF-induced S/B-conformational heterogeneity. B and S refers to B-type and stacked conformers, respectively.
The AF-induced S/B heterogeneity may permit specific macromolecular interactions, thereby influencing binding affinity and substrate selectivity in repair and translesion synthesis (TLS) pathways, respectively. Recent crystallographic studies showed low electron densities around a weakly bound AF moiety in the active site, giving credence to the solution/solid comparability of adduct structures (21, 22). Likewise, we have found that S conformeric AF adducts are more efficiently excised in the E. coli UvrABC nucleotide excision repair (NER) system (12). This study represents the first reported case of conformation-specific repair and provides molecular insights into the mechanisms of protein-DNA interactions involved in NER.
Despite the wealth of structural knowledge accumulated thus far, the factors defining carcinogen-induced conformational heterogeneity remain uncharacterized and its biological implications unclear. The objective of the present study was to gain thermodynamic and kinetic insights into the arylamine-induced S/B equilibrium using dynamic 19F NMR/CD and DSC measurements. Our results indicate that enthalpy-entropy compensation is important for modulating bulky adduct-induced conformational heterogeneity.
EXPERIMENTAL PROCEDURES
Caution: ABP and AF derivatives are suspected human mutagens/carcinogens and therefore must be handled with caution
Adduct Synthesis
Following the published procedures (12, 23), we prepared FABP- and FAF-modified 11-mer oligonucleotides d(5'-CCATCG*CAACC-3', in which G* is either an FABP- or FAF-modified dG adduct (Figure 1a). The HPLC system used for purification of the oligos consisted of a Hitachi EZChrom Elite unit (Ibaraki-ken, Japan) with a L2450 diode array as a detector and employed a Phenomenex Luna C18 column (10 × 150 mm, 2.5 µm) (Torrance, CA) with a 40-min gradient system involving 3 to 15% acetonitrile in pH 7.0 ammonium acetate buffer (0.10 M) with a flow rate of 2.0 mL/min. The modified oligos were characterized by ESI-TOF-MS analysis as reported previously (12)(Supporting Figures S1–S2) and annealed individually with appropriate complementary strand to produce the desired duplexes. An identical set of unmodified duplexes was also prepared as controls.
UV-Melting and Circular Dichroism (CD)
UV-melting data were obtained using a Beckman DU 800 UV/VIS spectrophotometer using the general procedures reported previously (12, 24). A typical melting experiment consisted of forward/reverse scans and was repeated three times. Thermodynamic parameters were calculated using the program MELTWIN® version 3.5. Circular Dichroism (CD). CD measurements were conducted on a Jasco J-810 spectropolarimeter using the published procedures (12, 25). Details on UV and CD are described in Supporting Information.
Dynamic 19F NMR
Duplex samples (20 ~ 40 ODS) were dissolved in 300 µL of a pH 7.0 buffer (100 mM NaCl, 10 mM Na3PO4, and 100 µM EDTA in 10% D2O/90% H2O) and filtered into through a Shigemi tube using a 0.2 µm membrane filter. All 1H and 19F NMR results were recorded using a dedicated 5-mm 19F/1H dual probe on a Bruker DPX400 Avance spectrometer operating at 400.0 and 376.5 MHz, respectively, as described previously (24, 26). Details on NMR are found in Supporting Information.
DSC Measurements
Measurements were performed using a VP-DSC Micro-calorimeter (Microcal Inc., Northampton, MA). Prior to temperature scanning, samples were degassed for at least 10 min under house vacuum in a closed vessel. Sample and reference solutions were loaded into the respective cells with a blunt-tip syringe. The cells were capped and external pressure (~3 atm) applied to prevent evaporation of the sample solution.
Raw data were collected as microwatts vs. temperature
Typically a 0.12 mM of duplex sample solution was scanned against a control buffer from 15 to 85°C at a rate of 0.75°C/min with at least 5 repetitions. Usually, the first scan showed deviations due to the conditioning of the DSC cells to the temperature and to contact of solution, and thus were excluded from further analysis. A buffer to buffer scan was subtracted from the sample scan, which was then normalized for the heating rate, i.e., each data point was divided by the corresponding heating rate. This results in base-corrected ΔCpex vs. temperature curves. Each transition showed negligible changes in heat capacity between the initial and final states, thus it was difficult to detect an appreciable ΔΔCpex for the melting transitions; subsequently, ΔΔCpex = 0 was assumed. The area of the resulting curve is proportional to the transition heat, which, when normalized for the number of the moles of the sample, is equal to the transition enthalpy, ΔH:
Tm was the temperature at half the peak area. The free energy change at the transition temperature or melting temperature, ΔGbo(Tm), was: ΔGbo(Tm)= −RTmln[4/Ct], where R is the gas constant 8.31451 Jmol−1K−1. The heat of binding at the transition temperature, ΔHbo(Tm), is then − ΔHco(Tm) and as a function of temperature was:
Thus, the free energy change for binding as a function of temperature was:
It should be noted that the parameters of ΔHbo(T) and ΔCp were model-independent while ΔGbo(T) assumed a two-state transition model. Finally, the change in the binding entropy as a function of temperature ΔSbo(T) was simply calculated as:
The uncertainties in the values of Tm, ΔHbo, ΔGbo and ΔSbo determined from DSC scans of the duplex represented only the random errors inherent in the DSC measurement. There were also possible systemic errors which can be combined with these random errors to obtain a combined standard uncertainty for the final reported values of Tm, ΔHbo, ΔGbo and ΔSbo. Estimates of these systemic standard uncertainties were 0.1 K for Tm, 0.02 kcal/mol for ΔGbo resulting from 3% uncertainty in the concentration determination, and 0.03 ΔHbo resulting from uncertainty in the concentration determination and the DSC power calibration.
RESULTS
Model Systems
In vivo activation of arylamines and heterocyclic amines followed by reactions with cellular DNA results in N-deacetylated dG adduct formation as the major and the most persistent lesion (27). Due to the flexibility around the glycosidyl bond, these adducts can exist in an equilibrium between B and S conformations, as illustrated by the AF example in Figure 1b (7). As model systems, we prepared fully paired fluorine-labeled 7-fluoro-2-aminofluorene (FAF)- and 4'-fluoro-4-aminobiphenyl (FABP)-modified 11-mer DNA duplexes [d(5'-CCATCG*CAACC-3'):d(3'-GGTTGCGATGG-5')] (G*=FAF and FABP) (Figure 1a). The modified oligonucleotides were characterized by mass spectrometry (Supporting Figures S1 and S2). This duplex sequence has been used previously for structural investigations of various bulky adducts derived from arylamines and polycyclic aromatic hydrocarbons (8, 25, 28). FAF and FABP are versatile fluorinated conformational probes for the potent carcinogens 2-aminofluorene (AF) and 4-aminobiphenyl (ABP), respectively (23, 26, 29–31).
UV Melting
The modified duplexes were significantly destabilized thermally and thermodynamically relative to the unmodified control in a wide range of concentrations, 0.3 ~ 10 µM (Supporting Figure S3). Adduct formation induced a major reduction in Tm of FAF and FABP at 0.1 mM by −8.5°C and −10.7°C, respectively (Supporting Table 1). Free energy differences (ΔG37°) were similarly decreased by −3.2 and −3.3 kcal/mol, respectively. The more robust thermal destabilization exhibited by FABP was unexpected because of its conformational flexibility and lesser impact on the duplex structure (detailed below).
19F NMR and ICD290–350 nm
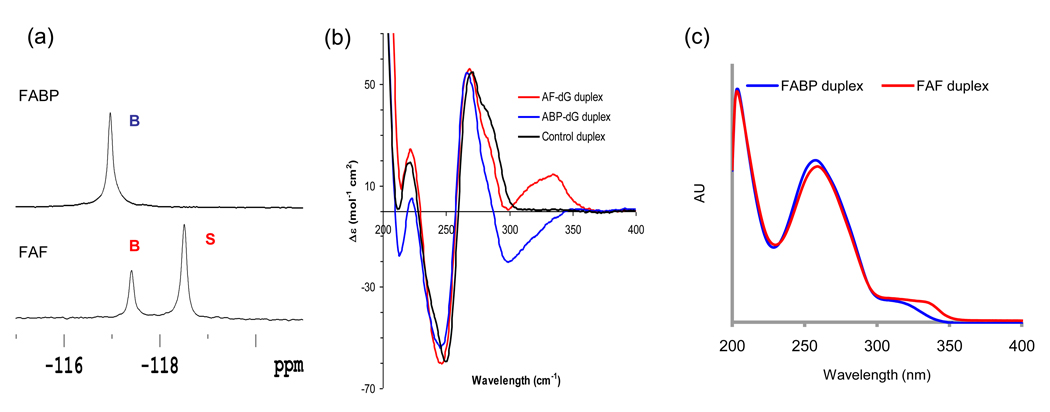
The conformation difference between FABP and FAF modified duplexes is clearly evidenced by their 19F NMR spectra obtained at 20°C (Figure 2a). The FABP-duplex exhibited a single homogeneous 19F signal at −116.8 ppm. This signal was assigned to a B conformer because of its relatively large H/D isotope effect (0.2 ppm) that indicated full exposure of the carcinogen to the solvent (23). The FAF duplex, on the other hand, yielded two signals at an approximate ratio of 34:66, favoring the S conformer. NOESY data (Supporting Figure S4) indicated that the two signals are gradually exchanged at ambient temperature. The peak at −117.4 ppm indicated a large H/D solvent effect (~0.2 ppm), whereas the upfield signal at −118.6 ppm showed a minimal effect (<0.02 ppm)(Supporting Figures S5–8). Consequently, these signals were designated as representing B and S conformers, respectively. The signal assignment was also verified by analyses of induced CD at 290–350 nm (ICD290–350 nm) (Figure 2b). We have previously shown that ICD290–350nm of a B conformer is mostly negative, while the W- and S-conformers are characterized by positive ICD290–360 nm values (25). Figure 2b indicates a clear conformational difference between the FABP and FAF adducts, where a negative ICD290–350 nm was observed for FABP and FAF exhibited a strong positive ICD290–350 nm. This observation is in contrast to the unmodified control which showed no noticeable ellipticities in the same region. For comparison, the UV spectra of FAF and FABP duplexes are shown in Figure 2c.
Fig. 2.
(a) 19F NMR Spectra of FABP and FAF-modified 11-mer duplexes at 20 ° C. (b) Overlays of (b) CD and (c) UV spectra for FABP and FAF-modified duplexes.
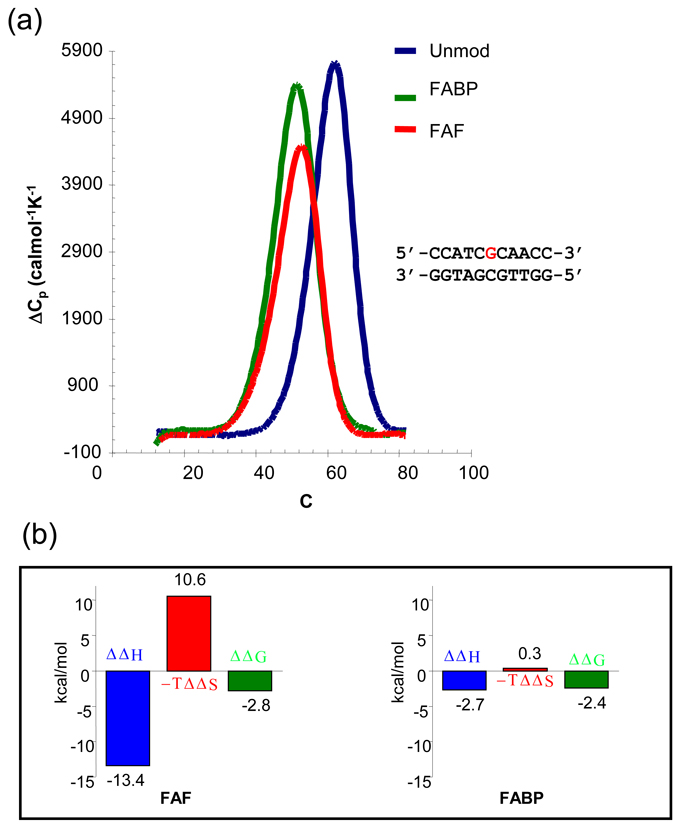
Microcalorimetry Data
To further investigate the thermodynamic impact of different carcinogens, the melting enthalpy of the duplexes with and without lesions was measured via Digital Scanning Calorimetry (DSC). In contrast to UV melting, DSC procedures are model-independent, thus yielding more accurate measurements of enthalpy and Tm (32). Figure 3 shows the DSC plots of excess heat capacity (Cpex) vs. temperatures at a constant oligo concentration of 0.12 mM. The Tm of both FAF- and FABP-duplexes was reduced by −9.1°C and −10.2°C, whereas ΔG37° were decreased by −2.4 and −2.8 kcal/mol, respectively. Consistent with the UV-melting results (Supporting Figure 3 and Table 1), both FABP and FAF lesions produced significantly lower thermal (Tm) and thermodynamic (ΔG) stabilities of the duplex (Table 1).
Fig. 3.
(a) DSC and (b) Thermodynamic plots of the FABP- and FAF-modified, and the control DNA duplexes.
Table 1.
Thermodynamic Parameters Derived from DSC Experiments
| CCATCGCAACC GGTAGCGTTGG | ||||
| −ΔH kcal/mol | −ΔS eu | −ΔG37kcal/mol | TmoC | |
| Control | 79.1 | 214.9 | 12.45 | 64.5 |
| FABP | 76.4 | 214.1 | 10.02 | 54.3 |
| FAF | 65.7 | 180.7 | 9.66 | 55.4 |
| −ΔΔH kcal/mol | −ΔΔS eu | −ΔΔG37 kcal/mol | ΔTmoC | |
| FABP | 2.7 | 0.7 | 2.43 | 10.2 |
| FAF | 13.4 | 34.2 | 2.79 | 9.1 |
Furthermore, the DSC results indicated that FABP modification produces a much smaller effect on the melting enthalpy, ΔH, (−2.7 kcal/mol) compared to FAF (−13.4 kcal/mol) (Figure 3, Table 1). Given similar overall duplex thermodynamic stability (ΔΔG), the large difference of enthalpy reduction between the FABP and FAF lesions must have originated from their differences in adduct conformation. For the FABP-duplex, the carcinogen is bound in a major grove external conformation to a minimally perturbed B-DNA helix with all base pairs essentially intact and the modified dG in the normal anti glycosidic configuration (i.e., B conformer). Unlike FAF, FABP lacks a methylene group, thus allowing greater conformational flexibility. The flexible FABP in the major groove maintains its minimal interaction with the duplex, resulting in a small change in melting enthalpy (ΔΔH 2.7 kcal/mol) and entropy (ΔΔS 0.7 eu) (Table 1). Consequently, a small enthalpy-entropy compensation was obtained. For example, the enthalpy decreases (ΔΔH 2.7 kcal/mol) observed for the FABP adduct represent 3.4% loss of total enthalpy; nearly all enthalpy loss contributes to the free energy loss (ΔΔG37° 2.4 kcal/mol), which represents almost 20% of the total free energy. The large thermal and thermodynamic destabilization observed for the FABP adduct is due to the mild interaction that lacks entropy compensation through adoption of a major groove binding B conformation.
In contrast, the flat FAF interacts strongly with the helical duplex, resulting in significant enthalpy reduction (ΔΔH 13.4 kcal/mol) with large entropy compensation (Figure 3). At 20°C, 66% of the FAF-duplex adopts the S conformation with both the modified dG and dC opposite the lesion being displaced in the major groove, while the FAF intercalated into the helix via a base-displacement. The large enthalpy reduction observed is a reflection of the ruptured Watson-Crick base pairs and loss of normal stacking in the helix. In addition, the effect of the lesion on the duplex is not limited to the lesion site; it also propagates into the immediately flanking and next nearest neighbor base pairs. As shown in Figure 3, the larger entropy compensation, TΔΔS, causes ΔΔG to be similar to ABP modification (−2.8 vs. −2.4 kcal/mol).
Dynamic 19F NMR
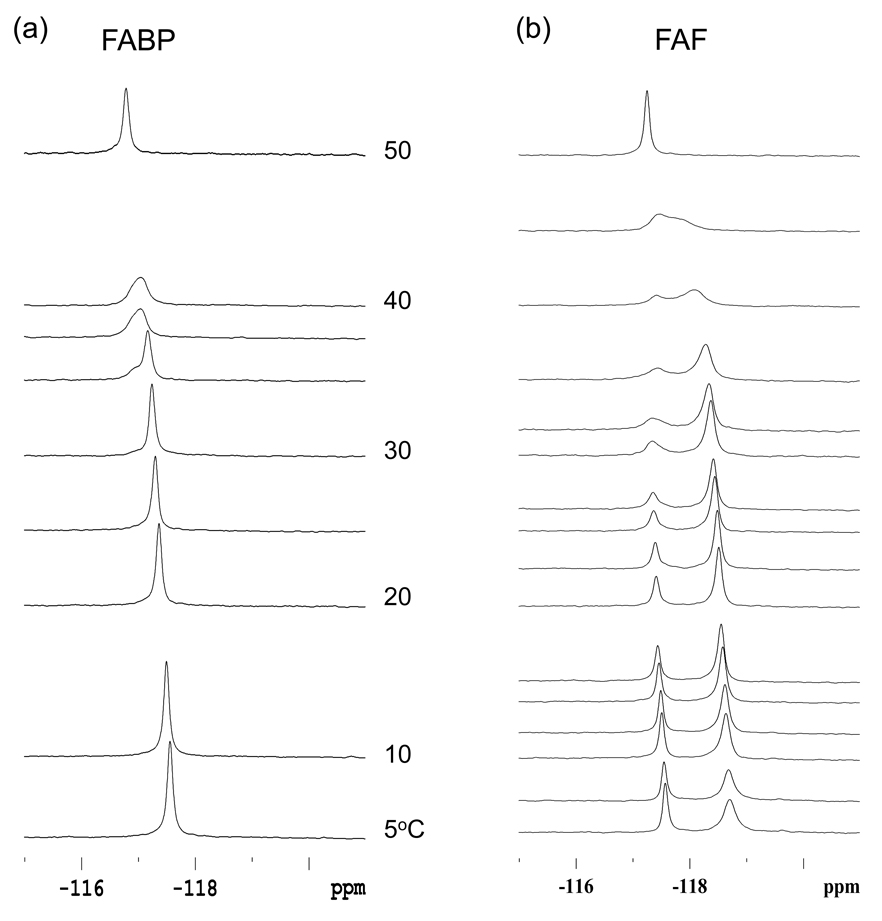
Although DSC is a powerful technique, the conformational heterogeneity at or near the duplex melting temperature (Tm) makes it difficult to separate the respective contributions of B and S conformers to the thermodynamic destabilization of the duplex. Dynamic 19F NMR, however, provides a unique way to investigate the conformational dynamic and thermodynamic properties of a lesion-containing duplex by providing simplified NMR spectra in the lower temperature range (5–40°C)(12, 23).
As shown in Figure 4a, the FABP-duplex exclusively exhibited B conformer signal throughout the entire temperature range (5–60°C), with little change among the chemical shifts. These results make sense because the fully-exposed characteristic of FABP would protect against substantial effects resulting from B conformer to random single strands conformational fluctuations (12). For FAF (Figure 4b), however, the S conformer population was favorably interchanged in a gradual manner until it coalesced around 45°C and eventually stabilized at near complete melting at 60°C (see Supporting Figure S9 for complete spectral series).
Fig. 4.
Dynamic 19F NMR of the FABP- and FAF-modified DNA duplexes as a function of temperatures.
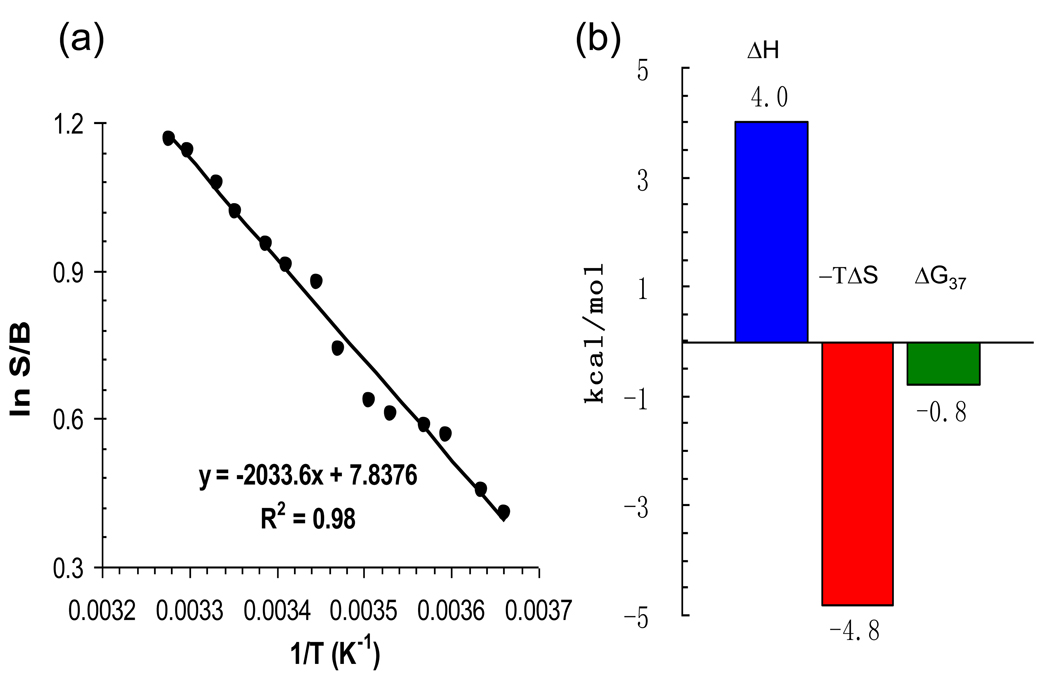
The excellent separation of the B and S 19F NMR signals allowed application of van’t Hoff analysis, an efficient approach to estimate the relative energy difference between the B and S adduct conformers. The lower temperature range (0~30°C) was used in order to minimize the interference of duplex melting on the conformational analysis (Figure 5). A matched NMR sample concentration (1 mM) from the DSC experiments was used. The van’t Hoff plot (lnS/B vs 1/T) indicated ΔHB/S = 4.1 kcal/mol enthalpy difference in favor of the major groove binding B conformer, and ΔSB/S = 15.6 cal mol−1 k−1 in favor of intercalated S conformer (Figure 5a). Therefore, the S conformer exhibited greater stability at a physiologic temperature due to the favorable entropy (Figure 5b). It can be concluded that the increased stability exhibited by the S-conformer in response to increasing temperatures is an entropy-driven event.
Fig. 5.
(a) van’t Hoff plot of the FAF-induced B/S conformational equilibrium as a function of 1/T (0 ~ 30°C). (b) Conformational thermodynamics of B–S equilibrium derived from van’t Hoff parameters.
The van’t Hoff data indicate that the free energy differences ΔGB/S(20°) = −0.39 kcal/mol favor S over B conformers at 20°C. Previous studies of FAF have indicated stacking and van der Waal dispersive forces (collectively known as 'hydrophobic effect') as well as electrostatic effects as responsible for shifting the balance from B to S. Our present data indicate that these factors may not be sufficiently responsible for the preference of S conformers at a physiological temperature. In regards to the S conformer, the possible gain in enthalpy from carcinogen stacking was offset by rupture of hydrogen bonds, unfavorable steric interactions, and loss of base stacking induced by base displacement as well as misplaced nucleotides at the lesion site (12). B conformer is favored enthalpically and would be expected to dominate the balance at lower temperatures. However, S-related entropy compensation would lead to dominant S conformers at higher temperatures. The larger entropy in S conformers was likely an effect of the increased conformational freedom of the modified dG and opposite dC that are displaced into the major groove and the reorientation of water molecules in the hydration shell of the hydrophobic carcinogen moiety.
Combining the conformational thermodynamic results derived from DSC melting and 19F NMR measurements, the destabilization effect of respective B and S conformers could be estimated. The ratio of B to S was approximately 25:75 at 30°C (Figure 4b) at which the duplex melting process begins. The melting enthalpy ΔH of the integration DSC profile included contributions from both conformers and their respective melting thermodynamic parameters are as follows:
These results indicated that the FAF in a B conformer also interacts considerably with the duplex, resulting in reduction of melting enthalpy (ΔΔH = 10.3 kcal/mol) and gain of entropy (ΔΔS = 22.9 cal/mol K), although to a lesser degree than that in an S conformer (ΔΔH = 14.4 kcal/mol, ΔΔS = 38.5 cal/mol K). As shown above, FABP produces minimal disturbances with duplex structures by adopting the B conformer state, resulting in nominal reduction of enthalpy and entropy compensation. The stronger interaction of the FAF B conformer, relative to FABP existing exclusively in B conformer state, may be a reflection of its larger ring size and more rigid nature. It is noteworthy that the interaction among the carcinogen, DNA duplex and solvent system may produce a more complex situation. The differences between B and S conformer heat capacity, ΔCp, might be significant because of considerable structural differences. Thus, the conformational enthalpy difference, ΔHB/S, and entropy difference, ΔSB/S, might be temperature-dependent.
Line Shape Analysis
A two-site dynamic exchange line shape was applied to calculate the kinetic parameters for the FAF-duplex (Table 2)(12, 23). The S/B interconversion energy barrier (ΔG‡) obtained at 20°C and using the Eyring equation, ΔG‡c = 4.58TC(10.32 + log TC/kC) cal/mol, was 13.6 kcal/mol. The exchange rate constant at TC (kC = 2.22Δν) was determined to be 921 s−1. The correlation time (τ = 1/κ) of the FAF-duplex, which signifies the amount of time the lesion spends in the B conformer state before changing to S, or vice versa, was 3.3 ms at 20°C. This value is comparable to the lifespan in G-tract G:C pairs, but is about 10-fold smaller than those for regular G:C Watson-Crick base pairs (33, 34).
Table 2.
Kinetic and Thermodynamic Parameters Derived from Dynamic 19F NMR Experiments
| B:S Ratio 20 ºC | 34%:66% |
| ΔG°20ºC | 0.39 kcal/mol |
| ΔH° | 4.04 kcal/mol |
| k30ºC | 300 s−1 |
| τ (1/k) | 3.33 ms |
| kC=2.22Δν | 921 s−1 |
| ΔG≠20ºC | 14.3 kcal/mol |
| ΔH≠ | 13.6 kcal/mol |
DISCUSSION
Our results showed a typical carcinogen-induced thermal (ΔTm) and thermodynamic (ΔΔG) destabilization upon FABP- and FAF-modification (24, 35). ΔΔG is considered to be a more reliable predictor for lesion-induced destabilization because of its focus on the behavior of the DNA duplex at physiologically relevant temperatures (25 or 37°C)(32).
The exclusive B conformer FABP interacts with the helix and causes minimal disturbance at the lesion site resulting in a small change in both enthalpy and entropy, both of which are compensated. The co-planar FAF, however, interacts significantly with the duplex, yielding an S/B conformational heterogeneity. Consequently, FAF-duplex formation resulted in a major reduction in enthalpy changes, the bulk of which was compensated for by favorable entropy gained from the conformational freedom of the displaced base and the rearranged water molecules in the hydration shell of the hydrophobic carcinogen at the lesion site. This enthalpy-entropy compensation appears to minimize sequence-dependent variations in duplex stability (ΔG37°C)(12, 24). The compensation effect could also explain why the B conformer is dominant at low temperatures, while the S conformer becomes favored at higher temperatures (12). Thus, favorability of the B conformer is enthalpic in origin at lower temperatures, but the equilibrium becomes entropically-driven at higher temperatures and favors the S conformer. This appears to be the case regardless of the nature of sequence contexts flanking the FAF-modified DNA duplexes (12, 24). A greater stacking ability of the hydrophobic FAF, in comparison to FABP, should play a role in shifting equilibrium toward the S conformer, but the stacking alone may not be sufficient to compensate for the loss of enthalpy resulting from ruptured hydrogen bonding, loss of base stacking and unfavorable steric strain at the lesion site (12). It should be noted that the larger amine adducts such as aminopyrene (16) and isoquinoline (19) favor the S conformation. It is the favorable entropy of the S over B conformer that defines the relative S/B population ratio at a given temperature.
Pronounced enthalpy-entropy compensation has widely been observed for various weakly interacting macromolecular settings, including conformational fluctuations occurring during the melting of the DNA helix and the binding of small ligands or drugs to DNA and/or proteins (34, 36, 37). It is difficult to dissect the myriad of forces that govern the kinetic and thermodynamic parameters involved in damaged DNA and proteins phenomena. The molecular contacts between lesion bound-DNA and nearby amino acid residues, such as those in the active site of a polymerase or a repair protein, can also alter the conformational dynamics at the lesion site (38, 39). Thus, protein enzymatic actions can magnify the enthalpic effects of the lesion while minimizing the entropy compensation; indeed, the enthalpy differences induced by adduct formation may serve as a better indicator of how enzymes perform replication and repair proteins recognize DNA damage (40). Other, more subtle contributory factors, such as changes in solvation characteristics around and within the protein-DNA assembly complex, can further complicate the analysis (36). For instance, the enhanced free energy differences which guide the competition between correct and incorrect base pairing during DNA replication could be large when considered as enthalpy differences, presumably accounting for the fidelity of a polymerase (31). These scenarios appear to be generally valid for many entropy-driven biological systems.
The AF-induced S/B equilibrium is analogous to the so-called “base flipping”, a mostly enzyme-induced mechanism that is frequently involved in precarious epigenetic control of gene expression as well as base excision repair (BER) of a damaged DNA (33). In the latter, upon recognition of DNA damage a glycosylase utilizes base flipping to form a catalytic package in the active site of the enzyme and then a glycosidic bond is broken to form an abasic site followed by insertion of the correct base. It has been shown that the enthalpy cost of removing a base from its helical stack is compensated for by the entropy gain from the increased conformational freedom gained upon base flipping (41, 42). Usually, the voided helix space created by base flipping is transiently occupied by nearby amino acids in the active site of the enzyme (43, 44). The AF-induced S/B equilibrium is different in that the duplex cavity created by the rotation of the gycosidyl bond of the modified dG at the lesion site is occupied by either the hydrophobic carcinogen (as with S conformer) or modified dG (as with B conformer)(12). The compensation effect observed for the S/B equilibrium, therefore, appears to be a consequence of the intrinsic ability of the carcinogen to influence its interaction with the helical DNA structure. The correlation time (τ = 1/κ) of the FAF-duplex was found to be 3.3 ms at 20°C. This value is comparable to the lifespan in G-tract G:C pairs, but is about 10-fold smaller than those for regular G:C Watson-Crick base pairs (33, 34). The amount of time the lesion spends in the B state before changing to S” includes an anti to syn conformational change of the glycosidic bond, which is not normal in unmodified DNA. Perhaps G tracts have some unusual properties, and the low correlation time for the adduct compared to normal G:C pairs may reflect the syn/anti transition. This situation is reminiscent of a model study conducted by Jiang et al. (45), in which the aromatic surrogate pyrene in a DNA strand opposite the uracil nucleotide acts both as a wedge to push the uracil away from the base stack in the free DNA and as a plug to hinder its reinsertion after base flipping, restoring the catalytic activity of mutant uracil DNA glycosylase. Using difluorophenyl as a model uracil, the same group has shown that the hydrophobic moiety is stabilized within the helix despite a rapid exchange rate (kC > 1200 s−1) between stacked and unstacked states, a range comparable to that observed for the S/B equilibrium (kC > 921 s−1)(45).
Base flipping is also observed in nucleotide excision repair (NER), the major pathway for repair of bulky DNA lesions (46). During prokaryotic NER, lesions are excised through an elaborate two-tier damage recognition process involving initial binding to UvrA and verification of the damage by UvrB (47, 48). The tandem detection by UvrAB mediates the subsequent formation of a stable pre-incision complex using a padlock-like mode of binding in which a beta-hairpin is inserted between the two strands of the DNA duplex in the vicinity of the DNA damage. Finally, a dual incision of the strand containing the lesion is carried out by UvrC. Correspondingly, Malta et al. (49) have shown that several bases around a cholesterol adduct, including the base immediately adjacent to the lesion on the 3'-side as well as its partner base on the complementary strand, are flipped out of the DNA duplex in NER. It has been shown that the yeast XPC orthologue Rad4 inserts a β-hairpin through the DNA duplex, causing the cyclobutane pyrimidine dimer (CPD) lesion to flip out of the double helix. The expelled nucleotides of the undamaged strand are recognized by Rad4, whereas the two CPD-linked nucleotides become disordered (50). Tubbs et al. (51) have recently shown that alkyltransferase-like proteins (ATLs) generate a stable complex involving an non-enzymatic flipping of the endogenous alkyguanine lesions into a bulky lesion for the NER pathway. Flipping patterns involving NER proteins appear to be dependent on the nature of lesions.
We have shown previously that the S-conformeric FAF lesion is more susceptible to repair than the B conformer in the E. coli UvrABC system (11). The molecular basis of this conformation-specific NER is not well understood, although the presence of Watson-Crick hydrogen base pairs at the lesion site and groove surface characteristics could be important (12). The influence of sequence context and the quality of Watson-Crick base pairs at or in the vicinity of the lesion are also important. For example, the AF adduct in the TG*T sequence context is incised by UvrABC proteins with consistently greater efficiency than in the CG*C context (52–54). Similar results have been obtained for nucleotide excision repair of oxidative DNA damage (8-oxo-dG and FAPY) (55, 56) and DNA modified by the 10S(+)-trans-anti-[BP]-N2-dG adduct (57), a major bulky adduct derived from a prominent environmental carcinogen. Results indicate a weakening of local base stacking interactions and poor quality of Watson-Crick base pairing at and in the vicinity of the lesion in the TG*T sequence context. A similar sequence effect has been observed in human NER for the BP-lesion, i.e., the ..CG*GC.. sequence context elicited NER efficiency 4 times greater than did the ..CG*C.. context, and a destabilized Watson-Crick hydrogen bonding in the CG*GC elicited the most robust NER responses (58). Taken together, these results indicate that the local distortion of base pairing interactions constitutes a recognition element for the NER repair apparatus.
In summary, combined calorimetric/CD/19F NMR studies provide valuable mechanistic insights into how arylamine carcinogens interact with the duplex at the lesion site. Our results also demonstrate how thermodynamic parameters contribute to conformational equilibrium of the AF-DNA adduct. The AF-induced S/B equilibrium strongly depends on not only the nature of the carcinogen’s structure and flanking sequences, but also its entropy compensation effect. A more complete understanding of these types of entropy-driven conformational heterogeneity requires additional investigations of the adducts in other DNA sequences, alone or in combination with appropriate repair proteins or polymerases. These investigations are in progress in our laboratory.
Supplementary Material
Abbreviations
- FAF
N-(2’-deoxyguanosin-8-yl)-7-fluoro-2-aminofluorene
- FABP
N-(2’-deoxyguanosin-8-yl)-4'-fluoro-4-aminobiphenyl
- CD
circular dichroism
- DSC
Digital Scanning Calorimetry
- 19F NMR
fluorine nuclear magnetic resonance spectroscopy
- ICD290–360 nm
induced circular dichroism at 290–360 nm.
Footnotes
We are grateful to the NIH (R01CA098296) for their financial support of this work. This research was also made possible in part by the use of the Research Core Facility supported by the NCRR/NIH (P20 RR016457).
Supporting Information Available: Experimental details, ESI-MS spectra (Figure S1–S2) and UV meling curves (Figure S3) of FABP- and FAF-modified oligonucleotides, NOESY/EXSY spectra of FAF-duplex (Figure S4), H/D isotope effects (Figures S5 and S7) and imino proton (Figures S6 and 7) spectra, full dynamic 19F NMR spectra (Figure S9), and UV melting thermodynamics (Table S1). This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Vogelstein B, Kinzler KW. p53 function and dysfunction. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- 2.Menendez D, Linga A, Resnick MA. The expanding universe of p53 targets. Nature Review Cancer. 2009;9:724–737. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 3.Greenbaltt MS, Bennett WP, Hollstein MH, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 4.Ljungman M. Targeting the DNA Damage Response in Cancer. Chem. Rev. 2009;109:2929–2950. doi: 10.1021/cr900047g. [DOI] [PubMed] [Google Scholar]
- 5.Berdis AJ. DNA Polymerases as Therapeutic Targets. Biochemistry. 2008;47:8253–8260. doi: 10.1021/bi801179f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hussain SP, Hollstein MH, Harris CC. p53 tumor suppressor gene: at the crossroads of molecular carcinogenesis, molecular epidemiology, and human risk assessment. Ann. N. Y. Acad. Sci. 2000;919:79–85. doi: 10.1111/j.1749-6632.2000.tb06870.x. [DOI] [PubMed] [Google Scholar]
- 7.Cho BP. Dynamic conformational heterogeneities of carcinogen-DNA adducts and their mutagenic relevance. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2004;22:57–90. doi: 10.1081/LESC-200038217. [DOI] [PubMed] [Google Scholar]
- 8.Patel DJ, Mao B, Gu Z, Hingerty BE, Gorin A, Basu AK, Broyde S. Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem Res Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- 9.Cho BP, Beland FA, Marques MM. NMR structural studies of a 15-mer DNA duplex from a ras protooncogene modified with the carcinogen 2-aminofluorene: conformational heterogeneity. Biochemistry. 1994;33:1373–1384. doi: 10.1021/bi00172a013. [DOI] [PubMed] [Google Scholar]
- 10.Eckel LM, Krugh TR. 2-Aminofluorene modified DNA duplex exists in two interchangeable conformations. Nat. Struct. Biol. 1994;1:89–94. doi: 10.1038/nsb0294-89. [DOI] [PubMed] [Google Scholar]
- 11.Meneni S, Shell SM, Zou Y, Cho BP. Conformation-specific recognition of carcinogen-DNA adduct in escherichia coli nucleotide excision repair. Chem Res Toxicol. 2007;20:6–10. doi: 10.1021/tx600273h. [DOI] [PubMed] [Google Scholar]
- 12.Meneni SR, Shell SM, Gao L, Jurecka P, Lee W, Sponer J, Zou Y, Chiarelli MP, Cho BP. Spectroscopic and theoretical insights into sequence effects of aminofluorene-induced conformational heterogeneity and nucleotide excision repair. Biochemistry. 2007;46:11263–11278. doi: 10.1021/bi700858s. [DOI] [PubMed] [Google Scholar]
- 13.Jain N, Reshetnyak YK, Gao L, Chiarelli MP, Cho BP. Fluorescence probing of aminofluorene-induced conformational heterogeneity in DNA duplexes. Chem Res Toxicol. 2008;21:445–452. doi: 10.1021/tx7003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro R, Ellis S, Hingerty BE, Broyde S. Effect of ring size on conformations of aromatic amine-DNA adducts: the aniline-C8 guanine adduct resides in the B-DNA major groove. Chem Res Toxicol. 1998;11:335–341. doi: 10.1021/tx970211q. [DOI] [PubMed] [Google Scholar]
- 15.Cho BP, Beland FA, Marques MM. NMR structural studies of a 15-mer DNA sequence from a ras protooncogene, modified at the first base of codon 61 with the carcinogen 4-aminobiphenyl. Biochemistry. 1992;31:9587–9602. doi: 10.1021/bi00155a011. [DOI] [PubMed] [Google Scholar]
- 16.Mao B, Vyas RR, Hingerty BE, Broyde S, Basu AK, Patel DJ. Solution conformation of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dC in a DNA duplex. Biochemistry. 1996;35:12659–12670. doi: 10.1021/bi961078o. [DOI] [PubMed] [Google Scholar]
- 17.Wang F, DeMuro NE, Elmquist CE, Stover JS, Rizzo CJ, Stone MP. Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for −2 bp deletions. J Am Chem Soc. 2006;128:10085–10095. doi: 10.1021/ja062004v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elmquist CE, Wang F, Stover JS, Stone MP, Rizzo CJ. Conformational differences of the C8-deoxyguanosine adduct of 2-amino-3-methylimidazo[4,5-f]quinoline (IQ) within the NarI recognition sequence. Chem Res Toxicol. 2007;20:445–454. doi: 10.1021/tx060229d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, Elmquist CE, Stover JS, Rizzo CJ, Stone MP. DNA sequence modulates the conformation of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme. Biochemistry. 2007;46:8498–8516. doi: 10.1021/bi700361u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown K, Hingerty BE, Guenther EA, Krishnan VV, Broyde S, Turteltaub KW, Cosman M. Solution structure of the 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine C8-deoxyguanosine adduct in duplex DNA. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8507–8512. doi: 10.1073/pnas.151251898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutta S, Li Y, Johnson D, Dzantiev L, Richardson CC, Romano LJ, Ellenberger T. Crystal structures of 2-acetylaminofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16186–16191. doi: 10.1073/pnas.0406516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu GW, Kiefer JR, Burnouf D, Becherel OJ, Fuchs RPP, Beese LS. Observing Translesion Synthesis of an Aromatic Amine DNA Adduct by a High-fidelity DNA Polymerase. J. Biol. Chem. 2004;279:50280–50285. doi: 10.1074/jbc.M409224200. [DOI] [PubMed] [Google Scholar]
- 23.Zhou L, Rajabzadeh M, Traficante DD, Cho BP. Conformational Heterogeneity of Arylamine-Modified DNA: 19F NMR Evidence. J Am Chem Soc. 1997;119:5384–5389. [Google Scholar]
- 24.Meneni SR, D'Mello R, Norigian G, Baker G, Gao L, Chiarelli MP, Cho BP. Sequence effects of aminofluorene-modified DNA duplexes: thermodynamic and circular dichroism properties. Nucleic Acids Res. 2006;34:755–763. doi: 10.1093/nar/gkj480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang F, Meneni S, Cho BP. Induced circular dichroism characteristics as conformational probes for carcinogenic aminofluorene-DNA adducts. Chem Res Toxicol. 2006;19:1040–1043. doi: 10.1021/tx0601253. [DOI] [PubMed] [Google Scholar]
- 26.Meneni S, Liang F, Cho BP. Examination of the Long-range Effects of Aminofluorene-induced Conformational Heterogeneity and Its Relevance to the Mechanism of Translesional DNA Synthesis. J. Mol. Biol. 2007;366:1387–1400. doi: 10.1016/j.jmb.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beland FA, Kadlubar FF. Handbook of Experimental Pharmacology. Heidelberg: Spring-Verlag; 1990. [Google Scholar]
- 28.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 29.Jain N, Li Y, Zhang L, Meneni SR, Cho BP. Probing the sequence effects on NarI-induced −2 frameshift mutagenesis by dynamic 19F NMR, UV, and CD spectroscopy. Biochemistry. 2007;46:13310–13321. doi: 10.1021/bi701386f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho BP, Zhou L. Probing the conformational heterogeneity of the acetylaminofluorene-modified 2'-deoxyguanosine and DNA by 19F NMR spectroscopy. Biochemistry. 1999;38:7572–7583. doi: 10.1021/bi990182d. [DOI] [PubMed] [Google Scholar]
- 31.Liang F, Cho BP. Probing the thermodynamics of aminofluorene-induced translesion DNA synthesis by differential scanning calorimetry. J Am Chem Soc. 2007;129:12108–12109. doi: 10.1021/ja075271p. [DOI] [PubMed] [Google Scholar]
- 32.Plum GE, Breslauer KJ, Roberts RW. Thermodynamics and Kinetics of Nucleic Acdi Association/Dissociation and Folding Processes. Amsterdam: Pergamon; 2002. [Google Scholar]
- 33.Priyakumar UD, MacKerell AD., Jr Computational approaches for investigating base flipping in oligonucleotides. Chem Rev. 2006;106:489–505. doi: 10.1021/cr040475z. [DOI] [PubMed] [Google Scholar]
- 34.Chen C, Russu IM. Sequence-dependence of the energetics of opening of at basepairs in DNA. Biophys J. 2004;87:2545–2551. doi: 10.1529/biophysj.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 36.Petruska J, Goodman MF. Enthalpy-entropy compensation in DNA melting thermodynamics. J Biol Chem. 1995;270:746–750. doi: 10.1074/jbc.270.2.746. [DOI] [PubMed] [Google Scholar]
- 37.Qu X, Ren J, Riccelli PV, Benight AS, Chaires JB. Enthalpy/Entropy Compensation: Influence of DNA Flanking Sequence on the Binding of 7-Amino Actinomycin D to Its Primary Binding Site in Short DNA Duplexes??AU - Qu, Xiaogang. Biochemistry. 2003;42:11960–11967. doi: 10.1021/bi0347813. [DOI] [PubMed] [Google Scholar]
- 38.Broyde S, Wang L, Zhang L, Rechkoblit O, Geacintov NE, Patel DJ. DNA adduct structure-function relationships: comparing solution with polymerase structures. Chem Res Toxicol. 2008;21:45–52. doi: 10.1021/tx700193x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guengerich FP. Interactions of carcinogen-bound DNA with individual DNA polymerases. Chem Rev. 2006;106:420–452. doi: 10.1021/cr0404693. [DOI] [PubMed] [Google Scholar]
- 40.Brabec V, Stehlikova K, Malina J, Vojtiiskova M, Kasparkova J. Thermodynamic properties of damaged DNA and its recognition by xeroderma pigmentosum group A protein and replication protein A. Arch Biochem Biophys. 2006;446:1–10. doi: 10.1016/j.abb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 41.O'Neil LL, Wiest O. Structures and energetics of base flipping of the thymine dimer depend on DNA sequence. J Phys Chem B. 2008;112:4113–4122. doi: 10.1021/jp7102935. [DOI] [PubMed] [Google Scholar]
- 42.Klimasauskas S, Szyperski T, Serva S, Wuthrich K. Dynamic modes of the flipped-out cytosine during HhaI methyltransferase-DNA interactions in solution. EMBO J. 1998;17:317–324. doi: 10.1093/emboj/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 44.Beuck C, Singh I, Bhattacharya A, Hecker W, Parmar VS, Seitz O, Weinhold E. Polycyclic aromatic DNA-base surrogates: high-affinity binding to an adenine-specific base-flipping DNA methyltransferase. Angew Chem Int Ed Engl. 2003;42:3958–3960. doi: 10.1002/anie.200219972. [DOI] [PubMed] [Google Scholar]
- 45.Jiang YL, McDowell LM, Poliks B, Studelska DR, Cao C, Potter GS, Schaefer J, Song F, Stivers JT. Recognition of an unnatural difluorophenyl nucleotide by uracil DNA glycosylase. Biochemistry. 2004;43:15429–15438. doi: 10.1021/bi0483864. [DOI] [PubMed] [Google Scholar]
- 46.Gillet LCJ, Scharer OD. Molecular Mechanisms of Mammalian Global Genome Nucleotide Excision Repair. Chem. Rev. 2005;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 47.Truglio JJ, Croteau DL, Van Houten B, Kisker C. Prokaryotic Nucleotide Excision Repair: The UvrABC System. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 48.Pakotiprapha D, Liu Y, Verdine GL, Jeruzalmi D. A structural model for the damage-sensing complex in bacterial nucleotide excision repair. J Biol Chem. 2009;284:12837–12844. doi: 10.1074/jbc.M900571200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malta E, Moolenaar GF, Goosen N. Base flipping in nucleotide excision repair. J Biol Chem. 2006;281:2184–2194. doi: 10.1074/jbc.M508901200. [DOI] [PubMed] [Google Scholar]
- 50.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 51.Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, McGown G, Thorncroft M, Santibanez-Koref MF, Millington C, Arvai AS, Kroeger MD, Peterson LA, Williams DM, Fried MG, Margison GP, Pegg AE, Tainer JA. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459:808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zou Y, Shell SM, Utzat CD, Luo C, Yang Z, Geacintov NE, Basu AK. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–12661. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jain N, Meneni S, Jain V, Cho BP. Influence of flanking sequence context on the conformational flexibility of aminofluorene-modified dG adduct in dA mismatch DNA duplexes. Nucleic Acids Res. 2009;37:1628–1637. doi: 10.1093/nar/gkn1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruan Q, Liu T, Kolbanovskiy A, Liu Y, Ren J, Skorvaga M, Zou Y, Lader J, Malkani B, Amin S, Van Houten B, Geacintov NE. Sequence context- and temperature-dependent nucleotide excision repair of a benzo[a]pyrene diol epoxide-guanine DNA adduct catalyzed by thermophilic UvrABC proteins. Biochemistry. 2007;46:7006–7015. doi: 10.1021/bi700294k. [DOI] [PubMed] [Google Scholar]
- 55.Kalam MA, Basu AK. Mutagenesis of 8-oxoguanine adjacent to an abasic site in simian kidney cells: tandem mutations and enhancement of G-->T transversions. Chem Res Toxicol. 2005;18:1187–1192. doi: 10.1021/tx050119r. [DOI] [PubMed] [Google Scholar]
- 56.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cai Y, Patel DJ, Geacintov NE, Broyde S. Dynamics of a Benzo[a]pyrene-derived Guanine DNA Lesion in TGT and CGC Sequence Contexts: Enhanced Mobility in TGT Explains Conformational Heterogeneity, Flexible Bending, and Greater Susceptibility to Nucleotide Excision Repair. J. Mol. Biol. 2007;374:292–305. doi: 10.1016/j.jmb.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kropachev K, Kolbanovskii M, Cai Y, Rodriguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, Geacintov NE. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions. J Mol Biol. 2009;386:1193–1203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.