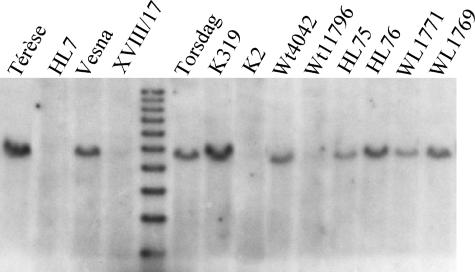Abstract
Genes in the TERMINAL FLOWER1 (TFL1)/CENTRORADIALIS family are important key regulatory genes involved in the control of flowering time and floral architecture in several different plant species. To understand the functions of TFL1 homologs in pea, we isolated three TFL1 homologs, which we have designated PsTFL1a, PsTFL1b, and PsTFL1c. By genetic mapping and sequencing of mutant alleles, we demonstrate that PsTFL1a corresponds to the DETERMINATE (DET) gene and PsTFL1c corresponds to the LATE FLOWERING (LF) gene. DET acts to maintain the indeterminacy of the apical meristem during flowering, and consistent with this role, DET expression is limited to the shoot apex after floral initiation. LF delays the induction of flowering by lengthening the vegetative phase, and allelic variation at the LF locus is an important component of natural variation for flowering time in pea. The most severe class of alleles flowers early and carries either a deletion of the entire PsTFL1c gene or an amino acid substitution. Other natural and induced alleles for LF, with an intermediate flowering time phenotype, present no changes in the PsTFL1c amino acid sequence but affect LF transcript level in the shoot apex: low LF transcript levels are correlated with early flowering, and high LF transcript levels are correlated with late flowering. Thus, different TFL1 homologs control two distinct aspects of plant development in pea, whereas a single gene, TFL1, performs both functions in Arabidopsis. These results show that different species have evolved different strategies to control key developmental transitions and also that the genetic basis for natural variation in flowering time may differ among plant species.
INTRODUCTION
For some fundamental aspects of plant biology, the genes involved have been identified through a molecular genetics approach using the model species Arabidopsis. From this basic information, comparative studies between species can begin, in particular to understand the genetic and molecular mechanisms responsible for the large diversity in plant morphology and to identify the genes involved in adaptive evolution (Cronk, 2001). In plants, the best example of such evolutionary developmental studies is the identification and analysis of MADS box genes involved in flower development in several plant species, including gymnosperms (reviewed by Ma and De Pamphilis, 2000). Isolation of putative orthologs in different species and studies of RNA and/or protein expression patterns provide insights into the conservation and diversification of gene function in plant development (Hofer and Ellis, 2002). For instance, the LEAFY (LFY) gene of Arabidopsis, which was isolated initially as FLORICAULA in snapdragon, is a key gene involved in floral development (Coen et al., 1990; Weigel et al., 1992). Orthologs of LFY have since been studied in numerous other species, including UNIFOLIATA in pea (Hofer et al., 1997). In certain cases, different regulator processes or new roles can be found. For example, UNIFOLIATA was proposed to regulate indeterminacy during both leaf and flower development. The function of LFY during leaf development was not described (Hofer et al., 1997).
Flowering time is a major adaptive trait in the life strategy of flowering plants, which have to synchronize their reproduction with favorable environmental conditions. After a vegetative phase, plants undergo the floral transition. The switch from the vegetative to the reproductive stage is controlled by physiological signals and genetic networks that integrate environmental (photoperiod and temperature) and endogenous (stage of the plant) conditions (Levy and Dean, 1998; Colasanti and Sundaresan, 2000). The molecular genetics of the long-day plant Arabidopsis enable the isolation and characterization of the genes that control flowering time (reviewed by Mouradov et al., 2002). In agronomic species, most of the genetic loci that control flowering time have been identified as quantitative trait loci (in maize or rice; Yano et al., 2001; Salvi et al., 2002) or as mutants (in pea; Murfet and Reid, 1993). Using a combination of map-based cloning and a candidate-gene approach, two quantitative trait loci, Hd1 and Hd3a, have been cloned in rice (Yano et al., 2000; Kojima et al., 2002) and one, VRN1, has been cloned in wheat (Yan et al., 2003). They correspond to genes that are similar to the Arabidopsis genes CONSTANS, FLOWERING LOCUS T (FT), and APETALA1, respectively.
Because pea is both a classic model species for plant development and an important crop in Europe, we used a molecular approach to study homologs of the snapdragon CENTRORADIALIS/Arabidopsis TERMINAL FLOWER1 (CEN/TFL1) genes in pea. CEN is involved in inflorescence architecture in snapdragon (Bradley et al., 1996). The cen mutation leads to the conversion of the indeterminate inflorescence to a terminal flower. Orthologs of CEN have been found in different species: TFL1 in Arabidopsis (Bradley et al., 1997), SELF PRUNING (SP) in tomato (Pnueli et al., 1998), CET in tobacco (Amaya et al., 1999), and LpTFL1 in Lolium perenne (Jensen et al., 2001). In Arabidopsis, tfl1 mutants have a terminal flower and flower earlier than the wild type (Bradley et al., 1997). This early-flowering phenotype was not observed in snapdragon. TFL1 may play a role in inflorescence meristem identity as well as in floral initiation control as a repressor of flowering. It was proposed that these two distinct roles are in fact one, with TFL1 controlling the length of both the vegetative and reproductive phases (Ratcliffe et al., 1998).
CEN and TFL1 are similar to a family of mammalian phosphatidylethanolamine binding proteins (PEBPs) also known as Raf-1 kinase inhibitor proteins. Crystallography analysis reveals that CEN may be involved in interaction with a kinase (Banfield and Brady, 2000). In tomato, SP was shown to interact with multiple proteins and was proposed to encode a modular protein with the potential to interact with a variety of signaling pathways (Pnueli et al., 2001). Expression analysis has revealed that genes closely related to TFL1 are expressed mainly in the shoot apical meristem in the region below the terminal meristem. CEN is induced during floral initiation (Bradley et al., 1996), whereas TFL1 expression also is found during the vegetative phase; this expression could explain the role of TFL1 in delaying flowering in Arabidopsis (Bradley et al., 1997). Analysis of mutants and sequencing of the entire Arabidopsis genome have revealed that the TFL1 genes belong to a small family (at least six genes) with functional divergence (Mimida et al., 2001). One of them, FT, has a TFL1-antagonistic role by promoting flowering in Arabidopsis (Kardailsky et al., 1999; Kobayashi et al., 1999). Studies of TFL1 homologs in other species may help us better understand the function and the evolution of the TFL1 family in flowering plants.
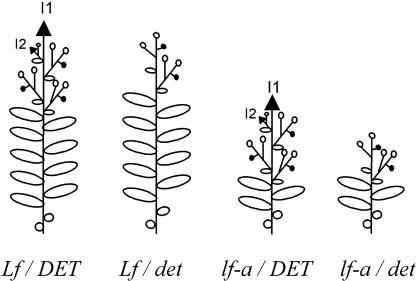
Floral initiation and development in pea have been studied for many decades (Murfet and Reid, 1993). Based on physiological and mutational analyses, a model for flowering that involves both a floral inhibitor and a stimulus has been developed (reviewed by Reid et al., 1996; Weller et al., 1997). The stimulus is specific to flowering and is under the control of GIGAS (Beveridge and Murfet, 1996). The synthesis of the floral inhibitor is controlled by different genes (STERILE NODE, HIGH RESPONSE, PHOTOPERIOD, DAY NEUTRAL, and EARLY) and is strongly regulated by photoperiod. The integration of the signals occurs in the apex and is controlled by the LATE FLOWERING (LF) gene. LF determines the node of flowering in pea for a given genetic background (Figure 1). Four natural and induced classes of alleles are known—Lf-d, Lf, lf, and lf-a—that in maximal inductive conditions confer minimum nodes of flowering of 15, 11, 8, and 5, respectively. The dominance order is Lf-d > Lf > lf > lf-a, with the lf-a allele being recessive (Murfet, 1975). LF is active in the shoot, and the different alleles determine the threshold of sensitivity of the apical meristem to flowering signals. Because mutations of LF lead to plants with an early phenotype, LF may be considered a repressor of flowering.
Figure 1.
Scheme of the Phenotypes of lf-a, det, and lf-a det in Pea.
Black arrows represent shoot apical meristems, open circles represent flowers, and closed circles represent stubs (terminal meristems with epidermal hairs). I1 and I2 indicate the primary and secondary inflorescence meristems, respectively.
Pea is an indeterminate-flowering plant, as is Arabidopsis. After floral initiation, the shoot apical meristem is converted to an inflorescence meristem (called I1; Figure 1). The I1 meristem grows indefinitely, and an axillary meristem in the leaf axil generates a secondary inflorescence, I2. Flowers (often two) arise laterally from I2. At the onset of senescence, the I2 meristem ceases growing and is converted to a stub, a terminal meristem with epidermal hairs (Singer et al., 1999). A pea mutant, known as determinate (det), produces a few axillary flowers and an apparent terminal flower (Reid and Murfet, 1984; Singer et al., 1990) (Figure 1). Scanning electron microscopy showed that this terminal flower actually arises from an axillary meristem and that the I1 meristem is converted to a stub as in the wild type (Singer et al., 1990) (Figure 1). A real terminal flower can be obtained in pea by crossing the det mutant with another mutant, vegetative1, which remains vegetative (Reid and Murfet, 1984; Singer et al., 1999).
Using degenerate primers, we isolated three TFL1 homologs in pea. Detailed analyses, by gene mapping, allele sequencing, and expression studies, revealed that two of these homologs correspond to important genes involved in flower initiation and development in pea: DET and LF. This study provides comparative information about the function and evolution of TFL1 genes in flowering plants.
RESULTS
Arabidopsis TFL1 Homologs Represent a Small Gene Family in Pea
To isolate TFL1-related sequences in pea, we designed several different degenerate primers corresponding to conserved domains identified from the alignment of published TFL1 homologs (Figure 2A) and other homologous EST sequences. Two different primer pairs successfully amplified fragments from pea genomic DNA. Two fragments of 450 bp (A) and 850 bp (B) obtained with the primer combination TFL1-3/TFL1-5, and a single fragment of 450 bp (C) obtained using the primer combination TFL1-1/TFL1-2, were isolated and sequenced. Each band yielded sequence similar to that of TFL1 and CEN. Additional sequences for each fragment were obtained by 3′ and 5′ rapid amplification of cDNA ends PCR on seedling or flower cDNA (see Methods). Complete sequences were obtained for transcripts corresponding to fragments A and C, and these genes were designated PsTFL1a and PsTFL1c, respectively. For the third gene, designated PsTFL1b, only a partial sequence was obtained.
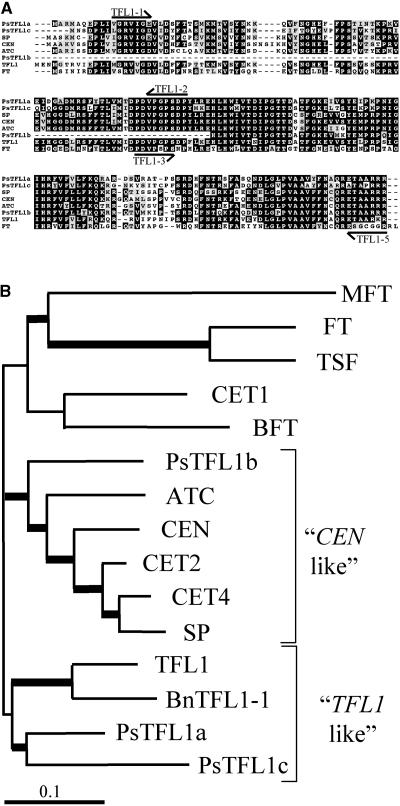
Figure 2.
Comparison of Pea TFL1 Homologs with TFL1 Related Genes.
(A) Alignment of the predicted amino acid sequences of PsTFL1a, PsTFL1b, and PsTFL1c, TFL1 (Bradley et al., 1997), SP (Pnueli et al., 1998), CEN (Bradley et al., 1996), ATC (Mimida et al., 2001), and FT (Kardailsky et al., 1999; Kobayashi et al., 1999). The alignment was performed with Multialign software (Corpet, 1988). Arrows represent the positions of the degenerate primers used to isolate the TFL1 homologs in pea.
(B) Phylogenic tree of TFL1-related proteins constructed using the NJ method with the program CLUSTAL W. Branches with a bootstrap value of >600 (of 1000) are shown with thick lines. In addition to the proteins shown in (A), TSF, BFT, MFT, CET1, CET2, and CET4 from tobacco (Amaya et al., 1999) and BnTFL1-1 from Brassica napus were included in the analysis.
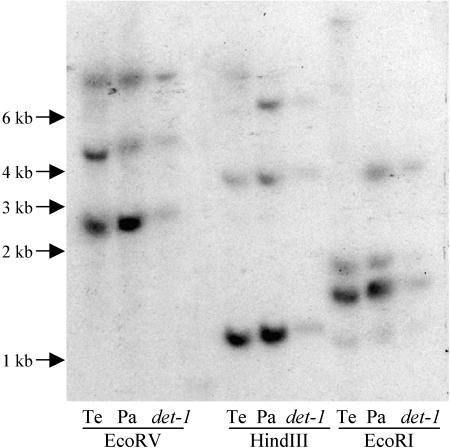
To evaluate the number of TFL1-related genes in pea, we made a DNA gel blot using PsTFL1a as a probe (Figure 3). Three hybridizing bands were seen in HindIII and EcoRV digests, and four bands were seen in digests with EcoRI. The additional band in the EcoRI digest can be explained by the presence of an EcoRI site in the PsTFL1b sequence. Therefore, we concluded that the three TFL1 homologs isolated probably represent the entire TFL1 family in pea.
Figure 3.
DNA Gel Blot of the Wild-Type Pea Lines Térèse and Paloma and the Mutant det-1 (JI 2121).
Genomic DNA was digested with EcoRV, HindIII, and EcoRI. The blot was hybridized with a probe corresponding to PsTFL1a. Hybridization and washing conditions were low stringency (temperature of 60°C, 2× SSC as wash salt). Sizes of the marker fragments are represented in kilobases. Pa, Paloma; Te, Térèse.
PsTFL1a and PsTFL1c are predicted to encode proteins of 174 and 173 amino acids, respectively, according to the computer software Eugène (Schiex et al., 2000). The predicted PsTFL1a and PsTFL1c proteins show 70% amino acid identity and 72 and 65% identity with TFL1, respectively. The PsTFL1b clone, which is incomplete at the 5′ end, covers 90 amino acids and shows 73% identity with TFL1 over this region. The protein sequence alignment in Figure 2A shows that the three predicted pea proteins contain large regions that are conserved across TFL1 homologs from other species. Intron/exon boundaries also are highly conserved across these genes (data not shown).
Previous studies have shown that despite their apparently similar functions, Arabidopsis TFL1 and Antirrhinum CEN are not particularly closely related (Mimida et al., 2001). Clustering of TFL1-related sequences based on amino acid similarity suggested the presence of several distinct groups (Figure 2B). Both PsTFL1a and PsTFL1c cluster with TFL1. However, PsTFL1b belongs to another group of genes that includes CEN, SP, and ATC. Although PsTFL1b is a partial sequence, the same results were obtained when the analysis was performed using only the C-terminal region. Other members of the Arabidopsis TFL1 family, such as FT, TSF, and BFT (Mimida et al., 2001), are more distant (Figure 2B).
The Map Locations of PsTFL1a and PsTFL1c Suggest That They May Be Candidate Genes for DET and LF, Respectively
To study the relationship between the TFL1-like sequences isolated and already known flowering loci in pea, we developed molecular markers corresponding to these sequences. We screened the parents of two different mapping populations for single nucleotide polymorphisms (SNP) and converted these to PCR-based cleaved amplified polymorphic sequence or derived cleaved amplified polymorphic sequence markers (see Methods).
PsTFL1a was mapped in the Térèse × K586 RIL population (Laucou et al., 1998) and was found to be located in group V between two RAPD markers, K3-3000 and D3-1100, a region shown previously to contain DET (Rameau et al., 1998). Analysis of an F2 population segregating for a det mutation showed no recombinant genes between DET and PsTFL1a among 120 individuals (data not shown). Thus, PsTFL1a is closely linked to DET, and in view of the phenotypic similarity between det mutants and Arabidopsis tfl1 mutants, we considered it to be a good candidate for the DET gene.
Because of the lack of SNP for PsTFL1b and PsTFL1c between Térèse and K586, these genes were mapped in another RIL population derived from a cross between the more distant lines JI 281 and JI 399 (Ellis et al., 1992). PsTFL1b maps on linkage group III close to the markers PsZF18 and C7/1 (Ellis et al., 1992; Laucou et al., 1998), a region containing no obvious candidate flowering loci. By contrast, PsTFL1c was located in linkage group II and segregated with the B5/9 marker in a region containing LF (Ellis et al., 1992; Laucou et al., 1998) and an important quantitative trait loci for flowering time (our unpublished results). This finding suggests that PsTFL1c could be a candidate gene for LF. Our subsequent analyses focused on PsTFL1a and PsTFL1c, because these potentially represented genes that regulate agronomically important flowering traits in pea.
det Mutants at the PsTFL1a Locus Carry Significant Mutations
To examine the relationship between PsTFL1a and DET, we next sequenced PsTFL1a from three different det mutants and their corresponding wild-type lines when available (Table 1). All three were found to contain SNP within the PsTFL1a gene.
Table 1.
Sequence Analysis of PsTFL1a in the det Mutants
| Mutant Line | Position | Nucleotide Substitution | Major Effect |
|---|---|---|---|
| det-1 | +202 (junction exon 1/intron 1) | TGGT → TGAT | Intron 1 is not spliced |
| det-2 | +12 (exon 1) | ATG → ATC | Met-4 → Ile |
| +139 (exon 1) | ACC → CCC | Thr-47 → Pro | |
| +401 (intron 2) | AACCA → AATCA | Silent | |
| +655 (exon 4) | CAA → CGA | Gln-127 → Arg | |
| +749 (exon 4) | GTT → GTG | Silent | |
| det-3 | +12 (exon 1) | ATG → ATC | Met-4 → Ile |
| +197 (exon 1) | ACA → ATA | Thr-66 → Ile | |
| +401 (intron 2) | AACCA → AATCA | Silent | |
| +584 (exon 4) | GAG → AAG | Glu-104→ Lys | |
| +749 (exon 4) | GTT → GTG | Silent |
The positions of the substitutions are labeled from the +1 of translation. For det-1, the sequence is compared with the progenitor line, Paloma, whereas for det-2 and det-3, the sequence is compared with the wild-type line Térèse. “Silent” indicates that the substitution has no effect at the amino acid sequence of the protein. Mutations in boldface are proposed to be responsible for the det phenotype (see details in the text).
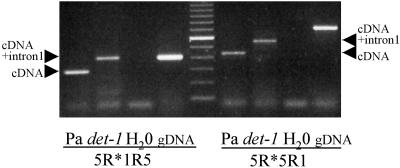
In the det-1 mutant line JI 2121 (Swiecicki, 1987), a point mutation was found at the first exon-intron junction, converting the consensus splicing donor motif TGGT to TGAT (Table 1). Using primer pairs surrounding the first and the second introns, reverse transcriptase–mediated (RT) PCR performed on root RNA from det-1 and the Paloma wild-type line confirmed that the first intron is not spliced out in PsTFL1a transcripts from det-1, whereas the second intron is spliced out normally in both the wild type and det-1 (Figure 4). Retention of the first intron in det-1 is predicted to result in the introduction of a stop codon, truncating PsTFL1a at amino acid 72.
Figure 4.
Analysis of the Nonsplicing of the First Intron in the det-1 Line.
PCR was performed on root cDNA from Paloma (Pa) and det-1 (JI 2121) with water (H2O) and genomic DNA (gDNA) as controls. The primer pairs 5R*1R5 and 5R*5R1 surrounded intron 1 and introns 1 and 2, respectively. Arrowheads indicate the size expected for the cDNA if the first intron is spliced (cDNA) or not spliced (cDNA+intron 1). A 100-bp size marker lane is shown in the center of the gel.
The det-2 line JI 1358 arises as a spontaneous mutant, for which no progenitor line is available (J. Hofer, personal communication). We found five SNP in PsTFL1a between the det-2 line and wild-type cv Térèse, some of which may represent natural polymorphism (Table 1). Of these, three are predicted to direct changes in the protein sequence. Two of these changes (Met-4 to Ile and Thr-47 to Pro) affect residues that are not notably conserved across the TFL1 family, whereas the Gln residue substituted for in the third change (Gln-127 to Arg) is invariant across all TFL1 sequences and is conserved even in more distantly related proteins such as FT and animal PEBPs. The high degree of conservation implies that mutation of this Gln-127 likely affects TFL1 function and could account for the det-2 phenotype.
The det-3 mutant was obtained by mutation of the SG line (Berdnikov et al., 1999), but this line has since been lost. Thus, as in the case of det-2, no progenitor line was available for det-3. Sequencing revealed five SNP between det-3 and wild-type cv Térèse (Table 1). Three of these are predicted to direct changes in the protein sequence. The first is Met-4 to Ile, which it shares in common with det-2. The second is Thr-66 to Ile. This residue is conserved perfectly across all known TFL1 sequences. Furthermore, the corresponding substitution in Arabidopsis TFL1 is the basis for the tfl1-14 mutant phenotype (Ohshima et al., 1997) and was shown to suppress interactions with putative partners (Pnueli et al., 2001). The third change occurs at position 104, where a conserved Glu is replaced by Lys.
Additional evidence for the importance of the Gln-127 and Thr-66 residues comes from a detailed analysis of the CEN (Banfield and Brady, 2000) and TFL1 (our unpublished data) protein structures. Thr-66 resides in the core of the protein, located on the central β-sheet F (as labeled for human PEBP; Banfield et al., 1998). The Thr side chain forms a hydrogen bond to the side chain of Gln-127. Therefore, these two residues (Thr-66 and Gln-127) map to the same region of the structure. Interestingly, two of the det mutants, det-2 and det-3, represent mutations in these residues. These mutations (Thr-66 to Ile in det-3 and Gln-127 to Arg in det-2) would disrupt the hydrogen bonding interaction between the two residues and likely would affect the structural integrity of this region. Because mutations that affect protein function have been mapped to this region in both Arabidopsis TFL1 and PsTFL1a, it follows that this must be an important part of the structure. However, this importance is likely to be structural (maintenance of protein fold) rather than directly functional (e.g., interaction with other proteins/small molecules).
Because three different det mutant alleles carry a substitution in the predicted amino acid sequence of PsTFL1a that is likely to have a significant effect on PsTFL1a function, we conclude that PsTFL1a corresponds to DET.
PsTFL1c Is Another Homolog of TFL1 and Corresponds to LF
We also examined the relationship between PsTFL1c and LF by sequencing PsTFL1c from a range of natural variant alleles and induced mutants at the LF locus (Table 2). A relatively large number of LF alleles are known and have been grouped into four phenotypic classes (Taylor and Murfet, 1993; Weller et al., 1997). Mutants in the lf-a class present the strongest phenotype and can flower as early as node 5. In four of six independent lf-a lines—HL7, XVIII/17, Wt11796, and K2—PsTFL1c could not be amplified from genomic DNA (data not shown), and a band corresponding to PsTFL1c was absent from these lines in DNA gel blot analyses (Figure 5). These lines were obtained from fast-neutron or γ-ray mutagenesis (Taylor and Murfet, 1993) and therefore likely carry large deletions in the region of PsTFL1c.
Table 2.
Results of Sequence Analysis of PsTFL1c in Several Pea Lines and Their Corresponding Mutants for LF
| Line | LF Class | Haplotype | Derived Mutant Line | LF Class | Haplotype |
|---|---|---|---|---|---|
| Vesna | Lf-d | A | XVIII/17 | lf-a | No |
| Torsdag | Lf | A | K319 | lf | C |
| K2 | lf-a | No | |||
| Wt4042 | Lf | A | Wt11796 | lf-a | No |
| HL75 | Lf | A | HL76 | lf-a | B |
| Porta | Lf | A | Wt11790 | lf | C |
| Wt11791 | lf | C | |||
| Paloma | Lf | A | Wt11795 | lf-a | E |
| WL1771 | Lf-d | D | WL1770 | Lf | D |
| WL1769 | lf | D | |||
| Champagne | Lf-d | A | |||
| Térèse | Lf | A | |||
| HL7 | lf-a | No |
Mutants and their progenitor lines are described by Taylor and Murfet (1993). The PsTFL1c sequences were grouped in five haplotypes (A to E) as described in Figure 6. “No” indicates that PsTFL1c could not be amplified by PCR and was not detected on a DNA gel blot (Figure 5). HL7 is a natural mutant; therefore, no progenitor line is available.
Figure 5.
DNA Gel Blot of Different Wild-Type and Corresponding Mutant Lines for LF.
Mutant lines are described in Table 2. The genomic DNA was digested with EcoRI, and the blot was hybridized with a PsTFL1c probe under high stringency.
In the remaining two lf-a mutants (HL76 and Wt11795), PsTFL1c was amplified and sequenced (Table 2, Figure 6). Compared with its isogenic Lf progenitor HL75, the lf-a line HL76 was found to carry a 6-bp deletion in the PsTFL1c coding region that is predicted to direct the replacement of three amino acids (Phe-Ala-Asp) at positions 147 to 149 with a single Tyr (haplotype B in Figure 6). This deletion maps to the α-C helix (as labeled for the human PEBP [Banfield et al., 1998]), and in the mutated form, it is highly unlikely that this helix would be able to form. Loss of this helix would have a seriously destabilizing effect on the protein's structure. Interestingly, this helix maps to the same region of the structure as the Thr-66/Gln-127 pair, again suggesting the importance of this area in protein stability and/or function. In the lf-a line Wt11795, a C-to-T substitution at position 1625 is responsible for the replacement of Pro-109 by Ser (haplotype E in Figure 6). This Pro is conserved in all TFL1-related plant sequences (Figure 2A) and forms an integral part of the proposed “putative ligand binding site” for these proteins (Banfield et al., 1998). Consequently, this mutation could affect protein function severely.
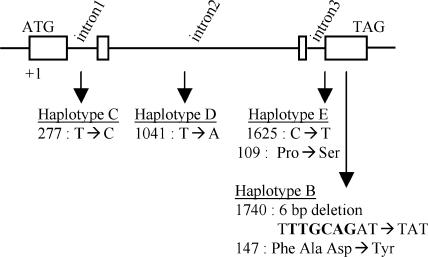
Figure 6.
Description of the Different Haplotypes Identified from the Sequence Analysis of PsTFL1c in Several Pea Lines and Their Corresponding Mutants for LF.
Mutant lines are described in Table 2. Scheme of the PsTFL1c genomic sequence. Boxes represent the coding sequence. Arrows indicate the positions of the substitutions relative to haplotype A. For each haplotype, the position and the nature of each substitution are described and the predicted effect on the amino acid sequence is given.
Our analysis of six independent lf-a mutants has shown in each case an important modification in the PsTFL1c sequence, either a large deletion or a functionally significant substitution mutation. Therefore, we conclude that PsTFL1c corresponds to LF.
The Three PsTFL1 Genes Present Different Patterns of Expression
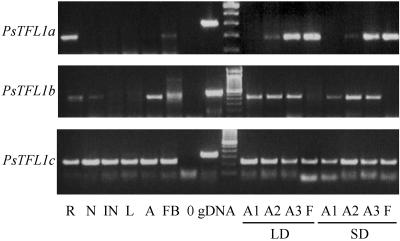
We next examined the presence/absence of transcript for each of the PsTFL1 genes by RT-PCR in different tissues at different stages. Specific intron-spanning primers were designed for each gene to control for contaminating genomic DNA. We analyzed a number of different tissues during the vegetative and reproductive phases in plants grown under either short-day or long-day conditions (Figure 7). The three genes showed distinct patterns of expression. PsTFL1a was expressed mainly in roots, in the apex after the floral transition, and in flower buds and flowers. Expression in roots is common for flowering genes such as LpTFL1 (Jensen et al., 2001), LUMINIDEPENDENS (Aukerman and Amasino, 1996), and GIGANTEA (Fowler et al., 1999). No function was proposed for the expression of these genes in roots. No expression of PsTFL1a was detected in the shoot apex before the floral transition. In both short-day and long-day conditions, a signal was detected in the apex after the floral transition (stage A2) and remained during the reproductive phase (stage A3). For PsTFL1b, expression was found in the apex during the vegetative and reproductive phases (Figure 7). Expression was found in roots and dormant nodes. No expression was detectable in flowers. As with PsTFL1a, no difference was observed between short-day and long-day conditions. PsTFL1c transcripts were present in all tissues studied (Figure 7), and no change in the expression pattern was detected during the flowering process.
Figure 7.
Expression Analysis of Pea TFL1 Homologs by RT-PCR.
Specific primers for PsTFL1a, PsTFL1b, and PsTFL1c were used to amplify the cDNA (see Methods). As controls, PCR was performed on genomic DNA (gDNA) and water (0). PCR was performed on cDNA obtained from different tissues: root (R), dormant node 4 (N), internode (IN), leaf (L), vegetative apex (A), flower bud (FB), flower (F), and shoot apex during the floral transition under short-day (SD) or long-day (LD) conditions at three different stages (A1, before the floral transition; A2, after the floral transition but before flowering; and A3, after flowering). These three stages are described in detail in Methods.
Intermediate Mutant Lines for LF Present Variations of the PsTFL1c Transcript Level
As shown previously, the stronger LF alleles (lf-a) carry deletions or substitutions of the PsTFL1c gene. Other alleles (lf, Lf, and Lf-d) were studied (Table 2). DNA gel blot analysis revealed no detectable structural change for some of these lines (K319 and WL1769; Figure 5). Sequence analysis showed no differences in the coding region. Only two silent substitutions in the introns were detected: one between lf mutants K319, Wt11790, and Wt11791 (haplotype C) and their wild-type progenitors (haplotype A) and another between the wild-type line WL1771, its derived mutant lines (haplotype D), and the other wild-type lines (haplotype A) (Table 2, Figure 6). Therefore, plants containing the Lf-d, Lf, or lf allele produce the same PsTFL1c protein.
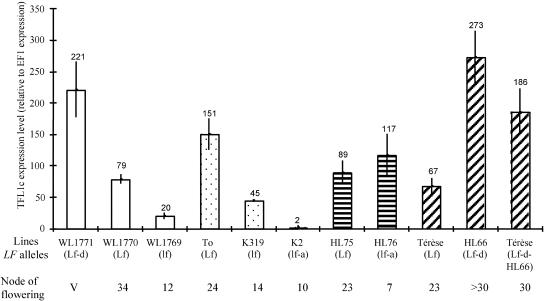
To determine whether differences between LF alleles occurred at the transcriptional level, PsTFL1c transcripts were analyzed by real-time PCR in the apices of plants with five expanded leaves (Figure 8). At this stage, plants have not initiated flowering, except those bearing the strong lf-a allele. In the mutant series WL1770 (Lf) and WL1769 (lf), derived from the WL1771 (Lf-d) progenitor line (Table 2), PsTFL1c was expressed sequentially higher. Between Lf-d and lf, there was a 10-fold difference (Figure 8). Therefore, for a given genetic background, there was a correlation between the expression of PsTFL1c and the node of flowering. A similar correlation was found in mutants K319 (lf) and K2 (lf-a) in the Torsdag (Lf) background. PsTFL1c transcript levels were threefold higher in Torsdag than in K319 (lf), whereas no significant expression was detected in K2, which carries a large deletion spanning the TFL1 gene (Figure 6).
Figure 8.
Analysis of the PsTFL1c Transcript Level in Different LF Lines by Real-Time PCR.
RNA was extracted from the apices of plants at the five-node stage. To compare the results, transcript levels also were evaluated for the elongation factor, E1Fα, which is supposed to be constant in our conditions. Numbers represent the differences in expression between PsTFL1c and EF1α. The real-time PCR experiment was repeated three times. For each LF line, the allele at the LF locus and the node of flowering (average of four different plants grown in short-day conditions) are given. V indicates that the plants did not flower after 40 nodes and remained vegetative.
These results suggested that the low level of PsTFL1c transcription was associated with early flowering. This correlation seemed to be contradicted by the analysis of HL76 (lf-a), in which a relatively high level of PsTFL1c transcript was associated with early flowering. The PsTFL1c transcript level was not significantly different between HL75 (Lf) and HL76 (lf-a) (Figure 8). This contradiction can be explained by the presence of a mutation in the predicted PsTFL1c protein of HL76 (Figure 6). For this line, protein sequence, not mRNA level, was the cause of the early-flowering phenotype. Because no change was detected in PsTFL1c transcript level between HL75 and HL76, this finding suggests that there is no feedback regulation for LF.
We noticed that within the same class of LF allele, there was variation in the level of PsTFL1c transcript. For example, for the Lf allele, the relative expression level varied from 67 (Térèse line) to 151 (Torsdag line). This variation could be a consequence of modulation resulting from different genetic backgrounds. To test this hypothesis, we introduced LF alleles in new genetic backgrounds by successive backcrosses. The Lf-d allele, carried by the HL66 line, was introduced into the Térèse background by seven successive backcrosses. In this new genetic background, the level of PsTFL1c transcript in Lf-d plants was 40% lower than that in HL66 but still significantly higher (threefold) than that in the nearly isogenic Lf line Térèse (Figure 8). From these results, we conclude that PsTFL1c transcription is determined by the LF allele but also by different genetic backgrounds.
DISCUSSION
The genetics and physiology of flowering in pea have been studied in detail (Reid et al., 1996; Weller et al., 1997). We used a candidate-gene approach to study the role of TFL1 homologs in the flowering process. Two of these homologs, PsTFL1a and PsTFL1c, were found to correspond to the pea genes DET and LF, respectively. Evidence for these correspondences and the conservation of TFL1 homolog gene function between Arabidopsis and pea are discussed below.
DET, an Arabidopsis TFL1 Homolog, Acts to Maintain the Fate of the Inflorescence Meristem in Pea
PsTFL1a mapped in the vicinity of DET. Sequencing of PsTFL1a in three independent det lines revealed mutations that would significantly modify PsTFL1a structure, leading to a nonfunctional protein. These results strongly indicate that PsTFL1a corresponds to DET. Further evidence that PsTFL1a corresponds to DET comes from mutant phenotype comparisons and gene interactions.
The phenotype of det mutants in pea is similar to those of tfl1 and cen. In all three species, indeterminate growth is changed to determinate growth during the reproductive phase (Singer et al., 1990; Bradley et al., 1996, 1997). This conversion was proposed to result from the acceleration of the reproductive phase and the conversion of the inflorescence to a floral meristem in Arabidopsis (Ratcliffe et al., 1998). In det mutants, the inflorescence meristem is converted to a stub (Singer et al., 1999). As with cen mutants in snapdragon, det mutants present no phenotype during the vegetative phase, whereas tfl1 mutants flower earlier in Arabidopsis. The absence of a vegetative phenotype may be explained by the fact that DET and CEN (Bradley et al., 1996) are expressed only after the floral transition, whereas TFL1 also is expressed during the vegetative phase (Bradley et al., 1997).
Interactions between TFL1 and other important regulatory genes are conserved between some species. In Arabidopsis, TFL1 antagonizes LFY, as does CEN with FLORICAULA in snapdragon. Double mutants (tfl1 lfy or cen flo) have a lfy or flo phenotype, respectively (Bradley et al., 1996, 1997). The same epistatic interaction is found in pea between DET and UNIFOLIATA, the LFY ortholog. The double mutant det uni has a uni phenotype (Singer et al., 1999).
LF, Another Arabidopsis TFL1 Homolog, Is a Repressor of Flowering
The results obtained demonstrate that PsTFL1c corresponds to LF. PsTFL1c was mapped to linkage group II in the vicinity of LF. Analysis of PsTFL1c genomic DNA sequence in the strongest lf-a mutants (plants presenting the earliest flowering phenotype) showed significant modifications: four lf-a mutants are complete deletion mutants lacking PsTFL1c and can be considered null alleles; two other lf-a mutants contained nonsilent changes that could modify the structure or function of PsTFL1c.
Comparison of the lf-a and tfl1 mutants reveals a similar early-flowering phenotype. In Arabidopsis, TFL1 acts to maintain the apical meristem during the vegetative stage and thereby control the length of the vegetative phase (Bradley et al., 1997; Ratcliffe et al., 1998). In pea, LF also can be considered a regulator of the length of the vegetative phase. Mutant lines for this gene have a lower flowering node and early flowering time (Taylor and Murfet, 1993). LF, like TFL1, acts as a floral repressor by lengthening the vegetative phase. In contrast to tfl1, no major floral phenotype was detected for mutants at the LF locus.
Natural and induced mutant alleles for LF have been grouped into four classes: Lf-d, Lf, lf, and lf-a (Taylor and Murfet, 1993). The strongest lf-a phenotype is the result of mutation or deletion of PsTFL1c. This loss of function is consistent with lf-a being a recessive mutation. Sequencing of the Lf-d, Lf, and lf alleles revealed no change in the predicted amino acid sequence of PsTFL1c (Table 2, Figure 6). We demonstrated that the flowering phenotype in these alleles could be explained by differences in PsTFL1c transcript levels during the vegetative phase in the shoot apex. When comparing series of alleles in the same genetic background, LF transcript levels were correlated with the flowering node in pea. Plants having the late-flowering Lf-d allele had a high level of LF transcript. The Lf-d plants behaved like 35S-TFL1 Arabidopsis plants, in which the upregulation of TFL1 is responsible for a delay in flowering and a highly branched architecture (Ratcliffe et al., 1998). In pea, the influence of flowering genes on branching is well known (Floyd and Murfet, 1986; Beveridge et al., 2003). In particular, the Lf-d allele, by delaying flower initiation, results in plants with a high number of aerial lateral branches. The high transcript levels in Lf-d plants are consistent with the dominance of this allele. Furthermore, a transcript dose effect also would explain the intermediate flowering node seen in heterozygous individuals (Murfet, 1975).
One hypothesis to explain the differences in transcript levels between the LF alleles is that important cis-regulatory elements are mutated. We have sequenced the untranslated region (200 bp 5′ and 260 bp 3′) and the introns in a series of different LF alleles (Table 2), and only two changes were detected. One substitution (haplotype D; Figure 6) seems to result from natural variation with no effect on flowering, because it is found in WL1771 (Lf-d), WL1770 (Lf), and WL1769 (lf) (Table 2). More interesting is the substitution in intron 1 (haplotype C; Figure 6), which is found in independent lf plants derived from different wild-type lines. lf lines Wt11790 and Wt11791 were obtained from the Lf line Porta (Murfet, 1991), whereas lf line K319 was obtained from the Lf line Torsdag (Uzhintseva and Sidorova, 1988). It is intriguing that identical alleles appeared independently in different mutagenesis programs. Important cis elements can be present in the introns, as was shown in the second intron of AGAMOUS (Lohmann et al., 2001). Modification of the expression of genes between different alleles has been demonstrated for important traits. In tomato, the fw2.2 alleles regulate fruit size through changes in transcript regulation rather than in the FW2.2 protein itself (Cong et al., 2002). In rice, Hd3, a FT homolog, shows different levels and timing of gene expression between different alleles (Kojima et al., 2002). In both cases, SNP between the alleles have been detected in putative regulatory regions. Further analysis will be required to prove the association between SNP and expression changes in the lf alleles.
Different Regulation between TFL1 Homologs in Pea and Arabidopsis
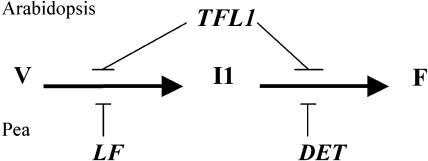
In pea, two TFL1 homologs have two distinct functions: LF is involved in the control of the vegetative phase by delaying floral initiation, the transition from the vegetative to the I1 inflorescence meristem, and DET is involved in the control of the floral phase by preventing the transition from the I1 inflorescence meristem to the flower (Figure 9). This regulation is different from that in Arabidopsis, in which only one TFL1 gene controls the length of both the vegetative and floral phases (Figure 9). To obtain the tfl1 phenotype in pea (early flowering and determinate growth), a det lf double mutant is necessary (Murfet, 1989). LF and DET are homologs and may derive from a common ancestor by duplication. In pea, the two genes may have evolved separately and become specialized for two distinct functions. The det lf-a double mutant has only additive effects and no extra phenotype (Murfet, 1989), which suggests nonredundant functions for DET and LF. It has been argued that subfunctionalization of duplicated genes is a mechanism whereby degenerative mutations can lead to the preservation of duplicated genes (Force et al., 1999; Lynch and Force, 2000). DET and LF may provide an example in which a gene that was expressed originally during both the vegetative and reproductive phases (such as TFL1 in Arabidopsis) diverged into two copies with a partitioning of gene expression patterns as predicted by the duplication/degeneration/complementation model. Like DET, CEN has only one function during the vegetative phase in snapdragon (Bradley et al., 1996), so it will be interesting to determine whether another TFL1 homolog exists in snapdragon that functions during the vegetative phase in a manner similar to LF.
Figure 9.
Model for the Control of Floral Initiation and Development by TFL1 Genes in Arabidopsis and Pea.
F, flower; I1, inflorescence meristem; V, vegetative phase.
In this study, we have demonstrated the important role played by a TFL1 homolog, LF, in the control of flowering time in pea. Detailed genetic analyses have enabled the identification of a few genes involved in the natural variation in flowering time among pea cultivars under controlled environmental conditions (Murfet, 1971b; Weller et al., 1997). LF was identified as a major contributor to natural variability, but other genes involved in the photoperiodic pathways also were shown to influence flowering time in pea, such as STERILE NODE and HIGH RESPONSE. These genes are implicated in the synthesis of a graft-transmissible floral inhibitor, which is perceived in the apex by LF (Murfet, 1971a). As a long-day flowering plant, pea has developed a strategy to control flowering in response to photoperiod and vernalization, with a central role for LF. Vernalization acts quantitatively by reducing the flowering node in two ways: by decreasing inhibitor production and by rendering the apex more sensitive to the flowering signal (Murfet and Reid, 1974; Reid and Murfet, 1975).
The strategy developed by Arabidopsis seems to be different, because TFL1 plays a minor role in the control of flowering time between ecotypes. In Arabidopsis, FLOWERING LOCUS C (FLC) and FRIGIDA (FRI) are central elements and are key components of the response to vernalization (Michaels and Amasino, 1999; Sheldon et al., 1999; Johanson et al., 2000). For instance, in natural Japanese Arabidopsis populations, the digenic interaction between FRI and FLC was the major genetic system controlling flowering time (Shibaike et al., 1999). FLC is a floral repressor, which must be repressed by vernalization for floral initiation to occur. FRI acts synergistically by maintaining high levels of FLC. Early-flowering ecotypes of Arabidopsis carry nonfunctional FRI and/or FLC alleles (Johanson et al., 2000) and consequently have low levels of FLC transcript, whereas late-flowering ecotypes have high levels of FLC expression (Rouse et al., 2002). This important role for FLC also is found in other Brassicaceae species (Tadege et al., 2001; Schranz et al., 2002). Thus, pea and Arabidopsis have developed different strategies to control flowering time, which may represent different strategies to respond to environmental conditions. In natural Arabidopsis populations, flowering time variation is explained mainly by response to vernalization, whereas in pea it seems to be explained mostly by response to photoperiod. Interestingly, in both cases, the transcript level of the repressor, FLC or LF, determines flowering time. Different alleles exist at the LF locus and are responsible for different LF transcript levels. The same results were found in Arabidopsis at the FLC locus, where different alleles confer different FLC transcript levels (Schlappi, 2001).
In conclusion, we have shown that TFL1 homologs in pea play an important role in floral initiation and development. LF is a major gene that controls flowering time by integrating different environmental and endogenous signals. Understanding the regulation of LF by genes involved in photoperiod response is an important next step for dissecting the molecular basis of flowering control in pea.
METHODS
Plant Material and Growth Conditions
det lines of pea (Pisum sativum) were obtained from the John Innes Pisum Germplasm collection. det-1 corresponds to JI 2121 and was obtained by mutation of cv Paloma (Swiecicki, 1987). det-2 (JI 1358) is a spontaneous mutant, and det-3 (JI 3100) was obtained by mutation of line SG (Berdnikov et al., 1999). Other pea cultivars were Térèse and Torsdag. The F2 and RI lines used for genetic mapping are described below. LF lines were provided by Ian Murfet (University of Tasmania, Hobart, Australia). Relevant information concerning the mutants and their progenitors is described by Taylor and Murfet (1993).
For the floral initiation experiments, the plants were grown in cabinets at 20°C during the day and 15°C during the night with illumination by mercury vapor lamps (135 mE·m−2·s−1) in a 1:1 mixture of sphagnum:clay under short-day (12 h of light) or long-day (18 h of light) conditions. For the long-day conditions, plants received 12 h of light from the mercury vapor lamps extended by 6 h of light from a series of 40-W incandescent/fluorescent bulbs. The apices were harvested at three different stages. The A1 stage was collected at least 10 nodes before plants flower. At this stage, the apices are still vegetative (Isabelle Lejeune, personal communication). The A2 stage was harvested just three nodes before flowering. At the A2 stage, the floral initiation has occurred but flowers are not opened. The A3 stage corresponds to the apices just after flowers open.
Cloning and Isolation of Genes
The TFL1 homologs were isolated in cv Térèse using degenerate primers. The PCR conditions were 35 cycles at 94°C for 60 s, 55°C for 60 s, and 72°C for 2 min. The primer combination TFL1-1 (5′-ATGGGGAGAGTGATA/T/CGGG/AGAA/TG-3′) and TFL1-2 (5′-TCACTAGGA/GCCA/TGGAACATCA/TGG-3′) gives a 450-bp band corresponding to PsTFL1c. Using the primer combination TFL1-3 (5′-GATGTTCCA/TGGA/TCCTAGTGAC/TCC-3′) and TFL1-5 (5′-CTTGCAGCA/GGTC/TC/TTCC/TCTC/TTG-3′), two bands of 400 and 800 bp were obtained corresponding to PsTFL1a and PsTFL1b, respectively.
The sequences of the genes were extended by 3′ rapid amplification of cDNA ends (RACE) PCR using the kit and the recommendations of the supplier (Life Technologies, Rockville, MD) on cDNA from seedlings and flowers. The following primers were used as gene-specific primers and nested gene-specific primers, respectively, for PsTFL1a (TFL1R1 [5′-TCAAACAAAGAGCGAGAGATTCAG-3′] and TFL1R2 [5′-AGACCA- TTTCAACACTCGTAG-3′]), for PsTFL1b (Tb31 [5′-GACAGATATACCAGGCACAAC-3′] and Tb32 [5′-GAAATATGAAATGCCACGTCC-3′]), and for PsTFL1c (TFL1-1 and TFL5R3 [5′-AGCCAAGGATTCAGATTCAAGG-3′]). The 5′ part of PsTFL1a was obtained by 5′ RACE PCR using the kit and the recommendations of the supplier (Life Technologies) on cDNA from flowers. The gene-specific primers used were TFLR3 (5′-CTACTTTGATACACACACGACAC-3′) and TFL5R1 (3′-GAATAGAAC- AAACACAAACCT-3′). The 5′ part of PsTFL1c was recovered by PCR walking on genomic DNA (Devic et al., 1997). Pea genomic DNA was digested by DraI, and adaptors were ligated to the digested DNA as described by Devic et al. (1997). The following specific primers were used to amplify the gene: TFL1cR8 (5′-TAACTGTAGAAGGAAAGGGTA-3′) and TFL1cR9 (5′-ATGCTTGCGGTAAAATAATCA-3′).
Mapping and Marker Development
Mapping was performed on two different mapping populations (recombinant inbred lines) obtained from the crosses Térèse × K586 (Laucou et al., 1998) and JI 281 × JI 399 (Ellis et al., 1992). Polymorphisms were sought between the parents in the mapping population to develop PCR markers such as cleaved amplified polymorphism sequence (CAPS) markers. For PsTFL1a, a single nucleotide polymorphism was detected between Térèse and K586. A derived CAPS (dCAPS) marker (Neff et al., 1998) was developed using the following primers (TFL2, 5′-GAACACTTG- CACTGGTAAATATAATAGA-3′; TFLR, 5′-TGTAGCATCTGTTGTTCC- TGG-3′) and the HinfI enzyme. For PsTFL1b and PsTFL1c, polymorphisms were found between JI 281 and JI 399 and CAPS markers were developed for PsTFL1b (primer pairs TFLb32 [5′-GAAATATGAAATGCC- ACGTCC-3′] and TFL1bR1 [5′-ACAAACTAGAACAACAACAACCC-3′] and restriction enzyme HhaI) and for PsTFL1c (primer pairs TFL1-1 and TFL1-2 and restriction enzyme HpyF44III).
DNA Extraction and Gel Blots
DNA was extracted from leaves according to the protocol described by Laucou et al. (1998). Ten micrograms of genomic DNA was digested with restriction enzymes and loaded on a 0.7% agarose gel. Blotting was performed according to the recommendation of the membrane supplier (Biotrans Nylon Membrane; ICN, Costa Mesa, CA). Hybridization was performed in Church and Gilbert (1984) buffer at 65°C overnight. Washing was done at 65°C according to Sambrook et al. (1989) (2× SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate] and 0.1% SDS for 30 min, 1× SSC and 0.1% SDS for 15 min, and 0.2× SSC and 0.1% SDS for 15 min).
RNA Extraction and Expression Analysis
RNA was extracted using the RNeasy Plant Mini Kit with a DNase treatment on an RNeasy column (Qiagen, Valencia, CA). Five micrograms of total RNA was used for cDNA synthesis. Before reverse transcription, total RNA was treated with amplification-grade DNase I (Life Technologies) according to the manufacturer's protocol. Reverse transcription was performed using 200 units of Superscript II RNase H− reverse transcriptase (Invitrogen, Carlsbad, CA) in the presence of 40 units of recombinant ribonuclease inhibitor (Life Technologies) with the AP primer. For PCR, cDNAs were resuspended in 100 μL of water, and 1 μL was used per reaction. Reverse transcriptase–mediated PCR was performed using specific primers for PsTFL1a (TFL5RACE [5′-TGAGTTGTACTCTTAAGTTCTTC-3′] and TFLaR5 [5′-AGGGCCAGGAACATCAGGGTC-3′]), PsTFL1b (TFL1bR1 and TFL1b31 [5′-GACAGATATACCAGGCACAAC- AG-3′]), and PsTFL1c (TFLcR2 [5′-AAATAAGCAGCAGCAACAGGG-3′] and TFLcR3 [5′-CAGACATTCCAGGGACAACAG-3′]) and the following program (40 cycles at 94°C for 30 s, 60°C for 60 s, and 72°C for 30 s). The PCR product was loaded on a 2% (w/v) agarose gel.
Real-time PCR was performed on a Roche Lightcycler using the FastStart DNA Master SYBR Green I kit (Mannheim, Germany) according to the manufacturer's protocol. Specific primers suitable for quantitative reverse transcriptase–mediated PCR were designed using LC Probe Design Report software (Roche) for EF1α (E1FF1 [5′-GATGCACCTGGACATCGTGACT-3′] and E1FR1 [5′-CTTAGGGGTGGTAGCATCCATCT-3′]) and PsTFL1c (cF3 [5′-CCACATTTGGAAAAGAGTTGACAAGC-3′] and cR3 [5′-GCGTCTTCTAGGAGCCGTTGC-3′]). The PCR program consisted of a first step of denaturation and Taq activation (95°C for 8 min) followed by 45 cycles of denaturation (94°C for 10 s), annealing (58°C for 7 s), and extension (72°C for 10 s). At the end, amplified products were denatured (95°C), renatured (65°C), and progressively denatured (step from 65 to 95°C over 30 min or 0.1°C/s for the fusion curve analysis). Both primers combinations (cR3/cF3 and E1FR1/E1FE1) were tested (fusion curve, linearity, and efficiency of the primers). The primer combinations for E1F and PsTFL1c have a PCR efficiency of 86 and 80%, respectively.
For PCR, cDNA was diluted 50 times and 5 μL was used as a template in a 20-μL reaction mix. PsTFL1c transcript level was estimated based on the level of the constitutive EF1α gene (Nesi et al., 2000). For each condition, the number of cycles necessary to reach a certain level (exit position) of fluorescence was evaluated for EF1α (nEF1α) and PsTFL1c (nPsTFL1c). The difference between the exit points of the two genes was calculated (d1 = nPsTFL1c − nEF1α). The value 2d1 represents the difference of copy number between PsTFL1c and the constitutively expressed gene, EF1α. Because EF1α was expressed at a higher level than PsTFL1c, the final ratio was calculated as follows: PsTFL1c level = (1/2dl) × 100,000% EF1α.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Catherine Rameau, rameau@versaille.inra.fr.
Accession Numbers
The sequence data described herein have been submitted to GenBank with accession numbers AY340579 for PsTFL1a, AY340580 for PsTFL1b, and AY343326 for PsTFL1c. Accession numbers for the sequences shown in Figure 2 are as follows: AB027506 (TSF), AB016880 (BFT), AF147721 (MFT), and BAA33415 (BnTFL1-1 from Brassica napus).
Acknowledgments
We thank Mike Ambrose, K. Sidorova, and Ian Murfet for providing the mutant lines. Thanks also are due to Bertrand Dubreucq for help and suggestions with the real-time PCR experiments, to Magali Goussot, Karine Haurogné, Xenie Johnson, and Nathalie Mansion for technical assistance, and to Patrick Grillot for his excellent management of the greenhouse. We also particularly thank Julie Hofer and Jim Weller for their critical comments and corrections on the manuscript and for their suggestions. Finally, we dedicate this article to Ian Murfet for his great contribution to the understanding of flowering genetics in pea. This work was supported by Genoplante (Project PEA-A) and the Human Frontiers in Science Program. M.J.B. is a Royal Society (UK) University Research Fellow.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015701.
References
- Amaya, I., Ratcliffe, O.J., and Bradley, D.J. (1999). Expression of CENTRORADIALIS (CEN) and CEN-like genes in tobacco reveals a conserved mechanism controlling phase change in diverse species. Plant Cell 11, 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman, M.J., and Amasino, R.M. (1996). Molecular genetic analysis of flowering time in Arabidopsis. Semin. Cell Dev. Biol. 7, 427–433. [Google Scholar]
- Banfield, M.J., Barker, J.J., Perry, A.C.F., and Brady, R.L. (1998). Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction. Structure 6, 1245–1254. [DOI] [PubMed] [Google Scholar]
- Banfield, M.J., and Brady, R.L. (2000). The structure of Antirrhinum CENTRORADIALIS protein (CEN) suggests a role as a kinase regulator. J. Mol. Biol. 297, 1159–1170. [DOI] [PubMed] [Google Scholar]
- Berdnikov, V.A., Gorel, F.L., Bogdanova, V.S., Kosterin, O.E., Trusov, Y.A., and Rpzov, S.M. (1999). Effect of a substitution of a short chromosome segment carrying a histone H1 locus on expression of the homeotic gene Tl in heterozygote in the garden pea Pisum sativum L. Genet. Res. 73, 93–109. [Google Scholar]
- Beveridge, C.A., and Murfet, I.C. (1996). The gigas mutant in pea is deficient in the floral stimulus. Physiol. Plant. 96, 637–645. [Google Scholar]
- Beveridge, C.A., Weller, J.L., Singer, S.R., and Hofer, J.M.I. (2003). Axillary meristem development: Budding relationships between networks controlling flowering, branching, and photoperiod responsiveness. Plant Physiol. 131, 927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, D., Carpenter, R., Copsey, L., Vincent, C., Rothstein, S., and Coen, E. (1996). Control of inflorescence architecture in Antirrhinum. Nature 379, 791–797. [DOI] [PubMed] [Google Scholar]
- Bradley, D., Ratcliffe, O., Vincent, C., Carpenter, R., and Coen, E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen, E.S., Romero, J.M., Doyle, S., Elliott, R., Murphy, G., and Carpenter, R. (1990). floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell 63, 1311–1322. [DOI] [PubMed] [Google Scholar]
- Colasanti, J., and Sundaresan, V. (2000). ‘Florigen’ enters the molecular age: Long-distance signals that cause plants to flower. Trends Biochem. Sci. 25, 236–240. [DOI] [PubMed] [Google Scholar]
- Cong, B., Liu, J., and Tanksley, S.D. (2002). Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl. Acad. Sci. USA 99, 13606–13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk, Q.C.B. (2001). Plant evolution and development in a post-genomic context. Nat. Rev. Genet. 2, 607–619. [DOI] [PubMed] [Google Scholar]
- Devic, M., Albert, S., Delseny, M., and Roscoe, T.J. (1997). Efficient PCR walking on plant genomic DNA. Plant Physiol. Biochem. 35, 109–116. [Google Scholar]
- Ellis, T.H., Turner, L., Hellens, R.P., Lee, D., Harker, C.L., Enard, C., Domoney, C., and Davies, D.R. (1992). Linkage maps in pea. Genetics 130, 649–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd, R.S., and Murfet, I.C. (1986). Branching in Pisum: Effect of the flowering and length genes. Pisum Newsl. 18, 12–15. [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.-l., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer, J., and Ellis, N. (2002). Conservation and diversification of gene function in plant development. Curr. Opin. Plant Biol. 5, 56–61. [DOI] [PubMed] [Google Scholar]
- Hofer, J., Turner, L., Hellens, R., Ambrose, M., Matthews, P., Michael, A., and Ellis, N. (1997). UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7, 581–587. [DOI] [PubMed] [Google Scholar]
- Jensen, C.S., Salchert, K., and Nielsen, K.K. (2001). A TERMINAL FLOWER1-like gene from perennial ryegrass involved in floral transition and axillary meristem identity. Plant Physiol. 125, 1517–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V.K., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., and Yano, M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105. [DOI] [PubMed] [Google Scholar]
- Laucou, V., Haurogné, K., Ellis, N., and Rameau, C. (1998). Genetic mapping in pea. 1. RAPD-based genetic linkage map of Pisum sativum. Theor. Appl. Genet. 97, 905–915. [Google Scholar]
- Levy, Y.Y., and Dean, C. (1998). The transition to flowering. Plant Cell 10, 1973–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann, J.U., Hong, R.L., Hobe, M., Busch, M.A., Parcy, F., Simon, R., and Weigel, D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and Force, A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H., and De Pamphilis, C. (2000). The ABCs of floral evolution. Cell 101, 5–8. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimida, N., Goto, K., Kobayashi, Y., Araki, T., Ahn, J.H., Weigel, D., Murata, M., Motoyoshi, F., and Sakamoto, W. (2001). Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 6, 327–336. [DOI] [PubMed] [Google Scholar]
- Mouradov, A., Cremer, F., and Coupland, G. (2002). Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 14 (suppl.), S111.–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfet, I.C. (1971. a). Flowering in Pisum: Reciprocal grafts between known genotypes. Aust. J. Biol. Sci. 24, 1089–1101. [Google Scholar]
- Murfet, I.C. (1971. b). Flowering in Pisum: Three distinct phenotypic classes determined by the interaction of a dominant early and a dominant late gene. Heredity 26, 243–257. [Google Scholar]
- Murfet, I.C. (1975). Flowering in Pisum: Multiple alleles at the lf locus. Heredity 35, 85–98. [Google Scholar]
- Murfet, I.C. (1989). Interaction of the det (determinate) mutant with other flowering genes. Pisum Newsl. 21, 44–47. [Google Scholar]
- Murfet, I.C. (1991). Early flowering mutants Wt11790 and Wt11791 result from mutation at the Lf locus. Pisum Genet. 23, 16–18. [Google Scholar]
- Murfet, I.C., and Reid, J.B. (1974). Flowering in Pisum: The influence of photoperiod and vernalising temperatures on the expression of genes Lf and Sn. Z. Pflanzenphysiol. 71, 323–331. [Google Scholar]
- Murfet, I.C., and Reid, J.B. (1993). Developmental mutants. In Peas: Genetics, Molecular Biology and Biotechnology, R. Casey and D.R. Davies, eds (Wallingford, UK: CAB International), pp. 165–216.
- Neff, M.M., Neff, J.D., Chory, J., and Pepper, A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 14, 387–392. [DOI] [PubMed] [Google Scholar]
- Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima, S., Murata, M., Sakamoto, W., Ogura, Y., and Motoyoshi, F. (1997). Cloning and molecular analysis of the Arabidopsis gene Terminal Flower 1. Mol. Gen. Genet. 254, 186–194. [DOI] [PubMed] [Google Scholar]
- Pnueli, L., Carmel-Goren, L., Hareven, D., Gutfinger, T., Alvarez, J., Ganal, M., Zamir, D., and Lifschitz, E. (1998). The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125, 1979–1989. [DOI] [PubMed] [Google Scholar]
- Pnueli, L., Gutfinger, T., Hareven, D., Ben-Naim, O., Ron, N., Adir, N., and Lifschitz, E. (2001). Tomato SP-interacting proteins define a conserved signalling system that regulates shoot architecture and flowering. Plant Cell 13, 2687–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameau, C., Dénoue, D., Fraval, F., Haurogné, K., Josserand, J., Laucou, V., Batge, S., and Murfet, I.C. (1998). Genetic mapping in pea. 2. Identification of RAPD and SCAR markers linked to genes affecting plant architecture. Theor. Appl. Genet. 97, 916–928. [Google Scholar]
- Ratcliffe, O.J., Amaya, I., Vincent, C.A., Rothstein, S., Carpenter, R., Coen, E.S., and Bradley, D.J. (1998). A common mechanism controls the life cycle and architecture of plants. Development 125, 1609–1615. [DOI] [PubMed] [Google Scholar]
- Reid, J.B., and Murfet, I.C. (1975). Flowering in Pisum: The sites and possible mechanisms of the vernalization response. J. Exp. Bot. 26, 860–867. [Google Scholar]
- Reid, J.B., and Murfet, I.C. (1984). Flowering in Pisum: A fifth locus, veg. Ann. Bot. 53, 369–382. [Google Scholar]
- Reid, J.B., Murfet, I.C., Singer, S.R., Weller, J.L., and Taylor, S.A. (1996). Physiological-genetics of flowering in Pisum. Cell Dev. Biol. 7, 455–463. [Google Scholar]
- Rouse, D.T., Sheldon, C.C., Bagnall, D.J., Peacock, W.J., and Dennis, E.S. (2002). FLC, a repressor of flowering, is regulated by genes in different inductive pathways. Plant J. 29, 183–191. [DOI] [PubMed] [Google Scholar]
- Salvi, S., Tuberosa, R., Chiapporina, E., Maccaferri, M., Veillet, S., van Benningen, L., Isaac, P., Edwards, K., and Phillips, R.L. (2002). Toward positional cloning of Vgt1, a QTL controlling the transition from the vegetative to the reproductive phase in maize. Plant Mol. Biol. 48, 601–613. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schiex, T., Moisan, A., and Rouzé, P. (2000). Eugène: An eucaryotic gene finder that combines several sources of evidence. In JOBIM'2000, O. Gascuel and M.F. Soagot, eds (Heidelberg, Germany: Springer-Verlag Heidelberg), pp. 111–125.
- Schlappi, M. (2001). RNA levels and activity of FLOWERING LOCUS C are modified in mixed genetic backgrounds of Arabidopsis thaliana. Int. J. Plant Sci. 162, 527–537. [Google Scholar]
- Schranz, M.E., Quijada, P., Sung, S.B., Lukens, L., Amasino, R., and Osborn, T.C. (2002). Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162, 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibaike, H., Ishiguri, Y., and Kawano, S. (1999). Genetic analysis of flowering time for eight natural populations of Arabidopsis thaliana (Brassicaceae) in Japan with special regard to the genes, FRI and FLC. Plant Spec. Biol. 14, 229–236. [Google Scholar]
- Singer, S., Sollinger, J., Maki, S., Fishbach, J., Short, B., Reinke, J., Fick, J., Cox, L., McCall, A., and Mullen, H. (1999). Inflorescence architecture: A developmental approach. Bot. Rev. 65, 385–410. [Google Scholar]
- Singer, S.R., Hsuing, L.P., and Huber, S.C. (1990). determinate (det) mutant of Pisum sativum L. (Leguminosae: Papilionoideae) exhibits an indeterminate growth pattern. Am. J. Bot. 77, 1330–1335. [Google Scholar]
- Swiecicki, W.K. (1987). determinate growth (det) in Pisum: A new mutant gene on chromosome 7. Pisum Newsl. 19, 72–73. [Google Scholar]
- Tadege, M., Sheldon, C.C., Helliwell, C.A., Stoutjesdijk, P., Dennis, E.S., and Peacock, W.J. (2001). Control of flowering time by FLC orthologues in Brassica napus. Plant J. 28, 545–553. [DOI] [PubMed] [Google Scholar]
- Taylor, S.A., and Murfet, I.C. (1993). Flowering in pea: A mutation from Lf-d to lf-a and a summary of induced Lf mutations. Pisum Genet. 25, 60–63. [Google Scholar]
- Uzhintseva, L.P., and Sidorova, K.K. (1988). Genetics of early flowering pea mutants. Pisum Newsl. 20, 39–40. [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. [DOI] [PubMed] [Google Scholar]
- Weller, J.L., Reid, J.B., Taylor, S.A., and Murfet, I.C. (1997). The genetic control of flowering in pea. Trends Plant Sci. 2, 412–418. [Google Scholar]
- Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., and Dubcovsky, J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100, 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., Baba, T., Yamamoto, K., Umehara, Y., Nagamura, Y., and Sasaki, T. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., Kojima, S., Takahashi, Y., Lin, H., and Sasaki, T. (2001). Genetic control of flowering time in rice, a short-day plant. Plant Physiol. 127, 1425–1429. [PMC free article] [PubMed] [Google Scholar]