Abstract
Over the past five years, it has become increasingly apparent to researchers that the initial promise and excitement of using gene replacement therapies to ameliorate folding diseases are still far from being broadly or easily applicable. Because a large number of human diseases are protein folding diseases (~30 to 50%), many researchers now realize that more directed approaches to target and reverse the fundamental misfolding reactions preceding disease are highly feasible and offer the potential of developing more targeted drug therapies. This is also true with a large number of so called “orphan protein folding diseases”. The development of a broad-based general screening array method using the chaperonin as a detection platform will enable us to screen large chemical combinatorial libraries for specific ligands against the elusive transient, primary reactions that often lead to protein misfolding. This development will provide a highly desirable tool for the pharmaceutical, academic and medical professions.
Introduction
Protein misfolding reactions are now recognized to be a major contributing factor in a number of Human disease states, including Amyotropic Laterial Sclerosis, Cystic Fibrosis, Alzheimer’s disease, Parkinson’s disease, cancers, and a host of many different amyloidosis and metabolic diseases. The molecular origins of a majority of protein folding diseases occur because missense mutations shift the equilibrium distribution from the native state to populate partially denatured or abnormally activated species. In addition to missense mutation induced changes, equilibrium shifts toward misfolded protein populations also can result from changes in the solution environment or through the simple overexpression or increased concentration of the native protein. One emerging strategy and potential therapy for ameliorating protein misfolding in disease states or increasing the lifetime of the native protein is to identify so-called “pharmacological chaperones” stabilize native folded distribution. Usually, these stabilizers are identified using small molecule combinatorial chemistry screens. Unfortunately, such screens are problematic because 1) the assay often has to be specifically tailored for each protein system and 2) some designed assays depend on extremely slow (timelines of hours to tens of hours to days) secondary reactions such as oligomer dissociation or protein aggregation. Typically, these assays are unable to detect the elusive transient misfolded or activated states that are generally considered to be the critical direct culprits in disease progression. With supporting data presented herein, we outline the development of a general and rapid screening method that relies on the high affinity nucleotide-free state of the GroEL to detect transient misfolded states. The advantage of this particular protein platform is that this form of GroEL is known to bind a very wide variety of aggregation-prone proteins be they associated with disease or even derived in equilibrium with natively folded proteins. We demonstrate how this chaperonin can serve as a screening platform to aid in the search for lead compounds and perhaps eventual strategies to stabilize proteins involved in protein folding diseases.
I) Rationale behind using the GroEL chaperonin as a thermodynamic sink: Lessons from in vivo and in vitro observations
The major role of molecular chaperones is to prevent or decrease molecular aggregation. The general protein binding function of chaperones promote proper folding, efficient cellular proteolysis and clearance, plays a role in maintaining activated states (a protein buffer), and also may engage in protection of proteins during oxidative stress conditions and in some cases promotes disaggregation.
It was recognized early on that chaperones can provide a general buffering effect against mutational stress. For example, it was noted that the GroE chaperonins could rescue and maintain properly folded states even in instances where missense mutants would lead to misfolding (1). Lindquist and colleagues have since discovered that removing this protective buffering function, in this case by decreasing Hsp90 interactions, uncovers genotypic variations that were suppressed by chaperone maintenance functions. In this role, chaperone proteins can play a role in broadening evolutionary selective advantages by expanding functional protein folded states (2,3). Recently, Tokuriki and Tawfik (4) have demonstrated that GroE chaperonin overexpression also allows for more rapid divergence of modified enzyme activities. This is thought to occur because the chaperonin allows folding of protein stability mutants. This interaction, in turn, will allow these enzymic systems to evolve by decreasing the selection stringency and expanding the fitness of the mutated enzyme through a greater conformational search space. Since enzymatic catalysis is now recognized by most to occur via multiple conformations (5), the act of removing structural constraints to folding and allowing for more expanded coupled comformational states appears to be advantageous during enzyme evolution.
Alternatively, the ability of post translational maturation chaperones, such as particular activated Hsp90 protein-cochaperone complexes, to fold and stabilize mutated or destabilized proteins appears to have deleterious effects in some disease states. For example, the broad recognition specificity of the Hsp90 chaperones appear to extend the lifetime of some activated oncogenic proteins such as kinases, steroid receptors and transcriptional factors. Intense medicinal chemistry based drug design efforts are being developed to inhibit the Hsp90 chaperoning abilities, leading to a selective destabilization of the activated proteins and enhancing clearance of those oncogenic proteins (6,7). An apparent decrease in Hsp90 activities by targeted chemical inhibition are also thought to result in a decrease in p35 activity, a kinase that is involved in aberrant tau phosphorylation (8) leading to neurodegeneration in Alzheimer’s patients.
It is well known that the addition of chaperone proteins to protein solutions in vitro extends the lifetime of protein activity. Most notably, Lorimer and colleagues were the first to note that proteins that were predominantly present in their native states will partition onto the high affinity state of the GroEL chaperonin (no nucleotide) if these native states were in equilibrium with a significant population of transiently unfolded states (9). Also, Martin et al., (10) noted that the chaperonin can play a protective role during heat stress. Later studies seem to indicate that the chaperonin can sequester misfolded proteins at elevated temperatures and can subsequently release the properly folded form once lower temperatures return (11). It remains to be determined if these in vitro effects have any relevance in vivo. However, it is important to note that the high affinity nucleotide free form of GroEL is very efficient at binding to proteins that explore transiently denatured or partially folded states. In conclusion, to quote Edward Eisenstein* (CARB, University of Maryland), the GroEL chaperonin can be thought of as a “Promiscuous Antibody”* that binds all types of protein folding intermediates if they expose a portion of their hydrophobic surfaces to the aqueous environment.
II) Significance of using “chaperonin sinks” to screen for small molecule stabilizers aimed at ameliorating protein folding diseases
Geneticist Susan Lindquist has estimated that missense mutants that result in protein misfolding may account for >30-50% of human disease conditions. Therefore, it is absolutely essential that we understand the basic physical properties that dictate these protein folding diseases. Fred Cohen emphasizes that “we’re doomed to failure if we don’t think of these conditions [protein folding diseases] as kinetic diseases.” Thus, it has been recognized by most in the field that the kinetic aspects of these protein folding disease states are very crucial to disease outcomes (12). An exciting new strategy for ameliorating protein folding diseases is based on identifying pharmacological chaperones that bind and stabilize native folds which prevent deleterious protein misfolding or through direct maintenance of the protein levels (13).
Thus far, a number of computationally intensive yet elegant in silico efforts has lead to some limited but notable in vitro successes. These select number of native-state stabilizers have been identified using computation free energy binding methods which either slow or prevent the key misfolding reactions for mutant forms of transthyretin (TTR) and superoxide dismutase (SOD) (14,15). Most notably, clinical trials have now been implemented to treat Gaucher’s disease and TTR amyloidosis based on lead compounds identified in various small molecule screens. For most folding diseases however, the search for potential pharmacological chaperones is hampered by the lack of rapid real time physiologically-relevant assays to monitor the initial misfolding reactions that precede deleterious aggregation reactions.
In these particular purified systems, indirect assays are employed to monitor the unfolding reaction and these assays are typically performed over very long time periods (tens of hours to days under denaturing conditions) because the competing back reactions to the native fold cannot be avoided. For example, monitoring the eventual dissociation of the missense ALS mutant A4V dimer of superoxide dismutase requires 20-40 hours of assay time (15). Likewise, monitoring the real time disappearance of TTR tetramers follows a time line measured in days even under denaturing conditions (16). To compound the problem, the monitoring processes are sometimes cumbersome (fluorescent dye binding assays, intrinsic fluorescence measurements etc) and are subject to misinterpretation due to fluorescence artifacts such as inner filter effects. Furthermore, these dye based assays are not universally used for large combinatorial chemical screens due to their inherent interferences with absorbing test compounds.
III) Early supporting data for the chaperonin sink
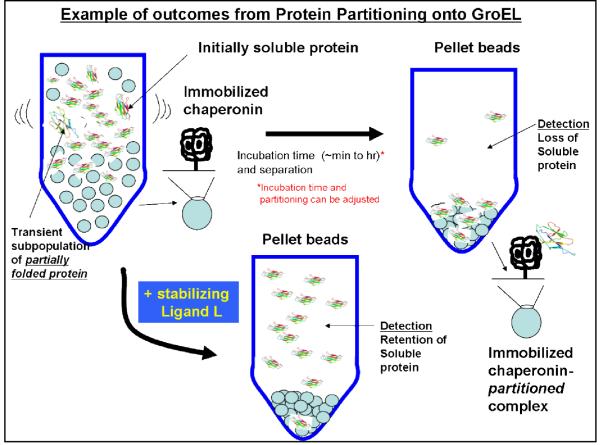
As stated above, most methods monitor downstream differences resulting from protein misfolding but are inherently indirect because they depend on late secondary reactions such as the slow disappearance of the oligomeric state or folded state rather than on the actual initial misfolding kinetic event(s). The key advantage of measuring the partition reactions of transient misfolded proteins onto the chaperonin (“the chaperonin sink” method) presented herein enables us to rapidly monitor misfolding reactions under reasonable physiological conditions. In most instances, the chaperonin sink partitioning rates are accelerated simply by increasing the chaperonin concentration whereby the rate eventually approaches an upper limit defined by the rate-limiting dissociation or transient unfolding rate constant (ref 27, see figure below).
The chaperonin sink approach can serve as a rapid direct detector of the existing partially folded states. Within a reasonable kinetic time frame, the presence of the chaperonin interferes with or even prevents the reverse refolding or reassociation reactions from occurring. The chaperonin detection system will also provide direct kinetic evidence that proposed pharmacological chaperones stabilize their respective target proteins via a kinetic stabilization mechanism (16). Most importantly however, this method can easily be scaled up for use as an essential and broad-based platform for combinatorial chemical screens to identify stabilizers of native protein folds. As indicated above, the partitioning of transiently misfolded forms that are in equilibrium with an initially stable form onto soluble GroEL has already been documented both in the Fisher laboratory as well as others (9,10, 17-19). Our laboratory has shown that inhibiting the specific reaction leading to rhodanese misfolding completely eliminates this chaperonin partitioning reaction, strengthening our hypothesis that we can expand this general chaperonin sink method to develop this assay into a new strategy to find small molecule stabilizers of the native fold.
Since numerous protein folding diseases result from the transient appearance of partially misfolded states, the high affinity form of the GroEL chaperonin can function as a general thermodynamic and kinetic “sink” to capture these transient misfolded forms. Specifically, we are developing a high-throughput screening system based on the chaperonin to identify potential stabilizing small molecule candidates that PREVENT the transient misfolded intermediate of the target protein from binding to the high affinity chaperonin. The measurable parameter in this assay is general because the read out for a positive hit is the increase in the amount of soluble protein in the particular test solution. Once identified, potential ligand/drugs can be tested and further developed with the aim to prevent protein folding diseases.
The preliminary work presented in this work will demonstrate the feasibility of using this GroEL capture system to rapidly detect partitioning of destabilized native proteins, disease variants, heterogenous misfolded populations of refolded proteins and transient forms of bacterial toxins. Our initial proof of concept indicates in some cases, the partitioning kinetics reveals a fast phase of partitioning onto the chaperonin platform. This rapid detection will most certainly aid in increasing the number of assays that one can perform as it applies to optimally designing high throughput systems to identify protein stabilizers.
Results
I) Proof of Principle Examples
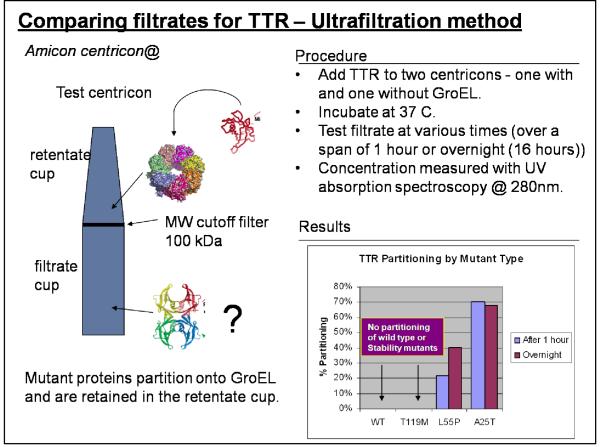
A) The Partitioning of mutant Transthyretins onto GroEL
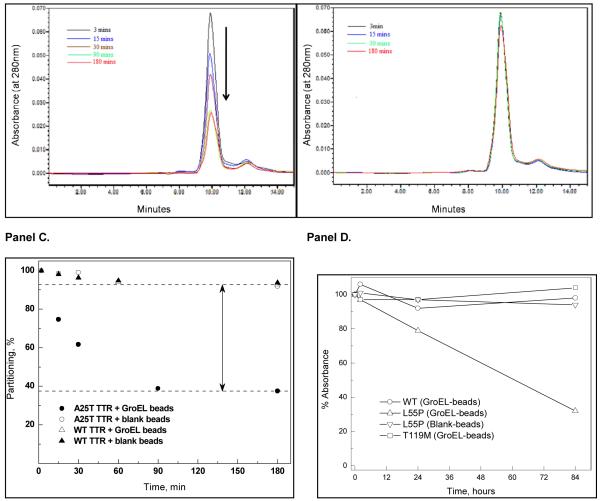
As mentioned previously, specific mutations of the serum protein, transthyretin (TTR) results in its dissociation and eventual amyloid formation leading in the deposition and accumulation of protein fibers, resulting in cardiomyopathies and neuropathies. The natively folded TTR and a stability mutant TTR (T119M) remain as stable tetramers under physiological conditions and these forms are not expected to readily partition onto GroEL. Conversely, due to their intrinsic instabilities, the two amyloidogenic mutant tetramers (L55P and A25T), are predicted to partition onto GroEL.
Transthyretin mutants partition onto immobilized GroEL
We used a series of purified TTR mutants, wild type and an enhanced stabilization mutant which were generously provided by Jeffery Kelly at Scripps. These assays were run either using the ultrafiltration method or by directly examining the loss of soluble TTR when the protein was incubated with immobilized GroEL beads. In the latter experiments, the soluble TTR could be rapidly evaluated after beads were spun down. We essentially demonstrated that the amyloid forming protein A25T transthyretin could rapidly partitions onto immobilized GroEL using either method. More importantly, we found that this partitioning reaction was fairly rapid compared with monitoring dissociation or fibril formation (16).
There are some drawbacks of separating GroEL from partitioned protein using the ultrafiltration procedure. When the protein samples within the filtrate cup were subjected to HPLC SEC analysis, in some instances, GroEL tetramer (802 kDa) was detected even though the MW cutoff was 100 kDa. From the accumulated data, we found that the failure rate of the ultrafiltration membrane exceeded manufacture specifications. Manufacturer specifications were listed at a less than 5% failure rate while we noted that this rate was much higher (~ 20%).
To confirm and improve on the test results from ultrafiltration, we also used an immobilized GroEL sepharose bead system to achieve easy and more rapid separation of the soluble protein fraction from the protein that partitions onto GroEL. This method was found to be superior to using free GroEL. It also lends itself to be more amenable to scale-up and automation of the screening system. The screening can be performed in multi-well plates into which a fixed quantity of beads has been incubated with the partially folded protein. The beads can then be easily separated from the free protein in solution which can be rapidly quantified by spectroscopic methods. Another advantage of using GroEL beads is that they can also be regenerated by treating them with a mixture of ATP and glycerol and reused.
In order to insure adequate mixing, samples were continuously rotated in small vials (37°C). We verified our initial observation that we could detect the misfolding of the TTR mutants particularly for A25T, much more rapidly than previously used mutant TTR oligomer dissociation/fibril formation detection kinetic assays (~ >100 hour at room temperature – (16)). Furthermore, since the chaperonin sink assay functions under physiological conditions, this strengthens its appeal as a general ligand screening assay. Also note that GroEL does not leach from the bead support as evidenced by the single HPLC peak attributed to TTR. The native TTR (right lower kinetic profiles) and a stabilization mutant T119M do not partition onto GroEL under these conditions. Interestingly the TTR missense mutant L55P is not as deleterious to patients as is A25T. The partitioning of this latter mutant was observed using a simple Bradford assay. Curiously under physiological temperatures (37°C), L55P also partitioned onto GroEL beads but at a much slower rate than A25T (Figure 4D). Again, as was observed using the previous ultrafiltration separation technique, the wild type and T119M did not partition on GroEL even during these longer incubation periods.
Figure 4.
Panel A. Using the chaperonin sink assay, a sample partitioning reaction is illustrated using HPLC SEC analysis. The A25T TTR decreases with increasing time during its incubation with GroEL immobilized on Beads (37°C). Panel B. In contrast, this same mutant shows no partitioning onto the bead support alone ). Panel C. The percentage of total protein that is being removed (partitioning) onto the GroEL bead supports is schematically shown for both the wild type TTR and the A25T mutant. Under these GroEL:TTR ratio conditions, the mutant TTR partitions onto GroEL within 90 min. The wild type TTR shows a very low partitioning onto GroEL beads ( ). Furthermore, this low partitioning extent is also observed with either the wild type or A25T protein. Panel D. Under the same conditions, the partitioning of L55P is significantly slower than that observed with the A25T TTR mutant. As expected, the wild type and T119M stability mutants do not show any partitioning onto the GroEL beads during this extended incubation period. Conditions can be adjusted to accelerate the partitioning onto GroEL.
). Furthermore, this low partitioning extent is also observed with either the wild type or A25T protein. Panel D. Under the same conditions, the partitioning of L55P is significantly slower than that observed with the A25T TTR mutant. As expected, the wild type and T119M stability mutants do not show any partitioning onto the GroEL beads during this extended incubation period. Conditions can be adjusted to accelerate the partitioning onto GroEL.
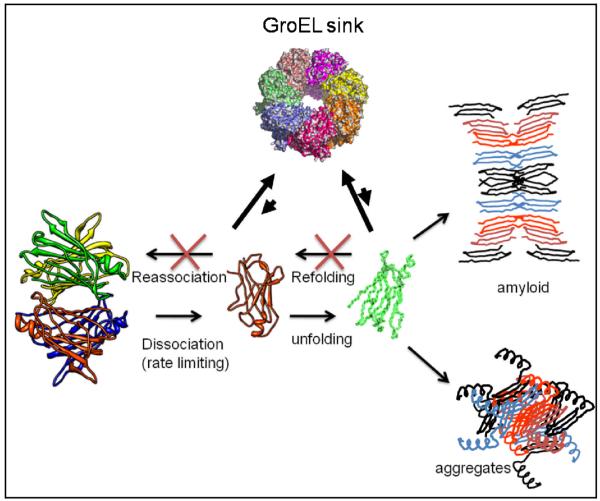
One likely explanation for the accelerated detection of A25T TTR misfolding in the presence of GroEL, particularly for an oligomeric protein, is the inherent ability of GroEL to bind and trap folding intermediates involved in refolding and reassociation reactions. From a detection standpoint, refolding and reassociation reactions compete with the dissociation and unfolding reactions leading to the natural delay in the appearance of measurable downstream aggregates. Since the chaperonin easily binds these folded intermediates, the end result is a kinetic sequestration of partially folded TTR forms (see below). These data also indicate that the partitioning reaction is dependent on the rate of dissociation and misfolding. L55P does not partition onto GroEL as rapidly as does the A25T mutant at 37°C under normal buffering conditions. This slow partitioning suggests that the dissociation/unfolding reactions of the L55P are slower than the A25T mutant. In this instance, the partitioning of L55P can be accelerated by including a mild denaturant such as urea, using a slightly elevated temperature (~ 40°C) or both.
B) β-2-microglobulin partitioning results
β-2-microglobulin is the primary protein involved in dialysis related amyloidosis. Dialysis patients are unable to secrete or degrade this protein. As this protein increases in concentration in the serum, it can cause amyloid plaques to develop in peripheral tissues. In addition, long term hemodialysis patients can develop cardiomyopathies due to amyloid deposition (20) as well as in the musculoskeletal system. Unlike most amyloid forming proteins, this protein does not have to exist as a mutant form in order to accumulate in the body as amyloid fibrils (21).
β-2- microglobulin and GroEL partitioning using ultrafiltration (UF)
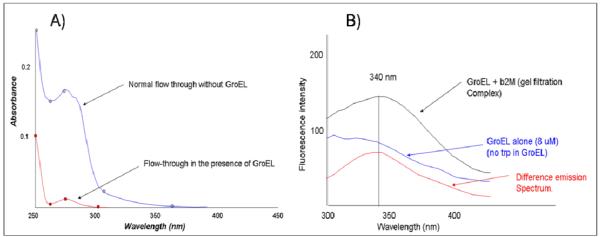
As described above, the partitioning reaction between β-2- microglobulin and soluble GroEL was assessed initially by ultrafiltration and confirmed by SEC analysis. In UF unit (MW 50kDa cutoff), the β-2- microglobulin was assayed alone while in the other unit, a 2:1 GroEL/β-2- microglobulin molar ratio mixture was incubated at 37°C. Over a span of 2 hours, the UF units were spun and the flow through (~200ul) was examined using absorbance spectroscopy and tryptophan fluorescence (Figure 6). Numerous control runs with GroEL alone (MW 802,000 Da) indicates that this large complex does not flow through the 50,000 MW cutoff filter. In the absence of GroEL, β-2- microgloblin readily flows through the ultrafiltration membrane while in the presence of GroEL, the flow through β-2-microgloblin concentration is significantly diminished (Figure 6 A). In both instances, the only protein detected in all of the test sample filtrate was β-2- microgloblin (~11 kDa). Separate SDS PAGE gels indicate that only β-2-microglobulin was observed. No GroEL (60 kDa subunit) could be detected in the analyzed filtrate. The complex between GroEL and β-2-microglobulin was also confirmed using gel filtration chromatography and tryptophan fluorescence. Using an S-300 sephadex fast flow matrix, an uncomplexed larger GroEL molecule elutes in the void volume while free β-2- microglobulin is retained in the matrix and elutes later. Since GroEL does not contain any tryptophans and β-2-microglobulin contains two trp residues, a complex between GroEL and β-2-microglobulin (b2M) was easily detected due to the increase in the tryptophan fluorescence emission spectrum contributed by β-2-microglobulin (excitation at 295 nm) (Figure 6B).
Figure 6.
Initially folded β-2- microglobulin can partition onto the GroEL chaperonin. In Panel A, the difference in a matched set of ultrafiltration experiments illustrate that the microglobulin is retained in the Amicon Centricons (50 kDa cutoff) that contain the chaperonin GroEL. A similar sample subjected to the same ultrafiltration procedure without GroEL is not retained. In Panel B, the fluorescence of a GroEL sample that has been incubated with β-2- microglobulin and isolated via gel filtration (S-300 column) shows a clear increase in tryptophan fluorescence compared with an equivalent amount of GroEL alone isolated in a separate gel filtration run. Since GroEL does not contain tryptophan, the total spectra and the difference spectra reveal the presence of tryptophan fluorescence indicating the contribution from GroEL bound β-2- microglobulin. A similar run with β-2- microglobulin alone did not show any co-migration with the GroEL peak.
C) Partitioning anthrax toxin pore transporter onto GroEL (verified by EM analysis)
The chaperonin/osmolyte system can capture and stabilize the anthrax toxin Pore complex for structural analysis
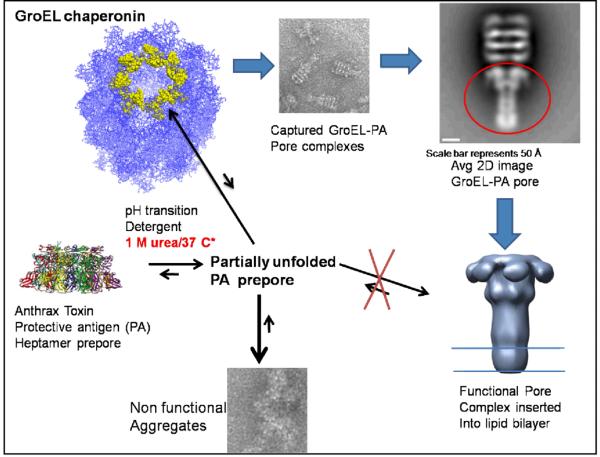
Our most spectacular demonstration of the utility of the chaperonin sink approach involves the capture of a membrane insertable form of the ~ 440 kDa anthrax toxin pore translocation complex (termed the Protective Antigen or PA) using GroEL (22). In the presence of non-denaturing concentrations of urea (~1 M), the refolded membrane insertable state of the protective antigen (PA) was captured and stabilized by GroEL. This captured complex was functional in translocating the lethal factor toxin component across a membrane bilayer. The formation of specific complexes between GroEL and the anthrax toxin pore allowed us to generate a low resolution structure (~23-25 Å) of this membrane protein using negative-stain Electron microscopy and single particle reconstruction.
We have demonstrated that we can capture and stabilize a bacterial toxin protein as it partially unfolds and refolds into a membrane insertable state. These data indicate that our GroEL/1 M urea osmolyte system can capture and stabilize a very large membrane protein complex while preventing its inappropriate aggregation even in the absence of lipids. This work also demonstrates that GroEL can bind extended protein surfaces of large structures so that they can be analyzed using single particle electron microscopy methods. It is conceivable that GroEL/osmolyte combinations can be used as a general tool to 1) increase the yields of correctly folded membrane proteins prior to membrane insertion and 2) more importantly, GroEL can serve as a large structural scaffold to facilitate membrane protein structure determination by electron microscopy. Currently we are exploring the general use of the GroEL chaperonin platform to capture other aggregation prone bacterial toxins as they transition from soluble to membrane insertable toxic states.
D.) Partitioning and Stabilization of the CFTR NBD using the Chaperonin Sink Approach
A majority of the individuals affected by cystic fibrosis carry a mutation, delta F508, in the nucleotide binding domain of the CFTR protein, resulting in the general misfolding of this important protein. The structure of the nucleotide domain region has been solved and it has been noted that this domain is stabilized by ATP (23). Upon removal of ATP, this domain misfolds and aggregates, and some researchers (24,25) have started to use this particular domain or the expressed protein to search for stabilizers of both the native and mutated NBD. The purified CFTR-NBD1 was a gift from the cystic fibrosis foundation and Phil Thomas of UT Southwestern.
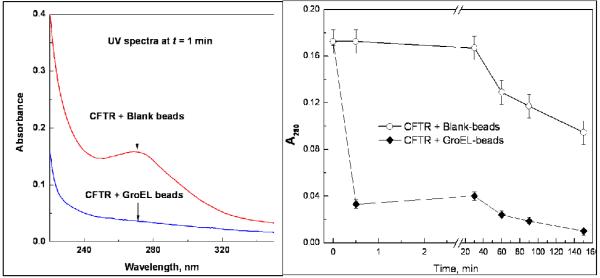
The data presented in Figure 8 indicates that the nucleotide free form of CFTR NBD1 readily partitions onto GroEL. Initial studies were carried out by incubating 1uM CFTR with a proper volume of oligomeric GroEL immobilized on sepharose beads (~ est. 5 uM final oligomer concentration based on labeling efficiency and wet bead volumes) to determine the extent of CFTR NBD partitioning. The concentration of CFTR in the reaction mixture was followed by measuring the UV absorbance of the protein at 280 nm, Bradford analysis, and by an immunodot blot assay. All three assays confirmed the partitioning of CFTR onto the chaperonin and illustrate the ease of using multiple detection schemes to follow the partitioning rates. As indicated in the right hand panel of Figure 8, the partitioning rate is very rapid (within 30sec mixing time at 37°C). As one would predict from a kinetic partitioning reaction, the CFTR-NBD1 partitioning rates decrease as the amount of GroEL present decreases (data not shown).
Figure 8.
After ATP is removed from the CFTR-NBD1, the protein domain was incubated with GroEL beads (~ estimated 5:1 GroEL oligomer to CFTR molar ratio). Within 1 min after mixing with the GroEL beads, CFTR-NDB1 levels decline dramatically. This decline was not observed for initially for CFTR samples that were incubated with blank beads.
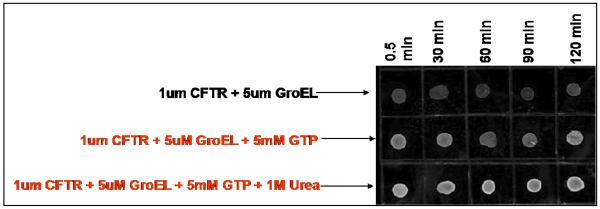
CFTR-NBD1 also binds GTP (26) and this interaction is known to stabilize this protein domain. However unlike ATP, GTP is not a factor in GroEL capture and release (details below). Upon the addition of GTP to the CFTR NBD, partitioning onto GroEL (as assessed by immunodot blot assays) is prevented (Figure 9). This partitioning was also prevented even when a low concentration of denaturant such as 1 M urea was present. These results indicate that GTP is still an excellent stabilizing ligand that prevents the CFTR NBD1 from unfolding even under mild denaturing conditions. As with any potential stabilizing ligand, one must rule out the possibility that the ligand (in this case GTP) inhibits CFTR binding to GroEL. This secondary screening control is necessary to rule out this possible outcome. If GTP were to interfere with the partitioning and capture of protein substrates, one would predict that the partitioning of a common test substrate, such as malate dehydrogenase, onto GroEL will also be affected in the presence of GTP. Malate dehydrogenase is slightly destabilized by low urea concentrations and resulting in facile partitioning onto GroEL (Figure 10 below). The inclusion of GTP into the partitioning reaction mixture does not interfere with capturing malate dehydrogenase as it partially unfolds (data not shown). With malate dehydrogenase as a test substrate, the partitioning rates remain the same regardless of the presence or absence of GTP. This outcome is not surprising since GTP neither binds to GroEL nor does it facilitate release and folding of proteins from GroEL (27).
Figure 9.
A stabilizing ligand prevents CFTR-NBD1 partitioning at 37°C. The addition of the nucleotide GTP inhibits the partitioning of CFTR-NBD1 onto GroEL immobilized on sepharose beads. The GroEL concentration was estimated as oligomer present with respect to the bead wet volume and the volume above the beads. Immunoblots were run at various time intervals to assess the amount of CFTR-NBD1 that remained in solution. The samples containing GTP showed significant amounts of CFTR that remained in solution. A standard immunoblot sample without GroEL present shows similar intensities to the samples above with GTP present. The addition of urea to the bottom panel examined the partitioning of CFTR-NBD1 in the presence of another destabilizing condition. Even with 1 M urea present, GTP still prevents the partitioning of CFTR-NBD1 onto GroEL.
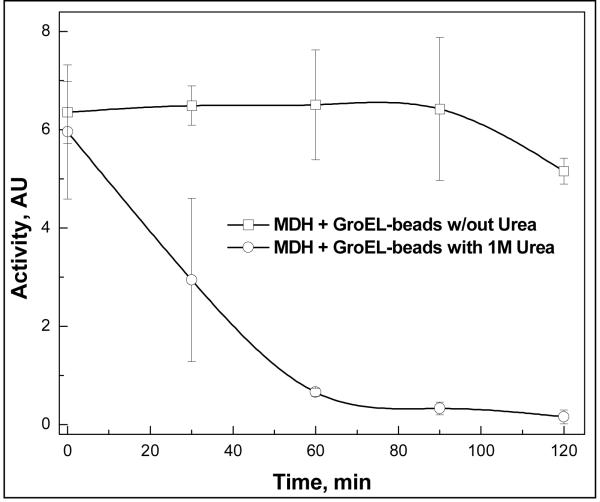
Figure 10.
Partitioning accelerated in the presence of 1 M Urea (37°C). Malate dehydrogenase (MDH) will partition onto GroEL beads if the proteins are incubated in 1 M urea ( ). In contrast, the partitioning reaction is much slower if 1 M urea is omitted from the incubation solution (
). In contrast, the partitioning reaction is much slower if 1 M urea is omitted from the incubation solution ( ).
).
In Figure 9, the blot intensity is a bit higher with GTP and urea present. The slightly lower signal with GTP alone appears to be the due to small loses of CFTR NBD on the sides of the incubating vials. Importantly, urea addition alone to CFTR in the presence of GroEL beads does not prevent its rapid partitioning so urea is not a stabilizer either by dot blot or spectroscopic analysis. In summary, we have demonstrated that the chaperonin platform can be used to screen for stabilizing conditions for the CFTR NBD, opening up the prospect that one could use the chaperonin system platform to screen for other pharmacological small molecule stabilizers of the CFTR NBD.
E.) Use of the chaperonin sink to identify Stabilizing solution conditions during folding or protein storage
During the folding of a Glutathione-S-Transferase (GST) chimera construct of human mitochondrial phosphoenolpyruvate carboxykinase (hmPEPCK) with GroEL, ATP and osmolytes (28-30), we noted that the activity of the PEPCK appears but then slowly disappeared if GroEL remains in contact with the folding protein. An additional bolus of ATP recovers some of this lost activity indicating that the refolded PEPCK was rebinding to the chaperonin. These results support the notion that in this case, the folded protein partitions back onto the GroEL chaperonin because the folded PEPCK is in equilibrium with its misfolded or partially folded form.
Based on these studies, we used GroEL as a thermodynamic and kinetic sink to help identify stabilizing conditions that would stabilize the refolded PEPCK and prevent its repartitioning back onto GroEL. We tested previous stabilization conditions and determined that raising the pH to 8.5 stabilized the refolded PEPCK. (see Figure 6 in 30). In addition to stabilizing PEPCK, we also demonstrated the utility of our chaperonin sink system by identifying stabilizing conditions against thermal denaturation of another protein, malate dehydrogenase (see Figure 8 in 30).
We have recently demonstrated that one of our natively folded test proteins, malate dehydrogenase, readily partitions onto immobilized GroEL when this protein is destabilized with 1 M urea. Here again, adding a stabilizing osmolyte such as sucrose or 0.5 M trehalose prevents this partitioning from occurring, demonstrating that the addition of a folding osmolyte with a destabilizing osmolyte such as 1 M urea results in an enhanced stabilization of the malate dehydrogenase fold and activity. This recapitulates the classic experimental demonstration presented by Bolen and colleague when they unequivocally showed that the stabilizing osmolyte trimethylamine-N-oxide (TMAO) at 1 M enhances the stability of proteins in the presence of low concentrations of urea (31).
F.) Methods for assessing remaining protein concentrations after partitioning
A particular strength of assessing protein stability with the chaperonin is that it is easy to quantitate the decrease in the soluble protein as a function of time. We have demonstrated that a number of facile and readily available techniques can be employed to monitor protein concentration changes. For example, we have used UV-vis spectroscopy, enzyme assays (Figure 8 in 30), protein fluorescence spectroscopy, Bradford analysis and western blotting to assess the remaining protein left in solution during a chaperonin partitioning experiment. Protein quantitation methods using dye based assays and immuno assays are useful in instances where absorbances of small molecule compounds from chemical screens will interfere with traditional spectroscopic measurements. The emphasis and attractive feature of the chaperonin sink assay is that one can chose from a broad spectrum of simple protein quantitation assays and one is not limited to just one detection system. Thus, all proteins involved in assay development can be easily monitored. In all cases presented thus far, multiple assays affirm that the partitioning reaction is easy to monitor.
II) Other General uses of the chaperonin partitioning platform
A) Screening for ligands and conditions with Native Proteins
The chaperonin platform can be used for virtually any protein that undergoes dynamic folding  unfolding transitions and these transition are common to most if not all mesophilic (normal temperature) proteins. This system will work with natively folded proteins (30) as well as specific missense disease proteins. Since this system is so broad based, our data indicates that this chaperonin array platform provides a solid foundation for drug identification and discovery for natively folded proteins. The key aspect that expands the utility of the platform for screening ligands of native proteins rests on the principle that natively folded proteins are always in equilibrium with partially folded species. Thus, one only has to perturb the unfolding equilibrium by subjecting the folded protein to very mild denaturation conditions (e.g. low denaturant concentrations or moderate temperature increase (37 to 45°C)). The former approach is particularly useful for proteins that do not exhibit a significant transient population of partially folded species that can readily interact with the high affinity GroEL chaperonin. To illustrate this point, it has been noted that folded malate dehydrogenase dimers do not readily partition onto GroEL at 37°C. However, since the GroEL oligomer is stable in low denaturant concentrations (in this instance, 1 M urea), simply incubating the high affinity GroEL mixture with active malate dehydrogenase with 1 M urea show a significant and accelerated partitioning of MDH onto GroEL. As was shown previously, this lost activity can be regained once ATP and osmolytes are added to the GroEL platform.
unfolding transitions and these transition are common to most if not all mesophilic (normal temperature) proteins. This system will work with natively folded proteins (30) as well as specific missense disease proteins. Since this system is so broad based, our data indicates that this chaperonin array platform provides a solid foundation for drug identification and discovery for natively folded proteins. The key aspect that expands the utility of the platform for screening ligands of native proteins rests on the principle that natively folded proteins are always in equilibrium with partially folded species. Thus, one only has to perturb the unfolding equilibrium by subjecting the folded protein to very mild denaturation conditions (e.g. low denaturant concentrations or moderate temperature increase (37 to 45°C)). The former approach is particularly useful for proteins that do not exhibit a significant transient population of partially folded species that can readily interact with the high affinity GroEL chaperonin. To illustrate this point, it has been noted that folded malate dehydrogenase dimers do not readily partition onto GroEL at 37°C. However, since the GroEL oligomer is stable in low denaturant concentrations (in this instance, 1 M urea), simply incubating the high affinity GroEL mixture with active malate dehydrogenase with 1 M urea show a significant and accelerated partitioning of MDH onto GroEL. As was shown previously, this lost activity can be regained once ATP and osmolytes are added to the GroEL platform.
The chaperonin platform enables researchers to rapidly test various potential ligand compounds on the target protein of interest. This system is useful as a primary or secondary screening system. In the latter instance, it is of interest to be able to easily test for potential interactions even using a single partitioning measurement. There are a number of advantages that the GroEL partitioning system brings to current ligand screening platforms (i.e. Thermofluor®) that are particularly pertinent. We are developing this system into a generic screening system product that other basic science and pharmaceutical interests will find appealing. In the latter case, the most important testing will come from the discovery and the verification/validation of drug-protein interactions.
Comparision with the current screening systems.
In contrast to dye based probes of thermal stability screens, the chaperonin does not perturb the intrinsic equilibrium between folding and misfolded states.
Non-two state denaturation, undetectable transitions and aggregation reactions present problems for data analysis of the thermal profiles. (chaperonin readily binds both partially folded monomeric or multimeric intermediates as well as aggregates so detection (decrease in detectable soluble protein) remains the same.
Potential Lead compounds are identified at physiological temperatures rather than at elevated temperatures.
Kinetics of misfolding does not depend on a thermal scan rate.
Chaperonin screening system is not as costly particularly since mass production of the chaperonin is facile (J. Seed, EdgeBiosystems).
Although the chaperonin based partitioning system requires a separation step, this is easily accomplished using immobilized GroEL onto solid support surfaces and beads. This construct will facilitate automation of the assay.
Signal is relatively constant for a stabilized protein (no spikes in data collection).
The binding of protein to GroEL is reversible.
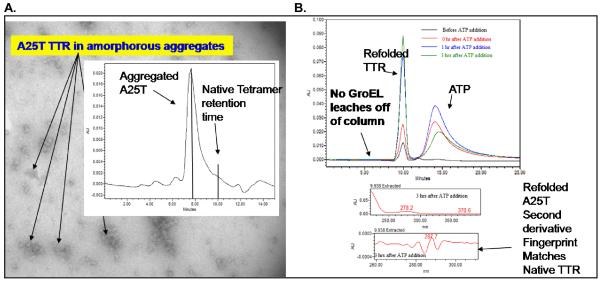
The last point is particularly useful for situations where a protein target needs to be recovered from bound GroEL. As shown below, one rescue an aggregated sample of the A25T mutant transthyretin to a refolding regime using GroEL (immobilized on Beads) ATP and the osmolyte glycerol (4 M). The refolding species in solution was periodically evaluated using SEC HPLC to test for homogeneity. Starting from aggregated A25T, the HPLC gel filtration profile indicates that the A25T mutant regained its proper molecular mass (compare Panel B chromatogram with chromatograms illustrated in panel A with aggregated A25T and wild type TTR). The correct folding of the mutant TTR was confirmed using second derivative UV-visible spectroscopy (see accompanying second derivative fingerprint spectra).
In addition to using the chaperonin system as a general use screening platform, one can also use this system to test potential lead compound under physiological conditions where proteins may have the tendency to interact with chaperone proteins in general. Having the possibility of employing an easy to use independent test for ligand interactions provides an additional assay to test for direct interactions. Secondary screens for stabilization will enhance compatibility issues in the presence of chaperone proteins and provide an additional evaluation of ligand candidates.
B) Role of GroEL Inhibiting or removing preformed protein aggregates in vitro
Does GroEL bind preformed protein aggregates? This is an important question considering the possibility that a protein may aggregate during its incubation with immobilized GroEL. To test this notion, the influence that the high affinity GroEL protein has on two common examples of aggregating protein reactions was examined. Of significance, both these aggregation conditions result in heterogenous populations of misfolded proteins. In one instance, we examined the consequences of adding the high affinity nucleotide free form of GroEL to an ongoing amyloid fibrillation reaction with carboxymethylated alpha lactalbumin. In the other example, we added the high affinity GroEL to a heterogenous mixture of refolded enzyme proteins that contained a substantial amount of misfolded protein to determine if GroEL could selectively bind to misfolded proteins while allowing folded active proteins to remain in solution .
i) Amyloid formation and inhibition by GroEL
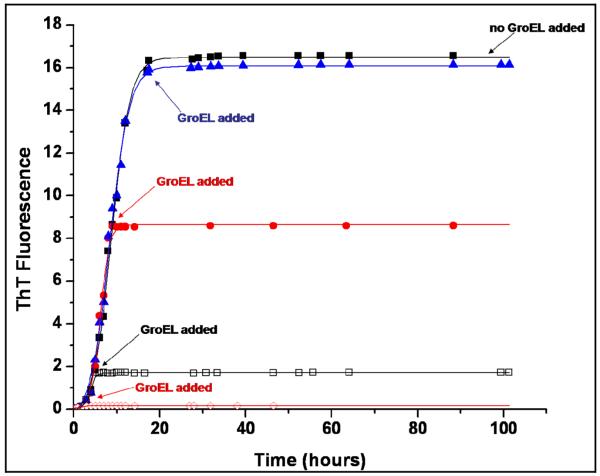
We examined the effect that GroEL would have on different phases of the carboxymethylated single disulfide alpha lactalbumin (1SS-α-LA) fibrillation/aggregation process (32). For these particular experiments, equimolar concentrations of GroEL were added to 1SS-α-LA samples at varying times after initiation of aggregation. The times selected for GroEL addition in this experiment corresponded to addition during the lag (GroEL addition at the 1 hour time point) and growth (GroEL addition at the 6, 10, and 17.5 hour time points) phases of 1SS-α-LA aggregation, as monitored by thioflavin T (ThT) fluorescence, and the resulting 1SS-α-LA aggregation profiles were then graphed as in the previous experiments and qualitatively compared to one another, as shown in Figure 12. It is observed from this data that the addition of GroEL to each sample resulted in a leveling off of the ThT fluorescence signal in that sample, regardless of the time at which GroEL addition occurred. In each of the four samples to which GroEL was added, the ThT signal showed no further increase over the time course of the experiment. Thus, all final ThT signal intensities in the samples containing GroEL remained below that measured for the equilibrium phase of the control sample, which was incubated in the absence of GroEL. Because ThT fluorescence intensity is a measure of fibrils/aggregates formed, this leveling off of the ThT signal indicates that further 1SS-α-LA aggregation in each GroEL-containing sample has been inhibited and thus kept at an amount of fibrils/aggregates that is below the amount formed in the control sample. This experiment shows that 1SS-α-LA aggregation was inhibited in each sample to which GroEL was added, and that this GroEL-induced inhibition of aggregation occurred irrespective of the aggregation phase during which the chaperonin was added. This result suggests that the addition of GroEL to 1SS-α-LA at any time during its aggregation process can halt further fibril/aggregate formation of the substrate protein, indicating that the high affinity chaperonin is effective in controlling 1SS-α-LA fibrillation/aggregation at many stages during the reaction.
Figure 12.
The effect of the GroEL chaperonin on 1SS-α-LA fibril/aggregate formation when added after the fibrillation/aggregation process has been initiated by incubation. Test samples of 1SS-α-LA (1mg/ml) were incubated at 37°C with stirring in sample buffer containing 10mM Mg+2 at pH 7.4. An equimolar concentration of GroEL was added to separate test samples of 1SS-α-LA at 1 hour ( ), 6 hours (□), 10 hours (
), 6 hours (□), 10 hours ( ), and 17.5 hours (
), and 17.5 hours ( ) after initiation of the fibrillation/aggregation process by incubation had begun. A control sample of aggregating 1SS-α-LA (■) was also tested and plotted as a comparison. Fibril/aggregate formation was monitored by an increase in thioflavin T (ThT) fluorescence intensity at 482nm and the symbols represent ThT fluorescence intensities determined experimentally. Error (average standard deviation of the mean) was ± 0.212 for the ThT intensity measurements in this experiment. ThT fluorescence intensity data is in arbitrary units and the lines were fitted to sigmoidal curves using Origin software.
) after initiation of the fibrillation/aggregation process by incubation had begun. A control sample of aggregating 1SS-α-LA (■) was also tested and plotted as a comparison. Fibril/aggregate formation was monitored by an increase in thioflavin T (ThT) fluorescence intensity at 482nm and the symbols represent ThT fluorescence intensities determined experimentally. Error (average standard deviation of the mean) was ± 0.212 for the ThT intensity measurements in this experiment. ThT fluorescence intensity data is in arbitrary units and the lines were fitted to sigmoidal curves using Origin software.
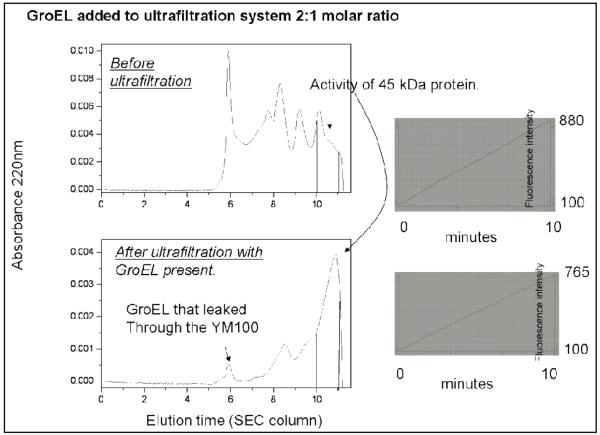
ii) Removing Misfolded Proteins from a Heterogeneous Refolding mixture
To test if the GroEL could be used as a general system to remove aggregated protein from folded conformers, a protease cathespin D containing 3 disulfides in its folded state was refolded in large scale (20 mL total volume) at concentration 5 μg/mL in 0.5 M Arginine for 7 days. A portion of this sample exhibited cathepsin activity. The heterogenous sample was concentrated to a final volume ~7 mL in a centriprep ultrafiltration device with a molecular mass cutoff of 10 kDa. Two aliquots of 200 μL from the resulting solution were mixed with refolding buffer, and one more aliquot of 200 μL was mixed with GroEL (final ratio “cathepsin : GroEL” was 2:1). One of the two samples with just refolding buffer (control) was subjected to HPLC in refolding buffer and collected fractions were assayed for cathepsin-activity (33). The other sample with just refolding buffer alone with the sample containing GroEL was subjected to ultra-filtration in microcon YM-100 and the flow through fractions from both samples were subjected to HPLC as well (Fig. 13). Collected fractions were assayed for APR-activity. Only the protein peak eluting between 10 and 10.5 min showed cathepsin activity. No significant activity was found before 10 min elution times for both samples. The most important result from this particular experiment indicates that the purified, active and most likely properly folded protein can be purified away from the hetereogeneous mixture by allowing the chaperonin system to aid in binding and removing approximately 75% (analyzed by peak areas) of the misfolded protein fractions. Of particular note is that the two prominent protein species that eluted prior to the cathespin activity (~10.5 min) perhaps representing dimer/ tetramer (?) peaks were substantially higher when the sample was simply injected onto the SEC column without treating the sample with GroEL. These comparative samples indicate that GroEL does not bind to the properly folded cathespin to any large extent and that a substantial clean up of the refolded sample could be possible when the refolded sample was incubated with GroEL. Furthermore, this experiment reaffirms the notion that GroEL does not bind to properly folded proteins. It should be noted that the cathespin is activated once the pH of the solution is lowered to pH 4.5. Incubating GroEL with active cathespin samples at pH 7.5 does not result in any observable GroEL cleavage.
Figure 13.
Comparison of gel filtration profiles of a poorly folded cathepsin D fraction before and after a single treatment with soluble GroEL. In this instance, GroEL does not bind to the properly folded cathepsin D and allowed this form to pass through the molecular mass cut off of the ultrafiltration unit. The cutoff removed larger aggregates while the smaller aggregates such as smaller dimers and tetramers (?) were diminished significantly. Control cutoff without GroEL showed substantially higher levels of dimers and tetramers (?) (between 8 and 10 min elution times) present. The cathepsin assays indicate (right hand side) that only a small portion of the folded cathepsin D (10-15%) was lost following the GroEL treatment and passage through the ultrafiltration device.
The advantage of using the GroEL bead for protein clean up procedures is obvious. For example, removing a large amount of misfolded conformers from heterogeneous mixtures has important utility in: 1) protein formulation, 2) target characterization and 3) crystallization efforts. The specificity in this approach comes from the inherent binding ability of the chaperonin for proteins that possess exposed hydrophobic surfaces. This appears to be the case both for individually misfolded proteins and for binding to preexisting aggregates.
Summary
The binding ability of the high affinity form of the chaperonin is a useful property that can be exploited to screen for potential ligand stabilizers of both mutant and natively folded proteins. We have demonstrated that this system can accelerate the time of detection due to its ability to facilitate kinetic partitioning of species that are only present in small quantities due to preexisting transient equilibria. This key property of the GroEL protein is extremely useful from a biotechnology standpoint. Furthermore, the ease at which one can reverse this binding using ATP and folding osmolytes (28-30) strengthens its utility as a recyclable and reusable platform system.
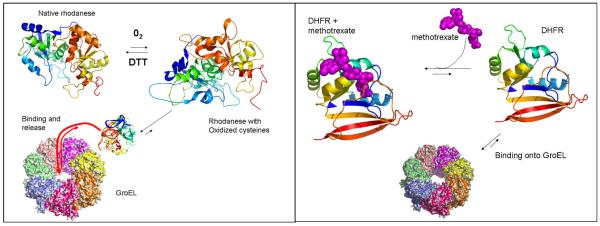
Figure 1.
Right panel The protein rhodanese partitions onto the chaperonin after oxidation of a number of critical cysteine residues. In the presence of DTT, this oxidation was reversed. However, in the presence of the high affinity chaperonin (no nucleotide), rhodanese partitioned onto the GroEL chaperonin. This partitioning was prevented once oxygen was removed from the system (17). Left panel, Lorimer and colleagues demonstrated that monomeric dihydrofolate reductase could partition onto GroEL from its native folded state and that this partitioning was prevented by providing a specific ligand for DHFR (9).
Figure 2.
The initial method for examining partitioning onto GroEL using the centricon ultrafiltration. The amount of the protein that passed through the filtration membrane was assessed by Bradford analysis, fluorescence and protein absorbance. Partitioning experiments were run in triplicate and protein concentrations showed a SD of approximately 5-10%. The two TTR mutants showed some significant partitioning onto the GroEL chaperonin and retention. In contrast, the wild type and stability mutant TTR isoforms readily passed through the ultrafiltration membrane.
Figure 3.
Our many test examples have shown that proper mixing with GroEL beads results in a partitioning of the target protein of interest. In the presence of a stabilizer, partitioning is prevented due to ligand binding and subsequent stabilization of the target protein (lower well). If stabilizers are absent (no binding), the target protein partitions onto immobilized GroEL beads and is removed from the sample solution (upper right well).
Figure 5.
Simplified kinetic scheme depicting the inhibition of reassocation and refolding reactions by the presence of the nucleotide free form of the GroEL chaperonin protein (GroEL sink). The actual partitioning reaction is definitely more complicated due to the presence of multiple forms (multiple landscape model) of folded, dissociated and unfolded forms of TTR. The reactions leading to fibril formation or general aggregation also illustrate the multiplicity of reactions that are present during misfolding reactions.
Figure 7. This montage shows the successful chaperonin capture and structural analysis of the anthrax toxin pore complex I while bound to the GroEL scaffold).
In the presence of the chaperonin sink, general aggregation of the anthrax toxin pore (protective antigen) is avoided, the anthrax toxin pore forms a complex with GroEL (visualized via Negative stain EM) and this functional toxin pore complex can be released from GroEL and insert into membrane bilayers (See more details in 22).
Figure 11.
Demonstration of Homogeneous folding of A25T TTR captured on GroEL beads: In Panel A, an A25T TTR sample aggregated upon storage (EM micrograph). The accompanying SEC HPLC chromatogram indicates degree of heterogeneity and lack of native tetramer species in the aggregated sample. The aggregated were disrupted and allowed to bind onto GroEL immobilized on a sepharose bead support system (GE Healthcare). The A25T TTR was released and refolded back into its tetrameric form as evidenced by the recovery of matching HPLC SEC retention times and second derivative UV-visible spectroscopy with wild type TTR tetramers. The refolded species was homogeneous. The refolding yield for this particular experiment was ~ 70%.
Acknowledgments
This research was supported in part by NSF MCB-0445936 (MTF), NIH R41 GM080074 (MTF), and a grant from the Kansas Technology Enterprise Corporation KTEC (MTF). IH was supported by the NSF grant. JH was specifically supported by a KUMC Medical Research Grant. ND and BK were supported by the KTEC and NIH grants. AAK was an undergraduate student supported by the NSF grant.
References
- 1.Van Dyk TK, Gatenby AA, LaRossa RA. Nature. 1989;342:451–453. doi: 10.1038/342451a0. [DOI] [PubMed] [Google Scholar]
- 2.Rutherford SL, Lindquist S. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 3.Queitsch C, Sangster TA, Lindquist S. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 4.Tokuriki N, Tawfik DS. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 5.Benkovic SJ, Hammes GG, Hammes-Schiffer S. Biochemistry. 2008;47:3317–3321. doi: 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]
- 6.Taldone T, Weilin S, Chiosis G. Bioorg. Med. Chem. 2009;17:2225–2235. doi: 10.1016/j.bmc.2008.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitesell L, Lindquist SL. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 8.Lou W, Dou F, Rodina A, Chip S, Kim J, Zhao Q, Moulick K, Aguirre J, Wu N, Greengard P, Chiosis G. Proc. Natl. Acad. Sci. USA. 2007;104:9511–9516. doi: 10.1073/pnas.0701055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viitanen PV, Donaldson GK, Lorimer GH, Lubben TH, Gatenby AA. Biochemistry. 1991;30:9716–9723. doi: 10.1021/bi00104a021. [DOI] [PubMed] [Google Scholar]
- 10.Martin J, Horwich AL, Hartl FU. Science. 1992;258:995–998. doi: 10.1126/science.1359644. [DOI] [PubMed] [Google Scholar]
- 11.Llorca O, Galán A, Carrascosa JL, Muga A, Valpuesta JM. J Biol Chem. 1998;273:32587–32594. doi: 10.1074/jbc.273.49.32587. [DOI] [PubMed] [Google Scholar]
- 12.Hammarstrom P, Wiseman RL, Powers ET, Kelly JW. Science. 2003;299:713–716. doi: 10.1126/science.1079589. [DOI] [PubMed] [Google Scholar]
- 13.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Ann. Rev. Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 14.Klabunde T, Petrassi HM, Oza VB, Raman P, Kelly JW, Sacchettini JC. Nat Struct Biol. 2000;7:312–321. doi: 10.1038/74082. [DOI] [PubMed] [Google Scholar]
- 15.Ray SS, Nowak RJ, Brown RH, Jr, Lansbury PT. Proc Natl Acad Sci U S A. 2005;102:3639–3644. doi: 10.1073/pnas.0408277102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson SM, Wiseman RL, Sekijima Y, Green NS, Adamski-Werner SL, Kelly JW. Acc. Chem. Res. 2005;38:911–921. doi: 10.1021/ar020073i. Review. [DOI] [PubMed] [Google Scholar]
- 17.Smith KE, Voziyan PA, Fisher MT. J. Biol. Chem. 1998;273:28677–28681. doi: 10.1074/jbc.273.44.28677. [DOI] [PubMed] [Google Scholar]
- 18.Walter S, Lorimer GH, Schmid FX. Proc Natl. Acad. Sci. U S A. 1996;93:9425–9430. doi: 10.1073/pnas.93.18.9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith KE, Fisher MT. J. Biol. Chem. 1995;270:21517–21523. doi: 10.1074/jbc.270.37.21517. [DOI] [PubMed] [Google Scholar]
- 20.Takayama F, Miyazaki S, Morita T, Hirasawa Y, Niwa T. Kidney Int Suppl. 2001;78:S172–6. doi: 10.1046/j.1523-1755.2001.59780172.x. [DOI] [PubMed] [Google Scholar]
- 21.Jahn TR, Parker MJ, Homans SW, Radford SE. Nat. Struct. Mol. Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 22.Katayama H, Janowiak BE, Brzozowski M, Jurcyk J, Falke S, Gogol EP, Collier RJ, Fisher MT. Nat. Struc. Mol. Biol. 2008;15:754–760. doi: 10.1038/nsmb.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis HA, Zhao X, Wang C, Sauder JM, Rooney I, Noland BW, Lorimer D, Kearins MC, Conners K, Condon B, Maloney PC, Guggino WB, Hunt JF, Emtage S. J Biol Chem. 2005;280:1346–1353. doi: 10.1074/jbc.M410968200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang XM, Wang XT, Yue H, Leung SW, Thibodeau PH, Thomas PJ, Guggino SE. J Biol Chem. 2003;278:51232–51242. doi: 10.1074/jbc.M309076200. [DOI] [PubMed] [Google Scholar]
- 25.Loo TW, Bartlett MC, Clarke DM. J Bioenerg Biomembr. 2005;37:501–507. doi: 10.1007/s10863-005-9499-3. [DOI] [PubMed] [Google Scholar]
- 26.Travis SM, Carson MR, Ries DR, Welsh MJ. J Biol Chem. 1993;268:15336–15339. [PubMed] [Google Scholar]
- 27.Horwich AL, Fenton WA. Prot. Sci. 1997;6:743–60. doi: 10.1002/pro.5560060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voziyan PA, Jadhav L, Fisher MT. J. Pharm. Sci. 2000;89:1036–1045. doi: 10.1002/1520-6017(200008)89:8<1036::aid-jps8>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 29.Voziyan PA, Johnston M, Chao A, Bomhoff G, Fisher MT. J. Struct. Funct. Genomics. 2005;31:1–6. doi: 10.1007/s10969-005-2646-6. [DOI] [PubMed] [Google Scholar]
- 30.Katayama H, McGill M, Kearns A, Brzozowski M, Degner N, Harnett B, Kornilayev B, Matkovic-Calogovic D, Holyoak T, Calvet JP, Gogol EP, Seed J, Fisher MT. J Struct Funct Genomics. 2009;10:55–65. doi: 10.1007/s10969-008-9053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang A, Bolen DW. Biochemistry. 1997;36:9101–9108. doi: 10.1021/bi970247h. [DOI] [PubMed] [Google Scholar]
- 32.Bomhoff G, Sloan K, Mclean C, Gogol EP, Fisher MT. Arch. Biochem Biophys. 2006;453:75–86. doi: 10.1016/j.abb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Yasuda Y, Kageyama T, Akamine A, Shibata M, Kominami E, Uchiyama Y, Yamamoto K. J. Biochem. 1999;125:1137–1143. doi: 10.1093/oxfordjournals.jbchem.a022396. [DOI] [PubMed] [Google Scholar]