Abstract
Single-molecule studies of biological macromolecules can benefit from new experimental platforms that facilitate experimental design and data acquisition. Here we develop new strategies to construct curtains of DNA in which the molecules are aligned with respect to one another and maintained in an extended configuration by anchoring both ends of the DNA to the surface of a microfluidic sample chamber that is otherwise coated with an inert lipid bilayer. This “double-tethered” DNA substrate configuration is established through the use of nanofabricated rack patterns comprised of two distinct functional elements: linear barriers to lipid diffusion that align DNA molecules anchored by one end to the bilayer and antibody-coated pentagons that provide immobile anchor points for the opposite ends of the DNA. These devices enable the alignment and anchoring of thousands of individual DNA molecules, which can then be visualized using total internal reflection fluorescence microscopy under conditions that do not require continuous application of buffer flow to stretch the DNA. This unique strategy offers the potential for studying protein−DNA interactions on large DNA substrates without compromising measurements through application of hydrodynamic force. We provide a proof-of-principle demonstration that double-tethered DNA curtains made with nanofabricated rack patterns can be used in a one-dimensional diffusion assay that monitors the motion of quantum dot-tagged proteins along DNA.
Introduction
Dynamic interactions between proteins and DNA underlie many biological processes and, as such, are the subject of intense investigation. Numerous laboratories are now tackling these problems using new optical microscopy-based methods that enable the direct visualization of DNA molecules or protein−DNA complexes at the single-molecule level in real time, and the information garnered from these experiments is being used to build detailed mechanistic models of many different types of reactions.1−4 Additionally, micro- and nanoscale devices, in combination with optical-based detection, are also becoming increasingly powerful tools for the manipulation and analysis of individual DNA molecules.5−7 One problem with many single-molecule techniques is that they are inherently designed to probe individual reactions, and as a consequence, it can be challenging to gather statistically relevant data. This difficulty is often compounded by the fact that these experiments are technically demanding. Therefore, it is advantageous to establish new experimental platforms that can increase throughput capacity of single-molecule imaging while at the same time making these approaches both easier and more readily applicable to biological reactions involving different types of DNA transactions.
In an effort to help make single-molecule techniques more accessible, we have sought to develop novel methods enabling high-throughput single-molecule imaging by integrating nanoscale engineering, microfluidics, and lipid bilayer-coated surfaces with single-molecule optical microscopy.8−10 A key aspect of these new experimental platforms is that they utilize inert lipid bilayers to passivate the fused silica surface of a microfluidic sample chamber. Artificial lipid membranes deposited on solid supports have proven to be useful for many types of biochemical studies.11−13 The chemical characteristics of the bilayer can be controlled through careful selection of the constituent lipids;11,14 they can be partitioned with chemical or mechanical barriers,15−17 and the distributions of molecules anchored to lipids can be manipulated using photochemical modulation, electrical fields, or hydrodynamic force.17−20 On the basis of these properties, we have demonstrated that artificial bilayers can be used in combination with manually etched microscale barriers to lipid diffusion to align hundreds of lipid-tethered DNA molecules; we refer to these aligned molecules as DNA curtains.10,21−24 Alignment of the lipid-tethered DNA is achieved by using hydrodynamic force to push the molecules into the leading edge of the diffusion barriers. More recently, we have employed electron beam (ebeam) lithography to fabricate diffusion barriers with nanoscale dimensions, which allows for much more precise control over both the location and lateral distribution of the DNA molecules within the curtains.8,9 These nanofabricated DNA curtains permit simultaneous visualization of thousands of individual DNA molecules that are perfectly aligned with respect to one another and offer the potential for massively parallel data acquisition from thousands of individual protein−DNA complexes in real time using a robust experimental platform amenable to a wide variety of biological applications. We have also shown that these DNA curtains are advantageous for studying protein−DNA interactions at the single-molecule level, and we have begun to apply these tools to biological systems such as nucleosomes and chromatin remodeling, homologous DNA recombination, and postreplicative mismatch repair.21−25
One drawback of our previous DNA curtain designs is that they require continuous application of a hydrodynamic force during data collection. This is because just one end of the DNA is anchored to the lipid bilayer. If buffer flow is terminated, the “single-tethered” DNA does not remain stretched, and as a consequence, it cannot be visualized along its full contour length because it drifts outside of the detection volume defined by the penetration depth of the evanescent field. The need for buffer flow is not problematic for many types of measurements; however, the hydrodynamic force exerted by the flowing buffer can potentially impact the behavior of proteins or protein complexes, and the magnitude of this impact is expected to scale in proportion to the hydrodynamic radius of the molecules under observation.26,27 The influence of buffer flow is especially apparent during measurements involving proteins that slide on DNA by one-dimensional diffusion, because an applied flow force can strongly bias the direction in which the proteins travel along the DNA.(26)
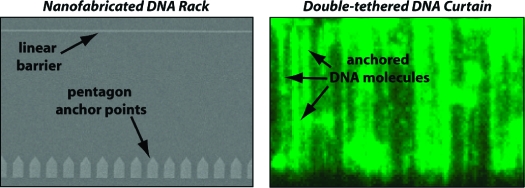
Here we have developed nanofabricated surface patterns for making curtains of aligned DNA molecules anchored by both ends to the surface of a microfluidic sample chamber. These patterns are termed DNA racks and are comprised of linear barriers to lipid diffusion, similar to our previous designs,8,9 along with arrays of metallic pentagons, which are coated with antibodies directed against a small molecule tag present at one end of the DNA. The lipid-tethered DNA molecules are first aligned along the linear barriers. The pentagons then serve as solid anchor points positioned at a defined distance downstream from the linear barriers. Once aligned at the linear barriers and anchored to the pentagons, the “double-tethered” DNA molecules are maintained in an extended state suspended above an inert lipid bilayer and remain confined within the detection volume defined by the penetration depth of the evanescent field. This anchoring strategy circumvents the requirement for continuous buffer flow and allows observation of long DNA molecules along their full contour length, even in the absence of an applied hydrodynamic force.
Materials and Methods
Electron Beam Lithography
Fused silica slides (G. Finkenbeiner, Inc.) were cleaned in NanoStrip solution (CyanTek Corp., Fremont, CA) for 20 min, then rinsed with acetone and 2-propanol, and dried with N2. The slides were spin-coated with a bilayer of polymethylmethacrylate (PMMA) [molecular weight of 25K, 3% in anisole, and molecular weight of 495K, 1.5% in anisole (MicroChem, Newton, MA)], followed by a layer of Aquasave conducting polymer (Mitsubishi Rayon). Each layer was spun at 4000 rpm for 45 s using a ramp rate of 300 rpm/s. Polygon patterns and linear barriers were written by Ebeam lithography using an FEI Sirion scanning electron microscope equipped with a pattern generator and lithography control system (J. C. Nabity, Inc., Bozeman, MT). After patterning, Aquasave was rinsed off with deionized water. Resist was developed using a 3:1 solution of 2-propanol and methyl isobutyl ketone (MIBK) for 1 min with ultrasonic agitation at 5 °C. The substrate was then rinsed in 2-propanol and dried with N2. A 15−20 nm layer of gold atop a 3−5 nm adhesion layer of either chromium (Cr) or titanium (Ti) was deposited using a Semicore electron beam evaporator. Alternatively, a 15−20 nm layer of Cr was deposited directly onto the fused silica without need for an adhesion layer. Liftoff was effected at 80 °C at a 9:1 methylene chloride:acetone ratio. Alternatively, barriers were made out of just a 15−20 nm layer of chromium, as previously described.8,9 Following liftoff, samples were rinsed with acetone to remove stray chromium flakes and dried with N2. Barriers were imaged using a Hitachi 4700 scanning electron microscope and a PSIA XE-100 scanning probe microscope in noncontact mode. Optical images of the barriers were taken with a Nikon Eclipse ME600 camera.
Nanoimprint Lithography
Nanoimprint masters were fabricated using electron beam lithography, liftoff, and inductively coupled plasma etching. Briefly, a bilayer of polymethylmethacrylate (PMMA, 25K and 495K) was spun onto a silicon wafer with a thin coating of silicon dioxide. Patterns were written by an FEI Sirion SEM outfitted with a Nabity Nanopattern Generation System and then developed in a 2-propanol/methyl isobutyl ketone mixture (3:1) at 5 °C in a bath sonicator. Samples were then rinsed with 2-propanol and dried with N2. A Semicore Ebeam evaporator was used to evaporate 20 nm Cr onto the masters. Liftoff was performed in acetone at 80 °C. The patterned masters were then plasma-etched to a depth of 100 nm in a mixture of C4F8 and O2 (9:1) for 90 s at a power of 300 W using an Oxford ICP etch tool. Nanoimprint masters were then coated with a fluorinated self-assembled monolayer (Nanonex, Princeton, NJ) to prevent adhesion between the master and resist.
To make nanoimprinted barriers, PMMA 35K (Microresist Technologies) was spin-coated on a fused silica microscope slide and baked on a hot plate for 5 min at 180 °C. Nanoimprint was performed in two stages: a 2 min preimprint phase with a pressure of 120 psi and pretemperature of 120 °C followed by a 5 min imprint phase with a pressure of 480 psi and temperature of 190 °C. This heated the PMMA well above its glass transition temperature and allowed it to conform to the mold. After imprinting had been conducted, a descum process was undertaken to remove ∼10 nm of residual PMMA. Descum was done in an inductively coupled plasma under CHF3 and O2 (1:1) and a power of 200 W for 40 s total (two iterations of 20 s). After descum, 15−20 nm of Cr was evaporated on the samples and liftoff was performed in a 9:1 mixture of methylene chloride and acetone at 65 °C for several hours, followed by bath sonication to remove stray metal flakes. Finally, nanopatterned slides were rinsed in acetone and dried with N2.
Lipid Bilayers and DNA Curtains
The flow cells were assembled from the nanopatterned fused silica slides. Inlet and outlet ports were made by boring through the slide with a high-speed precision drill press equipped with a diamond-tipped bit [1.4 mm outside diameter (Kassoy)]. The slides were cleaned by successive immersion in 2% (v/v) Hellmanex, 1 M NaOH, and 100% MeOH. The slides were rinsed with filtered sterile water between each wash and stored in 100% MeOH until they were used. Prior to assembly, the slides were dried under a stream of nitrogen and baked in a vacuum oven for at least 1 h. A sample chamber was prepared from a borosilicate glass coverslip (Fisher Scientific) and double-sided tape (∼25 μm thick, 3M). Inlet and outlet ports (Upchurch Scientific) were attached with hot-melt adhesive (SureBonder glue sticks, FPC Corp.). The total volume of the sample chambers was ∼4 μL. A syringe pump (Kd Scientific) and actuated injection valves (Upchurch Scientific) were used to control sample delivery, buffer selection, and flow rate. The flow cell and prism were mounted in a custom-built heater with computer-controlled feedback regulation to control the temperature of the sample between 25 and 37 °C (±0.1 °C), as necessary. After each use, the slides were soaked in MeOH to remove the ports and tape, rinsed with water, washed briefly (15−20 min) with Nanostrip, and rinsed with water. This procedure was sufficient to clean the slide surfaces for reuse, and each slide could be used multiple times without degrading the quality of the optical surface or the metallic patterns.
DNA curtains were constructed as described previously,8,9 with the exception of additional steps necessary for anchoring the second end of the DNA. All lipids were purchased from Avanti Polar Lipids, and liposomes were prepared as previously described. In brief, a mixture of DOPC (1,2-dioleoyl-sn-glycerophosphocholine), 0.5% biotinylated DPPE [1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl)], and 8−10% mPEG 550-PE {1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-550]}. The mPEG does not affect bilayer formation or assembly of the DNA curtains but rather serves to further passivate the surface against nonspecific adsorption of quantum dots (which we use in our studies of protein−DNA interactions). Liposomes were applied to the sample chamber in three injections of 200 μL followed by 5 min incubations. Excess liposomes were flushed away with 1 mL of buffer A, which contained 10 mM Tris-HCl (pH 7.8) and 100 mM NaCl, and the bilayer was incubated for an additional 30 min. Buffer A with 25 μg/mL anti-DIG Fab (Roche), anti-FITC (Invitrogen), or anti-BrdU IgG (Sigma) was then injected into the sample chamber and incubated for 30 min. The sample chamber was then flushed with 1 mL of buffer B, which contained 40 mM Tris-HCl (pH 7.8), 1 mM DTT, 1 mM MgCl2, and 0.2 mg/mL BSA, and incubated for an additional 5 min. One milliliter of buffer A containing neutravidin (330 nM) was then injected into the sample chamber and incubated for 20 min. The flow cell was then rinsed with 3 mL of buffer B to remove any unbound neutravidin. One milliliter of λ DNA (20 pM) labeled at one end with biotin and at the other end with either digoxigenin, FITC, or bromodeoxyuridine (as indicated) and prestained with 1−2 nM YOYO1 was injected into the sample chamber in five 200 μL aliquots, with a 2−3 min incubation period following each injection. The DNA was then aligned at the linear barriers using a flow rate of 0.02 mL/min, and this rate was then increased to 2−3 mL/min to anchor the second end of the DNA molecules.
DNA Substrates
The DNA substrates were made by ligating oligonucleotides to the 12-nucleotide overhangs at the end of the λ phage genome (48.5 kb). Ligation mixes (total volume of 1 mL) contained 4 nM λ DNA (Invitrogen), 1 μM biotinylated oligonucleotiode (5′-pAGG TCG CCG CCC [FITC]-3′ or 5′-pAGG TCG CCG CCC [DIG]-3′), 1 μM FITC-labeled oligonulceotide (or DIG-labeled oligonucleotide; 5′-pGGG CGG CGA CCT [BioTEG]-3′), and 1× ligase buffer (NEB). The reaction mix was warmed to 65 °C for 10 min and then cooled slowly to room temperature. After the mixture had cooled, ligase was added [T4 DNA ligase (400 units/μL) or Taq ligase (40 units/μL), NEB], and the mixture was incubated overnight at 42 °C. Reaction mixtures that included T4 ligase were then heat-inactivated at 65 °C for 10 min, and the ligated DNA products were purified over a Sephacryl S200HR column (GE Healthcare) run in 10 mM Tris-HCl (pH 7.8), 1 mM EDTA, and 150 mM NaCl. The purified DNA was stored at −20 °C.
For insertion of a DIG-labeled oligonucleotide complementary to a position 14711 bp from the biotinylated end of the λ DNA, 500 μL of λ DNA [200 pM, labeled at the ends with biotin and FITC (as described above)] was incubated for 2 h with the nicking enzyme Nt.BstNBI (50 units, NEB) at 55 °C in a buffer containing 10 mM Tris-HCl (pH 7.8), 10 mM MgCl2, and 150 mM NaCl. The enzyme was heat-denatured by incubation at 80 °C for 20 min. One millimolar oligonucleotide (5′-pTTC AGA GTC TGA CTT TT[DIG]-3′) was added to the solution, and the mixture was incubated at 55 °C for 20 min and then cooled slowly to room temperature over the course of 1 h. ATP was then added to a final concentration of 1 mM along with 2000 units of T4 ligase, and the reaction mixture was incubated at room temperature for 90 min. The ligase was then heat-denatured by incubation at 65 °C for 20 min.
TIRFM and One-Dimensional Diffusion Assays
The basic design of the microscope used in this study has been previously described.(10) In brief, the system is built around a Nikon TE2000U inverted microscope with a custom-made illumination system. For this study, a 488 nm, 200 mW diode-pumped solid-state laser (Coherent, Sapphire-CDHR) was used as the excitation source. The laser was attentuated as necessary with a neutral density filter and centered over the DNA curtain by means of a remotely operated mirror (New Focus). The beam intensity at the face of the prism was typically ∼10−15 mW. TIRFM images were collected using a 60× water immersion objective lens (Nikon, 1.2 NA Plan Apo) and a back-illuminated EMCCD detector (Photometrics, Cascade 512B). Anti-Flag-labeled quantum dots (QDs, Invitrogen) were prepared as described previously.(21) Mlh1 (250 nM) and anti-FLAG QDs (500 nM) were incubated at a 1:2 protein:QD ratio in 20 mM Tris (pH 7.8) and 100 mM NaCl for more than 30 min on ice. The QD-tagged Mlh1 was diluted to a final concentration of 10 nM in buffer containing 40 mM Tris (pH 7.8), 1 mM DTT, 1 mM ATP, 50 mM NaCl, 1 mM MgCl2, and 0.4 mg/mL BSA and then injected into a sample chamber containing preassembled double-tethered DNA curtains. Diffusion coefficients were calculated as previously described.(21)
Results
Design Elements and Nanofabrication of the DNA Rack
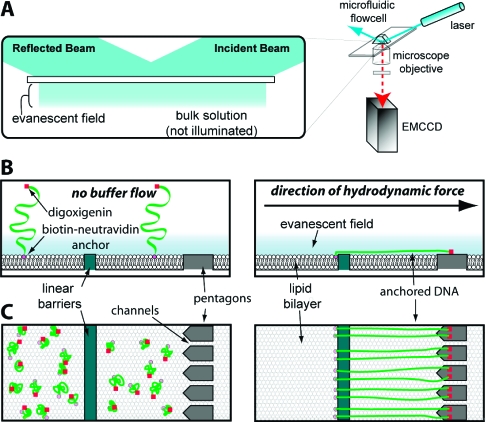
Here we expand on our previous work and demonstrate the development of new nanofabricated barrier patterns, termed DNA “racks”, which can be used to make DNA curtains where both ends of the DNA molecules are anchored to the flow cell surface. The rack patterns utilize a combination of two distinct functional elements, and an overview of the general design is presented in Figure 1A,C. In principle, one end of the DNA is first anchored via a biotin−neutravidin interaction to a supported lipid bilayer coating the surface of the fused silica sample chamber (Figure 1B,C), as previously described.8,9 In the absence of a hydrodynamic force, the molecules are randomly distributed on the surface and lie primarily outside of the detection volume defined by the penetration depth of the evanescent field (∼150−200 nm). Application of buffer flow pushes the DNA through the sample chamber while the biotinylated DNA ends remain anchored within the mobile bilayer. The first pattern elements are linear barriers to lipid diffusion,15−17 which are oriented perpendicular to the direction of buffer flow at strategic locations in the path of the DNA (Figure 1B,C); these linear barriers are designed to halt the forward movement of the lipid-tethered DNA molecules through the sample chamber, causing them to accumulate at the leading edge of the barriers where they then extend parallel to the surface.8,9 The second elements of the pattern are a series of arrayed pentagons positioned at a defined distance behind the linear barriers. The distance between the linear barriers and the pentagons is optimized for the length of the DNA to be used for the experiments (Figure 1B,C, and see below). The channels between the adjacent pentagons are intended to minimize accumulation of lipid-tethered DNA molecules between the linear barriers and the pentagon arrays. This design feature takes advantage of our previous observation that geometric barrier patterns can be used to direct the movement of DNA by making use of barrier edges that are not perpendicular to the direction of buffer flow.(9) Any DNA molecules anchored to the bilayer between the two barrier elements are expected to slide off the angled edges of the pentagons and should be funneled through the channels. The pentagons are also designed to present a large, exposed surface that can be nonspecifically coated with antibodies directed against small molecule haptens [either digoxigenin (DIG), fluorescein isothiocyanate (FITC), or bromodeoxyuridine (BrdU) (see below)], which are covalently linked to the ends of the DNA opposite the ends bearing the biotin tag (Figure 1B). When the DNA molecules are aligned along the linear barriers and stretched perpendicular to the surface, the hapten-tagged DNA ends should bind the antibody-coated pentagons (Figure 1C). In this scenario, the DNA molecules should remain stretched parallel to the surface even when no buffer is being pushed through the sample chamber (Figure 1C).
Figure 1.
Schematic of DNA rack design. Panel A shows a diagram of the total internal reflection fluorescence microscope (TIRFM) used to image single molecules of DNA. For imaging with the TIRFM, the long DNA molecules (48 kb) used in these studies must be extended parallel to the surface of the sample chamber to remain confined within the evanescent field. Panels B and C depict a cartoon illustration of the bilayer on the surface of a fused silica slide, and a single barrier set comprised of a linear barrier and a series of aligned pentagons separated by nanochannels. Also depicted is the response of tethered DNA molecules to the application of a hydrodynamic force. The magenta circles are the biotinylated ends, and the red squares are the hapten (digoxigenin, FITC, or BrdU)-labeled ends of the DNA. The top and bottom parts of panels B and C depict views from the side and above, respectively. In the absence of buffer flow, the DNA molecules are tethered to the surface but are not confined within the evanescent field, nor are they aligned at the barrier. As depicted in panel C, when flow is applied, the DNA molecules are dragged through the bilayer until they encounter the linear diffusion barrier, at which point they will align with respect to one another and the DIG-labeled ends become anchored to the antibody-coated pentagons. DNA located between the linear barriers and the pentagons passes through the nanochannels and goes to the next available linear barrier in the pattern.
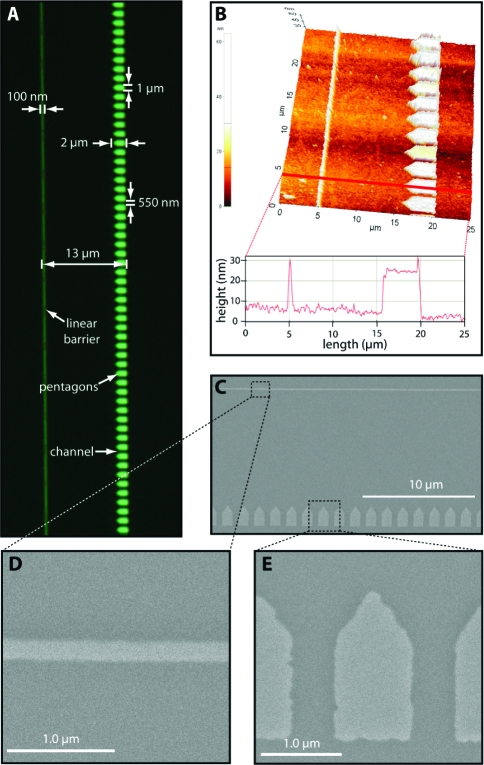
To achieve these desired design features, chromium (Cr) or gold (Au) barrier patterns were made on a 1 in. × 3 in. fused silica slide glass by either ebeam lithography as previously described8,9 or nanoimprint lithography28,29 (see Materials and Methods). For most of the work described below, each slide contained 16 total rack patterns, arranged in a 4 × 4 array at the center of the slide. Each rack pattern was 260 μm in length with a distance of 13 μm between the linear barriers and the back of pentagons. The center-to-center distance between each of the rack patterns within the 4 × 4 array was 500 μm in the x-direction (parallel to the direction of buffer flow) and 370 μm in the y-direction (perpendicular to the direction of buffer flow). The total area encompassed by the 4 × 4 array of rack patterns was 1513 μm × 1370 μm (2.073 mm2). Figure 2 shows the characterization of a typical DNA rack pattern made by ebeam lithography. Figure 2A shows an optical image of the overall pattern design; Figure 2B shows measurements of these parameters using atomic force microscopy (AFM), and Figures 2C, D, and E show characterization of the patterns with scanning electron microscopy (SEM). As demonstrated from these images, the height of the pattern elements was typically on the order of 20 nm and the width of the channels between adjacent pentagons was 500 nm, and as indicated above, the optimal distance between the leading edge of the linear barriers and the back of the pentagon array was ∼13 μm. This distance was specifically selected for use with λ phage DNA, a commercially available linear DNA substrate that is 48502 bp with a fully extended contour length of approximately 16.5 μm (see below). The separation distance of 11−13 μm between the leading edge of the linear barrier and the front and rear edges of the pentagons corresponds to ∼65−80% mean extension of the λ DNA substrate, depending upon the precise anchoring position on the pentagon surface.
Figure 2.
Nanofabrication and characterization of the DNA rack elements. Panel A shows an optical image of a single barrier set collected at 100× magnification, and relevant pattern dimensions are indicated. An AFM image of a rack pattern is shown in panel B highlighting the height of the linear barriers and the pentagons, as well as the distance between these two barrier elements. An SEM image of another rack pattern with a single linear barrier and the arrayed pentagons is shown in panel C, and details of the different barrier elements are shown in panels D and E.
Assembly and Characterization of Double-Tethered DNA Curtains
The overall design of the DNA rack relies upon the selective but nonspecific adsorption of antibodies to the relatively large exposed surface of the metallic pentagons. The bilayer will coat the fused silica surface but will not coat the metallic patterns, leaving the nanofabricated patterns exposed to solution.8,9,16 Antibodies injected into the sample chamber should not adsorb to the inert lipid bilayer, nor should they adsorb to the fused silica slide glass that is protected by the bilayer. The antibodies can nonspecifically adsorb to the exposed surfaces of both the linear barriers and the pentagons, because neither of these are protected by the bilayer and both are made from the same material. However, the larger exposed surface area of the pentagons should ensure adsorption of more antibody relative to the smaller area encompassed by the linear barriers. Each pentagon has a surface area of 1.55 μm2, and there are 167 pentagons for each 260 μm long linear barrier. This corresponds to a total surface area of 372 μm2 for the pentagons compared to just 36 μm2 for an entire linear barrier. In addition, any antibodies bound to the linear barriers will not interfere with the overall anchoring strategy, because any DNA anchored to the linear barriers through a hapten−antibody interaction would not be confined to the region encompassed by the evanescent field once buffer flow was terminated and would not be visible with the TIRFM (see below).
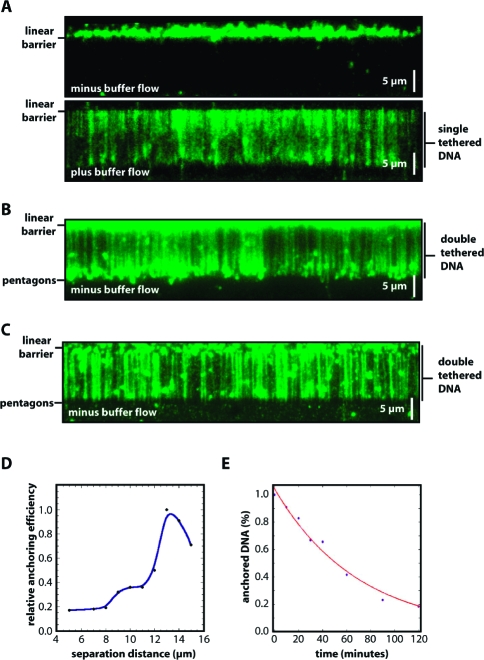
To assemble the double-tethered DNA curtains, the surface of the flow cell was first coated with a lipid bilayer, as previously described,8,9 with the exception that BSA was omitted from all buffers used prior to deposition of the antibodies. Omission of BSA was essential to ensure that the exposed pentagon surfaces were not passivated prior to the addition of the antibodies. Once the bilayer was assembled on the surface, anti-DIG Fab (Roche) or anti-FITC IgG (Invitrogen) was injected into the sample chamber and allowed to adhere nonspecifically to the exposed metal barriers. Following a brief incubation, the free antibody was rinsed from the flow cell and replaced with buffer containing 0.2 mg/mL BSA, which served as a nonspecific blocking agent to passivate any remaining exposed surfaces. Neutravidin was then injected into the sample chamber, where it could bind to the biotinylated lipids. DNA labeled at one end with biotin and at the other end with DIG (see Materials and Methods) was stained with the fluorescent intercalating dye YOYO1 and then injected into the sample chamber. Following injection of the DNA, the sample chamber was incubated briefly without buffer flow to allow the biotinylated ends to adhere to the surface of the bilayer, and then buffer flow was applied to push the DNA molecules into the linear barriers. The anchored DNA molecules were then imaged with the TIRFM in the presence and absence of buffer flow. As previously reported, single-tethered DNA curtains assembled with linear barriers could only be viewed when buffer was flowing, but the molecules drifted out of the evanescent field and disappeared from view when flow was terminated (Figure 3A).8,9 In contrast, when the DNA curtains were made using the new rack patterns, the anchored molecules remained fully extended and visible by TIRFM even when no buffer was flowing through the sample chamber (Figure 3B,C). Any DNA molecules that were anchored to the surface by just one end (either via the biotin or DIG tag) cannot be viewed along their contour length when buffer flow was terminated. Rather, the bilayer-tethered ends of these molecules remain visible, causing a high background of fluorescence signal upstream of the linear barrier (i.e., outside the area of observation). This does not happen at the leading edge of the pentagons, because fewer DNA molecules become anchored to the relatively small area of bilayer between the linear barrier and pentagon array, and also because any DNA molecules that do happen to become anchored in the region are free to pass through the gaps between the adjacent pentagons. The fact that the single-tethered molecules disappear from view in the absence of buffer flow can be used to take advantage of the spatially selective illumination geometry provided by TIRF illumination and ensures that any single-tethered DNA molecules will not comprise imaging of the double-tethered DNA.
Figure 3.
Curtains of double-tethered DNA. Panel A shows a typical example of a single-tethered curtain, in the absence (top) and presence (bottom) of buffer flow. Panels B and C show examples of double-tethered DNA curtains in the absence of any buffer flow, illustrating that the DNA molecules remain extended even in the absence of continual hydrodynamic force. The Cr barrier patterns in panel B were made by ebeam lithography, and those in panel C were made by nanoimprint lithography. Panel D shows the relative anchoring efficiency of the second DNA end as a function of the separation distance between the linear barrier and the polygon array, revealing a peak tethering efficiency corresponding to 13 μm, which corresponds to ∼80% extension of the λ DNA relative to its full contour length (48502 bp, ∼16.5 μm). Panel D shows the percent of digoxigenin-labeled DNA that remains anchored after defined time intervals, revealing a half-life of 69 min.
Several experimental parameters were assessed to optimize assembly of the double-tethered DNA curtains. The relative anchoring efficiency was tested for rack patterns made with variable spacing between the linear barriers and pentagon anchor points (ranging from 5 to 15 μm). As expected, the most efficient anchoring occurred with pattern distances corresponding to the length of the flow-stretched λ DNA [∼13 μm (Figure 3D)].(10) Control experiments with DNA lacking the hapten tag or flow cells not treated with antibodies verified that both the DIG label and the Fab fragments were necessary for efficiently anchoring the two ends of the DNA. Approximately 95% of the anchored DNA could be attributed to specific antibody−hapten interactions, with the remaining 5% due to nonspecific anchoring of the DNA ends to the antibody-coated pentagons (data not shown). The identity of the antibody−hapten pair was also investigated, and double-tethered DNA curtains could be made using substrates that were end-labeled with a single digoxigenin, FITC, or BrdU, and pentagons coated with the corresponding anti-DIG, anti-FITC, or anti-BrdU antibodies, respectively. The number of double-tethered DNA molecules anchored to the surface decreased slowly over time, with a half-life of >1 h for the DIG-labeled molecules (Figure 3E), in good agreement with the expectations for a high-affinity antibody−hapten interaction. The half-life of the FITC-anchored DNA was comparable to that of the DIG-anchored DNA (not shown). However, the BrdU-anchored DNA molecules (labeled with either one or three BrdU tags) had a much stronger tendency to dissociate from the pentagons, displaying a half-life on the order of just a few minutes (data not shown). Finally, the double tethering procedure worked equally well with barriers made or chromium (Cr) or gold (Au), and neither of these materials interfered with acquisition of the fluorescent images. The Cr patterns appeared dark against the background (Figure 3B), because the Cr blocks the incident light from the laser beam, whereas Au barriers appeared bright against the darker background, presumably because a surface plasmon (SP) is established within the thin Au layer and either the SP is more intense than the surrounding evanescent field on the fused silica or additional light is scattered due to the intrinsic roughness of the gold surface.30,31 This allowed the Au patterns to be directly imaged with the TIRFM, enabling the pattern elements to serve as fiduciary markers on the sample chamber surface (see below and Figure 4C).
Figure 4.
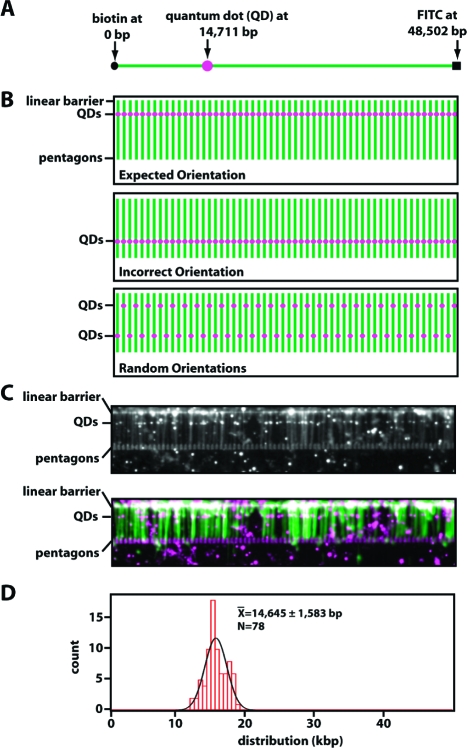
Defined orientation of the DNA molecules. Panel A shows a schematic diagram of the DNA substrate labeled at each end with either biotin or FITC, as indicated, and labeled at an internal position with digoxigenin (DIG). The internal DIG tag was located 14711 bp from the biotinylated end of the DNA and was labeled with an anti-DIG-coated quantum dot (QD). Panel B shows a schematic representation of the expected location of the fluorescent QD (magenta dots) if the DNA molecules (green lines) are in the expected orientations (top panel) or the incorrect orientation (bottom panel) or randomly distributed between the two possible orientations (bottom panel). Panel C shows an example of a double-tethered DNA curtain labeled at the internal position with the anti-DIG-coated QDs; the top panel shows a black and white image, and the bottom panel shows a pseudocolored version of the same image (the DNA is green and the QDs are magenta). Fluorescent points outside of the nanofabricated pattern represent QDs that are nonspecifically adsorbed to the bilayer. These are easily distinguished from those bound to DNA because they do not colocalize with the DNA molecules and are also free to diffuse in two dimensions, whereas those immobilized on the DNA are not. The patterns used in this image were made from Au (15−20 nm) with a thin (3−5 nm) Ti adhesion layer and appear bright against a darker background. The location of each of the DNA-bound QDs was determined by fitting the images to two-dimensional Gaussian functions, as described previously,(23) and the position data were then plotted as a histogram in panel D.
Defined Orientation of the Double-Tethered DNA Curtains
The differential chemistries used to tag the two ends of the DNA were selected such that the molecules within the double-tethered curtains should be aligned in a defined orientation. That is, the biotinylated end should be anchored at the edge of the linear barriers, and the hapten-tagged end should be anchored to the downstream pentagons. This orientation specificity can be tested using an asymmetrically labeled DNA substrate tagged with a fluorescent quantum dot [QD (Figure 4A)]. If the DNA molecules were aligned as expected, then a QD located at a single specific site within the DNA should appear as a fluorescent “line” spanning the DNA curtain (Figure 4B, top panel). The line of QDs should be oriented perpendicular to the long axis of the extended DNA molecules and should coincide with the known location of the engineered tag. If the DNA were in the opposite orientation from that which was expected, then the QDs would be found at an incorrect location within the double-tethered curtain (Figure 4B, middle panel). Finally, if the DNA molecules were randomly oriented, then the QDs should reveal both possible orientations of the DNA molecules (Figure 4B, bottom panel). To confirm that the DNA was oriented correctly, the molecules were labeled at one end with FITC, labeled at the other with biotin, and labeled with DIG at a position 14711 bp from the biotin tag (see Materials and Methods). The internal DIG labels were made using an oligonucleotide replacement strategy in which the DNA was nicked at specific sites with a nicking endonuclease and short ssDNA fragments flanked by the resulting nicks were replaced with a DIG-tagged oligonucleotide (see Materials and Methods). Experiments with DNA curtains anchored by a single end confirmed that the QDs were present at a single location within the DNA molecules as dictated by the sequences of the oligonucleotides.(25) The double-tethered curtains were then assembled using pentagons coated with anti-FITC antibodies, and the DIG tags were labeled by injecting anti-DIG-coated QDs into the sample chamber. As shown in Figure 4C, the DNA-bound anti-DIG QDs were aligned with one another, and the mean position of the QD tags was found to be 14645 ± 1585 bp (N = 78) from the biotinylated DNA ends, which coincided to within 66 bp of the expected location (Figure 4D). No QD tags were observed at other locations on the DNA. These results confirmed that all of the DNA molecules anchored to the surface via the nanofabricated rack patterns were aligned in the same orientation, as defined by the distinct function groups engineered at the opposing ends of the DNA molecules.
Visualizing Proteins Sliding on the Double-Tethered DNA Curtains
The primary motivation for development of these double-tethered DNA curtains was to facilitate visualization of passive diffusion of proteins along DNA.(26) For example, in previous work, we have demonstrated that the Msh2−Msh6 protein complex can diffuse in one dimension (1D) along duplex DNA.(21) The Msh2−Msh6 protein is an essential component of the postreplicative mismatch repair machinery and is responsible for locating and initiating repair of biosynthetic DNA replication errors. Our initial studies with the Msh2−Msh6 complex relied upon DNA molecules that were tethered by both ends to neutravidin nonspecifically absorbed to a fused silica surface.(21) With this approach, we would typically obtain only 10−30 DNA molecules per flow cell that were suitable for making diffusion measurements, often limiting data collection to just one molecule per field of view. Moreover, these double-tethered DNA molecules were randomly distributed on the flow cell surface and had to be manually located by scanning the entire surface of the flow cell, which made these 1D diffusion measurements technically demanding and time-consuming. As shown above, using the engineered surfaces with DNA rack patterns, we can now visualize thousands of DNA molecules per flow cell, and on the order of 50−100 in each field of view. Here we demonstrate that these anchored curtains of DNA are suitable for studying protein−DNA interactions.
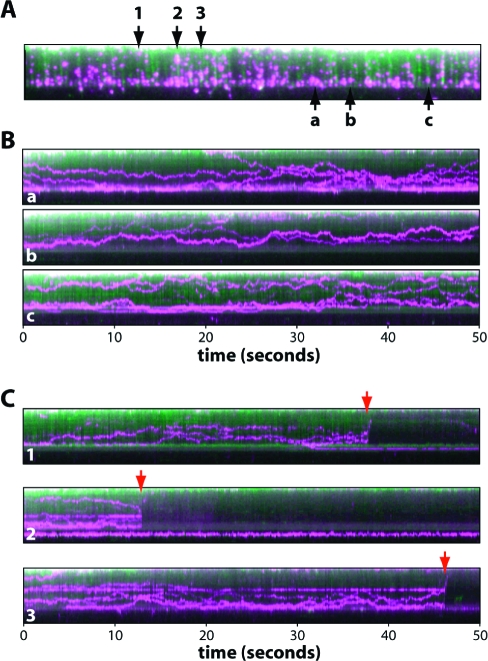
To determine whether the double-tethered DNA curtains made using the nanofabricated rack patterns could be used to study 1D diffusion of proteins, we tested the mismatch repair protein Mlh1,(32) which was labeled with a single QD via a FLAG epitope tag; complete analysis of QD-tagged Mlh1 will be presented elsewhere.(33) The labeled proteins were injected into a flow cell containing a curtain of YOYO1-stained double-tethered DNA molecules, and videos were collected over a 60 s period. As shown in Figure 5A, we could readily detect binding of Mlh1 to the DNA molecules within the double-tethered curtains. There were 79 DNA molecules and 235 DNA-bound proteins observed in this single field of view (Figure 5A), illustrating the dramatic improvement of this approach compared to our previous technique for making double-tethered DNA molecules.21,22 The region of the bilayer below the pentagons was devoid of QDs, indicating that adsorption of the QD-tagged Mlh1 complex to the bilayer was minimal. The proteins that were bound to the double-tethered DNA curtain diffused rapidly in one dimension along the DNA molecules, exhibiting a mean diffusion coefficient of 0.14 ± 0.13 μm2/s, and this motion was revealed in kymograms made from representative examples of molecules found within the DNA curtains (Figure 5B; detailed analysis of this diffusive behavior will be presented elsewhere(33)). This experiment provides direct evidence that the double-tethered DNA curtains assembled using nanofabricated rack patterns can be used to visualize the lateral motion of fluorescently tagged proteins along the DNA molecules.
Figure 5.
Using DNA racks to visualize 1D protein diffusion. Panel A shows an example of a double-tethered DNA curtain bound by QD-labeled Mlh1. The DNA is colored green, and the proteins are colored magenta. This image represents a single 100 ms image taken from a 1 min video (not shown). Panel B shows three representative kymograms (designated a−c) made from individual DNA molecules from within panel A. Panel C shows examples of DNA molecules taken from panel A that broke during the course of DNA collection (numbered 1−3), demonstrating that both the DNA and the bound proteins diffuse rapidly away from the surface and out of the evanescent field, this confirming that the QD-tagged proteins are not adsorbed to the bilayer.
To verify that the lipid bilayers remained inert when used with the DNA rack devices, we assessed whether the protein or DNA molecules remained bound to the surface after breaking the DNA. If either the proteins or the DNA were nonspecifically linked to the lipid bilayer that coated the sample chamber surface, then they should remain within the evanescent field and in the field of view even if the DNA breaks or detaches from one of its anchor points. In contrast, if neither the proteins nor the DNA interacts nonspecifically with the bilayer, then detachment of the DNA should cause the molecules to retract away from the surface and they should rapidly disappear from view.8−10,21−23,25 As shown in Figure 5C, when the DNA molecules randomly detached from the surface during the course of an observation, the proteins almost immediately disappeared from view as they drifted outside the detection volume defined by the penetration depth of the evanescent field. Taken together, these data demonstrate that the fluorescently tagged proteins were bound to the DNA, that the DNA molecules interacted only with the sample chamber surface through their anchored ends, and that neither the QD-tagged proteins nor the DNA interacted nonspecifically with the lipid bilayer.
Discussion
Single-molecule imaging offers many unique opportunities to probe biological reactions in ways not possible through conventional biochemical or biophysical approaches. To facilitate these studies, we are developing new techniques for controlling the spatial organization of surface-anchored DNA molecules within the confines of a microfluidic sample chamber. Our unique approaches are based upon surface engineering techniques that enable us to control the distribution of DNA substrates with micro- and nanoscale precision.8,9 Here we build upon our previous work by using direct-write electron beam or nanoimprint lithography to fabricate patterns comprised of linear diffusion barriers followed by pentagonal anchor points, which together provide different functional features enabling DNA curtains to be anchored by both ends on the surface of a microfluidic sample chamber. We refer to these patterned surfaces as DNA racks because the molecules are stretched out and anchored to the flow cell surface where they can be viewed with the TIRFM. We have demonstrated that these rack patterns can be used along with wide-field TIRF microscopy to visualize hundreds of individual, perfectly aligned DNA molecules, all of which are arranged in the same orientation and anchored by both ends to the sample chamber surface. Although we have used pentagons as anchor points in this study, it is likely that other shapes would function just as well. An important aspect of these devices is that the fused silica surface is coated with a supported lipid bilayer, and the DNA molecules are suspended above this bilayer, ensuring that they are maintained within an inert microenvironment compatible with a range of biological molecules. As with our previous nanofabricated devices, this new approach is relatively simple and robust, the flow cells are reusable, the barriers themselves are uniform, and they do not compromise the optical quality of the fused silica or interfere with signal detection. Other procedures for anchoring numerous, long DNA molecules to surfaces have been described.34−39 However, none of these methods offers the ability to pattern thousands of DNA molecules all aligned in the same orientation using an experimental platform that is compatible with protein biochemistry and single-molecule fluorescence imaging. One drawback of our approach is that we have limited control over the tension of the DNA, and the tension cannot be varied during the course of an experiment, as can be done with techniques such as laser tweezers. However, it may be possible to integrate a multiplexed optical trap into our DNA curtain experiments, which would provide a means for independently controlling the tension on the DNA molecules within the curtain.
As a first conceptual demonstration, we have shown that the curtains of double-tethered DNA can be used to image 1D diffusion of DNA-binding proteins. Our previous approach to these experiments relied upon DNA that was randomly anchored to a surface via biotin−neutravidin interactions, which typically yielded no more than one to three molecules of DNA per field of view, with just ∼10−30 double-tethered DNA molecules present over the entire surface of the flow cell.21,22 In addition, the molecules were randomly oriented on the flow cell surface, making it difficult to locate and compare different DNA molecules. While this original approach proved to be useful for our initial studies, it was tedious and challenging to collect sufficient data for thorough analysis. As demonstrated here, we are now able to visualize on the order of 100 DNA molecules per field of view, thousands of molecules are present on the surface of a typical flowcell, and all of these DNA molecules are aligned in the exact same orientation. This novel approach will make 1D diffusion measurements and molecule-to-molecule comparisons more straightforward in future work.
Acknowledgments
We thank members of our laboratories for insightful discussions and for carefully reading the manuscript. We thank Aaron Plys and Eric Alani (Cornell University, Ithaca, NY) for providing the Mlh1 protein. This research was funded by the Initiatives in Science and Engineering grant (ISE, awarded to E.C.G. and S.W.) program through Columbia University, and by National Institutes of Health (NIH) Grant GM074739 and an NSF PECASE Award to E.C.G. T.F. was supported in part by a National Science Foundation Graduate Research Fellowship. J.G. was supported by an NIH training grant for Cellular and Molecular Foundations of Biomedical Sciences (T32GM00879807). This work was partially supported by the Nanoscale Science and Engineering Initiative of the National Science Foundation under NSF Award CHE-0641523 and by the New York State Office of Science, Technology, and Academic Research (NYSTAR). E.C.G. is an HHMI Early Career Scientist.
Funding Statement
National Institutes of Health, United States
References
- Bustamante C.; Bryant Z.; Smith S. B. Nature 2003, 421, 423–7. [DOI] [PubMed] [Google Scholar]
- Cairns B. R. Nat. Struct. Mol. Biol. 2007, 14, 989–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J.; van Holde K. Mol. Cell 2006, 24, 317–29. [DOI] [PubMed] [Google Scholar]
- van Oijen A. M. Biopolymers 2007, 85, 144–53. [DOI] [PubMed] [Google Scholar]
- Riehn R.; Austin R. H.; Sturm J. C. Nano Lett. 2006, 6, 1973–6. [DOI] [PubMed] [Google Scholar]
- Riehn R.; Lu M.; Wang Y. M.; Lim S. F.; Cox E. C.; Austin R. H. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 10012–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegenfeldt J. O.; Prinz C.; Cao H.; Huang R. L.; Austin R. H.; Chou S. Y.; Cox E. C.; Sturm J. C. Anal. Bioanal. Chem. 2004, 378, 1678–92. [DOI] [PubMed] [Google Scholar]
- Fazio T.; Visnapuu M. L.; Wind S.; Greene E. C. Langmuir 2008, 24, 10524–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visnapuu M. L.; Fazio T.; Wind S.; Greene E. C. Langmuir 2008, 24, 11293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granéli A.; Yeykal C.; Prasad T. K.; Greene E. C. Langmuir 2006, 22, 292–9. [DOI] [PubMed] [Google Scholar]
- Chan Y. H.; Boxer S. G. Curr. Opin. Chem. Biol. 2007, 11, 581–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez T. D.; Nelson W. J.; Boxer S. G.; Kam L. Langmuir 2005, 21, 11963–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd D. L.; Ragains J. R.; Skehel J. J.; Harrison S. C.; van Oijen A. M. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 15382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackmann E. Science 1996, 271, 43–8. [DOI] [PubMed] [Google Scholar]
- Cremer P. S.; Boxer S. G. J. Phys. Chem. B 1999, 103, 2554–9. [Google Scholar]
- Groves J. T.; Boxer S. G. Acc. Chem. Res. 2002, 35, 149–57. [DOI] [PubMed] [Google Scholar]
- Groves J. T.; Ulman N.; Boxer S. G. Science 1997, 275, 651–3. [DOI] [PubMed] [Google Scholar]
- Tanaka M.; Hermann J.; Haase I.; Fischer M.; Boxer S. G. Langmuir 2007, 23, 5638–44. [DOI] [PubMed] [Google Scholar]
- Groves J. T.; Boxer S. G.; McConnell H. M. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 13390–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson B. L.; Nye J. A.; Groves J. T. Langmuir 2008, 24, 6189–93. [DOI] [PubMed] [Google Scholar]
- Gorman J.; Chowdhury A.; Surtees J. A.; Shimada J.; Reichman D. R.; Alani E.; Greene E. C. Mol. Cell 2007, 28, 359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granéli A.; Yeykal C.; Robertson R. B.; Greene E. C. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 1221–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad T. K.; Robertson R. B.; Visnapuu M. L.; Chi P.; Sung P.; Greene E. C. J. Mol. Biol. 2007, 369, 940–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad T. K.; Yeykal C.; Greene E. C. J. Mol. Biol. 2006, 363, 713–28. [DOI] [PubMed] [Google Scholar]
- Visnapuu M. L.; Greene E. C. Nat. Struct. Mol. Biol. 2009, , in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J.; Greene E. C. Nat. Struct. Mol. Biol. 2008, 15, 768–74. [DOI] [PubMed] [Google Scholar]
- Tafvizi A.; Huang F.; Leith J. S.; Fersht A. R.; Mirny L. A.; van Oijen A. M. Biophys. J. 2008, 95, L01–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. Y.; Krauss P. R.; Renstrom P. J. Science 1996, 272, 85–7. [Google Scholar]
- Gao H.; Tan H.; Zhang W.; Morton K.; Chou S. Y. Nano Lett. 2006, 6, 2438–41. [DOI] [PubMed] [Google Scholar]
- Knoll W.; Rothenhäusler B. Nature 1988, 332, 615–7. [Google Scholar]
- Kim G.; Campbell C. T. Biomaterials 2007, 28, 2380–92. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A.; Erie D. A. Annu. Rev. Biochem. 2005, 74, 681–710. [DOI] [PubMed] [Google Scholar]
- Gorman J.; Plys A.; Alani E.; Greene E. C., manuscript in preparation.
- Lebofsky R.; Bensimon A. Briefings Funct. Genomics Proteomics 2003, 1, 385–96. [DOI] [PubMed] [Google Scholar]
- Kabata H.; Kurosawa O.; Arai I.; Washizu M.; Margarson S. A.; Glass R. E.; Shimamoto N. Science 1993, 262, 1561–3. [DOI] [PubMed] [Google Scholar]
- Guan J.; Lee L. J. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 18321–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Blainey P. C.; Schroeder C. M.; Xie S. X. Nat. Methods 2007, 4, 397–9. [DOI] [PubMed] [Google Scholar]
- Assi F.; Jenks R.; Yang J.; Love C.; Prentiss M. J. Appl. Phys. 2002, 92, 5584–6. [Google Scholar]
- Lin J.; Qi R.; Aston C.; Jing J.; Anantharaman T. S.; Mishra B.; White O.; Daly M. J.; Minton K. W.; Venter J. C.; Schwartz D. C. Science 1999, 285, 1558–62. [DOI] [PubMed] [Google Scholar]