Abstract
Chromosomal processes related to formation and function of meiotic chiasmata have been analyzed in Sordaria macrospora. Double-strand breaks (DSBs), programmed or γ-rays-induced, are found to promote four major events beyond recombination and accompanying synaptonemal complex formation: (1) juxtaposition of homologs from long-distance interactions to close presynaptic coalignment at midleptotene; (2) structural destabilization of chromosomes at leptotene/zygotene, including sister axis separation and fracturing, as revealed in a mutant altered in the conserved, axis-associated cohesin-related protein Spo76/Pds5p; (3) exit from the bouquet stage, with accompanying global chromosome movements, at zygotene/pachytene (bouquet stage exit is further found to be a cell-wide regulatory transition and DSB transesterase Spo11p is suggested to have a new noncatalytic role in this transition); (4) normal occurrence of both meiotic divisions, including normal sister separation. Functional interactions between DSBs and the spo76-1 mutation suggest that Spo76/Pds5p opposes local destabilization of axes at developing chiasma sites and raise the possibility of a regulatory mechanism that directly monitors the presence of chiasmata at metaphase I. Local chromosome remodeling at DSB sites appears to trigger an entire cascade of chromosome movements, morphogenetic changes, and regulatory effects that are superimposed upon a foundation of DSB-independent processes.
Keywords: Spo76/Pds5p, Spo11p, meiosis, bouquet, chromosome stability, recombination
The central unique event of meiosis, reductional segregation of homologs at division I, is mediated by chiasmata. These observable connections between homologs correspond to sites of crossing over at the DNA level and arise during meiotic prophase via a complex series of chromosomal and nuclear changes that extend well beyond the process of DNA recombination and are both intricate and poorly understood.
Recombination involves a series of local biochemical changes that begin with programmed double-strand breaks (DSBs) at early prophase and occupy most of prophase (review in Keeney 2001). Formation of chiasmata requires two other types of local changes (e.g., Blat et al. 2002). First, crossing over must occur not only within the DNA, but also between the underlying chromatid axes at corresponding positions. Second, because only one chromatid of each replicated homolog is involved, sister chromatids must be differentiated and separated locally at both the DNA and axis levels. The recombination complexes seen associated with their underlying chromatid axes during early prophase (e.g., Moens et al. 2002) likely mediate spatial, temporal, and functional linkage between events at the DNA and axis levels along the chiasma formation pathway.
This progression of local changes occurs in close temporal coordination with a series of global changes in chromosome structure. One obvious meiotic structural feature is the synaptonemal complex (SC), a close-packed array of transverse filaments that links homolog axes at a distance of 100 nm all along their lengths. The SC appears, persists, and then disappears during mid-prophase, concomitant with intermediate and late stages of the recombination process (Roeder and Bailis 2000; Hunter and Kleckner 2001).
Meiotic prophase also involves dramatic chromosome movements as implied by dynamic changes in chromosome disposition within the nucleus. One type of movement is homolog juxtaposition. During the first part of prophase, homologs identify one another and come progressively closer together until finally they are completely joined by the SC. Homologs can identify one another independently of recombination (Dernburg et al. 1998; McKim and Hayashi-Hagihara 1998; Cha et al. 2000; Nabeshima et al. 2001). In other organisms, recombination is required for pairing and SC formation: SC is absent when programmed DSBs are absent and is restored by exogenous DSBs (e.g., Celerin et al. 2000; Romanienko and Camerini-Otero 2000). Thus, recombination may play broader roles in homolog juxtaposition (Goldman and Lichten 2000; Peoples et al. 2002) as well as in nucleating installation of the SC between already juxtaposed axes (Zickler and Kleckner 1999).
A second type of chromosome movement involves formation and elimination of the bouquet configuration (e.g., for reviews, see Zickler and Kleckner 1998; Scherthan 2001). During late leptotene/zygotene, immediately preceding and during SC formation, chromosome ends become clustered in a limited region of the nuclear envelope, opposite the spindle pole body/centrosome in organisms with defined microtubule organizing centers. Then, at early-mid-pachytene, after SCs are complete, the chromosomes open up out of this bouquet and chromosome ends become dispersed throughout the nuclear periphery. The basis for, and significance of, this striking, uniquely meiotic feature remains essentially unknown.
The presence of chiasmata also confers important requirements on the meiotic process with respect to events that occur after they have formed. Chiasmata ensure reductional segregation at division I because they constitute physical connections between the homologs, a role analogous to that provided by sister centric-region connections during mitosis (e.g., Paliulis and Nicklas 2000; Lee and Orr-Weaver 2001). Because of these connections, bipolar orientation of homologs results in tension on their kinetochores, which stabilizes this appropriate configuration. A chiasmate connection requires the combined effects of local crossing over, maintenance of axis continuity, and cohesion between sister chromatids all along their lengths. Then, actual separation and poleward movement of homologs requires release of sister-chromatid connections, distal to the position of the most centromere-proximal chiasmata on each arm. Programed release of sister connections thus plays as crucial a role for division I as for mitosis (for review, see Lee and Orr-Weaver 2001; Nasmyth 2001).
Full elucidation of meiosis requires understanding these individual component processes and delineation of whether and how they are interrelated. This is a challenging task because nearly all processes are linked to one another in one or more ways, as illustrated by the fact that mutations in meiotic chromosomal components almost always confer pleiotropic effects. To make matters worse, even a single protein may carry out multiple, sequential and mechanistically distinct roles (e.g., Zickler and Kleckner 1999; Roeder and Bailis 2000).
The current study examines the basic events of meiosis with respect to their dependence on DSBs and with respect to the roles of two important, evolutionarily conserved molecules, Spo76/Pds5p and Spo11p. Spo76/BIMD/Pds5p is a basic chromosome structure component that occurs all along the chromosome axes. This cohesin-related protein has been recruited from the mitotic cycle (Hartman et al. 2000; Tanaka et al. 2001) and functionally adapted for use in the meiotic program (van Heemst et al. 1999, 2001; Wang et al. 2002). Spo11p is the topoisomerase-related transesterase that catalyzes DSB formation (Klapholz et al. 1985; for review, see Keeney 2001). Spo11 protein also has noncatalytic functions, for example, in S-phase entry in budding yeast (Cha et al. 2000).
We report here the molecular identification of the Sordaria SPO11 gene, cytological localization of Spo11p, detailed analysis of 12 spo11 mutants, and analysis of double mutants carrying spo11 mutations in combination with a unique SPO76 allele, spo76-1 (van Heemst et al. 1999). We find that SPO11 is one of several previously isolated “ASY” suppressor genes in which mutations alleviate the spo76-1 sporulation defect (Huynh et al. 1986). To probe the roles of DSBs (and thus recombination) directly, we also determined the effects of γ-irradiation on spo11 and spo11 spo76-1 phenotypes. Because of unique advantages of Sordaria as an experimental system, we have been able to examine the entire spectrum of chromosomal and nuclear processes from early prophase to telophase II. These studies provide new informations about homolog juxtaposition, the nature of the leptotene/zygotene transition, the bouquet stage, and separation of homologs at anaphases I and II. They reveal also new roles for both Spo11p and Spo76p during mid-prophase.
Results
ASY1 is Sordaria SPO11
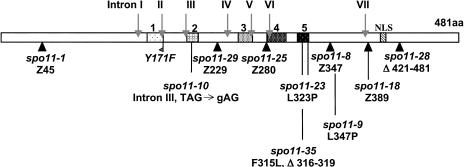
Sordaria macrospora SPO11 encodes a predicted protein of 481 amino acids (Fig. 1; Materials and Methods). Identities with other Spo11/Top6A family members are very high in conserved regions I-V (Fig. 1; Bergerat et al. 1997) and significant throughout: 58% with Neurospora crassa, 33%-35% with human and mouse, and 28% with budding yeast. SPO11 is ASY1: All 10 asy1 suppressors of spo76-1 are complemented by SPO11+, and corresponding mutations, which include deletion, missense, and nonsense alleles, can be identified through the entire coding region (Fig. 1). A complete deletion mutant (spo11Δ) and a mutant altered at the active site tyrosine invariant among all Spo11 proteins (spo11-Y171F) were constructed molecularly (Fig. 1; Materials and Methods).
Figure 1.
Sordaria Spo11p. Indicated are the five conserved regions (rectangles with different motifs), intron location, nuclear localization signal (NLS), and sites of 11 spo11 mutations. (black triangles) Predicted stop codons; (flag) tyrosine spo11-Y171F mutation; (Δ) deletion.
All spo11 mutants show wild-type growth rates and none is UV or X-ray sensitive (data not shown). All exhibit the same basic meiotic phenotypes (below). In mutant spo11-10, 80% of nuclei show mutant phenotypes whereas 20% exhibit normal meiosis except for reduced crossovers. spo11-10 is a point mutation in the 3′ acceptor site of intron III (gAG instead of TAG) and, by reverse transcriptase PCR (RT-PCR), at least some mutant transcripts are correctly spliced.
Spo11p is chromatin associated during early-mid-prophase
Chromosomal localization of Spo11p was assessed via a Spo11 protein tagged at its C terminus by Green Fluorescent Protein (GFP). The tagged construct has no effect on wild-type growth or sporulation, efficiently complements all spo11 mutants, and gives the same chromosomal staining pattern in both spo11Δ and wild type, which thus likely accurately reflects that of wild-type Spo11p. Patterns seen by GFP fluorescence were confirmed with anti-GFP antibodies.
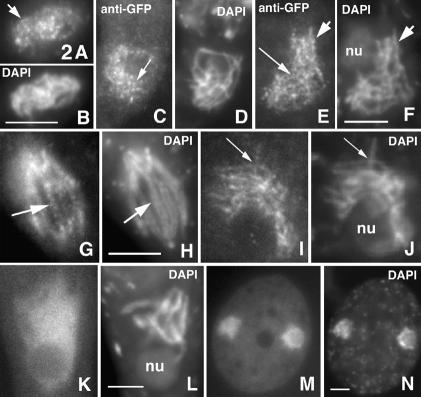
Spo11-GFP first appears, as chromatin-associated foci, during karyogamy, when the two haploid nuclei have fused but their nucleoli have not (and thus after S phase; Fig. 2A,B). At early leptotene, when homologs are still well separated, nuclei exhibit numerous foci located throughout the chromatin (Fig. 2C,D). Thereafter, foci are seen in rows (Fig. 2E,F), which persist during presynaptic alignment (Fig. 2G,H), suggestive of association with chromosome axes. Then, Spo11p stains as irregular, discontinuous lines along all synapsing chromosomes with few foci remaining on the chromatin (Fig. 2I,J). Lines remain visible during early pachytene but from mid- to end pachytene, Spo11p is seen as a bright diffuse signal spread over the entire nucleus (Fig. 2K,L), and disappears during the diffuse stage. Spo11p reappears in the nuclei of newly membranated ascospores (Fig. 2M,N) but is gone in mature ascospores. No staining was seen in vegetative nuclei.
Figure 2.
Spo11p localization. Spo11-GFP staining (left) and corresponding DAPI (right). Karyogamy nucleus (A,B) and early leptotene nucleus (C,D; double-stained with anti-GFP antibody) showing chromatin-associated Spo11-GFP foci (arrows). (E,F) Mid-leptotene nucleus (anti-GFP antibody); foci are mainly in rows (arrows). (G,H) Leptotene nucleus with clear presynaptic alignment and Spo11-GFP foci in rows (arrow). (I,J) Spo11-GFP stains as discontinuous narrow lines along all chromosomes of late leptotene and bouquet nuclei (arrow). (K,L) Late pachytene nucleus with diffuse Spo11p signal. (M,N) Ascospore with two nuclei resulting from the second postmeiotic division stained by Spo11-GFP and DAPI. Bars, 5 µm.
Sordaria SPO11 mutations abrogate recombination initiation, crossover, and chiasma formation
RecA homolog Rad51p marks the sites of meiotic DSBs (e.g., Bishop 1994). When analyzed with antibodies against Rad51p, wild-type nuclei exhibit maximum abundance of foci from late leptotene through early zygotene (58-45 foci; n = 70 nuclei). Rad51 foci are rare in all spo11 mutants (e.g., 4% of spo11Δ nuclei show one dot; n = 250).
A subset of DSBs are processed into crossovers, seen at diplotene as chiasmata. Wild-type diplotene nuclei exhibit 21 ± 3 chiasmata over the seven bivalents (n = 50). In spo11Δ, chiasmata are reduced ∼500-fold (two seen among 50 diplotenes). The other spo11 mutants exhibit similar reductions except spo11-10, where 80% of nuclei have no chiasmata and ∼20% show 8-14 chiasmata (n = 50).
Crossing over is also strongly reduced. In the interval between yas1 and pam2, two spore color markers that map far from their centromere on each arm of chromosome 4, the frequency of exchanges in wild-type meiosis, measured by tetrad analysis, is 44 ± 5% (n = 2000). Among rare viable ascospores emerging from spo11 homozygous crosses, yas1-pam2 crossover frequencies were ∼10-fold reduced: 2.1% (spo11Δ; 210 spores analyzed), 4.2% (spo11-8; 203 spores), and 6.3% (spo11-1; 70 spores). Lesser reduction in crossovers as compared with chiasmata could reflect underestimation of wild-type crossover frequencies due to the large yas1-pam2 map distance or an increased probability of spore viability when a crossover is present. Residual crossovers (and a 10-fold reduction) were also observed in Arabidopsis spo11-1 mutants (Grelon et al. 2001).
Many viable ascospores (67%) issued from the above spo11 crosses are aneuploid, as indicated by spore color (black instead of mutant) and subsequent tests. Among 200 potentially aneuploid spo11Δ ascospores tested (Materials and Methods), 95.4% showed no crossover and cosegregation of both parental chromosomes 4, a configuration that implies aberrant segregation at division I, with or without aberrant segregation at division II. The remaining 4.6% of ascospores had a [(yas1 +)(+ +)(+ pam2)] genotype that implies defective segregation not only at division I, but also at division II (plus crossing over between one of the tester markers and the centromere). As second division missegregation cannot be detected in the majority aneuploid class, its frequency could be occurring at even higher levels. Second division missegregation is not straightforwardly predicted by absence of chiasmata.
SPO11 is dispensable for basic chromosome organization and condensation but is required for spatial juxtaposition of homologs and synapsis/SC formation
In Sordaria, karyogamy is immediately followed by a classical prophase program. We have examined the effects of all 12 spo11 mutations on prophase events. Ascus growth (10 µm after karyogamy to 150 µm at the end of prophase), nuclear volumes, and nucleolar changes were similar in mutants and wild type and thus were used as references for staging.
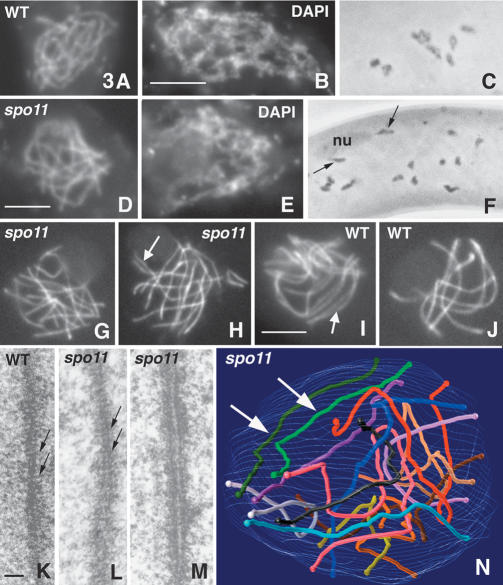
During wild-type meiosis, as revealed by staining with DAPI and hematoxylin, chromosomes emerge as smooth, discrete units due to chromatin compaction during leptotene (Fig. 3A) and remain as such through pachytene. From leptotene onward, homologs become progressively juxtaposed (Fig. 2H) and, by pachytene, seven fully synapsed pairs are always seen. Chromosomes exit pachytene into the diffuse stage, characterized by chromatin decondensation (Fig. 3B), and then re-emerge at diplotene as seven compact units with homologs connected by chiasmata (Fig. 3C). Compaction continues into metaphase I. spo11 mutants exhibit the same progression as wild type (Fig. 3D-F) except that diplotene nuclei exhibit 14 individual chromosomes not connected by chiasmata (Fig. 3F).
Figure 3.
Prophase phenotypes of wild type and spo11 mutants. Nuclei are stained by DAPI (A,B,D,E) and by hematoxylin (C,F). (A-C) Wild-type. (A) Leptotene. (B) Diffuse stage. (C) Diplotene nuclei. (D-F) Corresponding spo11Δ nuclei. (F) Note similar chromosome condensation at diplotene despite no chiasmata among the 14 univalents; arrows point to the two widely separated homologs attached to the nucleolus (nu). (G-J) Chromosome axes are visualized by Spo76-GFP. (G,H) Early and mid-prophase nuclei of spo11Δ. Note partial alignment of one pair of homologs (arrow in H). (I,J) Wild-type nuclei at equivalent ascus sizes. (I) Zygotene; arrow points to aligned segments at the same distance as aligned segment noted in H. (J) Pachytene with seven synapsed bivalents. (K-M) Electron micrographs of thin sections of AEs from wild-type (K) and spo11-1 (L,M) nuclei. Note striations (arrows) in wild-type and mutant AEs. (M) Late prophase spo11 AE shows thicker, separated sister axes. (N) Three-dimensional representation of a reconstructed spo11-1 nucleus (from 56 sections/pictures); arrows indicate an aligned homolog pair. Bars: LM pictures, 5 µm; EM pictures, 0.1 µm.
Prophase axis structure and the process of homolog pairing and synapsis were further studied at higher resolution. Chromosome axes, stained with Spo76-GFP (van Heemst et al. 1999), were visualized by confocal microscopy; the presence of Spo76-GFP did not affect spo11 mutant phenotypes. Axial elements (AEs) and SCs were visualized by ultrastructural studies. Complete three-dimensional reconstructions were made of 25 serially sectioned nuclei of spo11-1 and spo11-9, both null alleles by phenotype; results were compared with 85 wild-type serially sectioned nuclei. Sordaria is an especially favorable system for analysis of meiotic pairing because continuous AEs emerge very early in prophase and their chromosome identities can be assigned by length differences and centromere index (Zickler 1977).
From early to mid-prophase, wild type and spo11 mutants are indistinguishable with respect to axis development and organization. In all 12 mutants, as in wild type, Spo76p first appears at the telomeres at late karyogamy and then is seen all along the chromosome axes (Fig. 3G-J) until the diffuse stage (for wild type, see also van Heemst et al. 1999). In wild type and mutants, AEs are continuous along each of the 14 chromosomes by midleptotene, at which point both AE ends are anchored within the nuclear envelope. Total spo11 AE lengths, calculated from the 25 reconstructed nuclei, match perfectly the lengths of wild-type AEs at the corresponding stages: for younger nuclei, 73-78 and 72 ± 5 µm, and for later stages, 82-92 and 90 ± 6 µm, respectively. Interestingly, even though spo11 homologous AEs were generally far apart (below), they always had exactly the same lengths. spo11 AEs also exhibit the same regular striations as wild-type AEs (Fig. 3K,L). Mutant AEs remain normal through the time when wild-type AEs become the lateral elements of SCs. At a stage corresponding to late pachytene in wild type (by ascus and nucleolus criteria), mutant AEs become thicker and double (Fig. 3M). Similar late AE doubling is also seen in Sordaria asynaptic mutants from three other genes and in situations where synapsis is absent naturally, for example, in unsynapsed regions of mammalian X and Y chromosomes (Zickler and Kleckner 1999), and thus could be a consequence of absence of synapsis.
All spo11 mutants are severely defective in both spatial juxtaposition and synapsis of homologs (Fig. 3, cf. G,H and wild type in I,J). In wild type, by late leptotene, homologs are coaligned at a distance of 200-400 nm (“presynaptic alignment”; Zickler 1977). SC is then installed, linking homologs at a distance of 100 nm. In spo11 mutants, complete coalignment of all homologs at a distance of 400 nm or less was never observed. In confocal series of Spo76-GFP stained nuclei (n = 350) and in each of the 25 serially sectioned nuclei discussed above, only one to three homologs were found coaligned along various portions of their lengths (Fig. 3H), with occasional alignment along entire chromosome lengths (Fig. 3N). These configurations (Fig. 3G,H) remain unchanged through stages corresponding to zygotene and pachytene in wild type (Fig. 3I,J). Neither synapsis, indicated in wild type by merger of Spo76-GFP-stained homologs (Fig. 3J), nor SC in EM sections are seen, except for rare late nonhomologous SC (data not shown).
spo11 mutants show normal bouquet formation and delayed bouquet exit
In wild-type meiosis, specific recognition and juxtaposition of homologs is accompanied by global chromosome movements mediated by chromosome ends. All chromosome ends (attached to the nuclear envelope at early leptotene, above) migrate into a relatively limited area of the nuclear envelope such that, by the time chromosomes are presynaptically coaligned at late leptotene, most or all ends are present in a “loose bouquet” (Fig. 4A). Afterwards, in a rather transient stage that is concomitant with SC formation, all chromosome ends converge into a highly polarized organization to give a classical “tight bouquet” configuration (Fig. 4B). All 12 spo11 mutants exhibit bouquet formation with the same progressive series of spatial changes seen in wild type (Fig. 4C,D). Telomere clustering is then released (Fig. 4E) at a stage corresponding to mid-pachytene in wild type (Fig. 4F). Because mutant nuclei exhibit only very limited pairing of homologs (above), we can conclude that bouquet formation requires neither DSBs nor prominent interactions between homologs. Conversely, the failure of homolog juxtaposition in spo11 mutants cannot be attributed to a failure to make a bouquet.
Figure 4.
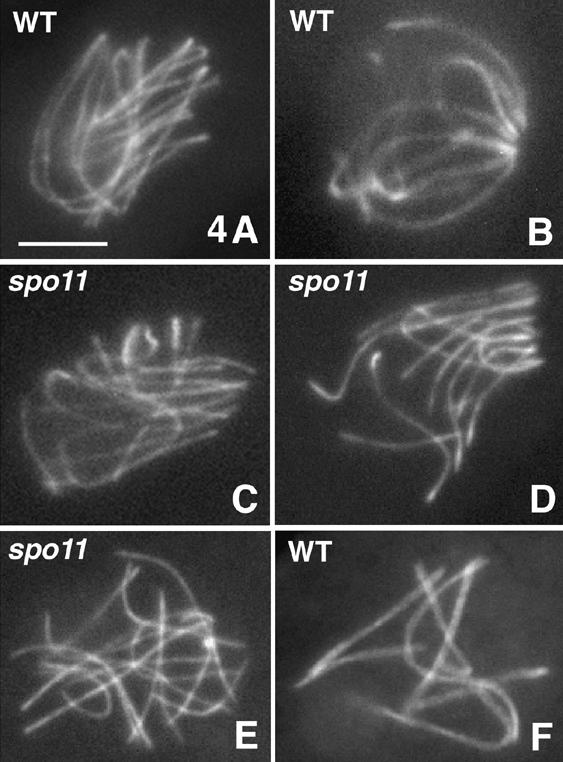
Wild-type and spo11 bouquet. Chromosome axes are stained by Spo76-GFP. Wild-type late leptotene (A; note aligned homologs) and early pachytene nucleus (B) with respectively loose and tight bouquets. spo11Δ early (C) and later (D) bouquet. Telomere clustering releases at late prophase in both spo11-Y171F (E) and wild type (F; note the seven synapsed bivalents). Bar, 5 µm.
spo11 mutants are, however, defective in exiting the bouquet. Wild-type nuclei with a bouquet configuration never exceed 5%-10% of prophases (three experiments; n = 1050, 1016, and 875). Dramatically higher frequencies are seen in all spo11 mutants, implying that this stage is substantially prolonged relative to other prophase stages. Interestingly, unlike all other analyzed phenotypes, the extent of this prolongation is allele specific. The most pronounced effect occurs in spo11-Y171F, with ∼60% bouquet nuclei, versus ∼40% in spo11Δ and ∼22% in spo11-10 (three experiments with, respectively, n = 655, 395, and 972; 1071, 754, and 850; and 347, 321, and 552 nuclei analyzed; these differences are statistically significantly different from one another: P < 0.0001). We further find that mutant bouquet-staged nuclei have the same size as their wild-type counterparts and that mutant asci with bouquet release configurations exhibit the same progressive increase in size, with the same correspondence between size and ends configuration, as wild type. These findings imply that arrest at the bouquet stage in spo11 mutants affects ascus development, as well as the state of the chromosomes, and thus is a cell-wide phenomenon.
spo11 mutants exhibit aberrant homolog segregation at metaphase I, extra SPB duplication after division I, and delayed sister arm separation at anaphase II
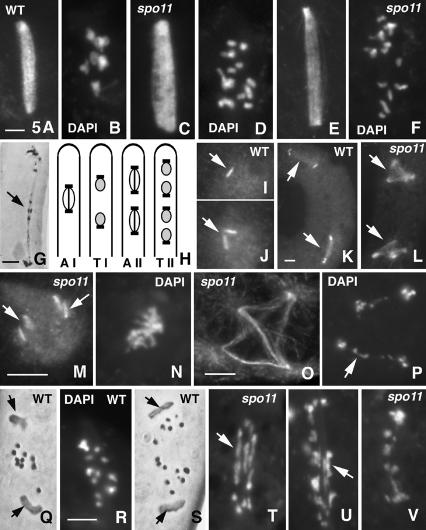
Spindle morphogenesis was analyzed in spo11 mutants by immunostaining of microtubules and spindle pole bodies (SPBs), using α-tubulin and MPM-2 antibodies respectively. Two experiments were made with n = 754 and 850 asci for spo11Δ plus 655 and 972 asci for spo11-Y171F (representing all stages from prophase through sporulation) and compared with wild type (n = 1016 and 875). In mutants, as in wild type, SPBs separate at prometaphase; chromosomes then attach to a short spindle that elongates as cells enter metaphase I (MI; Fig. 5, cf. A,B and C,D) and further elongates during anaphase A and anaphase B (Fig. 5E,F). Most chromosomes move efficiently to the poles, with the 14 pairs of sister chromatids variably distributed due to absence of chiasmata (Fig. 5F). Ten percent of anaphase I (AI) nuclei (n = 130) exhibit one or several laggard chromosomes (Fig. 5G). Also, the proportion of asci with M/AI spindles is higher in spo11 mutants than in wild type (5% and 5.5% compared with 1.6% and 1.4% of the total; P < 0.0003), implying delayed progression through this stage.
Figure 5.
Division I and II in wild-type and spo11 asci. Spindles and SPBs are stained with anti-α-tubulin and anti-MPM2 antibodies, respectively. Wild-type MI spindle (A) and corresponding seven bivalents (B) stained by DAPI. (C-G) spo11Δ nuclei. MI spindle (C) and corresponding DAPI showing 14 univalents (D). (E,F) AI spindle and DAPI. (G) Late AI (hematoxylin) showing laggard chromosomes (arrow). (H) Drawing of wild-type asci at anaphase I (AI), telophase I (TI), anaphase II (AII), and telophase II (TII). (I) One of the two wild-type TI nuclei with one SPB (arrow). (J) Wild-type prophase II nucleus showing duplicated SPBs (arrow). (K) Wild-type ascus with two division II bipolar spindles (arrows) and four SPBs. (L) Corresponding spo11Δ ascus with two tetrapolar division II spindles (arrows). (M) One of the two spo11Δ prophase II nuclei with four SPBs (arrows; cf. J). (N) Corresponding DAPI. (O) Tetrapolar spo11Δ AII spindle with clear astral microtubules emanating from the four poles and crossed intranuclear microtubules. (P) Corresponding DAPI: Both segregating sets of chromosomes show laggards (arrow). (Q) Wild-type MII stained with hematoxylin: Three chromosomes show separating chromatids. Arrows point to the two SPBs. (R) Wild-type early AII stained by DAPI; note that hematoxylin and DAPI give similar chromosome pictures. (S) Wild-type AII (hematoxylin); 14 sister chromatids separate and segregate regularly (arrows point to SPBs). (T) In contrast, spo11 MII show elongated chromosomes (arrow). (U,V) The two spo11 AII nuclei from one ascus; both show stretched chromosomes (arrow in U) and a mixture of chromosomes and chromatids (smaller units). Bars, 5 µm.
In wild type, two nuclei are formed at telophase I (Fig. 5H). SPB duplication (Fig. 5I,J) and metaphase II (MII) spindle assembly then occurs in each nucleus (Fig. 5H,K). In spo11 mutants, a striking aberrancy is observed during prophase II: 40%-50% of the nuclei (n = 119) behave like wild type (Fig. 5K); the other 50%-60% undergo an extra round of SPB duplication (Fig. 5L-N) and the resulting MII spindles are tri- or tetrapolar (Fig. 5L,O,P), instead of bipolar (Fig. 5K).
Wild-type sister chromatids separate at AII, yielding four nuclei that are regularly positioned along the length of the ascus (Fig. 5H). Division II occurs efficiently in all spo11 asci, regardless of spindle configuration. However, a second striking aberrancy is seen in the course of this division: Although chromatids segregate as seven regular units in wild-type MII (Fig. 5Q) and AII (Fig. 5R,S), mutant chromosomes exhibit a highly stretched form at MII (Fig. 5T) and AII (Fig. 5U,V), on both bi- and tri-/tetrapolar spindles. This mutant configuration suggests that sister centromeres have undergone bipolar attachment, with concomitant sister centromere separation whereas separation of sisters along their arms appears severely impeded or delayed (arrow, Fig. 5T,U). This defect fits with the aberrant meiosis II segregation of sisters identified by genetic analysis (above). The proportion of mutant asci with division II spindles is higher than in wild type (4% and 4.1% compared with 0.7% and 1% of the total; P < 0.002), implying delayed progression through this stage.
Postmeiotic mutant mitoses are almost entirely normal with respect to SPB, spindle morphology, and chromosome segregation, despite highly variable chromosome numbers. Sister chromatids usually segregate regularly (1-3 laggards seen in 10% of nuclei; n = 270). Ascospore production is reduced in all 12 mutants (15%-25% of wild type; n = 400) due to aneuploidy plus defective placement of ascospore walls resulting from aberrant division II spindle placement (Thompson-Coffe and Zickler 1993).
Ionizing radiation can efficiently restore recombination, chiasmata, pairing, synapsis, bouquet exit, and sporulation in all spo11 mutants
To critically assess which spo11 defects are specifically attributable to the absence of DSBs and to determine the effectiveness of an exogenous DSB in substituting for a programmed Spo11-mediated DSB, we subjected young fruiting bodies of spo11Δ and spo11-Y171F to γ rays and examined meiotic prophase and later stages. Sporulation was restored most efficiently at 200 Gy. Spectacularly, whereas eight-spored asci were never produced in unirradiated mutants (Fig. 6A), 15%-22% of mature asci from irradiated spo11Δ and spo11-Y171F strains were eight-spored (Fig. 6B; n = 400). Furthermore, all eight ascospores were viable (30 asci tested), implying full restoration of regular meiosis.
Figure 6.
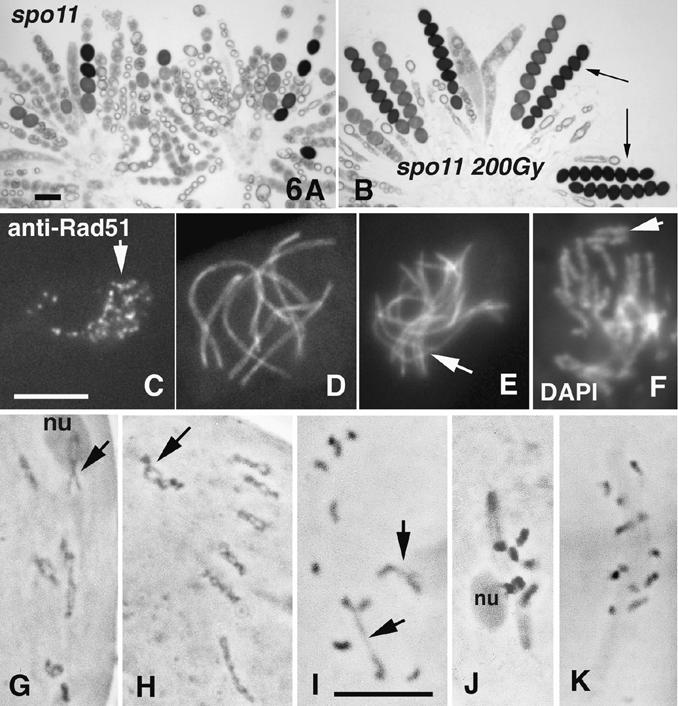
Phenotypes of irradiated spo11 mutants. (A) spo11Δ asci (from one perithecium) form only few ascospores. (B) After γ irradiation, eight-spored asci are regularly formed (arrows). (C-J) γ-Irradiated spo11Δ asci. (C) Leptotene nucleus with numerous Rad51 foci (arrow); anti-Rad51 antibody staining. (D,E) Spo76-GFP staining. (D) Restored pachytene nucleus with seven bivalents (cf. Fig. 3H). (E) Nucleus with only partial synapsis (arrow). (F) DAPI-stained nucleus showing broken chromosomes (arrow; cf. their sizes and chromosomes in D). (G-K) Chromosomes are stained by hematoxylin. (G-I) Restored diplotene nuclei with clear chiasmata. (G) All bivalents exhibit at least one chiasma. (H) The seven bivalents show several chiasmata each. Arrows point to the nucleolar organizer bivalent 2 (nu), exhibiting two (G) and four (H) chiasmata. (I) Nucleus with only two chiasmata (arrows). (J) MI spindle with seven bivalents; the nucleolar bivalent (left) is still attached to the nucleolus (nu). (K) Control unirradiated spo11Δ MI nucleus with 14 univalents. Bars, 5 µm.
Irradiated asci were examined for occurrence of prophase events (n = 420 for spo11Δ and 384 for spo11-Y171F). About half of leptotene and zygotene nuclei exhibited Rad51 foci, ranging from 1 to more than 60 (Fig. 6C); thus, irradiation produces DSBs. High levels of restoration were also observed for essentially all other spo11-abrogated processes. (1) Bouquet nuclei were rarer in irradiated nuclei (10%-15%) than in unirradiated nuclei (40%-60%, above), suggesting that this stage is no longer prolonged. (2) Among mid-prophase nuclei, homologous synapsis was fully restored in ∼20% (Fig. 6D); ∼30% exhibited synapsis of only some homologs (Fig. 6E). The remainder showed broken chromosomes (Fig. 6F) and complicated aggregates of nonhomologous synapsis. (3) Among 80 analyzed diplotene nuclei, 18% showed at least one chiasma per bivalent (Fig. 6G), with many having the wild-type number or more (Fig. 6H). An additional 22% showed only one to three chiasmata (Fig. 6I). The remaining 60% contained no chiasmata. (4) Normal divisions were restored in 20% of asci (Fig. 6J). In control experiments, no Rad51 foci, no synapsis and no chiasmata (Fig. 6K) were observed (n = 150 for each feature). The frequency of nuclei with at least one chiasma per bivalent (18%) corresponds to the number of eightviable-spored asci (15%-22%). Thus, if a Sordaria spo11Δ nucleus acquires a sufficient number of DSBs, it will proceed to give a genetically normal meiotic outcome.
spo11 mutations ameliorate spo76-1 prophase chromosome destabilization
A hallmark of spo76-1 meiosis is that, at the time when synapsis and early recombination nodules are initiated, AEs are widely split (arrows in Fig. 7A) and sometimes broken, with SC present only in regions where AEs remain single (van Heemst et al. 1999). Earlier, at leptotene, AEs are continuous and unsplit. In striking contrast, in 20 serially sectioned spo11 spo76-1 nuclei, AEs were continuous and single throughout prophase, though somewhat less electron dense (Fig. 7B) than AEs in either wild type or spo11 single mutants (Fig. 3K,L). No SCs were seen. Analogously, light microscopy of spo11 spo76-1 nuclei show no evidence of sister chromatid separation before the diffuse stage (Fig. 7C,D) and no synapsis (Fig. 7E). Thus, spo11 mutations fully suppress the sister individualization and AE fracturing defects of spo76-1 but abrogate SC formation. Absence of SC in double mutants is an expected consequence of the absence of DSBs (above). Amelioration of sister cohesion and axis integrity, was not anticipated.
Figure 7.
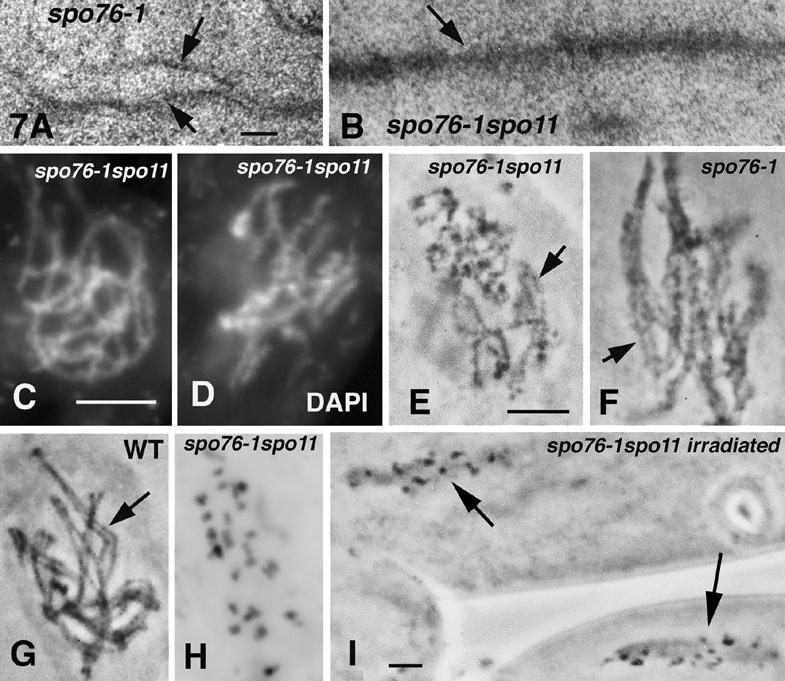
spo11-1 spo76-1 phenotypes. (A) Electron micrograph of a spo76-1 AE; note clear progressive AE splitting (arrows) from right to left. (B) spo76-1 spo11 longitudinally sectioned AE (arrow). (C,D) Early and late prophase nuclei of spo11-1 spo76-1 stained by DAPI: Chromosomes are more kinky and diffuse at later stages (D) than at early leptotene (cf. chromosomes of single spo11 mutants, Fig. 3D). (E-I) Hematoxylin staining. spo11 spo76-1 chromosomes are unpaired (E) and show similar kinkiness (arrow) as paired spo76-1 chromosomes (F; arrow). (G) Wild-type paired chromosomes are smooth and straight (arrow). (H) spo11 spo76-1 onset of AI with 28 chromatids. (I) Two asci of irradiated spo11 spo76-1: All nuclei arrest at AI. Bars: LM picture, 5 µm; EM pictures, 0.1 µm.
spo11-Y171F confers the same effects as other spo11 alleles, including spo11Δ. The same effects are also conferred by mutations in four other suppressor ASY genes, each of which shows restoration of eight-viable-spored asci by γ rays and thus presumably also abrogates DSB formation (data not shown). Moreover, when spo11 spo76-1 double mutants are irradiated, they exhibit again a spo76-1 phenotype at mid-prophase. These results suggest that DSBs are responsible for the spo76-1 axis fracturing and sister individualization at the chromatin level.
Light microscopy further shows that mid-prophase spo76-1 chromosomes are diffuse and kinky (Fig. 7F) when compared with wild type (Fig. 7G). Also, at diplotene, spo76-1 nuclei always exhibit 28 chromatids, implying a further loss of sister cohesion during the diffuse stage. Mid-prophase spo11 spo76-1 chromosomes are less diffuse than those of spo76-1, but more than those of wild type or spo11 (cf. Figs. 7D and 3A,D) and are still almost as kinky as those of spo76-1 (Fig. 7, cf. E and F). At diplotene, 63% of double mutant nuclei (n = 70) display a mixture of chromatids and chromosomes whereas the remaining 37% exhibit 28 chromatids (Fig. 7H). Thus, spo11 mutations suppress these additional spo76-1 defects only partially.
Meiosis I arrest of spo76-1 is dependent on DSBs
spo76-1 nuclei enter MI with 28 individual chromatids and then permanently arrest, with elongated spindles. The 37 extragenic suppressors of spo76-1, including all of the asy1/spo11 alleles, were identified on the basis that they eliminate this arrest (Huynh et al. 1986). We now find that suppression of metaphase arrest is conferred equivalently by all 12 spo11 mutants: In all double mutants, all asci from each fruiting body (n = 200) progress through both meiotic divisions. Thus, arrest is suppressed in nuclei having either a mixture of single chromatids and sister pairs or the same 28 single chromatids seen in spo76-1 single mutant. Furthermore, after γ-ray treatment of spo11Δ spo76-1, and spo11-Y171F spo76-1 fruiting bodies, 55% of irradiated asci now show permanent cell cycle arrest at MI as in spo76-1 (Fig. 7I; n = 450). The remaining 45% of asci form abnormal ascospores or aborted asci with condensed nuclei, presumably because they were not in a susceptible stage. Thus, DSBs, Spo11-mediated or exogenous, are required for MI arrest of spo76-1.
Discussion
In Sordaria, as in other organisms, meiotic recombination initiates via programmed DSBs catalyzed by the evolutionarily conserved transesterase protein Spo11. The current study shows that DSBs promote a cascade of chromosome-related processes, some not previously recognized, and provide new information about these processes and about the roles of Spo11p and Spo76p for meiotic chromosomes.
DSBs promote the chromosome movements required for presynaptic alignment
Some organisms are known to exhibit DSB-independent pairing during meiotic prophase (Dernburg et al. 1998; McKim and Hayashi-Hagihara 1998; Cha et al. 2000; Nabeshima et al. 2001). Partial homolog pairing occurs also in Sordaria spo11 mutants. It occurs before the bouquet (e.g., Fig. 3N), and thus is not a general consequence of telomere clustering. In contrast, in other organisms, DSBs are required for SC formation (e.g., Celerin et al. 2000; Romanienko and Camerini-Otero 2000; Grelon et al. 2001; Lichten 2001). Our data show that DSBs are required for juxtaposition of homologs into presynaptic alignment, which does not occur in Sordaria spo11 mutants, including spo11-Y171F, but which is restored after γ irradiation. Because homologs are generally widely separated in early wild-type and mutant prophases, this result implies that DSBs promote the long-range chromosomal movements that bring homologs close together along their entire lengths. These findings are in accord with DSB-mediated homolog juxtaposition seen in budding yeast (Goldman and Lichten 2000; Peoples et al. 2002). DSBs might exert their effects indirectly. However, a direct effect appears more likely because studies in plants and animals suggest that presynaptic alignment is mediated by interaxis linkages that include RecA homologs (e.g., Zickler and Kleckner 1999; Moens et al. 2002). We propose that a DSB located within an axis-associated post-DSB recombinosome on one homolog interacts stably with an homologous sequence in a partner chromatin loop and then reels the partner axis into juxtaposition. DSB-promoted presynaptic alignment is probably not sufficient to explain the role of DSBs in promoting SC formation, which likely involves additional processes specifically related to crossovers (Zickler and Kleckner 1998).
Bouquet exit is a cell-wide transition modulated by DSBs
Sordaria spo11 mutants remain in the bouquet configuration longer than wild type, as also seen in budding yeast (Trelles-Stricken et al. 1999). The current study provides two further pieces of information. First, exogenous DSBs restore timely release of the bouquet, showing that exit from the bouquet stage is directly or indirectly promoted by DSBs. Second, delayed bouquet exit reflects a delay in overall cellular progression, and not simply a defect in telomere movement.
Defective bouquet exit has also been observed in yeast rad50S, Caenorhabditis elegans syp-1, and mouse Atm-/- mutants (Trelles-Stricken et al. 1999; Scherthan et al. 2000; MacQueen et al. 2002), all of which make DSBs but are defective in later stages of recombination and in SC. The latter two studies proposed that synapsis/SC formation is required for bouquet exit. We suggest, alternatively, that exit from the bouquet is mediated by regulatory processes that sense progression of recombination-related interactions beyond the DSB stage (Hunter and Kleckner 2001) and then direct cell-wide progression via signal transduction mechanisms.
Bouquet exit is delayed longer in spo11-Y171F, where the protein is present but catalytically defective, than in spo11Δ, where Spo11p is absent. The same difference is also seen in budding yeast (O. Nanassy and N. Kleckner, unpubl.). Thus, Spo11p may negatively regulate bouquet exit via a noncatalytic function. A negative role for Spo11p during yeast S phase was similarly defined (Cha et al. 2000). Factors that oppose progression are often used to create barriers, which can then be eliminated in an appropriately regulated way. Perhaps Spo11p first promotes DSBs and then, in its negative role, helps to couple bouquet exit to post-DSB recombinational steps. Spo11p is axis-associated in mouse (Romanienko and Camerini-Otero 2000) and, in Sordaria, Spo11-GFP forms lines along chromosomes during the bouquet stage. Thus, Spo11p might mediate its negative role by binding along chromosome axes.
Our results also have important implications for the role of the bouquet stage in meiosis. It is often suggested that the bouquet promotes homolog pairing. In wild-type Sordaria, DSB-independent recognition and substantial DSB-mediated pairing occurs before the bouquet stage. However, bouquet formation could be important for complete alignment. Also, because DSB-mediated control is exerted specifically on “exit” from the bouquet, rather than “entry”, exit could be the functionally critical aspect. Perhaps the accompanying chromosome dispersal destabilizes inappropriate nascent recombinational interactions that have created interlocks or ectopic and/or homoeologous linkages (e.g., Goldman and Lichten 2000; Niwa et al. 2000).
Meiotic DSBs destabilize intersister connections and axis continuity at the leptotene/zygotene transition in spo76-1
In spo76-1, AEs form normally at leptotene and then fracture and split at zygotene. We show here that destabilization of spo76-1 AEs is dependent on DSBs. We infer that DSBs also tend to destabilize wild-type axes and that spo76-1 chromosomes, being defective, are sensitized to such effects. A functional rationale for DSB-promoted axis destabilization can be suggested. Chiasmata are sites not only of crossing over at the DNA level but also of exchange at the axis level; furthermore, these changes involve only one chromatid of each homolog, implying that chiasma formation involves local differentiation and separation of sister chromatids. Axis destabilization and sister separation are precisely the DSB-promoted defects observed in spo76-1.
spo76-1 destabilization occurs well after DSB formation (above), during a period which corresponds to the time of the next major biochemical transition of recombination, onset of stable strand exchange, as defined in budding yeast; crossover control may also occur at this point (Hunter and Kleckner 2001). Interestingly, the number and distribution of spo76-1 AE fracturing events, determined in EM three-dimensional reconstructions (van Heemst et al. 1999) correspond to the number and distribution of crossover-designation events predicted for spo76-1. In wild type, 21 chiasmata occur per meiosis. In spo76-1, given a 30% reduction in Rad51 foci (van Heemst et al. 1999), only ∼15 crossover designations would be expected and each of the seven spo76-1 bivalents shows one to two disruptions (∼10-15 per nucleus). Furthermore, just as every wild-type bivalent always exhibits at least one chiasma, every spo76-1 bivalent exhibits at least one axis disruption. Thus, axis fracturing could be triggered at sites of axis-associated recombination complexes as part of the crossover-designation process.
Our finding further suggests that a fundamental role of Spo76p during prophase is to oppose the local effects of DSB-mediated axis destabilization. Destabilization both along axes and between sister chromatids could be driven by expansion of chromatin loops (Blat et al. 2002). If so, Spo76p could act as a spring-clip, enforcing axis integrity in opposition to chromatin expansion forces. The roles of other meiotic axis components can be interpreted similarly: Mutants of budding yeast and C. elegans defective for meiotic cohesin Rec8 show early centromere separation at/before diplotene (Klein et al. 1999; Pasierbek et al. 2001). Also, C. elegans rec8 mutants exhibit chromosome fragmentation at diakinesis that is suppressed by absence of SPO11 (Pasierbek et al. 2001); and a yeast mutant lacking axis component Red1p exhibits sister splitting at/before pachytene (Bailis and Roeder 1998).
Meiotic DSBs are required for regular progression through both meiotic divisions
In Sordaria, as in spo11 mutants of other organisms, the two meiotic divisions occur efficiently but with random segregation of homologs at meiosis I due to absence of chiasmata (e.g., Jiao et al. 1999; Buonomo et al. 2000; Shonn et al. 2000; Grelon et al. 2001; Sharif et al. 2002). Sordaria spo11 mutants also exhibit additional defects at division II: (1) An extra round of SPB duplication is seen in 50% of nuclei at prophase II; (2) sister chromatids separation is delayed at AII; and (3) both divisions are prolonged. Division I is also prolonged in budding yeast spo11Δ, where it reflects “spindle checkpoint” sensing of the absence of chiasmata (Shonn et al. 2000). The three Sordaria defects are restored by exogenous DSBs, and thus are direct or indirect consequences of the absence of DSBs. Sordaria spo11 mutants also exhibit a tendency for missegregation of sister chromatids at division II, which fits with the defective sister separation seen cytologically and the division II missegregation found in Schizosaccharomyces pombe rec12 (spo11 homolog) mutants (Sharif et al. 2002).
In wild-type meiosis, sister-arm connections are eliminated by the end of AI (e.g., Lee and Orr-Weaver 2001). Thus, the aberrantly retained sister connections seen in Sordaria spo11 mutants at division II could reflect a failure of normal sister arm separation during division I. Three considerations support this possibility. First, budding yeast spo11 mutants exhibit, at division I, persistence of both sister connectedness at telomeres and securin Pds1, whose release is required for cohesin cleavage (Buonomo et al. 2000; Shonn et al. 2000). Second, the extra round of SPB duplication seen at prophase II in Sordaria can be a hallmark of defective sister separation: The budding yeast gene that mediates cohesin cleavage was named ESP1 because corresponding mutants exhibit “Extra Spindle Pole bodies” during mitosis (Baum et al. 1988). Third, because a division I bivalent maintains its normal segregation behavior when micro-manipulated onto a division II spindle (Paliulis and Nicklas 2000), it would not be surprising if unreleased division I sister connections in spo11 mutants were maintained into division II.
In wild-type meiosis, sister separation at division I appears to result from the combination of two successive effects: programmed cleavage of sister chromatid cohesion followed by mechanical disruption of residual linkages by mechanical forces of poleward movement in regions distal to any chiasma(ta) (Lee and Orr-Weaver 2001). In spo11 mutants, because chiasmata are absent, the latter forces are not imposed and “residual” linkages will thus never be eliminated. This effect could explain the retention of sister-arm connections into division II when DSBs are absent. Alternatively, or in addition, the DSB-mediated tendency for sister separation at mid-prophase (above) may help to prepare sister linkages for elimination at division I in wild-type meiosis, with spo11 mutants lacking this additional meiosis-specific “assistance”. Finally, the spindle checkpoint likely delays programmed cohesin cleavage in a spo11 mutant (Shonn et al. 2000), thus potentially exacerbating other defects.
Meiotic DSBs trigger arrest in spo76-1
DSBs are required for the permanent MI arrest seen in Sordaria spo76-1. One possible explanation is that residual lesions remain at DSB sites and are sensed by a DNA damage checkpoint, which triggers arrest. On the other hand: (1) spo76-1 diplotene chromosomes are all single chromatids and no broken chromatids are seen; (2) Rad51 foci appear on spo76-1 chromosomes at leptotene and disappear at pachytene, suggesting that recombination can proceed beyond the DSB stage at all sites (van Heemst et al. 1999; this work); (3) in other organisms, defects in meiotic recombination and SC status at mid-prophase result in arrest at the end of prophase, not at MI (Roeder and Bailis 2000). A second possible explanation is that spo76-1 arrest is triggered by the spindle checkpoint. However, in budding yeast, the spindle checkpoint confers only a delay in mitotic progression, not permanent arrest (Nasmyth 2001).
These considerations raise a general question: Given that connections between homologs play such an important role during meiosis, why does there appear to be no mechanism that specifically senses the absence of such connections, as seen by the fact that DSB-negative mutants still carry out division I despite the absence of chiasmata? We would like to propose that there is such a mechanism, and that it is set up in response to DSBs as part of the general cascade of DSB-triggered, chiasma-related processes. This mechanism would be absent in spo11 and other DSB-negative mutants, as well as spo11 spo76-1, but present in spo76-1 and γ-irradiated spo11 spo76-1, thus triggering arrest. The most intriguing possibility would be direct monitoring of tension on/at chiasmata at MI.
General implications
Meiotic DSBs promote not only recombination, SC formation, and homolog segregation but an entire cascade of downstream processes, all related to formation or function of chiasmata. In contrast, basic organization and morphogenesis along the chromosomes, some early homolog recognition, bouquet formation and exit, plus the mechanics of the two meiotic divisions, are all independent of DSBs. Thus, the DSB cascade has likely been superimposed upon a foundation of more basic processes. Exogenous DSBs substitute efficiently for programmed Spo11-mediated DSBs for many events of this cascade (Thorne and Byers 1993; Dernburg et al. 1998; Celerin et al. 2000; Malkova et al. 2000; Romanienko and Camerini-Otero 2000; this work). However, programmed DSBs may be required for regulatory features, for example, that ensure at least one DSB and/or at least one crossover per bivalent or crossover interference.
The meiotic program must integrate two antithetical activities. DNA DSBs create lesions that could potentially destroy the genome but ultimately create the substrates for chiasmata required for regular chromosome partitioning and viable gametes (Lichten 2001). The current results suggest that the same antithetical requirements may exist at the structural level: Destabilization of chromosome axis integrity must be permitted locally at recombination sites while being maintained globally via the action of axis-related proteins such as Spo76/Pds5.
Materials and methods
Cloning and sequencing of SPO11
A SPO11-containing cosmid clone was identified by PCR in a S. macrospora indexed genomic cosmid library (Pöggeler et al. 1997) using primers based on Neurospora crassa SPO11 (Munich Information Center for Protein Sequences; code 13e11 290, contig 9a3). Mutants were sequenced on both strands, with gene-specific primers (Sigma Genosys). Sequencing used the BigDyeTM Terminator, Cycle Sequencing Kit (Applied) and a 310 DNA Sequencer (Applied) or Thermo Sequenase (USB kit). Total RNA, enriched for meiotic transcripts, was obtained by guanidine thiocyante extraction of young fruiting bodies. cDNAs were made using Gene racer KIT (Invitrogen) and directly sequenced.
Strains, plasmids and transformations of Sordaria
The homothallic S. macrospora wild-type strain, from which all mutants are derived, is “St. Ismier” strain FGSC 4818. spo11Δ, Y171F, and GFP alleles were constructed by standard methods. In spo11Δ, a hygromycin resistance cassette replaces the SPO11 coding region (3 bp upstream of the ATG to 192 bp downstream from the stop codon). In SPO11-GFP, GFP coding sequence (pEGFP-1; Clontech) was fused just after C-terminal amino acid 481. Strains carrying mutant alleles substituted for wild-type SPO11 were obtained by transformation with selection for hygromycin resistance (spo11Δ) or cotransformation with a second plasmid-encoding phleomycin resistance (spo11-Y171F) plus confirmation by Southern blotting. GFP alleles were integrated ectopically by selection for linked hygromycin resistance (van Heemst et al. 1999).
Genetic analysis
The Sordaria life cycle provides a powerful tool for analysis of aneuploid genotypes, by vegetative outgrowth and back-crossing (Moreau et al. 1985). Wild-type ascospores are black; yas1 and pam2 ascospores are yellow and pink, respectively. The frequency of exchanges in the pam2-yas1 interval of chromosome 4 was estimated by the frequency of (++) over (++) and (+pam2) genotypes. The frequency of disomy for chromosome 4 was analyzed in viable ascospores issued from the following crosses: pam2 spo11Δ × yas1 spo11Δ, pam2 spo11-1 × yas1 spo11-1, and pam2 spo11-8 × yas1 spo11-8, according to Moreau et al. (1985). The dose of irradiation (200 Gy) that gave optimal restoration of spores is precisely that predicted to give at least one crossover per bivalent (details available upon request).
Cytology
Asci were processed for immunofluorescence as described (Thompson-Coffe and Zickler 1993). Primary antibodies were MPM-2 (kindly provided by P.N. Rao) at 1:300, anti-Rad51 (Oncogene) at 1:400, anti-GFP (Roche) at 1:500, and anti-α-tubulin (Amersham) at 1:500. Secondary antibodies were FITC anti-mouse and anti-rabbit or CyTM3 anti-rabbit (Jackson) at 1:100 and 1:4000. Controls included primary or secondary antibodies alone. Chromatin was visualized with DAPI (0.5 µg/mL). All samples, including EGFP, were observed on a Zeiss Axioplan microscope with a CCD Princeton camera and a Zeiss LSM 510 Confocal Laser Scanning Microscope (LSM software). For light and electron microscopy, asci were fixed and analyzed as in Zickler (1977). Three-dimentional rendering used Vector Works and Zoom software.
Acknowledgments
This paper is dedicated to the memory of Ahn Dao Huynh. We thank Stefanie Pöggeler for cosmid identification help. This work, A.S., and S.T. were supported by grants from the Centre National de la Recherche Scientifique (UMR 8621) to D.Z. and from the National Institutes of Health (GM25326) to N.K. S.G. was supported by MURST-CNR program lex 488/92 Cluster 02.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.275203.
References
- Bailis J.M. and Roeder, G.S. 1998. Synaptonemal complex morphogenesis and sister-chromatid cohesion require Mek1-dependent phosphorylation of a meiotic chromosomal protein. Genes & Dev. 12: 3551-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P., Yip, C., Goetsch, L., and Byers, B. 1988. A yeast gene essential for regulation of spindle pole duplication. Mol. Cell Biol. 8: 5386-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat A., de Massy, B., Gadelle, D., Varoutas, P.C., Nicolas, A., and Forterre, P. 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386: 414-417. [DOI] [PubMed] [Google Scholar]
- Bishop D.K. 1994. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79: 1081-1092. [DOI] [PubMed] [Google Scholar]
- Blat U., Protacio, R.U., Hunter, N., and Kleckner, N. 2002. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111: 1-12. [DOI] [PubMed] [Google Scholar]
- Buonomo S.B., Clyne, R.K., Fuchs, J., Loidl, J., Uhlmann, F., and Nasmyth, K. 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103: 387-398. [DOI] [PubMed] [Google Scholar]
- Celerin M., Merino, S.T., Stone, J.E., Menzie, A.M., and Zolan, M.E. 2000. Multiple roles of Spo11 in meiotic chromosome behavior. EMBO J. 19: 2739-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha R.S., Weiner, B.M., Keeney, S., Dekker, J., and Kleckner, N. 2000. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes & Dev. 14: 493-503. [PMC free article] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald, K., Moulder, G., Barstead, R., Dresser, M., and Villeneuve, A.M. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387-398. [DOI] [PubMed] [Google Scholar]
- Goldman A.S.H. and Lichten, M. 2000. Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc. Natl. Acad. Sci. 97: 9537-9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M., Vezon, D., Gendrot, G., and Pelletier, G. 2001. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20: 589-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T., Stead, K., Koshland, D., and Guacci, V. 2000. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 151: 613-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. and Kleckner, N. 2001. The single-end invasion: An asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106: 59-70. [DOI] [PubMed] [Google Scholar]
- Huynh A.D., Leblon, G., and Zickler, D. 1986. Indirect intergenic suppression of a radiosensitive mutant of Sordaria macrospora defective in sister-chromatid cohesiveness. Curr. Genet. 10: 545-555. [DOI] [PubMed] [Google Scholar]
- Jiao K., Bullard, S.A., Salem, L., and Malone, R.E. 1999. Coordination of the initiation of recombination and the reductional division in meiosis in Saccharomyces cerevisiae. Genetics 152: 117-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52: 1-53. [DOI] [PubMed] [Google Scholar]
- Klapholz S., Waddell, C.S., and Esposito, R.E. 1985. The role of the SPO11 gene in meiotic recombination in yeast. Genetics 110: 187-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Mahr, P., Galova, M., Buonomo, S.B., Michaelis, C., Nairz, K., and Nasmyth, K. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91-103. [DOI] [PubMed] [Google Scholar]
- Lee J.Y. and Orr-Weaver, T.L. 2001. The molecular basis of sister-chromatid cohesion. Annu. Rev. Cell. Dev. Biol. 17: 753-777. [DOI] [PubMed] [Google Scholar]
- Lichten M. 2001. Meiotic recombination: Breaking the genome to save it. Curr. Biol. 11: R253-R256. [DOI] [PubMed] [Google Scholar]
- MacQueen A.J., Colaiacovo, M.P., McDonald, K., and Villeneuve, A.M. 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes & Dev. 16: 2428-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A., Klein, F., Leung, W.Y., and Haber, J.E. 2000. HO endonuclease-induced recombination in yeast meiosis resembles Spo11-induced events. Proc. Natl. Acad. Sci. 97: 14500-14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K.S. and Hayashi-Hagihara, A. 1998. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: Evidence that the mechanism for initiating meiotic recombination is conserved. Genes & Dev. 12: 2932-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens P.B., Kolas, N.K., Tarsounas, M., Marcon, E., Cohen, P.E., and Spyropoulos, B. 2002. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115: 1611-1622. [DOI] [PubMed] [Google Scholar]
- Moreau P.J.F., Zickler, D., and Leblon, G. 1985. One class of mutants with disturbed centromere cleavage and chromosome pairing in Sordaria macrospora. Mol. Gen. Genet. 198: 189-197. [Google Scholar]
- Nabeshima K., Kakihara, Y., Hiraoka, Y., and Nojima, H. 2001. A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J. 20: 3871-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. 2001. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35: 673-745. [DOI] [PubMed] [Google Scholar]
- Niwa O., Shimanuki, M., and Miki, F. 2000. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 19: 3831-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliulis L.V. and Nicklas, R.B. 2000. The reduction of chromosome number in meiosis is determined by properties built into the chromosomes. J. Cell Biol. 150: 1223-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch, M., Melcher, M., Schleiffer, A., Schweizer, D., and Loidl, J. 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes & Dev. 15: 1349-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples T.L., Dean, E., Gonzalez, O., Lambourne, L., and Burgess, S.M. 2002. Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes & Dev. 16: 1682-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler S., Nowrousian, M., Jacobsen, S., and Kück, U. 1997. An efficient procedure to isolate fungal genes from an indexed cosmid library. J. Microbiol. Methods 29: 49-61. [Google Scholar]
- Roeder G.S. and Bailis, J.M. 2000. The pachytene checkpoint. Trends Genet. 16: 395-403. [DOI] [PubMed] [Google Scholar]
- Romanienko P.J. and Camerini-Otero, R.D. 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6: 975-987. [DOI] [PubMed] [Google Scholar]
- Scherthan H. 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell. Biol. 2: 621-627. [DOI] [PubMed] [Google Scholar]
- Scherthan H., Jerratsch, M., Dhar, S., Wang, Y.A., Goff, S.P., and Pandita, T.K. 2000. Meiotic telomere distribution and Sertoli cell nuclear architecture are altered in Atm- and Atm-p53-deficient mice. Mol. Cell Biol. 20: 7773-7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif W.D., Glick, G.G., Davidson, M.K., and Wahls, W.P. 2002. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome 1: 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonn M.A., McCarroll, R., and Murray, A.W. 2000. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 289: 300-303. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Hao, Z., Kai, M., and Okayama, H. 2001. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 20: 5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Coffe C. and Zickler, D. 1993. Cytoskeletal interactions in the ascus development and sporulation of Sordaria macrospora. J. Cell Sci. 104: 883-898. [Google Scholar]
- Thorne L.W. and Byers, B. 1993. Stage-specific effects of X-irradiation on yeast meiosis. Genetics 134: 29-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken E., Loidl, J., and Scherthan, H. 1999. Bouquet formation in budding yeast: Initiation of recombination is not required for meiotic telomere clustering. J. Cell Sci. 112: 651-658. [DOI] [PubMed] [Google Scholar]
- van Heemst D., James, F., Poggeler, S., Berteaux-Lecellier, V., and Zickler, D. 1999. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell 98: 261-271. [DOI] [PubMed] [Google Scholar]
- van Heemst D., Kafer, E., John, T., Heyting, C., van Aalderen, M., and Zickler D. 2001. BimD/Spo76 is at the interface between cell cycle progression, chromosome morphogenesis and recombination. Proc. Natl. Acad. Sci. 98: 6267-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.W., Read, R.L., and Norbury, C.J. 2002. Fission yeast Pds5 is required for accurate chromosome segregation and for survival after DNA damage or metaphase arrest. J. Cell Sci. 115: 587-598. [DOI] [PubMed] [Google Scholar]
- Zickler D. 1977. Development of the synaptonemal complex and the 'recombination nodules' during meiotic prophase in the seven bivalents of the fungus Sordaria macrospora. Chromosoma 61: 289-316. [DOI] [PubMed] [Google Scholar]
- Zickler D. and Kleckner, N. 1998. The leptotene-zygotene transition of meiosis. Ann. Rev. Genet. 32: 619-697. [DOI] [PubMed] [Google Scholar]
- ____. 1999. Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33: 603-754. [DOI] [PubMed] [Google Scholar]