Abstract
The human Y-family DNA polymerases, Polι, Polη, and Polκ, function in promoting replication through DNA lesions. However, because of their low fidelity, any involvement of these polymerases in DNA synthesis during base excision repair (BER) would be highly mutagenic. Mechanisms, therefore, must exist to exclude their participation in BER. Here, we show that although Polι, Polη, and Polκ are all able to form a covalent Schiff base intermediate with the 5′-deoxyribose phosphate (5′-dRP) residue that results from the incision of DNA at an abasic site by an AP endonuclease, they all lack the ability for the subsequent catalytic removal of the 5′-dRP group. Instead, the covalent trapping of these polymerases by the 5′-dRP residue inhibits their DNA synthetic activity during BER. The unprecedented ability of these polymerases for robust Schiff base formation without the release of the 5′-dRP product provides a means of preventing their participation in the DNA synthetic step of BER, thereby avoiding the high incidence of mutagenesis and carcinogenesis that would otherwise occur.
Keywords: Y-family DNA polymerases, base excision repair, abasic site, 5′-doexyribose phosphate, 5′-dRP lyase
Abasic (AP) sites, resulting from spontaneous hydrolysis of the N-glycosylic bond, arise in DNA relatively frequently, as hydrolytic depurination causes the loss of ∼9000 bases per day from the DNA of a mammalian cell (Nakamura et al. 1998). AP sites are also formed in DNA as intermediates in the removal of damaged bases by base excision repair (BER; Wallace 1997). A plethora of endogenous and exogenous agents yield modified or damaged bases. For example, the DNA bases cytosine and 5-methyl cytosine are subject to hydrolytic deamination to uracil and thymine, respectively, and oxygen-free radicals produced during normal cellular metabolism are an important source of endogenous damage to DNA bases that results in oxidized purines such as 8-hydroxyguanine, and ring-saturated derivatives of pyrimidines such as thymine glycol (Wallace 1994, 1997; Seeberg et al. 1995). The different types of modified or damaged bases are removed by specific DNA glycosylases by cleavage of the N-glycosylic bond between the base and the deoxyribose moiety of the nucleotide. The resulting AP site is then acted on by a class II AP endonuclease, which cleaves the phosphodiester backbone on the 5′ side of the AP site, leaving a 3′-hydroxyl group and 5′-baseless deoxyribose-5′-phosphate (5′-dRP) residue (Wallace 1994, 1997; Seeberg et al. 1995). The 5′-dRP residue is then removed by a dRP lyase, such as that contained in DNA polymerase (Pol) β (Matsumoto and Kim 1995; Piersen et al. 1996), and the resulting single nucleotide gap is filled in by Polβ (Wilson 1998). Because of the high frequency with which endogenous base modifications, base damages, and AP sites are generated, the repair synthesis step of BER needs to be fairly accurate to keep the incidence of mutations low.
Polβ is especially adapted to function in BER because it can efficiently catalyze both the removal of the 5′-dRP group via β-elimination and the subsequent single-nucleotide gap-filling reaction. Limited proteolysis studies have revealed two independently folded domains in Polβ, the 8-kD N-terminal portion, which contains the 5′-dRP lyase activity, and the 31-kD C-terminal portion, which contains the polymerase activity. Lys 72 in the conserved helix–turn–helix motif of the 8-kD domain acts as the nucleophile that initiates the 5′-dRP removal reaction (Prasad et al. 1998b; Deterding et al. 2000). First, Lys 72 initiates an attack on the C1 position of the 5′-dRP group. Next, a transient covalent intermediate is formed in which the DNA substrate is linked covalently to the enzyme as a Schiff base that can be stabilized by reduction with sodium borohydride. Finally, the 5′-dRP-group is removed from the DNA and released from Polβ (Matsumoto et al. 1998; Prasad et al. 1998a). Polβ-null mice cells are hypersensitive to the cytotoxic effects of the alkylating agent methyl methanesulphonate (MMS), indicating an in vivo requirement for Polβ in the repair of alkylated bases (Sobol et al. 1996). Only the 5′-dRP lyase activity of Polβ, however, is critical for resistance to MMS, as Polβ-null mice cells stably transfected with a Polβ minigene carrying mutations in the 5′-dRP lyase active site still retain their sensitivity to MMS, whereas transfection with a Polβ minigene carrying mutations in the DNA polymerase active site complement the MMS sensitivity of Polβ-null mice cells (Sobol et al. 2000). Thus, in mammalian cells, Polβ is primarily responsible for the rate-limiting removal of the 5′dRP-group, whereas the other DNA polymerases can functionally substitute for the polymerase activity of Polβ in BER.
The human Y-family DNA polymerases, Polη, Polι, and Polκ, promote replication through DNA lesions that present a block to the normal replication machinery. Polη replicates through UV-induced thymine–thymine (TT) dimers with the same efficiency and accuracy as it replicates through undamaged Ts (Johnson et al. 1999b, 2000c; Washington et al. 2000), and genetic studies in yeast have implicated Polη in the error-free bypass of cyclobutane dimers formed at TC and CC sites (Yu et al. 2001). Because of the involvement of Polη in the error-free bypass of cyclobutane pyrimidine dimers, its mutational inactivation in humans causes an increase in the incidence of UV mutagenesis and leads to the cancer-prone syndrome, the variant form of xeroderma pigmentosum (Johnson et al. 1999a; Masutani et al. 1999). Polη can efficiently replicate through other DNA lesions as well such as 7,8-dihydro-8-oxoguanine (8-oxoG) and O6-methyl guanine (m6G; Haracska et al. 2000a,b). In contrast to Polη, the ability of which to replicate through DNA lesions derives both from its ability to proficiently incorporate nucleotides opposite the lesion site and to proficiently extend from the inserted nucleotide, the bypass of certain DNA lesions requires the sequential action of two DNA polymerases, wherein one inserts the nucleotide opposite the lesion site and the other extends from the inserted nucleotide (Prakash and Prakash 2002). Polι is able to incorporate nucleotides opposite the 3′-T of a (6–4) T–T photoproduct or opposite an AP site; but it does not extend from the nucleotide opposite these lesions sites (Johnson et al. 2000b). Polκ, on the other hand, is unable to insert nucleotides opposite DNA lesions, as, for example, opposite the 3′-T of a TT dimer or opposite an m6G lesion, but it can efficiently extend from the nucleotides opposite these lesion sites (Haracska et al. 2002a; Washington et al. 2002). The Y-family polymerases possess an active site that is more tolerant of geometric distortions in the DNA than the replicative DNA polymerases that are unable to bypass DNA lesions. The cost of the ability of Polη, Polι, and Polκ to promote lesion bypass, however, is borne by their low fidelity. Steady-state kinetic analyses have indicated that Polη and Polκ misincorporate nucleotides with a frequency of ∼10-2 to 10-3 and ∼10-3 to 10-4, respectively (Washington et al. 1999; Johnson et al. 2000a,c). The fidelity of Polι is much poorer than that of Polη, particularly for incorporating nucleotides opposite the template pyrimidines (Johnson et al. 2000b; Tissier et al. 2000).
Previously, a role for human Polι has been implicated in BER (Bebenek et al. 2001). However, considering that Polι is an extremely low-fidelity DNA polymerase, DNA synthesis by this enzyme in BER would be very highly mutagenic. Here, we address whether the human Y-family DNA polymerases Polη, Polι, and Polκ can, in fact, play a role in BER, or whether these low-fidelity polymerases are somehow excluded from the DNA synthesis step of BER. Interestingly, we found that although each of these human polymerases can form a covalent Schiff base complex with the 5′-dRP group in DNA, they are unable to carry out the subsequent removal of the 5′-dRP group. Furthermore, and in striking contrast to Polβ, the DNA synthetic activity of Polι, Polη, and Polκ is inhibited upon their binding to the 5′-dRP group. From these and other observations presented here, we conclude that nonproductive binding to the 5′-dRP group represents a means to exclude these DNA polymerases from the DNA synthetic step of BER.
Results
Human DNA polymerases ι, η, and κ form a covalent complex with the 5′-dRP group
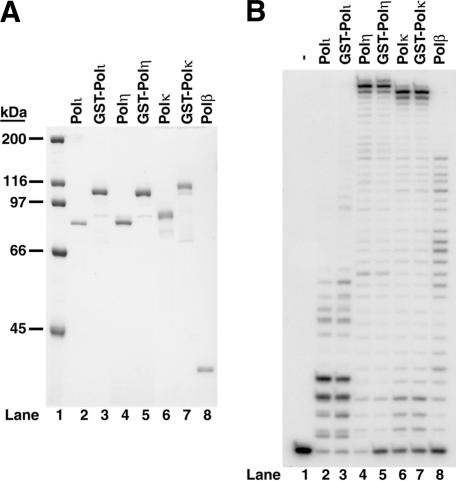
We purified human Polι, Polη, and Polκ as native as well as N-terminal GST-fusion proteins (Fig. 1A), and first examined their DNA synthesis activities. As shown in Figure 1B, the DNA synthesis activity of the GST-fusion polymerases were nearly identical to the corresponding native enzymes. Thus, the GST-fusion does not interfere with the activity of Polι, Polη, or Polκ.
Figure 1.
Purity and DNA synthetic activity of human Polι, Polη, Polκ, and Polβ. (A) Purified native and N-terminal GST-fusion Polι, Polη, Polκ, and native Polβ were analyzed on an 8% denaturing polyacrylamide gel and stained with Coomassie blue. (Lane 1) Molecular weight standards containing 500 ng of protein in each band; (lane 2) 300 ng of Polι; (lane 3) 500 ng of GST-Polι; (lane 4) 300 ng of Polη; (lane 5) 500 ng of GST-Polη; (lane 6) 500 ng of Polκ; (lane 7) 500 ng of GST-Polκ; (lane 8) 300 ng of Polβ. (B) The DNA synthetic activity of purified GST-fusion and native Polι, Polη, Polκ, and native Polβ was assayed using a 75-nt template oligonucleotide primed with a 40-nt 5′-32P-labeled oligonucleotide (S1 substrate) in the presence of all four deoxynucleotides (100 μM) in buffer A containing 40 mM Tris-HCl (pH 7.5), 8 mM MgCl2, 1 mM dithiothreitol, 100 μg/mL bovine serum albumin, and 10% glycerol. The DNA substrate (20 nM) was incubated with 2 nM of DNA polymerase at 37°C for 10 min. The reaction products were analyzed on an 8 M urea, 8% polyacrylamide gel, and the DNA bands were visualized by autoradiography.
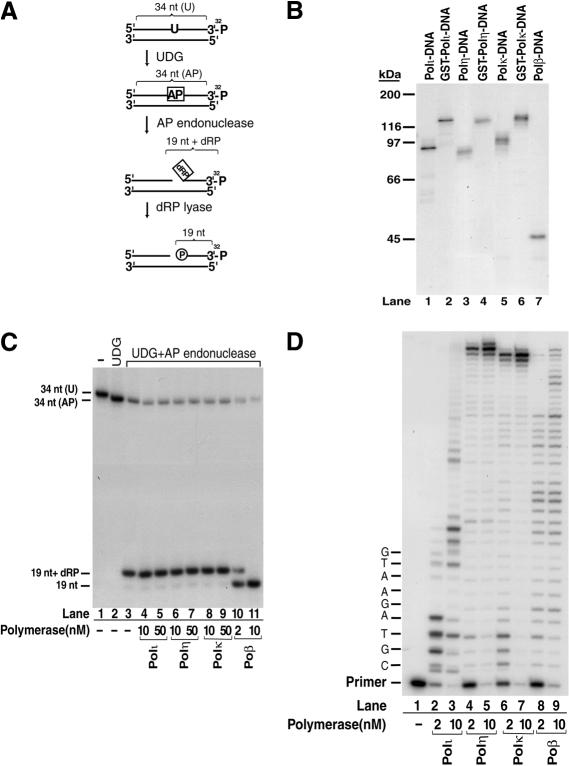
Next, we examined whether Polι, Polη, and Polκ, can form a Schiff-base covalent complex with the 5′-dRP-containing DNA substrate. A Schiff base can be formed between a properly positioned N-terminal α NH2 group (Dodson et al. 1993) or an ε NH2 group (Matsumoto et al. 1998; Prasad et al. 1998a) of a protein and the –CHO group of an open abasic residue in DNA, and the covalent linkage between DNA and protein can be trapped by the reduction of this Schiff base with sodium borohydride (Piersen et al. 1996). As outlined in Figure 2A, we generated a DNA substrate containing a 5′-dRP residue at an internal nick by the sequential treatment of a single-uracil-containing DNA duplex with uracil DNA glycosylase and a class II AP endonuclease. Figure 2B shows that, similarly to Polβ, Polι, Polη, and Polκ are active in the borohydride trapping assay. Because of the cross-linking of the polymerase to the 5′-dRP-containing DNA, the trapped products exhibit a slower mobility on an SDS–polyacrylamide gel than the unmodified polymerases (cf. Figs. 2B and 1A). We were able to cross-link the GST-fusion proteins as well as the native polymerases to the DNA, which indicates that the GST-fusion part is not required for the Schiff base formation. In conclusion, Polι, Polη, and Polκ are able to react with the 5′-dRP residues in DNA to form a Schiff-base covalent complex.
Figure 2.
Covalent binding of human Y-family DNA polymerases to the 5′-dRP residue in DNA. (A) Schematic representation of reactions to generate a 5′-dRP-group-containing DNA substrate. The initial substrate (S2 substrate) was generated by annealing a single-uracil-containing 3′-end-labeled 34-nt oligonucleotide [34 nt (U)] to its complementary oligonucleotide. Treatment with UDG generated an abasic-site-[34 nt (AP)]-containing DNA, which was incised at the 5′ side of the abasic site by a class II AP endonuclease, generating the 5′-dRP-containing substrate (19 nt + dRP). Removal of the 5′-dRP residue by dRP lyase will result in a 3′-32P-labeled 19-nt DNA fragment. (B) Trapping of polymerase–DNA covalent complexes with sodium borohydride (NaBH4). In 10 μL of reaction mixture, the 3′-32P-labeled 5′-dRP-containing DNA (100 nM), shown in A, was incubated with 50 nM each Polι (lane 1), GST-Polι (lane 2), Polη (lane 3), GST-Polη (lane 4), Polκ (lane 5), GST-Polκ (lane 6), or Polβ (lane 7) in buffer B containing 50 mM HEPES (pH 7.5), 10 mM MgCl2, 20 mM KCl, and 2 mM dithiothreitol, in the presence of 20 mM NaBH4 for 30 min on ice. Reactions were terminated by the addition of SDS-containing loading buffer, samples were resolved on an 8% SDS-PAGE gel, and products were visualized by autoradiography. (C) 5′-dRP lyase activity. The 5′-dRP-containing DNA substrate (20 nM) generated as shown in A was incubated in buffer B at 37°C for 10 min with Polι (lanes 4,5), Polη (lanes 6,7), Polκ (lanes 8,9), and Polβ (lanes 10,11), which were present at two different concentrations as indicated, followed by stabilization of the reaction product by incubation in the presence of 340 mM NaBH4 at 0°C for 30 min. The products were analyzed by a denaturing 16% polyacryamide gel and visualized by autoradiography. (D) DNA synthesis activity. S1 DNA substrate (20 mM) containing a 5′-32P-labeled primer was incubated with the DNA polymerase added at two different concentrations (2 nM or 10 nM), as indicated, in the presence of all four deoxynucleotides (100 μM) in buffer B at 37 °C for 10 min.
Lack of 5′-dRP lyase activity in human DNA polymerases ι, η, and κ
Although Schiff base formation is a prerequisite for a nonhydrolytic dRP lyase reaction, it does not necessitate a strong subsequent β-elimination reaction. To test whether the Schiff base formed by Polι, Polη, and Polκ with DNA was an intermediate in the 5′-dRP excision reaction, we examined whether Polι, Polη, and Polκ could excise the 5′-dRP group. We directly compared the 5′-dRP lyase (Fig. 2C) and DNA polymerase (Fig. 2D) activities of Polι, Polη, and Polκ at different enzyme concentrations, and as a control, we used Polβ. Whereas Polβ displayed a strong activity for removing the 5′-dRP group from the AP endonuclease-incised oligonucleotide (19 nt + dRP band), thereby generating the 19-nt product that lacks the 5′-dRP group and therefore has a faster mobility (Fig. 2C, lanes 10,11), we were unable to detect any 5′-dRP lyase activity associated with Polι, Polη, or Polκ (Fig. 2C, lanes 4–9). Despite the fact that at 2 nM concentration, Polι, Polη, and Polκ had as robust a DNA polymerase activity as Polβ (Fig. 2D), neither Polι, Polη, nor Polκ removed any 5′-dRP group even at as high a concentration as 50 nM. With Polβ, however, as little as 2 nM enzyme removed 70% of the 5′-dRP group, and 10 nM Polβ could remove all of the 5′-dRP groups from the DNA. Thus, although the human Y-family DNA polymerases are all able to form a covalent complex with the 5′dRP-group in DNA, the resulting polymerase–DNA complexes do not act as intermediates for the 5′-dRP excision reaction.
To further verify the absence of a 5′-dRP lyase activity in DNA polymerases ι, η, and κ, we examined the kinetics of borohydride cross-linking with these enzymes. Depending on whether the polymerase–5′-dRP–DNA complex is an intermediate in the dRP lyase reaction or not, the borohydride cross-linking should occur with different kinetics, and therefore, the amount of the cross-linked product should change if the polymerase were first preincubated with the 5′-dRP substrate for varied intervals before the addition of borohydride. For an enzyme such as Polβ, which has a robust 5′-dRP lyase activity, the cross-linked protein–DNA complex would diminish following preincubation with the enzyme because the enzyme would have removed the 5′-dRP group from the DNA. However, for an enzyme that has no 5′-dRP lyase activity, as our results indicate for Polι, Polη, and Polκ, preincubation of these enzymes with the 5′-dRP substrate should cause no decrease in the amount of cross-linked protein–DNA complex. Whereas Polι, Polη, and Polκ, as well as Polβ, rapidly formed a complex with the 5′-dRP substrate, as even without any preincubation, we were able to trap their complex with borohydride (Fig. 3, lane 1), striking differences were observed between Polβ and the other polymerases upon their preincubation with the 5′-dRP substrate before borohydride addition. Thus, after preincubation for 10 min, the intensity of the Polβ–5′-dRP complex diminished, while the intensity of the Polι–5′-dRP, Polη–5′-dRP, and Polκ–5′-dRP–DNA complexes increased (Fig. 3, cf. lanes 1 and 2). With a 30-min preincubation, the amounts of Polι, Polη, and Polκ covalent complexes showed a further increase, but no cross-linked product could be seen with Polβ (Fig. 3, lane 3). These results show that in contrast to Polβ, which has a potent 5′-dRP lyase activity, Polι, Polη, and Polκ did not excise the 5′-dRP group after complex formation with the 5′-dRP in DNA, and instead, with these DNA polymerases, the amount of cross-linked product increased with the increase in preincubation time. These are the results expected for an enzyme that has no 5′-dRP lyase activity but can form a covalent complex with the 5′-dRP group in DNA.
Figure 3.
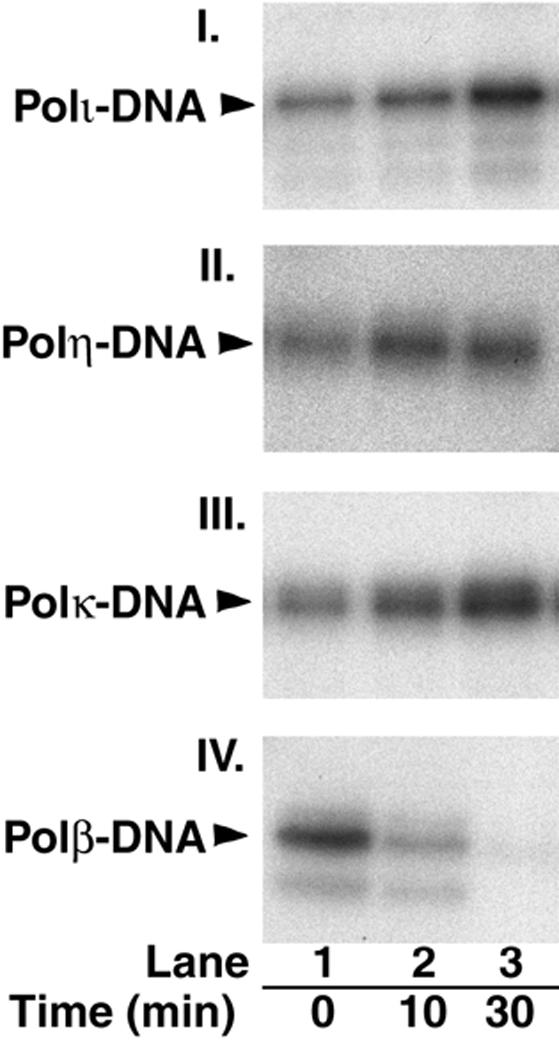
Time course of polymerase–DNA cross-linking by borohydride. Polι (panel I), Polη (panel II), Polκ (panel III), or Polβ (panel IV; each at 10 nM) were mixed at 0°C with the internal 5′-dRP-containing 3′-32P-labeled DNA (50 nM) shown in Figure 2A, followed by incubation at 37°C for 0, 10, or 30 min (lanes 1–3, respectively) before the addition of 20 mM NaBH4 and further incubation at 0°C for 30 min. After the addition of SDS-containing loading buffer, the samples were resolved on an 8% SDS–polyacrylamide gel, and the cross-linked polymerase–DNA products, indicated by arrows on the left of panels, were analyzed by autoradiography.
Exclusion of DNA Polymerases ι, η, and κ from DNA synthesis at an incised abasic site
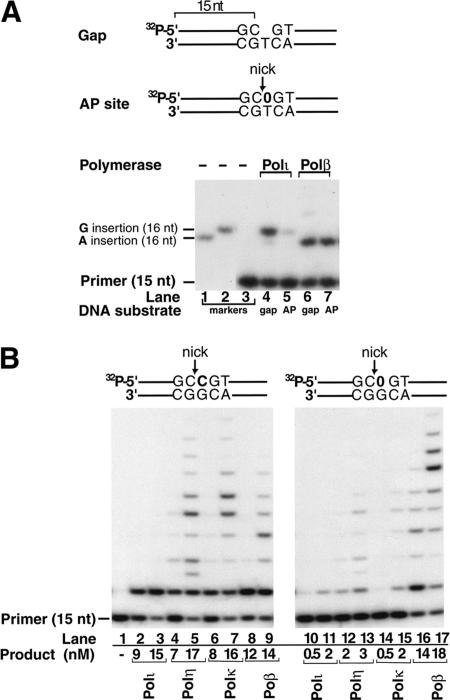
The ability of Polι, Polη, and Polκ, to form a covalent complex with the 5′-dRP group without carrying out the subsequent lyase reaction raised the possibility that binding to the 5′-dRP group precludes these polymerases from carrying out the DNA synthesis step during BER. To test this possibility, first we compared the DNA synthetic activity of Polι and Polβ on two DNA substrates, one of which was incised at the AP site and the other had a single nucleotide gap (Fig. 4A). To examine DNA synthesis, and to determine which nucleotide is inserted opposite the template residue T, which is situated opposite the 5′-incised AP site or opposite the single nucleotide gap, we monitored the extension of the 5′-32P-labeled 15-nt standing start primer, the 3′-end of which is situated right before the gap or the AP site, in the presence of all four deoxynucleotides. We compared the electrophoretic mobilities of the products of DNA synthesis with 16-nt oligonucleotide markers containing the 15-nt primer plus a G or an A residue at position 16, on a 20% polyacrylamide gel (Fig. 4A, lanes 1,2). As expected, whereas Polβ inserted an A opposite the template T (Fig. 4A, lanes 6,7), Polι inserted predominantly a G opposite the template T (Fig. 4A, lanes 4,5). In striking contrast to Polβ, which displayed nearly identical DNA synthetic activity on the two DNA substrates (Fig. 4A, lanes 6,7), the DNA synthetic activity of Polι was severely inhibited on the nicked AP-site-containing DNA (Fig. 4A, cf. lanes 4 and 5). Thus, not only does Polι have an extremely low fidelity in single nucleotide gap filling, it is also very poor in carrying out the DNA synthesis reaction on the 5′-nicked AP-site-containing DNA substrate.
Figure 4.
Exclusion of Y-family DNA polymerases from DNA synthesis on a 5′-dRP-containing DNA substrate. (A) The DNA synthetic activity of Polι and Polβ on a DNA substrate containing a single nucleotide gap or a 5′-dRP residue at the nick. Polι (2 nM; lanes 4,5) or Polβ (2 nM; lanes 6,7) was incubated with the DNA substrate (20 nM) at 37°C for 10 min in the presence of all four deoxynucleotides in buffer B. On the schematic representation of DNA substrates (shown on top), the AP site is indicated by a 0. The gapped substrate (S3–; lanes 4,6) and the substrate nicked at the AP site (S4–; lanes 5,7) contained a template T opposite both the gap and the AP site, and the 15-nt primer was 5′-32P-labeled. The nucleotide inserted opposite the T was determined by comparing the products of DNA synthesis reactions to 16-nt 5′-32P-labeled oligonucleotide markers containing the 15-nt primer (lane 3) plus an A (lane 1) or a G (lane 2) at their 3′-end. The reaction products were resolved on an 8 M urea, 20% polyacrylamide gel, and the DNA bands were visualized by autoradiography. (B) DNA synthetic activity of Polι, Polη, Polκ, and Polβ on DNA substrates containing a nick 5′ to a C or a nick at an AP site. DNA polymerases, each at 2 nM or 5 nM concentrations, were incubated with the DNA substrate (20 nM) at 37°C for 20 min in the presence of all four deoxynucleotides in buffer B. The DNA substrates nicked 5′ to a C (S5 substrate; lanes 1–9) or nicked 5′ to an AP site (S6 substrate; lanes 10–17) are shown schematically. The amount of reaction product was calculated from the intensity of bands generated by DNA synthesis divided by the intensity of all the bands.
To test if the 5′-dRP group inhibits the DNA synthesis activity of all three Y-family DNA polymerases, we compared the activity of Polι, Polη, Polκ, and Polβ on two DNA substrates containing a 5′-C or a 5′-dRP residue at the nick (Fig. 4B). Compared with DNA synthesis on the nicked DNA substrate without the 5′-dRP residue (Fig. 4B, lanes 2–7), the DNA synthesis activities of Polι, Polη, and Polκ were severely inhibited on the DNA substrate containing the 5′-dRP moiety (Fig. 4B, lanes 10–15), whereas no such inhibition was observed with Polβ (Fig. 4B, cf. lanes 8,9 and 16,17). Thus, relative to primer extension on the nicked DNA substrate lacking the 5′-dRP residue, the inhibition of the DNA synthetic activity on the 5′-dRP-containing DNA substrate was ∼10-fold for Polι, ∼5-fold for Polη, and ∼ 10-fold for Polκ, whereas Polβ was ∼20% more efficient at synthesizing DNA on the 5′-dRP substrate than on the nicked DNA substrate lacking this lesion. In conclusion, binding of Polι, Polη, and Polκ to the 5′-dRP group in DNA severely inhibits their DNA synthetic activity.
Discussion
Translesion DNA synthesis is one of the main roles of human Polη, Polι, and Polκ (Prakash and Prakash 2002). These DNA polymerases, however, replicate DNA with a low fidelity, and the fidelity of Polι is the poorest, particularly for incorporating nucleotides opposite pyrimidine templates. Therefore, to prevent increased mutagenesis, it is critical that the DNA synthetic activity of these polymerases be restricted. Because of the preponderance of a variety of base damages, and because of the high incidence of AP sites, which are generated as a result of spontaneous base loss and as intermediates in BER, it is particularly important that high-fidelity DNA synthesis prevail during BER.
We show here that Polη, Polι, and Polκ can each form a Schiff base with 5′-dRP-containing DNA. These enzyme–DNA complexes, however, are not intermediates of a 5′-dRP lyase reaction, as we found no 5′-dRP lyase activity in our purified Polι, Polη, and Polκ preparations, which are fully active in the DNA synthesis reaction. Thus, in contrast to Polβ, where as little as 2 nM of the enzyme removed ∼70% of the 5′-dRP group from the substrate, Polι, Polη, and Polκ had no 5′-dRP lyase activity, even at a concentration as high as 50 nM. The absence of a 5′-dRP lyase activity from Polι, Polη, and Polκ was further supported from studies in which these enzymes were preincubated with the 5′-dRP DNA substrate prior to borohydride reduction of the polymerase–DNA complexes. Although preincubation with Polβ before the addition of borohydride decreased the Polβ–DNA cross-linked product, consistent with a robust 5′-dRP lyase activity in Polβ, such preincubation with Polι, Polη, and Polκ resulted in an increase in the amount of Pol–DNA cross-linked product. The persistence of 5′-dRP residues, even after prolonged preincubation with Polι, Polη, and Polκ, has provided unambiguous evidence for the lack of any 5′-dRP lyase activity in these enzymes.
The ability of Polι, Polη, and Polκ to form a covalent protein–DNA Schiff base intermediate, but not to promote the subsequent 5′-dRP lyase reaction, could arise from the differences in the chemistry for the two reactions. When the environment of a particular –NH2 group in a protein lowers its pKa, it becomes a good nucleophile at physiological pH and is able to form a Schiff base with the –CHO group at the C1 position of an AP site. However, for the protein to display a strong 5′-dRP lyase activity, at this intermediate stage, the pKa of the imine group needs to be high enough for it to be protonated and to carry out an efficient β-elimination reaction; otherwise, the 5′-dRP group removal will be highly inefficient, occurring via a spontaneous β-elimination reaction. Thus, Schiff base formation is a prerequisite for the lyase activity, but it is not sufficient for the subsequent β-elimination reaction to occur.
Covalent binding to the 5′-dRP residue in DNA inhibits the DNA synthetic activity of Polι, Polη, and Polκ. This inhibition of synthesis may arise because covalent binding to the 5′-dRP group results in a physical orientation so that the polymerase can no longer access the 3′-primer end; alternatively, a transient conformational change may occur in the polymerase that inhibits its binding to the 3′-primer end or inhibits its catalytic activity. Because the covalent complexes of Polι, Polη, and Polκ with the 5′-dRP-containing DNA can be stabilized by borohydride, structural studies of such complexes could help unravel the mechanism of inhibition of their DNA synthetic activity.
Previously, a 5′-dRP lyase activity has been reported for human Polι (Bebenek et al. 2001; Prasad et al. 2003). This observation is inconsistent with our results. In these studies, however, no convincing evidence was provided to establish that the lyase activity is, in fact, intrinsic to Polι. The dRP lyase activity was inferred to be intrinsic to Polι because a covalent Polι–DNA Schiff-base intermediate could be trapped by reduction with sodium borohydride (Bebenek et al. 2001), and this site in Polι has been recently mapped to a 40-kD domain spanning residues Met 79 to ∼460 (Prasad et al. 2003). Our results confirm the formation of a covalent Polι–DNA Schiff-base intermediate, but we have found no evidence for a 5′-dRP lyase activity in Polι. We have examined many different highly purified native Polι and GST-Polι preparations that support robust DNA synthesis, but even with long incubation periods and with high enzyme concentrations, we have found no evidence of any significant 5′-dRP lyase activity in Polι. Moreover, as we show here, Polη and Polκ also form a Schiff base intermediate, but they have no 5′-dRP lyase activity. And, for Polη and Polκ also, we expect the region corresponding to Polι's 40-kD domain to be involved in Schiff base formation. In view of our observations, the formation of a Schiff base intermediate cannot be taken as evidence to support the existence of a 5′-dRP lyase activity in Polι. Furthermore, in the absence of additional supporting evidence, the coelution of Polι's polymerase activity with 5′-dRP lyase cannot be considered definitive, as that could be fortuitous (Prasad et al. 2003). Although we cannot be sure of the source of the 5′-dRP lyase activity that has been reported for Polι, any number of factors, as, for example, the presence of an extraneous peptide or a protein in the Polι preparation, could have been the source of that activity. Because of the extreme lability of a 5′-terminal abasic site, the removal of a 5′-dRP group can occur spontaneously in the absence of β-elimination catalysts (Bailly and Verly 1989), and basic proteins such as histones and polyamines stimulate this reaction by inducing β-elimination (Bailly and Verly 1988). Moreover, even short basic peptides such as Lys–Trp–Lys and Lys–Tyr–Lys, can promote this reaction (Helene et al. 1982).
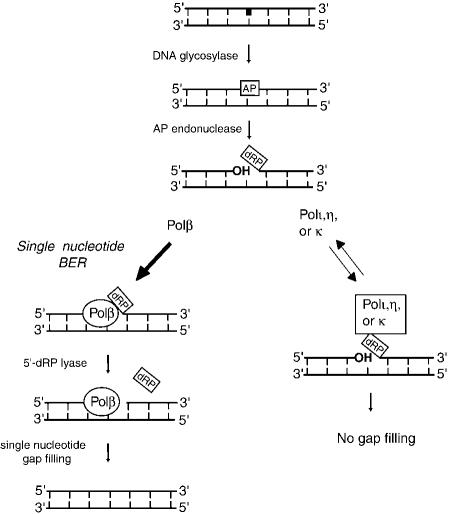
In summary, with respect to their interaction with a 5′-dRP group in DNA, all three human polymerases, Polι, Polη, and Polκ, behave very similarly. They all form a covalent Schiff base intermediate with the 5′-dRP group, but have no detectable 5′-dRP lyase activity (Fig. 5). To our knowledge, such robust Schiff base formation without the release of any significant 5′-dRP product is unprecedented, as all the other DNA polymerases that yield a covalent Schiff base intermediate also exhibit 5′-dRP lyase activity (Matsumoto et al. 1998; Pinz and Bogenhagen 2000; Garcia-Diaz et al. 2001). Our observation that the DNA synthetic activity of Polι, Polη, and Polκ is greatly reduced on a 5′-dRP-containing DNA substrate strongly supports the inference that covalent binding to the 5′-dRP residue represents a means by which these low-fidelity polymerases are excluded from the DNA synthetic step of BER (Fig. 5), thereby avoiding the high incidence of mutagenesis and carcinogenesis that would otherwise result.
Figure 5.
A model for the exclusion of Y-family polymerases from DNA synthesis during BER. An AP site is generated in DNA either by the action of a DNA glycosylase on the damaged base (shown in bold) or via spontaneous hydrolysis of purines (data not shown). Nicking of the phosphodiester backbone on the 5′-side of the AP lesion by a class II AP endonuclease generates a 3′-OH terminus and a 5′-dRP moiety, which is removed by the potent 5′-dRP lyase activity of Polβ, followed by single nucleotide gap filling by this enzyme (left). The thick arrow on the left denotes that because of its high affinity for the lesion site, Polβ would be the predominant enzyme to carry out the reactions shown. The low-fidelity Y-family polymerases, however, would be excluded from the DNA synthesis reaction during BER because of their unproductive binding to the 5′-dRP residue, which inhibits their ability to synthesize DNA on such a substrate (right). The bidirectional thin arrows on the right signify the less efficient and transient binding of Y-family polymerases to the lesion site. Dissociation of these polymerases from the 5′-dRP residue would free up the residue for removal by the 5′-dRP lyase activity of Polβ and for repair synthesis, as is shown on the left.
Materials and methods
DNA substrates
Oligonucleotides were synthesized by Midland Certified Reagent Co. The S1 substrate, used for the DNA synthesis reactions shown in Figures 1B and 2D, was generated by annealing a 75-nt oligonucleotide template, 5′-AGCTACCATGCCTGCC TCAAGAATTCGTAAGATGCCTACACTGGAGTACCGGA GCATCGTCGTGACTGGGAAAAC-3′ to the 5′-32P-labeled, 40-nt oligonucleotide primer, 5′-GTTTTCCCAGTCACGAC GATGCTCCGGTACTCCAGTGTAG-3′. For the single-uracil-containing S2 substrate, shown schematically in Figure 2A, the 34-nt 3′-32P-labeled oligonucleotide, 5′-CTGCAGCTGATGC GCUGTACGGATCCCCGGGTAC-3′, was annealed to the 34 nt template oligonucleotide, 5′-GTACCCGGGGATCCGTAC GGCGCATCAGCTGCAG-3′. The 5′-dRP-containing DNA substrate, used for the experiments shown in Figures 2B and C and 3, was generated by treatment of the S2 substrate with uracil DNA glycosylases (UDG; Trevigen) followed by treatment with human APE1, a class II AP endonuclease (Trevigen), as described below. For the DNA polymerase reactions shown in Figure 4A, the DNA substrates were generated by annealing three oligonucleotides together; the 34-nt template oligonucleotide, 5′-GTACCCGGGGATCCGTACTGCGCATCAGCTGC AG-3′; the 5′-32P-labeled 15-nt oligonucleotide primer, 5′-CT GCAGCTGATGCGC-3′; and either the 18-nt oligonucleotide, 5′-pGTACGGATCCCCGGGTAC-3′ (S3 substrate); or the 19-nt oligonucleotide, 5′-pUGTACGGATCCCCGGGTAC-3′ (S4 substrate). The DNA substrates shown in Figure 4B were generated by annealing three oligonucleotides together, the 34-nt template oligonucleotide used also for generating the S2 substrate; the 5′-32P-labeled 15-nt oligonucleotide primer used also for generating the S3 and S4 substrates; and either the 19-nt oligonucleotide 5′-pCGTACGGATCCCCGGGTAC-3′ (S5 substrate), or the 19-nt oligonucleotide used for generating the S4 substrate (S6 substrate). The AP site in the S4 and S6 substrates was generated by the removal of uracil by UDG treatment as described below.
Purification of DNA polymerases
Human Polι, Polη, and Polκ in fusion with glutathione S-transferase (GST) were expressed in yeast strain BJ5464 and bound to a glutathione-Sepharose 4B column as described (Haracska et al. 2001a,b, 2002b). GST-Polι, GST-Polη, and GST-Polκ were eluted from one-half of the glutathione-Sepharose 4B beads with 20 mM reduced glutathione in elution buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, 0.01% Nonidet P-40, and 10% glycerol. The other half of the glutathione-Sepharose 4B beads were incubated with PreScission protease (Amersham), which cleaves the GST fusion from the native proteins, releasing the native Polι, Polη, and Polκ in elution buffer. Purified proteins were concentrated by using a Microcon 30 (Amicon) and frozen at -70°C. Human Polβ purified as described (Rajendran et al. 1998) was kindly provided by W. Bujalowski (University of Texas Medical Branch, Galveston, TX).
DNA polymerase assays
The DNA polymerase reaction mixture (10 μL) contained 20 nM DNA substrate; 100 μM dGTP, dATP, dCTP, and dTTP; in either buffer A containing 40 mM Tris-HCl (pH 7.5), 8 mM MgCl2, 1 mM dithiothreitol, 100 μg/mL bovine serum albumin, and 10% glycerol (Fig. 1B); or in buffer B containing 50 mM HEPES (pH 7.5), 10 mM MgCl2, 20 mM KCl, and 2 mM dithiothreitol (Figs. 2D, 4). Polymerase reactions were started by the addition of Polι, Polη, Polκ, or Polβ in concentration as indicated in the figure legends. After incubation at 37°C for 10 or 20 min as indicated in the figure legends, the reactions were terminated by the addition of 40 μL of formamide containing loading buffer (95% formamide, 20 mM EDTA, 0.3% bromphenol blue, and 0.3% cyanol blue). The reaction products were resolved on 8% or 20% polyacrylamide gels containing 8 M urea, as indicated in the figure legends. Quantitation of the products was done using a Molecular Dynamics STORM PhosphorImager and ImageQuant software.
Preparation of 5′-dRP-containing DNA
The incised AP-site-containing DNA was generated by first pretreating the DNA duplex, which contained a single U in the 3′-32P-labeled DNA strand, with UDG (2 units/1 pmole DNA) in 50 mM HEPES (pH 7.5) at 30°C for 10 min, followed by treatment with human APE1, a class II AP endonuclease (2 units/1 pmole DNA) in buffer B at 30°C for 10 min, which incises the phosphodiester backbone on the 5′-side of the AP site, leaving a 3′-OH and a 5′-dRP residue. Owing to the labile nature of the AP site, the 5′-dRP-containing DNA substrate was prepared just before use.
Trapping of polymerase–DNA complexes by sodium borohydride
DNA polymerases were mixed on ice in buffer B with DNA substrate containing a 5′-dRP residue followed by immediate addition of 20 mM sodium borohydride. The reaction with sodium borohydride was continued for 30 min on ice. In the experiment shown in Figure 3, the polymerase was first preincubated with the DNA substrate at 37°C for the indicated time before the addition of sodium borohydride. The reactions were terminated by the addition of sodium dodecyl sulfate (SDS)-containing loading buffer, and the cross-linked polymerase-3′-32P-labeled DNA complexes were resolved on an 8% SDS–polyacrylamide gel. Products were visualized by autoradiography.
5′-dRP lyase assay
Reaction mixtures (10 μL) contained the DNA polymerases at two different concentrations, as indicated in the figure legends, and the 5′-dRP-containing DNA substrate (20 nM) in buffer B. Reactions were incubated at 37°C for 10 min. Thereafter, the reaction product was stabilized by the addition of 2M sodium borohydride to a final concentration of 340 mM, followed by incubation at 0°C for 30 min. Formamide-containing gel loading buffer (40 μL) was then added, and the reaction products were resolved on 16% polyacrylamide gels containing 8 M urea followed by visualization by autoradiography.
Acknowledgments
We thank M.L. Dodson for discussions and W. Bujalowski for human Polβ. This work was supported by NIH grant GM19261 and NIEHS grant ES012411.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1146103.
References
- Bailly V. and Verly, W.G. 1988. Possible roles of β-elimination and δ-elimination reactions in the repair of DNA containing AP (apurinic/apyrimidinic) sites in mammalian cells. Biochem. J. 253: 553-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1989. The multiple activities of Escherichia coli endonuclease IV and the extreme lability of 5′-terminal base-free deoxyribose 5-phosphates. Biochem. J. 259: 761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K., Tissier, A., Frank, E.G., McDonald, J.P., Prasad, R., Wilson, S.H., Woodgate, R., and Kunkel, T.A. 2001. 5′-Deoxyribose phosphate lyase activity of human DNA polymerase ι in vitro. Science 291: 2156-2159. [DOI] [PubMed] [Google Scholar]
- Deterding L.J., Prasad, R., Mullen, G.P., Wilson, S.H., and Tomer, K.B. 2000. Mapping of the 5′-2-deoxyribose-5-phosphate lyase active site in DNA polymerase β by mass spectrometry. J. Biol. Chem. 275: 10463-10471. [DOI] [PubMed] [Google Scholar]
- Dodson M.L., Schrock, R.D., and Lloyd, R.S. 1993. Evidence for an imino intermediate in the T4 endonuclease V reaction. Biochemistry 32: 8284-8290. [DOI] [PubMed] [Google Scholar]
- Garcia-Diaz M., Bebenek, K., Kunkel, T.A., and Blanco, L. 2001. Identification of an intrinsic 5′-deoxyribose-5-phosphate lyase activity in human DNA polymerase λ. J. Biol. Chem. 276: 34659-34663. [DOI] [PubMed] [Google Scholar]
- Haracska L., Prakash, S., and Prakash, L. 2000a. Replication past O6-methylguanine by yeast and human DNA polymerase η. Mol. Cell. Biol. 20: 8001-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Yu, S.-L., Johnson, R.E., Prakash, L., and Prakash, S. 2000b. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat. Genet. 25: 458-461. [DOI] [PubMed] [Google Scholar]
- Haracska L., Johnson, R.E., Unk, I., Phillips, B., Hurwitz, J., Prakash, L., and Prakash, S. 2001a. Physical and functional interactions of human DNA polymerase η with PCNA. Mol. Cell. Biol. 21: 7199-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001b. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc. Natl. Acad. Sci. 98: 14256-14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Prakash, L., and Prakash, S. 2002a. Role of human DNA polymerase κ as an extender in translesion synthesis. Proc. Natl. Acad. Sci. 99: 16000-16005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L., Unk, I., Johnson, R.E., Phillips, B.B., Hurwitz, J., Prakash, L., and Prakash, S. 2002b. Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol. Cell. Biol. 22: 784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helene C., Toulme, J.-J., Behmoaras, T., and Cazenave, C. 1982. Mechanisms for the recognition of chemically-modified DNA by peptides and proteins. Biochimie 64: 697-705. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Kondratick, C.M., Prakash, S., and Prakash, L. 1999a. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285: 263-265. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Prakash, S., and Prakash, L. 1999b. Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, Polη. Science 283: 1001-1004. [DOI] [PubMed] [Google Scholar]
- ____. 2000a. The human DINB1 gene encodes the DNA polymerase Polθ. Proc. Natl. Acad. Sci. 97: 3838-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Washington, M.T., Haracska, L., Prakash, S., and Prakash, L. 2000b. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406: 1015-1019. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Washington, M.T., Prakash, S., and Prakash, L. 2000c. Fidelity of human DNA polymerase η. J. Biol. Chem. 275: 7447-7450. [DOI] [PubMed] [Google Scholar]
- Masutani C., Kusumoto, R., Yamada, A., Dohmae, N., Yokoi, M., Yuasa, M., Araki, M., Iwai, S., Takio, K., and Hanaoka, F. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399: 700-704. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y. and Kim, K. 1995. Excision of deoxyribose phosphate residues by DNA polymerase β during DNA repair. Science 269: 699-702. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Kim, K., Katz, D.S., and Feng, J. 1998. Catalytic center of DNA polymerase β for excision of deoxyribose phosphate groups. Biochemistry 37: 6456-6464. [DOI] [PubMed] [Google Scholar]
- Nakamura J., Walker, V.E., Upton, P.B., Chiang, S.-Y., Kow, Y.W., and Swenberg, J.A. 1998. Highly sensitive apurinic/pyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 58: 222-225. [PubMed] [Google Scholar]
- Piersen C.E., Prasad, R., Wilson, S.H., and Lloyd, R.S. 1996. Evidence for an imino intermediate in the DNA polymerase β deoxyribose phosphate excision reaction. J. Biol. Chem. 271: 17811-17815. [DOI] [PubMed] [Google Scholar]
- Pinz K.G. and Bogenhagen, D.F. 2000. Characterization of a catalytically slow AP lyase activity in DNA polymerase γ and other family A DNA polymerases. J. Biol. Chem. 275: 12509-12514. [DOI] [PubMed] [Google Scholar]
- Prakash S. and Prakash, L. 2002. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes & Dev. 16: 1872-1883. [DOI] [PubMed] [Google Scholar]
- Prasad R., Beard, W.A., Chyan, J.Y., Maciejewski, M.W., Mullen, G.P., and Wilson, S.H. 1998a. Functional analysis of the amino-terminal 8-kDa domain of DNA polymerase β as revealed by site-directed mutagenesis. J. Biol. Chem. 273: 11121-11126. [DOI] [PubMed] [Google Scholar]
- Prasad R., Beard, W.A., Strauss, P.R., and Wilson, S.H. 1998b. Human DNA polymerase β deoxyribose phosphate lyase. J. Biol. Chem. 273: 15263-15270. [DOI] [PubMed] [Google Scholar]
- Prasad R., Bebenek, K., Hou, E., Schock, D.D., Beard, W.A., Woodgate, R., Kunkel, T.A., and Wilson, S.H. 2003. Localization of the deoxyribose phosphate lyase active site in human DNA polymerase ι by controlled proteolysis. J. Biol. Chem. 278: 29649-29654. [DOI] [PubMed] [Google Scholar]
- Rajendran S., Jezewska, M.J., and Bujalowski, W. 1998. Human DNA polymerase β recognizes single-stranded DNA using two different binding modes. J. Biol. Chem. 273: 31021-31031. [DOI] [PubMed] [Google Scholar]
- Seeberg E., Eide, L., and Bjorås, M. 1995. The base excision repair pathway. Trends Biochem. Sci. 20: 391-397. [DOI] [PubMed] [Google Scholar]
- Sobol R.W., Horton, J.K., Kuhn, R., Gu, H., Singhal, R.K., Prasad, R., Rajewsky, K., and Wilson, S.H. 1996. Requirement of mammalian DNA polymerase-β in base-excision repair. Nature 379: 183-186. [DOI] [PubMed] [Google Scholar]
- Sobol R.W., Prasad, R., Evenski, A., Baker, A., Yang, X.-P., Horton, J.K., and Wilson, S.H. 2000. The lyase activity of the DNA repair protein β-polymerase protects from DNA-damage-induced cytotoxicity. Nature 405: 807-810. [DOI] [PubMed] [Google Scholar]
- Tissier A., McDonald, J.P., Frank, E.G., and Woodgate, R. 2000. Polι, a remarkably error-prone human DNA polymerase. Genes & Dev. 14: 1642-1650. [PMC free article] [PubMed] [Google Scholar]
- Wallace S.S. 1994. DNA damages processed by base excision repair: Biological consequences. Int. J. Radiat. Biol. 66: 579-589. [DOI] [PubMed] [Google Scholar]
- ____. 1997. Oxidative damage to DNA and its repair. In Oxidative stress and the molecular biology of antioxidant defenses (ed. J.G. Scandalios), pp. 49-90. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Washington M.T., Johnson, R.E., Prakash, S., and Prakash, L. 1999. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J. Biol. Chem. 274: 36835-36838. [DOI] [PubMed] [Google Scholar]
- ____. 2000. Accuracy of thymine–thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl. Acad. Sci. 97: 3094-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington M.T., Johnson, R.E., Prakash, L., and Prakash, S. 2002. Human DINB1-encoded DNA polymerase κ is a promiscuous extender of mispaired primer termini. Proc. Natl. Acad. Sci. 99: 1910-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.H. 1998. Mammalian base excision repair and DNA polymerase β. Mutat. Res. 407: 203-215. [DOI] [PubMed] [Google Scholar]
- Yu S.-L., Johnson, R.E., Prakash, S., and Prakash, L. 2001. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol. 21: 185-188. [DOI] [PMC free article] [PubMed] [Google Scholar]