Abstract
Monoclonal B cell lymphocytosis (MBL) is a pre-clinical hematologic syndrome characterized by small accumulations of CD5+ B lymphocytes. Most MBL share phenotypic characteristics with chronic lymphocytic leukemia (CLL). While some MBL progress to CLL, most MBL have apparently limited potential for progression to CLL, particularly those MBL with normal absolute B cell counts (“low count” MBL). Most CLL are monoclonal and it is not known whether MBL are monoclonal or oligoclonal; this is important because it is unclear whether MBL represent indolent CLL or represent a distinct pre-malignant precursor prior to the development of CLL. We used flow cytometry analysis and sorting to determine immunophenotypic characteristics, clonality, and molecular features of MBL from familial CLL kindreds. Single cell analysis indicated 4 of 6 low count MBL consisted of two or more unrelated clones; the other 2 MBL were monoclonal. 87% of low count MBL clones had mutated immunoglobulin genes, and no immunoglobulin heavy chain rearrangements of VH family 1 were observed. Some MBL were diversified, clonally related populations with evidence of antigen-drive. We conclude that while low count MBL share many phenotypic characteristics with CLL, many MBL are oligoclonal. This supports a model for step-wise development of MBL into CLL.
Keywords: Monoclonal B Lymphocytosis, Chronic Lymphocytic Leukemia, B Cell Repertoire, Human B Cells
Introduction
Chronic lymphocytic leukemia (CLL) is a malignant lymphoproliferative disorder characterized by the progressive clonal expansion and accumulation of CD5+ B cells. Although the cause of CLL is unknown, risk factors for the development of CLL have been identified including advanced age, male gender, Caucasian ethnicity, and a family history of CLL or other lymphoproliferative disorder (1-3). Because the average age at diagnosis of CLL in the United States is 72 years , previous studies investigated whether normal CD5+ B cells undergo clonal expansion with increasing age (4, 5). Studies in older subjects showed that normal CD5+ B cells are typically polyclonal. Furthermore, CD5+ B cells are predominantly antigen naïve, and somatic mutations of immunoglobulin heavy (IGVH) and light chains are typically absent (6). These observations contrast with CLL, where virtually all cases are monoclonal and approximately half of CLL cases show somatic mutation of rearranged immunoglobulin heavy chain genes (IGVH) (7, 8).
Monoclonal B cell lymphocytosis (MBL) is a preclinical hematologic syndrome where small B cell clones (defined as a clone size less than 5,000 cells / μL (9)) with an abnormal immunophenotype are present in the peripheral blood. Most MBL have a typical chronic lymphocytic leukemia (CLL) immunophenotype: CD5+, CD19+, CD20lo, CD23+, CD79blo, sIglo; though other, less common, phenotypes have also been reported (10-12). Using 6 or 8 color flow cytometry, MBL is observed in approximately 3 - 12% of adults over age 50 in the general population (12-14). MBL is significantly more common among unaffected first-degree relatives from families with two or more individuals with CLL (familial CLL), with observed frequencies of 13% and 18% in two different familial CLL cohorts (15, 16).
Given the immunophenotypic similarity between MBL and CLL, and the fact that MBL is more prevalent among CLL kindreds than in the general population, MBL had been hypothesized to be a precursor state for CLL. A recent report by Rawstron et al (2008) showed that MBL can progress to CLL, but the rate of progression to need for CLL-specific therapy is low, approximately 1.1% per year (17). This low observed rate of progression was expected, given that MBL is at least 100 times more common in the general population than CLL. Further, a recent study by Landgren et al (2009) showed that MBL precedes virtually all cases of CLL (18). Both of these studies evaluated IGVH gene usage and mutation status, and both studies found that MBL are monoclonal with predominantly mutated IGVH (>80% in both series as compared to approximately 50% in CLL), with the distribution of rearranged IGVH similar to mutated CLL.
Dagklis et al (2008) recently reported a population-based screen for MBL of 1725 individuals in a small village in Italy (19). The authors identified MBL in 89 of the 1725 study subjects (5.2%). They observed that the majority of the subjects with MBL had very small MBL clones (average of 34 MBL lymphocytes / μL), and all but 3 subjects had normal absolute B cell counts. They termed the presence of small MBL clone size in combination with a normal total B cell count “low count” MBL. Within this group of low count MBL, they noted that 6 of 86 (7.0%) were polyclonal based on flow cytometric evaluation of surface κ/λ staining. Using bulk cell preparations, they were able to clone and sequence the IGVH in only 51 cases, and again found a predominance of somatically mutated IGVH.
While initial studies suggest that CLL-like MBL are predominantly monoclonal (12, 17-19), the clonality of MBL is yet to be established using adequately sensitive techniques. Because most MBL do not progress to CLL, it remains unclear whether MBL is a monoclonal, biologically indolent form of CLL, or whether MBL is a transitional state between normal CD5+ B cells and CLL. Therefore, we investigated the clonality of MBL by sequencing the rearranged immunoglobulin genes of single MBL cells. Similar to CLL lymphocytes, we show that flow cytometry-enriched MBL cells from the unaffected kindred of CLL families commonly have mutated immunoglobulin genes, and carry deletions of 13q14.3. However, unlike CLL, MBL are frequently oligoclonal rather than monoclonal. Our results support the hypothesis that CLL-like MBL are pauci-clonal CD5+ B cells with potential for progression to CLL.
Materials and Methods
MBL Subjects
Research subjects were identified through patients with familial CLL at Duke University Medical Center and the Durham Veterans Affairs Medical Center. A diagnosis of CLL was based upon standard criteria (9, 20). Familial CLL was defined as a family with 2 or more first or second degree relatives with CLL. Eighty-eight first and second degree relatives without CLL from 10 pedigrees with familial CLL were enrolled. Peripheral blood samples were collected for complete blood counts and MBL screening by flow cytometry. Participants identified as having MBL were asked to provide additional blood for sorting MBL and further phenotypic analysis. All subjects gave written informed consent. This study was approved by the Institutional Review Boards at Duke University and the Durham VA Medical Centers.
Flow Cytometry
MBL Identification
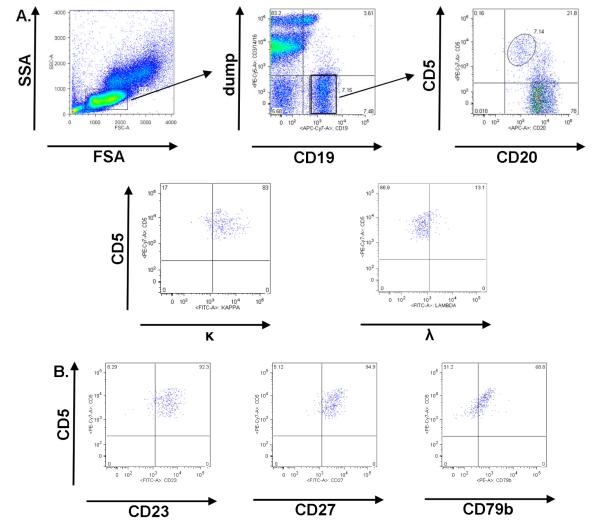
MBL screening was adapted from Rawstron et al (13). Venous blood for flow cytometry was collected in a CPT™ cell preparation tube (Becton Dickinson; Franklin Lakes, NJ), and PBMC were purified by density centrifugation. 2.5 × 105 PBMC were incubated with CD79b PE, CD3 PE-Cy5, CD14 PE-Cy5, CD16 PE-Cy5, CD5 PE-Cy7, CD20 allophycocyanin (APC), CD19 APC-Cy7 combined with one each of the following, CD27, CD23, Kappa or Lambda FITC. PE-Cy5 conjugated CD3, CD14, and CD16 were combined as a “dump” channel to exclude T cells, monocytes, NK cells and granulocytes. Isotype controls matched to test antibody isotype, concentration, and fluorescent conjugate were used for each test antibody. A table of reagents used is provided in Supplemental Table 1. Flow cytometry was performed on a Becton Dickinson ARIA flow cytometer. Instrument QC/QA using AlignFlow Plus flow cytometry alignment beads (Invitrogen) was performed daily by the Duke Human Vaccine Institute Flow Cytometry and Cell Sorting Facility. Compensation was performed by staining Quantum Simply Cellular anti-Mouse IgG beads (Bangs Laboratories, Inc) to saturation and using unstained blank beads and stained beads as negative and positive, respectively. Additionally, quantitative beads were used in each experiment to normalize MFI between experimental samples. Analysis of flow cytometry results was performed using FlowJo software (Tree Star Inc; Ashland, OR). MBL were defined as CD19+, CD5+, CD20lo, CD79blo B cells that were CD23+ (Figure 1).
Figure 1. MBL Identification by flow cytometry.
In A, the MBL flow cytometric gating strategy from PBMC is shown. MBL gating steps included a forward by side scatter plot, with low forward and side-scatter events gated, a CD19 by CD3 / CD14 / CD16 (dump antibodies to exclude T cells, monocytes, NK cells and granulocytes) plot, with CD19+ and dump negative events gated, and a CD20 by CD5 plot, with CD5+CD20lo MBL gated. In this sample, immunoglobulin light chain κ restriction was observed. Further confirmation of MBL was performed and indicated that MBL were (B) CD23+, CD27+, and CD79blo.
MBL Immunophenotyping and FACS
Approximately 40 mL of heparinized venous blood was collected for MBL characterization. The sample was incubated with RosetteSep™ Human B Cell Enrichment Cocktail (Stem Cell Technologies, Vancouver, British Columbia, Canada) and purified by Ficoll-Hypaque density centrifugation to enrich B cells. 2.5 × 105 B cells were aliquoted and fluorescently stained as described above, except the following antibodies were substituted in different tubes for the FITC- and PE-conjugated antibodies: CD27 FITC, CD23 FITC, Igκ FITC, Igλ FITC, and IgM FITC; CD79b PE and IgD PE. Flow cytometry and data analysis were performed as described above. MBL (CD19+, CD5+, CD20lo) were sorted as single cells into Promega 1x PCR buffer with 20ng rRNA (20 ul/well; 1 cell/well) in 96 well plates for Ig gene analysis. MBL were also sorted in bulk (2.5 - 5.0 × 105 cells) into 100 uL of HBSS for FISH.
Immunoglobulin Gene Analysis
Single MBL cells were amplified, sequenced and analyzed as previously described (21). In brief, 50 cycles of whole genome amplification (WGA) using 15-base oligomers was performed. Amplification of the rearranged immunoglobulin heavy and light chains were performed using nested PCR of WGA DNA. PCR products were sequenced on an ABI 3730 automated sequencer (PE Applied Biosystems, Foster, CA), the immunoglobulin gene sequences were aligned using Sequencher 4.8 (Gene Codes, Ann Arbor, MI), and immunoglobulin gene sequences were compared with those in the online sequence directory IMGT (22). β-globin was used as a positive control for PCR amplification. Our group has shown that polymerase errors are rare using this methodology; in a prior study only 2 unique mutations in β-globin were identified among PCR products from 90 whole-genome amplified single cells (21).
FISH
MBL were FACS purified as described above. The MBL suspension was loaded into a cytospin apparatus and brought to a final volume of 150 uL by adding PBS. MBL cells were spun onto untreated glass slides at 500 rpm for 5 minutes. The slides were air dried and stored at room temperature.
FISH was performed in the Duke Molecular Diagnostics Laboratory using a standard methodology for the analysis of clinical CLL samples. Fluorescent probes to detect deletion of 13q14.3 (D13S319), 13q34, 17p13.1 (TP53) and 11q22.3 (ATM) and a chromosomal enumeration probe (12p11.1-q11) to detect trisomy 12 (Vysis; Des Plaines, IL) were used. Following fixation of cells on slides, denaturation of MBL DNA, and fluorescent probe labeling of MBL chromosomes, specimens were examined on a fluorescence microscope to detect chromosomal abnormalities. For each sample, 100 adequate cells were examined and the percentage of MBL cells with each chromosomal abnormality was recorded.
Results
MBL Identification
MBL were detected in 11 of 88 (12.5%) first and second degree relatives from 10 familial CLL kindreds. All MBL cases had a typical CLL immunophenotype: CD5+, CD19+, CD20lo, CD23+, CD27+, CD79b+, sIglo. Sample MBL flow cytometry plots are shown in Figure 1. The mean age among participants with MBL was 67 years (range 43 to 94). Five subjects with MBL were male, and six were female. The 11 subjects with MBL were from 5 different families. The MBL subjects were predominantly siblings (9 subjects) of a family member with CLL, one MBL subject was a parent of a CLL patient, and one was a niece of a patient with CLL. Six participants with “low count” MBL provided additional blood for detailed MBL characterization. Demographic data from these 6 subjects with MBL are shown in Table 1. We observed significant variability in the size of the MBL clone as a percentage of the total B cell compartment. Nonetheless, the absolute number of MBL cells was small. All six participants had < 150 MBL cells / μL of blood (Table 1), and an absolute B cell count < 600 B cells / μL. One subject (a 7th MBL subject) was excluded from additional analysis because the absolute size of the MBL clone was approximately 4,000 MBL cells / μL. Among the 4 other MBL subjects who did not provide an additional sample, one lived at a distance too great to allow for timely processing of blood samples, one refused participation, one was lost to follow-up, and one died from an unrelated illness.
Table 1.
MBL subjects and characteristics
| Familial MBL Participant |
Patient characteristics |
MBL numbers | Immunophenotype | FISH | ||||
|---|---|---|---|---|---|---|---|---|
| Sex | Age (yrs) |
% MBL of CD19+ B cells |
MBL / μL | IgD | IgM | κ/λ ratioa |
||
| 0209 | F | 59 | 7% | 47 | + | − | 6.6 | Normal |
| 0226 | F | 73 | 58% | 140 | − | − | 0.8 | Biallelic del 13q14 (85%) Monoallelic del 13q14 (15%) |
| 0504 | F | 71 | 26% | 62 | + | − | 1.1 | Monoallelic del 13q14 (9%) |
| 0602 | M | 94 | 52% | 10 | + | + | 1.6 | Monoallelic del 13q14 (42%) |
| 1107 | F | 82 | 4% | 60 | + | − | 23.5 | Monoallelic del 13q14 (38%) |
| 1109 | M | 73 | 25% | 27 | − | − | 1.1 | Normal |
Ratio of mean fluorescence intensity of surface immunoglobulinκ to λ
MBL Immunoglobulin Gene Analyses
Single MBL cells were sorted by flow cytometry for immunoglobulin heavy and light chain sequence analysis to determine the clonality of MBL in these subjects. These results are summarized in Table 2. We observed two or more unrelated MBL clones in 4 of 6 participants. Individual cells with unique immunoglobulin sequences could be non-MBL B cells from the FACS procedure, so we did not consider unique single cells in clonality assessments. Because prior reports have shown that clonally-related B cells are not identified in the normal B CD5+ cell compartment (4, 5), we believe that identification of two or more clonally identical cells within the MBL population identifies an abnormal clonal expansion. Three different general patterns of clonality were observed. (i) Two subjects were monoclonal (0226 and 1107). (ii) Two subjects showed a predominant clone with a second, smaller clone comprising less than 10% of the total MBL population (0504 and 0602). (iii) The final two subjects were oligoclonal, with 3 or more clones (0209 and 1109). In participant 0209, three unique clones were observed: a mutated VH4-59 clone (38% of all MBL cells), a mutated VH4-34 clone (5%), and an unrelated, unmutated VH4-34 clone (57%). The two VH4-34 clones in subject 0209 appear unrelated because they do not share a D-J segment and contain 26 unique, unshared mutations in their V regions. Analysis of immunoglobulin genes in single cells from subject 1109 identified 4 unique MBL clones. A predominant MBL clone with rearranged VH3-30 was identified. Additional minor clones with rearrangements of VH3-15, VH4-61*02 and VH4-61*04 were also identified.
Table 2.
Immunoglobulin heavy and light chain gene usage in MBL single cells
| Familial MBL subject |
VH | VL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| #of cells seq. |
V | D | J | Somati c Muta- tionsa (%) |
Produc tiveb |
# of cells seq. |
V | J | Somati c Muta- tions (%) |
Produc tive |
|
| 209 | 36 | 4-34 | 3-22 | 1 | N (1.0) | P | 35 | λ2-14 | λ3 | N (1.7) | P |
| 29 | κ2-29 | κ2 | N (0.0) | NP | |||||||
| 22 | 4-59 | 2-8 | 4 | Y (3.6) | P | 6 | κ3-20 | κ2 | N (1.8) | P | |
| 6 | λ10-54 | λ3 | N (1.7) | P | |||||||
| 4 | 4-34 | 2-2 | 4 | Y (9.2) | P | 2 | κ1-5 | κ1 | Y (5.9) | P | |
| 4 | κ4-1 | κ1 | Y (5.2) | NP | |||||||
| 0226 c | 30 | 3-15 | 5-24 | 4 | Y (8.3) | P | 30 | λ1-51 | λ2 | N (1.1) | P |
| 27 | κ2-24 | κ5 | N (0.0) | P | |||||||
| 3 | 3-33 | 3-10 | 5 | Y (6.9) | NP | 3 | λ1-51 | λ2 | N (1.1) | P | |
| 3 | κ2-24 | κ5 | N (0.0) | P | |||||||
| 0504 | 53 | 3-23 | 3-22 | 3 | Y (4.1) | P | 48 | λ3-21 | λ3 | Y (5.2) | P |
| 2 | 3-7 | 5-24 | 4 | Y (3.1) | P | 2 | λ4-69 | λ3 | N (1.1) | P | |
| 2 | κ2-40 | κ1 | N (0.0) | P | |||||||
| 2 | κ4-1 | κ4 | N (0.0) | NP | |||||||
| 0602 | 35 | 3-07 | 3-10 | 5 | Y (3.1) | P | 29 | λ1-51 | λ2 | N (1.0) | P |
| 6 | κ4-1 | κ4 | N (0.0) | P | |||||||
| 4 | 3-07 | 2-21 | 3 | Y (5.9) | P | 3 | κ1-5 | κ2 | Y (4.5) | P | |
| 1107 | 42 | 3-07 | 4-17 | 5 | N (1.7) | P | 35 | κ1-33 | κ2 | Y (3.0) | P |
| 31 | κ4-1 | κ2 | Y (3.0) | NP | |||||||
| 1 | 3-30 | 5-5 | 4 | Y (2.4) | P | 1 | κ1-16 | κ2 | Y (2.3) | P | |
| 1 | 3-23 | 1-26 | 4 | N (0.3) | P | 1 | κ1-5 | κ4 | N (0.3) | P | |
| 1109 c | 25 | 3-30 | 5-5 | 3 | Y (7.0) | P | 25 | λ10-54 | λ3 | N (1.7) | P |
| 8 | κ2-29 | κ3 | N (0.0) | NP | |||||||
| 7 | 4-61 | 2-2 | 6 | Y (3.4) | P | 5 | κ3-20 | κ5 | Y (3.7) | P | |
| 5 | 4-34 | 2-15 | 6 | Y (9.4) | NP | 4 | κ3-20 | κ5 | Y (3.7) | P | |
| 5 | 4-61 | 6-13 | 4 | Y (2.4) | P | 3 | κ2-24 | κ4 | N (0.7) | P | |
| 4 | 3-15 | 5-24 | 4 | Y (2.7) | P | 4 | λ2-14 | λ1 | Y (2.7) | P | |
| 3 | κ1-33 | κ2 | N (0.0) | NP | |||||||
| 3 | κ2-30 | κ2 | N (0.0) | NP | |||||||
| 2 | 3-23 | 2-15 | 4 | Y (8.0) | P | 1 | κ3-11 | κ4 | N (1.7) | P | |
| 1 | 3-7 | 2-21 | 1 | N (1.3) | P | 1 | κ1-5 | κ2 | N (0.7) | P | |
Percent deviation from germline immunoglobulin sequence. A sequence was designated as mutated if the experimentally obtained immunoglobulin sequence deviated ≥ 2% from germline.
P: Immunoglobulin gene rearrangement predicted to be productive; NP: non-productive
Single cell analysis from subjects 0226 and 1109 identified single cells with identical light chains but different immunoglobulin heavy chains (subject 0226: VH3-15 and VH3-33; subject 1109: VH4-61 and VH4-34). In both cases one heavy chain rearrangement is predicted to be productive and the other is predicted to be non-productive. As such, these cells are likely derived from a single clone wherein both heavy chain loci are recombined, possibly due to B cell receptor revision events, with one productive rearrangement and the other non-productive. However, since both heavy chains were not amplified from any single cell in either subject, these heavy chain rearrangements are listed separately.
In approximately 3% of IGVH mutated CLL, clonally related subgroups of leukemic cells with progressive IGVH gene mutations are identified (21). We identified progressive immunoglobulin gene mutation in one of the MBL subjects (Figure 2). The VH3-07, D3-10 clone from subject 0602 consisted of related subgroups of MBL that shared the same immunoglobulin gene rearrangements and some somatic mutations but also had several partially shared mutations. MBL immunoglobulin gene rearrangements from subject 0602 were arranged into a genealogical tree in which sequentially related clones were ordered based on sequence homology with the germline sequence and subsequent contiguous clones with progressive somatic mutations. MBL clones arranged in the genealogical tree furthest away from the putative germline immunoglobulin sequence had evidence of antigen-driven immunoglobulin sequence changes (23). We and others have suggested that similar clonally-related subgroups from CLL and MBL represent antigen-driven refinement of the BCR (21, 24-27). We believe that evidence of oligoclonal diversification, such as that observed in participant 0602, favors an antigen-driven process for some subjects with MBL.
Figure 2. MBL subject with intraclonal diversification.
In A, genealogical analysis of related MBL subgroups based on sequences from 37 single MBL cells from subject 0602, and analysis of the rearranged immunoglobulin VH3-07 heavy and VL1b light chains from each MBL cell subgroup. Letters A — L represent individual subgroups of MBL cells with a related immunoglobulin heavy chain gene and light chain rearrangement. The number of individual cells identified from each subgroup is shown in orange to the right of the subclone sequence. GL represents the germline VH3-07 sequence. Empty circles with dashed lines (representing multiple potential clones) and circles with dashed lines labeled H1, H2 and H3 (individual clones) represent hypothetical transitional MBL clones that were not observed in the analysis. In B, antigen drive analysis with p values calculated using the method of Lossos et al (2000) (23). In brief, the ratio of amino acid replacement (R) to synonomous (S) nucleotide base changes is compared in both the framework (Fr) and complementarity determining region (CDR) to determine if the observed mutations are random or antigen driven (21).
MBL Immunophenotype
As previously stated, all MBL showed a typical CLL immunophenotype. The MBL subjects were analyzed for light chain restriction. Similar to CLL, MBL show low expression of surface immunoglobulin. Three MBL expressed sIg κ based on a κ:λ mean fluorescence intensity ratio of > 3:1, and the remainder showed low sIg expression with a normal κ:λ ratio.
The majority of normal CD5+ B cells express both IgM and IgD and have not undergone isotype switching (4, 6). CLL B cells express low levels of surface immunogloubulin, typically of the IgM subtype. A small proportion of CLL lymphocytes can isotype switch to non-IgM subtypes (28). Analysis of sIg isotype by flow cytometry in the 6 MBL subjects tested showed 1 MBL expressed both surface IgM and IgD, 3 expressed IgD only, and 2 did not express IgD or IgM. The observation that 5 of 6 MBL have switched to a non-IgM subtype is an intriguing observation because these immunoglobulin isotypes are less common in clinical CLL.
MBL Chromosomal Analyses
Somatic deletion of chromosome 13q14.3 is observed in over 50% of CLL cases. When observed as a sole abnormality, this is associated with biologically indolent disease and improved clinical outcomes (29). Therefore, we hypothesized that deletion of the 13q14.3 locus would be observed in MBL cells. MBL cells were sorted in aliquots of 2.5 – 5.0 × 104 MBL for interphase cytogenetic analysis to determine whether MBL have chromosomal aberrations. Mono- or biallelic deletions of chromosome 13q14.3 were identified in 4 of 6 subjects tested (Table 1). Trisomy 12 and chromosomal deletions of 11q or 17p were not identified. The percentage of MBL with 13q14.3 deletions ranged from 9% of MBL cells in participant 0504 to 100% of MBL cells in participant 0226. This result suggests that loss of the micro-RNA cluster on chromosome 13q14.3 (30) is an early, but not required event in the development of MBL.
Discussion
MBL share many phenotypic characteristics with CLL and are precursors for CLL (11, 17, 18). Prior studies of MBL and MBL progression to CLL have primarily focused on subjects with MBL populations that are > 1,000 MBL cells / uL (26, 31, 32). Because the absolute number of MBL lymphocytes has prognostic significance (17), biological differences likely exist between MBL with high cell counts and MBL with low cell counts. We therefore limited this study to subjects with relatively small MBL populations (< 150 MBL cells / uL) and tested the hypothesis that “low count” MBL are heterogeneous and thus may have different capacities for transformation to CLL. We found that many MBL are oligoclonal based on FISH and single cell immunoglobulin sequence analyses, and that some MBL showed oligoclonal diversification suggestive of antigen drive.
Our finding of unrelated, separate MBL clones in 4 of 6 participants may be the most striking difference between MBL and CLL observed in this study. Biclonal CLL is rarely observed, and to our knowledge, there are no published reports of tri-clonal CLL (33). This suggests that during CLL leukemogenesis, a single dominant CLL clone typically arises and either obscures or eliminates other MBL clones. One potential limitation of our data is that these subjects with MBL, all of whom have a family history of CLL, may have an inherited predisposition that favors formation and maintenance of MBL clones. The recent report by Dagklis et al (19) of an unselected population-based screen for MBL showed that 28% of identified MBL subjects clustered in 5 main families, also suggesting an inherited basis for MBL. Whether oligoclonal MBL are observed in MBL cases that are not associated with familial CLL remains an open question, though both Nieto et al (14 of 73) and Dagklis et al (6 of 89) observed oligoclonal MBL by surface κ:λ expression in population based cohorts (14, 19).
An important question raised by our results is whether multiple MBL clones are driven by tonic stimulation by one or multiple antigens. Because IGVH mutated and unmutated CLL are reactive to different types of antigens (34), the finding of unrelated IGVH mutated and unmutated clones in participant 0209 suggests that there may be unique antigens for each clone. Similarly, it is unlikely that a single antigen generated the 4 unique MBL clones observed in subject 1109.
Prior reports have stressed the diagnostic utility of light chain restriction in the identification of MBL (10, 12, 17). Because (i) surface immunoglobulin is weakly expressed, (ii) CLL-like MBL have an abnormal immunophenotype, and (iii) as we report here, MBL with a small absolute clone size is commonly oligoclonal, we propose that demonstration of a restricted κ:λ ratio is not required for the identification of CLL-like MBL, especially when single cell immunoglobulin gene sequence data are available. For other “non-CLL like” MBL in which the surface immunophenotype is shared with normal B cell subsets (e.g. CD5- or CD20bright) (12, 19), demonstration of a skewed κ:λ ratio is necessary for identification.
Although 14 of 16 (87%) MBL clones identified in the 6 study subjects had mutated IGVH chains, unmutated immunoglobulin heavy chain rearrangements were observed in two cases (subjects 0209 and 1107; Table 2). We found rearrangement of VH3 and VH4 immunoglobulin genes in all isolated MBL cells. The VH3 and VH4 immunoglobulin heavy and light chain genes used by MBL are also commonly used in CLL (7). Our results are concordant with recent reports by Rawstron et al (2008), Dagklis et al (2008), and Landgren et al (2009) (17-19). In Rawstron et al, 35 of 40 (88%) MBL showed a mutated IGVH; rearrangements of VH3-07, VH3-23, and VH4-34 comprised over 25% of all identified sequences (17). Dagklis et al showed 36 of 51 (71%) “low count” MBL had a mutated IGVH and rearrangements of VH4-59 and VH4-61 were most common (19). Finally, Landgren et al showed that 27 of 35 (77%) MBL that had progressed to CLL expressed IGVH genes that were mutated and rearrangements of VH3-23 and VH4-34 were most common (18). Unlike the data presented here, in these previous reports, all MBL were monoclonal based on immunoglobulin gene sequencing. Analysis of IGVH failed in 22% of subjects in Landgren and 39% in Dagklis, presumably because both reports used bulk preparations of B cells for IGVH sequencing and only those MBL that were predominantly monoclonal and had largely replaced the B cell compartment could be characterized (18, 19). Our detailed single cell analysis methods reveal the previously unappreciated diversity of some MBL.
Unlike in CLL in which VH 1-69 is the most commonly expressed IGVH, rearrangements of VH family 1 only occur in approximately 10-15% of normal CD5+ B cells, and VH1-69 rearrangements are rare (5, 7, 19, 35-37). We did not observe any VH1 rearrangements in this cohort of MBL. Among the subjects in the three previously discussed manuscripts, only Landgren et al identified a single unmutated VH1-69 (17-19). As such, MBL may use a more normal, albeit restricted, distribution of VH families than CLL. Alternatively, when rearrangements of VH 1-69 do occur, these cells may have enhanced malignant potential and quickly progress to overt leukemia, and are thus unlikely to be observed as MBL. This same rationale may explain the observation that MBL more commonly show somatically mutated IGVH genes than CLL. Whether the restricted BCR usage in MBL reflects antigen selection or a restricted pool of precursor CD5+ lymphocytes remains unclear. Among the heavy chain genes expressed in MBL, we did not observe any stereotyped mutations (7, 36).
We and others have observed that in clinical CLL, somatic deletion of 13q14.3 correlates with mutated IGVH (8, 38). While the number of MBL analyzed in our study was limited, the same correlation was observed in this dataset. The absence in our cohort of MBL with other chromosomal abnormalities typically associated with CLL emphasizes the indolent nature of most MBL and suggests that subsequent acquisition of additional chromosomal abnormalities may portend progression to CLL. This hypothesis is supported by the report of Rawstron et al (17), where deletions of 11q and 17p were only identified in MBL subjects with an absolute lymphocytosis and were not observed in subjects with low count MBL. A prior report by Ng et al (39) suggested that deletion of 13q14.3 may be more commonly observed in cases of familial CLL (11 of 13, 85%). Finally, we would emphasize that even in the absence of monoclonality, the finding of acquired chromosomal aberrations in MBL suggests that these populations are intrinsically abnormal.
In summary, we determined the characteristics of MBL lymphocytes from subjects ascertained from familial CLL kindreds. Using highly sensitive single cell sorting techniques, we found that MBL with cell counts < 150 MBL cells / uL were commonly multi-clonal, typically had mutated rearrangements of the IGVH, commonly showed non-IgM immunoglobulin isotypes, and frequently had loss of chromosome 13q14.3. These MBL appear indolent on this basis. The determinants of progression from MBL to CLL remain unclear. Future longitudinal analysis of this cohort will determine whether clonal expansion is observed in MBL, and if new MBL clones will emerge.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Andy Rawstron for his thoughtful review of this manuscript. We thank the study participants for their willingness to participate in this study, and the hematologyoncology physicians, nurses and physician assistants for their special help. Flow Cytometry was performed in the Duke Human Vaccine Institute Flow Cytometry Core Facility that is supported by the National Institutes of Health award AI-51445.
Grant Support: MC Lanasa is a fellow of the Leukemia and Lymphoma Society of America. This research is funded by the Bernstein Fund for Leukemia Research, the VA Research Service, and a grant from the National Institutes of Health (NCI R03 CA128030).
References
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005 Feb 24;352(8):804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Dighiero G, Hamblin TJ. Chronic lymphocytic leukaemia. Lancet. 2008 Mar 22;371(9617):1017–1029. doi: 10.1016/S0140-6736(08)60456-0. [DOI] [PubMed] [Google Scholar]
- 3.Goldin LR, Pfeiffer RM, Li X, Hemminki K. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood. 2004 Sep 15;104(6):1850–1854. doi: 10.1182/blood-2004-01-0341. [DOI] [PubMed] [Google Scholar]
- 4.Fischer M, Klein U, Kuppers R. Molecular single-cell analysis reveals that CD5-positive peripheral blood B cells in healthy humans are characterized by rearranged Vkappa genes lacking somatic mutation. J Clin Invest. 1997 Oct 1;100(7):1667–1676. doi: 10.1172/JCI119691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geiger KD, Klein U, Brauninger A, Berger S, Leder K, Rajewsky K, et al. CD5-positive B cells in healthy elderly humans are a polyclonal B cell population. Eur J Immunol. 2000 Oct;30(10):2918–2923. doi: 10.1002/1521-4141(200010)30:10<2918::AID-IMMU2918>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 6.Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998 Nov 2;188(9):1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, Allen SL, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998 Oct 15;102(8):1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinberg JB, Volkheimer AD, Chen Y, Beasley BE, Jiang N, Lanasa MC, et al. Clinical and molecular predictors of disease severity and survival in chronic lymphocytic leukemia. Am J Hematol. 2007 Dec;82(12):1063–1070. doi: 10.1002/ajh.20987. [DOI] [PubMed] [Google Scholar]
- 9.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008 Jun 15;111(12):5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marti GE, Rawstron AC, Ghia P, Hillmen P, Houlston RS, Kay N, et al. Diagnostic criteria for monoclonal B-cell lymphocytosis. Br J Haematol. 2005 Aug;130(3):325–332. doi: 10.1111/j.1365-2141.2005.05550.x. [DOI] [PubMed] [Google Scholar]
- 11.Marti G, Abbasi F, Raveche E, Rawstron AC, Ghia P, Aurran T, et al. Overview of monoclonal B-cell lymphocytosis. Br J Haematol. 2007 Dec;139(5):701–708. doi: 10.1111/j.1365-2141.2007.06865.x. [DOI] [PubMed] [Google Scholar]
- 12.Ghia P, Prato G, Scielzo C, Stella S, Geuna M, Guida G, et al. Monoclonal CD5+ and CD5- B- lymphocyte expansions are frequent in the peripheral blood of the elderly. Blood. 2004 Mar 15;103(6):2337–2342. doi: 10.1182/blood-2003-09-3277. [DOI] [PubMed] [Google Scholar]
- 13.Rawstron AC, Green MJ, Kuzmicki A, Kennedy B, Fenton JA, Evans PA, et al. Monoclonal B lymphocytes with the characteristics of “indolent” chronic lymphocytic leukemia are present in 3.5% of adults with normal blood counts. Blood. 2002 Jul 15;100(2):635–639. doi: 10.1182/blood.v100.2.635. [DOI] [PubMed] [Google Scholar]
- 14.Nieto WG, Almeida J, Romero A, Teodosio C, Lopez A, Henriques AF, et al. Increased frequency (12%) of circulating chronic lymphocytic leukemia-like B-cell clones in healthy subjects using a highly sensitive multicolor flow cytometry approach. Blood. 2009 Jul 2;114(1):33–37. doi: 10.1182/blood-2009-01-197368. [DOI] [PubMed] [Google Scholar]
- 15.Rawstron AC, Yuille MR, Fuller J, Cullen M, Kennedy B, Richards SJ, et al. Inherited predisposition to CLL is detectable as subclinical monoclonal B-lymphocyte expansion. Blood. 2002 Oct 1;100(7):2289–2290. doi: 10.1182/blood-2002-03-0892. [DOI] [PubMed] [Google Scholar]
- 16.Marti GE, Carter P, Abbasi F, Washington GC, Jain N, Zenger VE, et al. B-cell monoclonal lymphocytosis and B-cell abnormalities in the setting of familial B-cell chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2003 Mar;52(1):1–12. doi: 10.1002/cyto.b.10013. [DOI] [PubMed] [Google Scholar]
- 17.Rawstron AC, Bennett FL, O’Connor SJ, Kwok M, Fenton JA, Plummer M, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008 Aug 7;359(6):575–583. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 18.Landgren O, Albitar M, Ma W, Abbasi F, Hayes RB, Ghia P, et al. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009 Feb 12;360(7):659–667. doi: 10.1056/NEJMoa0806122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dagklis A, Fazi C, Sala C, Cantarelli V, Scielzo C, Massacane R, et al. The immunoglobulin gene repertoire of low-count chronic lymphocytic leukemia (CLL)-like monoclonal B lymphocytosis is different from CLL: diagnostic implications for clinical monitoring. Blood. 2009 Jul 2;114(1):26–32. doi: 10.1182/blood-2008-09-176933. [DOI] [PubMed] [Google Scholar]
- 20.Cheson BD, Bennett JM, Grever M, Kay N, Keating MJ, O’Brien S, et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996 Jun 15;87(12):4990–4997. [PubMed] [Google Scholar]
- 21.Volkheimer AD, Weinberg JB, Beasley BE, Whitesides JF, Gockerman JP, Moore JO, et al. Progressive immunoglobulin gene mutations in chronic lymphocytic leukemia: evidence for antigen-driven intraclonal diversification. Blood. 2007 Feb 15;109(4):1559–1567. doi: 10.1182/blood-2006-05-020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefranc MP, Giudicelli V, Kaas Q, Duprat E, Jabado-Michaloud J, Scaviner D, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2005 Jan 1;33:D593–597. doi: 10.1093/nar/gki065. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lossos IS, Tibshirani R, Narasimhan B, Levy R. The inference of antigen selection on Ig genes. J Immunol. 2000 Nov 1;165(9):5122–5126. doi: 10.4049/jimmunol.165.9.5122. [DOI] [PubMed] [Google Scholar]
- 24.Gurrieri C, McGuire P, Zan H, Yan XJ, Cerutti A, Albesiano E, et al. Chronic lymphocytic leukemia B cells can undergo somatic hypermutation and intraclonal immunoglobulin V(H)DJ(H) gene diversification. J Exp Med. 2002 Sep 2;196(5):629–639. doi: 10.1084/jem.20011693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbasi F, Longo NS, Lipsky PE, Raveche E, Schleinitz TA, Stetler-Stevenson M, et al. B-cell repertoire and clonal analysis in unaffected first degree relatives in familial chronic lymphocytic leukaemia kindred. Br J Haematol. 2007 Oct 17; doi: 10.1111/j.1365-2141.2007.06857.x. [DOI] [PubMed] [Google Scholar]
- 26.Rawstron AC, Bennett F, Hillmen P. The biological and clinical relationship between CD5+23+ monoclonal B-cell lymphocytosis and chronic lymphocytic leukaemia. Br J Haematol. 2007 Dec;139(5):724–729. doi: 10.1111/j.1365-2141.2007.06863.x. [DOI] [PubMed] [Google Scholar]
- 27.Degan M, Bomben R, Bo MD, Zucchetto A, Nanni P, Rupolo M, et al. Analysis of IgV gene mutations in B cell chronic lymphocytic leukaemia according to antigen-driven selection identifies subgroups with different prognosis and usage of the canonical somatic hypermutation machinery. Br J Haematol. 2004 Jul;126(1):29–42. doi: 10.1111/j.1365-2141.2004.04985.x. [DOI] [PubMed] [Google Scholar]
- 28.Efremov DG, Ivanovski M, Batista FD, Pozzato G, Burrone OR. IgM-producing chronic lymphocytic leukemia cells undergo immunoglobulin isotype-switching without acquiring somatic mutations. J Clin Invest. 1996 Jul 15;98(2):290–298. doi: 10.1172/JCI118792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000 Dec 28;343(26):1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 30.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002 Nov 26;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fung SS, Hillier KL, Leger CS, Sandhu I, Vickars LM, Galbraith PF, et al. Clinical progression and outcome of patients with monoclonal B-cell lymphocytosis. Leuk Lymphoma. 2007 Jun;48(6):1087–1091. doi: 10.1080/10428190701321277. [DOI] [PubMed] [Google Scholar]
- 32.Shanafelt TD, Kay NE, Call TG, Zent CS, Jelinek DF, LaPlant B, et al. MBL or CLL: which classification best categorizes the clinical course of patients with an absolute lymphocyte count >or= 5 × 10(9) L(-1) but a B-cell lymphocyte count <5 × 10(9) L(-1)? Leuk Res. 2008 Sep;32(9):1458–1461. doi: 10.1016/j.leukres.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang H, Cerny J. Molecular characterization of chronic lymphocytic leukemia with two distinct cell populations: Evidence for separate clonal origins. Am J Clin Pathol. 2006 Jul;126(1):23–28. doi: 10.1309/0YYF-17GF-KFJF-NP5G. [DOI] [PubMed] [Google Scholar]
- 34.Herve M, Xu K, Ng YS, Wardemann H, Albesiano E, Messmer BT, et al. Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J Clin Invest. 2005 Jun;115(6):1636–1643. doi: 10.1172/JCI24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghia P, Stamatopoulos K, Belessi C, Moreno C, Stella S, Guida G, et al. Geographic patterns and pathogenetic implications of IGHV gene usage in chronic lymphocytic leukemia: the lesson of the IGHV3-21 gene. Blood. 2005 Feb 15;105(4):1678–1685. doi: 10.1182/blood-2004-07-2606. [DOI] [PubMed] [Google Scholar]
- 36.Murray F, Darzentas N, Hadzidimitriou A, Tobin G, Boudjogra M, Scielzo C, et al. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood. 2008 Feb 1;111(3):1524–1533. doi: 10.1182/blood-2007-07-099564. [DOI] [PubMed] [Google Scholar]
- 37.Brezinschek HP, Foster SJ, Brezinschek RI, Dorner T, Domiati-Saad R, Lipsky PE. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(-)/IgM+ B cells. J Clin Invest. 1997 May 15;99(10):2488–2501. doi: 10.1172/JCI119433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krober A, Seiler T, Benner A, Bullinger L, Bruckle E, Lichter P, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002 Aug 15;100(4):1410–1416. [PubMed] [Google Scholar]
- 39.Ng D, Toure O, Wei MH, Arthur DC, Abbasi F, Fontaine L, et al. Identification of a novel chromosome region, 13q21.33-q22.2, for susceptibility genes in familial chronic lymphocytic leukemia. Blood. 2007 Feb 1;109(3):916–925. doi: 10.1182/blood-2006-03-011825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.