KIF9B localizes to the axoneme and basal body and is needed for flagella assembly, whereas KIF9A localizes only to the axoneme and controls flagella motility without affecting their structure.
Abstract
Numerous eukaryote genome projects have uncovered a variety of kinesins of unknown function. The kinesin 9 family is limited to flagellated species. Our phylogenetic experiments revealed two subfamilies: KIF9A (including Chlamydomonas reinhardtii KLP1) and KIF9B (including human KIF6). The function of KIF9A and KIF9B was investigated in the protist Trypanosoma brucei that possesses a single motile flagellum. KIF9A and KIF9B are strongly associated with the cytoskeleton and are required for motility. KIF9A is localized exclusively in the axoneme, and its depletion leads to altered motility without visible structural modifications. KIF9B is found in both the axoneme and the basal body, and is essential for the assembly of the paraflagellar rod (PFR), a large extra-axonemal structure. In the absence of KIF9B, cells grow abnormal flagella with excessively large blocks of PFR-like material that alternate with regions where only the axoneme is present. The functional diversity of the kinesin 9 family illustrates the capacity for adaptation of organisms to suit specific cytoskeletal requirements.
Introduction
Kinesins are usually plus end–directed microtubule motors that move vesicles, organelles, or chromosomes (Sharp et al., 2000; Hirokawa and Noda, 2008). Members of this superfamily are defined by a conserved motor domain that binds to microtubules and transforms the chemical energy of nucleotide triphosphate into mechanical force, resulting in motility. Kinesins have been grouped into families depending on the position of their motor domain, the type and number of subunits composing their active form, and their motility. Recent classifications of superfamilies are based on the large datasets available from genome projects (Miki et al., 2005; Wickstead and Gull, 2006).
Cilia and flagella perform essential functions such as motility, sensing, or morphogenesis. Their conserved architecture is a cylinder of nine doublet microtubules that form the outer circumference of the axoneme. At least five kinesin superfamilies are limited to flagellated species (kinesin 2, 9, 13, and probably 16 and 17). Kinesin 2 and 13 participate in flagellum formation by controlling intraflagellar transport (IFT) and microtubule depolymerization (Scholey, 2008). KLP1 (kinesin-like protein 1) is the founding member of the kinesin 9 family (KIF9) that is characterized by a specific neck domain, which is downstream from the catalytic core domain (Miki et al., 2005). First described in the green algae Chlamydomonas reinhardtii, CrKLP1 (C. reinhardtii KLP1) is localized to the central pair of singlet microtubules within the axoneme (Bernstein et al., 1994) and is involved in motility, possibly by regulating flagellar dynein activity (Yokoyama et al., 2004).
Trypanosoma brucei, the protist that causes sleeping sickness, is an amenable model to study the formation and function of flagella (Ralston and Hill, 2008). Its motile flagellum contains a classically structured “9 + 2” axoneme and is attached along the cell body for most of its length. It also possesses a paraflagellar rod (PFR), which is a large, lattice-like structure that runs parallel to the axoneme from where it emerges from the cell body to the distal tip. The PFR is composed of two major proteins, PFR1 and PFR2, and contains at least 20 other proteins (Portman et al., 2009). Association of the PFR within the flagellum is required for cell motility (Bastin et al., 1998) and is essential for survival at the bloodstream stage of the parasite (Branche et al., 2006; Broadhead et al., 2006).
In this study, we demonstrate that the two members of the T. brucei kinesin 9 family, KIF9A and KIF9B, are strongly associated with the flagellar skeleton and participate in flagellar motility. However, their individual contributions are distinct because only inhibition of KIF9B effects construction of the PFR, thus revealing the first kinesin involved in the formation of an extra-axonemal structure.
Results and discussion
Trypanosome KIF9 proteins display different characteristics and locations
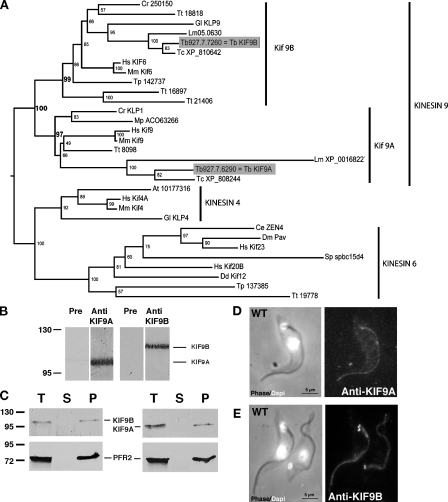
Searching the T. brucei genome database (http://www.genedb.org/genedb/tryp/blast.jsp) with the CrKLP1 protein sequence (P46870) identified two candidate members for the KIF9 family, which were termed KIF9A (NCBI Protein Database accession no. XP_846252) and KIF9B (NCBI Protein Database accession no. XP_846346). Reciprocal Blastp analysis showed that both KIF9A and KIF9B sequences recognized the CrKLP1 (expectancy [e]: KIF9A-CrKLP1 = 9 e − 66; KIF9B-CrKLP1 = 9 e − 67) as well as members of the KIF9 family from numerous flagellated species. Phylogenetic analyses demonstrated the existence of two subfamilies of KIF9 in all flagellated species analyzed (Fig. 1 A). The KIF9A family includes CrKLP1 and human KIF9, whereas the KIF9B family includes the so-called KIF6 human protein. Nevertheless, the kinesin 9 gene family was clearly separate from the kinesin 6 family (Fig. 1 A). Trypanosome KIF9A and KIF9B possess the typical kinesin motor domain and ATP-binding domain signatures (P-loop, Switch1, and Switch2). Trypanosome KIF9A is characterized by a unique 35–amino acid insertion in its N-terminal domain, whereas KIF9B is marked by at least seven insertions in its C-terminal domain (Fig. S1 A).
Figure 1.
Characterization of the kinesin 9 family. (A) Phylogenetic tree constructed with kinesin 4, 6, and 9 protein sequences (348 positions), identifying two groups within the kinesin 9 family, KIF9A and KIF9B. (B) Western blot on T. brucei whole cell extracts (107 cells/lane) probed with preimmune sera (Pre), anti-KIF9A (left; 1:500) or anti-KIF9B (right; 1:500) antisera. Data were reproduced at least three times (all five mouse sera gave the same result). (C) Western blot on T. brucei cell extracts fractionated in detergent (107 cells/lane) and reproduced five times. T, total; S, supernatant (soluble fraction); P, pellet. L8C4, recognizing the PFR2 protein, was used as control. (D and E) IFA staining on WT detergent-extracted cells reproduced at least 10 times. (left) Combined phase-contrast and DAPI (white) images; (right) IFA with anti-KIF9A (D) or anti-KIF9B (E) antibodies. Values on blots are given in kilodaltons.
To determine the cellular location of KIF9A and KIF9B, a fragment of each of the divergent C-terminal domains (KIF9A, amino acids 479–891; KIF9B, amino acids 490–1,041; Fig. S1 A) was expressed as His-tagged fusion proteins in Escherichia coli and used to immunize mice. The resulting polyclonal sera recognized a single band on trypanosome whole cell extracts, 100 kD for KIF9A (calculated 96.9 kD) and 115 kD for KIF9B (calculated 113.3 kD), which is in contrast to preimmune sera that gave no detectable signal (Fig. 1 B and Fig. S1 B). Both proteins were tightly associated with the cytoskeletal fraction, as shown after removal of the membrane by detergent treatment (Fig. 1 C). This is in agreement with the presence of KIF9A in a proteomic analysis of the structural components of flagella purified upon detergent and high salt treatment (Broadhead et al., 2006).
Indirect immunofluorescence assays (IFAs) of detergent-extracted cytoskeletons revealed the presence of KIF9A and KIF9B on the flagellum, but KIF9B also localized to the basal body (BB; Fig. 1, D and E; and Fig. S1 G). The preimmune sera did not produce any signal (unpublished data). These results were confirmed by IFA on nonextracted cells using KIF9A (Fig. S1 C) or in live cells by expression of a GFP∷KIF9B fusion protein (Fig. S1 H). Double staining with L8C4 (PFR marker; Kohl et al., 1999) or MAb25 (axoneme marker; Pradel et al., 2006) showed that both KIF9A and KIF9B are associated with the axoneme and not the PFR. The initiation of PFR construction happens only after the flagellum has emerged from the cell body (Fig. S1, D, E, I, and J). Double IFA with BB markers showed that KIF9A staining was absent from the BB area in contrast to KIF9B (Fig. S1, F and K). Unfortunately, immunogold electron microscopy using antibodies against KIF9A, KIF9B, or GFP in the case of GFP∷KIF9 failed to produce a significant signal, whereas a clear positive signal was obtained for the control L3B2 monoclonal antibody (Kohl et al., 1999).
The axonemal localization and the association with the cytoskeleton are similar to CrKLP1 (Bernstein et al., 1994), but KIF9B localization at the BB was more surprising. Dual localization to the axoneme and BB is typical for kinesin 2 and IFT proteins, but these are not tightly linked to the flagellar skeleton (Cole et al., 1998; Absalon et al., 2008a). Consistently, movement of GFP∷KIF9B within the flagellum could not be detected (Video 1), which is in contrast to the GFP∷IFT52 control (Absalon et al., 2008a).
Silencing of KIF9A and KIF9B causes different motility defects
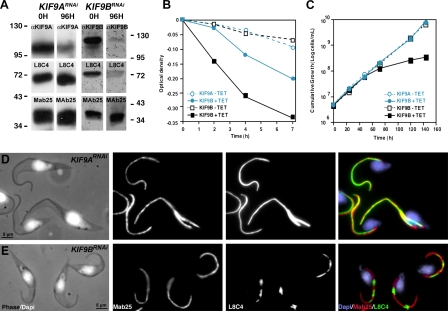
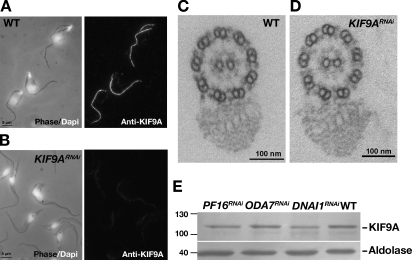
Plasmids allowing tetracycline-inducible expression of KIF9A or KIF9B double-stranded RNA were used to generate KIF9ARNAi or KIF9BRNAi cell lines. Western blotting demonstrated knockdown efficiency in both cases as well as absence of cross RNAi (Fig. 2 A; and Fig. S2, A and D). Observation of live KIF9ARNAi cells revealed a significant reduction in flagellum and cell movement, resulting in cell sedimentation (Fig. 2 B and Video 2), which is in agreement with the proposed function for KLP1 in C. reinhardtii (Yokoyama et al., 2004). The motility phenotype was more pronounced in KIF9BRNAi cells, leading to cell paralysis (Fig. 2 B and Video 3) and growth arrest (Fig. 2 C) in contrast to KIF9ARNAi. This growth phenotype could be a result of extremely reduced motility, as it has been shown that motility contributes to the completion of cytokinesis (Branche et al., 2006; Broadhead et al., 2006; Ralston et al., 2006). Because kinetics of RNAi can be variable from one target to another, KIF9ARNAi cells were induced for up to 8 d, but this did not modify the intensity of the motility phenotype (Fig. S2, B and C). IFA with the anti-KIF9A antibody demonstrated the efficiency of silencing at the cellular level (Fig. 3, A and B). Electron microscopy analysis of cross sections of KIF9ARNAi flagella failed to show any striking modifications: the nine doublets, the central pair, the outer or inner dynein arms, and the PFR all appear normal, as was the orientation of the central pair relative to the PFR (Fig. 3, C and D), which is a common feature of many motility mutants in trypanosomes (Branche et al., 2006; Gadelha et al., 2006; Ralston et al., 2006). KIF9A could be affected in mutants lacking some of the flagellar substructures. Therefore, we examined the abundance and location of KIF9A by Western blotting (Fig. 3 E) or IFA (not depicted) in various RNAi mutants where dynein arm (DNAI1RNAi or ODA7RNAi) or central pair (PF16RNAi) components were affected. No significant modifications were observed.
Figure 2.
Both KIF9 proteins participate in cell motility, but only KIF9B is involved in PFR assembly. (A) Western blot of total protein samples (107 cells per lane) from noninduced (0 h) and 96 h–induced KIF9ARNAi cells (left) and KIF9BRNAi cells (right), with the experiment performed at least three times. The same membrane was cut into three parts and probed with anti-KIF9A antibody (left), anti-KIF9B antibody (right), or L8C4 and MAb25. Values on blot are given in kilodaltons. (B) Sedimentation assay showing the mean of five reproducible experiments. Trypanosomes were incubated in cuvettes, and optical density was measured before and after mixing. KIF9ARNAi (blue) and KIF9BRNAi (black) were induced for 4 d or 3 d, respectively (solid lines). Control cells were grown without tetracycline (dashed line). (C) Cumulative growth curves of KIF9ARNAi and KIF9BRNAi mutants under noninduced conditions (dashed line) and induced conditions (solid lines), representative of four experiments. (D and E) IFA on 72 h–induced KIF9ARNAi (D) and KIF9BRNAi cells (E) performed at least 20 times. Combined phase-contrast and DAPI images (left), MAb25 (middle left), L8C4 (middle right), and combined DAPI, MAb25, and L8C4 images are shown (right).
Figure 3.
Characterization of the KIF9ARNAi mutant. (A and B) IFA on WT (A) or KIF9ARNAi (B) detergent-extracted trypanosomes. The anti-KIF9A antibody (1:800) labels the flagellum only in noninduced cells. (C and D) Transmission electron micrographs showing cross sections of flagella from WT (C) or 96 h–induced KIF9ARNAi cells (D). Representative images from 20 sections of WT and 20 sections of KIF9ARNAi. (E) Western blot on whole cell extracts of control or the indicated T. brucei axoneme mutants (5 × 106 cells/lane) induced for 3 d. The blot was probed with the anti-KIF9A antibody (1:500) and an antialdolase antibody (1:3,000) as a loading control. The experiment was reproduced twice. Values on blots are given in kilodaltons.
In Western blots, the MAb25 signal, an axoneme marker, remained unchanged in both KIF9ARNAi and KIF9BRNAi cells. However, an unexpected decrease in the amount of PFR2 was observed in KIF9BRNAi-induced cells (Fig. 2 A). By IFA, MAb25 showed the expected labeling from the proximal to the distal tip of the flagellum in both cases. L8C4 showed a uniform labeling of the flagellum on KIF9ARNAi cells (Fig. 2 D) but not in the KIF9BRNAi mutants where it produced a discontinuous signal, alternating regions of intense labeling with regions where the signal was negative (Fig. 2 E). This observation was reproduced using antibodies that recognize both PFR1 and PFR2 proteins (Fig. S3 A) or a component of the distal PFR domain (Fig. S3 B), indicating global defects in PFR organization.
KIF9BRNAi cells fail to assemble a normal PFR
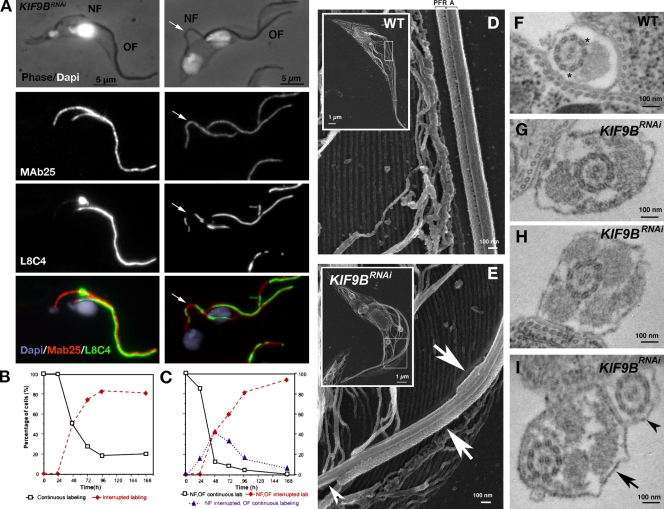
When a new flagellum is built, the PFR is only added to the axoneme as the flagellum exits the flagellar pocket and the two structures are built almost simultaneously (Sherwin and Gull, 1989). The PFR abnormalities in KIF9BRNAi cells could result from either its partial assembly or postassembly destabilization. Cells were carefully analyzed at early time points of RNAi induction, revealing that only the new flagellum showed discontinuous labeling, whereas the old flagellum was normal (Fig. 4 A and Fig. S2 F). We quantified the emergence of the PFR-related phenotype in cells with either one or two flagella at various time points after RNAi-mediated knockdown (n > 100). In cells with one flagellum, an irregular PFR labeling was first detected after 24 h. The abundance of these cells then increased steadily to reach a plateau at ∼80% (Fig. 4 B). In biflagellated cells (Fig. 4 C), the proportion of cells with both new and old normal PFR labeling decreased rapidly: they were replaced by siblings with an abnormal new flagellum (up to 42% at 48 h). After 24 h, most cells possessed two flagella that were both affected. This can be explained if one daughter cell inherits the flagellum with a normal PFR structure at cytokinesis, and the other daughter exhibits the abnormal PFR. Further division of the cell with abnormal PFRs results in irregular labeling of both the old and new flagellum.
Figure 4.
Characterization of the KIF9BRNAi mutant. KIF9B is involved in PFR assembly. (A) IFA on 48 h–induced KIF9BRNAi cells. (left) Cell with one nucleus, an old flagellum (OF), and a short new flagellum (NF) are shown. The old flagellum shows a normal axoneme/PFR labeling, whereas the new flagellum shows a normal axoneme but a disrupted PFR. (right) Cells with two nuclei and two flagella exhibit the same phenotype. Detached flagella are shown by arrows. The experiment was reproduced more than three times, and at least 100 biflagellated cells were analyzed. (B and C) KIF9BRNAi cells with a normal PFR labeling (black lines) are progressively replaced by cells with an interrupted labeling (red lines) both in uniflagellated (B) and biflagellated cells (C). In biflagellated cells, the phenotype is first visible in the new flagellum (purple line). For both populations, at least 100 cells were analyzed per induction time. (D and E) Scanning electron micrograph of a WT cell (D) and a 72 h–induced KIF9BRNAi cell (E) extracted with cold Triton X-100. The white rectangle indicates the position of the magnified area. Representative images were chosen from at least 20 cells. (F–I) Transmission electron micrographs showing cross sections of flagella from WT (F) or 72 h–induced KIF9BRNAi cells (G–I). Representative images were chosen from >50 sections of WT and 89 sections of KIF9BRNAi. Asterisks indicate doublets 4–7, arrows point toward excessive PFR, and arrowheads show a naked axoneme.
PFR structure in KIF9BRNAi cells
Although both old and new flagellum of wild-type (WT) trypanosomes have a constant diameter of ∼300 nm all along their length, induced KIF9BRNAi cells exhibit a highly variable diameter (150–450 nm) along the length of the flagellum (Fig. S3, D and E). To better visualize flagellar structure, cells were treated with cold triton to remove the pellicular and flagellar membrane (Absalon et al., 2008b). In WT trypanosomes, the characteristic lattice-like PFR structure connected to the axoneme is clearly visible with a constant diameter of ∼300 nm (Fig. 4 D). In contrast, the flagellum of induced KIF9BRNAi cells shows regions with one or more PFR-like structures (Fig. 4 E, arrows), alternating with portions containing only the axoneme (Fig. 4 E, arrowhead). The locally excessive PFR material is still connected to the axoneme but is not resistant to extraction with 1M NaCl (Fig. S3, F and G), indicating that these segments are not as stably connected to the axoneme as in WT cells.
In transmission electron microscopy of WT cells, the PFR has a diameter of ∼150–200 nm and is anchored to the axoneme via connections to doublets 4–7 (Fig. 4 F, asterisks). In KIF9BRNAi cells, several situations are encountered: a naked axoneme (36%), one PFR-like structure (15%), or two or more PFR-like structures (49%; n = 89). Several connections were visible between the multiple PFR-like structures and the axoneme (Fig. 4, G–I). The axoneme appeared normal, with the 9 + 2 organization and the dynein arms, but the orientation of the central pair could not be determined as a result of the presence of multiple PFR-like structures. In addition to these structural modifications, induced KIF9BRNAi trypanosomes display a flagellum that is partially detached from the cell body (Fig. S3, H and I), which is in contrast to KIF9ARNAi cells. The flagellum of cells with normal PFR labeling was always attached, whereas ∼35% of cells with interrupted PFR labeling exhibited a detached flagellum (Fig. S3 J). This could be a consequence of the motility defect as described in other mutants with decreased flagellar beating (Absalon et al., 2007; Li and Wang, 2008).
Separate functions for KIF9A and KIF9B in flagellum beating and construction
Our data demonstrate that the two kinesin 9 proteins are performing distinct functions in the trypanosome flagellum. KIF9A is required for motility but does not play a role in PFR assembly. In C. reinhardtii, CrKLP1 regulates flagellar dynein activity (Yokoyama et al., 2004) and interacts with Hydin, a central pair protein essential for motility in algae (Lechtreck and Witman, 2007) and trypanosomes (Dawe et al., 2007). It has been shown recently that Hydin is also essential for ciliary motility and has a similar effect on central pair structure in mice (Lechtreck et al., 2008). In contrast to algae or ciliates, the central pair does not twist during beating in trypanosomes (as in humans), and this functional difference could be associated with discrete structural variations of the axoneme.
KIF9B displays a conserved motor domain in its N-terminal region and a divergent C-terminal region with multiple insertions. We postulate that these insertions could be interacting either directly with PFR proteins or with proteins ensuring the junction between axoneme and PFR. During PFR assembly, PFR1 and PFR2 subunits are actively transported to their assembly site at the distal tip of the growing flagellum (Bastin et al., 1999a,b). This transport does not require preassembly, as shown in the mutant snl-1, where in the absence of PFR2, PFR1 is still transported to the distal tip (Bastin et al., 1999a). This transport could be ensured by the IFT machinery as reported for radial spokes or dynein arms in the C. reinhardtii axoneme (Qin et al., 2004). This is supported by the absence of a PFR in mutants where IFT has been abolished (Kohl et al., 2003). In this case, KIF9B would be deposited on the growing axoneme by the IFT machinery and would serve as a linker protein with the growing PFR structure or in the incorporation of PFR proteins in the PFR structure. However, the PFR is added to the axoneme only after the emergence of the flagellum from the flagellar pocket, i.e., when the axoneme is already 2-µm long. Therefore, the absence of PFR in IFT mutants could be caused by the lack of an axoneme that is needed as support for the PFR.
The intriguing localization of KIF9B to the BB region could also be significant for the role of this protein in flagellar assembly and motility. BBs appear ninefold symmetric but have asymmetric accessory structures. KIF9B could be important in either recognizing or determining this asymmetry, thereby allowing proper PFR assembly at defined microtubule doublets 4–7, whereas its absence would lead to patches of PFR that assemble with random radial orientation.
KIF9B could be involved in a specific machinery constructing the PFR, associating with kinesin 2, like kinesin 2 and osm-3 do in sensory cilia of Caenorhabditis elegans (Snow et al., 2004). KIF9B could transport PFR and associated proteins to the tip for assembly, allowing coordinated elongation of PFR and axoneme. In the absence of KIF9B, kinesin 2 still drives IFT and axoneme formation, whereas PFR elongation fails with proteins being dropped along the way. However, no movement of KIF9B could be detected in live cells, and the protein is tightly attached to the axoneme. Using its polar movement, KIF9B could translocate PFR material from one KIF9B molecule to its neighbor toward the distal tip. The extremely divergent C-terminal domain of KIF9B could be used as an adapter to interact with different partners according to the cell type where KIF9B is expressed. In mammals, KIF9A and KIF9B are expressed in a variety of tissues, such as testis, brain, and lung, but their function is unknown. Expression in testis could be related to the presence of extra-axonemal structures in the flagella of spermatozoa (Escalier, 2003), and investigation of their function in the construction of these elements holds promises. In conclusion, this work illustrates the diversification of the KIF9 family with contribution at the axoneme and the PFR level. Although sharing the motor domain, they have diversified their C-terminal tail to suit specific cytoskeletal requirements.
Materials and methods
Trypanosome cell lines and cultures
All cell lines used for this study were derivatives of strain 427 of T. brucei and cultured in SDM79 medium with hemin and 10% fetal calf serum. Cell lines KIF9BRNAi and KIF9ARNAi express double-stranded RNA from two tetracycline-inducible T7 promoters facing each other in pZJM vector (Wang et al., 2000) transformed in 29–13 cells that express the T7 RNA polymerase and the tetracycline repressor (Wirtz et al., 1999). RNAi was induced by addition of 1 µg tetracycline per milliliter of medium, and fresh tetracycline was added at each cell dilution.
The GFP∷KIF9B fusion contains 1–1,725 bp of KIF9B gene in frame behind the GFP gene in the vector pPCPFReGFP. The fusion gene has been cloned between PFR1 and PFR2 intergenic sequences. The vector was linearized by SphI and integrated in the endogenous locus, resulting in a reconstructed complete GFP∷KIF9B fusion. The plasmid was electroporated in WT cells, and resistant clones were selected by addition of 1 µg/ml puromycin.
For recombinant protein expression, the less-conserved C-terminal domains of the proteins (KIF9A, 479–891; KIF9B, 491–1,041) were chosen. Corresponding DNA fragments were cloned in frame with a hexahistidine tag (pET22b vector; EMD). After induction (with 0.5 mM IPTG for 2 h at 25°C), bacteria were lysed (BugBuster; EMD) in the presence of protease inhibitors (Roche) and lysozyme (Roche) followed by ultrasound sonication (three times for 2 min). The recombinant protein was purified by passage on a nickel nitrilotriacetic acid resin (EMD) and used for immunization of two groups of five mice (10 µg protein and complete Freund’s adjuvant) followed by two boosts with 10 µg protein in incomplete Freund’s adjuvant. 4 wk after the last injection, the mice were exsanguinated, and the serum was used for IFA and Western blotting. Primer sequences are available upon request.
RT-PCR
Total RNA was extracted from cells grown with or without tetracycline for the indicated periods of time and purified using Trizol. DNA was eliminated by DNase treatment, and RNA purity was confirmed by conventional PCR. After primer calibration and determination of optimal conditions, semiquantitative RT-PCR was performed using 50–100 ng template total RNA and 0.8 µM primers. Amplification was performed using the Superscript OneStep RT-PCR (Invitrogen) enzyme mix. Quantification was performed from ethidium bromide gels using the Image Quant software (version 5.2; Molecular Dynamics).
Immunofluorescence
Trypanosomes were settled onto poly-L-lysine–coated slides, and cytoskeletons were prepared by extraction with 1% NP-40 in 100 mM Pipes, pH 6.9, 2 mM EGTA, 1 mM MgSO4, and 0.1 mM EDTA. After fixation in methanol at −20°C for 5 min followed by rehydration (PBS for 10 min), the slides were incubated with primary antibodies. As PFR markers, we used the monoclonal antibodies L8C4 (Kohl et al., 1999), L13D6 (Kohl et al., 1999), 2E10 (Ismach et al., 1989), and ROD-1 (Woods et al., 1989). Monoclonal antibodies MAb25 and MAb22 were used as markers for the axoneme (MAb25; Pradel et al., 2006) and the BB (MAb22; Bonhivers et al., 2008).
Subclass-specific secondary antibodies coupled to Alexa Fluor 488 (Invitrogen), Cy3 (Jackson ImmunoResearch Laboratories, Inc.), or Cy5 (Jackson ImmunoResearch Laboratories, Inc.) were used for double labeling. Slides were stained with DAPI for visualization of kinetoplast and nuclear DNA content and mounted with antifade Vectashield medium (Vector Laboratories).
IFA was performed as described previously (Absalon et al., 2007). The primary antibodies L8C4 (Kohl et al., 1999), MAb25 (Pradel et al., 2006), L13D6 (Kohl et al., 1999), 2E10 (Ismach et al., 1989), and ROD-1 (Woods et al., 1989) were used.
For IFA with the anti-KIF9A and the anti-KIF9B antibodies, cells were treated with 1% Triton X-100 for 10 min at 4°C and fixed for 15 min at room temperature in 4% paraformaldehyde and 0.1% glutaraldehyde. After blocking by incubation for 45–60 min in PBS containing 1% BSA, the slides were incubated for 45–60 min with the antisera diluted 1:200 in PBS/0.1% BSA and treated as described previously (Absalon et al., 2007). Slides were mounted in Vectashield medium.
Slides were observed at room temperature with a microscope (DMR; Leica) using a 100× NA 1.3 Plan Fluotar objective, and images were captured with a camera (Cool Snap HQ; Roper Industries). Alternatively, slides were viewed on a microscope (DMI4000; Leica) using a 100× NA 1.4 Plan Apo objective, and images were acquired with a camera (Retiga-SRV; Q-Imaging). Images were analyzed using the IPLab Spectrum software (version 3.9; BD) or ImageJ (National Institutes of Health) and processed with Photoshop (CS2; Adobe). For live cell imaging, cells were filmed at room temperature on a microscope (DMI4000) using the aforementioned settings.
Electron microscopy
For transmission electron microscopy, WT and induced KIF9BRNAi samples were washed in 0.1 M cacodylate buffer, pH 7.2, and fixed in 2.5% glutaraldehyde, 2% paraformaldehyde, and 0.5% tannic acid solution for 2 h at room temperature. Fixed samples were washed three times in 0.1 M cacodylate buffer, pH 7.2, and postfixed in 1% 0.1 M osmium-cacodylate solution, pH 7.2, for 2 h in the dark under agitation. Samples were stained en block with 2% aqueous uranyl solution at 4°C overnight, gradually dehydrated in ethanol, and embedded in Spurr resin followed by polymerization for 3 d at 70°C. For detergent extraction of cytoskeletons, cells were treated with 100 mM Pipes, 2 mM EGTA, and 1 mM MgSO4 containing 1% Nonidet P40 solution for 10 min (Sherwin and Gull, 1989). Washed samples were fixed in 2.5% glutaraldehyde, 0.5% tannic acid solution for 2 h at room temperature, and processed as described for whole cells (see previous paragraph). 60-nm-thick sections were cut with an Ultracut E microtome (Reichert S; Leica) and picked up on copper rhodium–coated grids. Grids were analyzed on an electron microscope (JEM 1010; Jeol), and images were captured with a digital camera (C4742-95; Hamamatsu Photonics). Fixation, embedding, and sectioning for transmission electron microscopy of whole cells from WT and induced KIF9BRNAi samples were performed as described previously (Branche et al., 2006).
For scanning electron microscopy, cells were washed in PBS, fixed for at least 1 h with 2.5% glutaraldehyde in PBS or 0.2 M cacodylate buffer, washed, and postfixed in 1% osmium tetroxide. After dehydration, samples were critical point dried (K850 [Emitech] or UnionCPD30 [Balzers]) and coated with gold (JFC-1200 [Jeol] or Ion Beam Coater 681 [Gatan]). Samples were visualized with a scanning microscope (JM6700 F; Jeol). For detergent-extracted cytoskeleton preparations, cells were treated with 1% Triton X-100 at 4°C in PBS for 10 min to strip the plasma membrane. After two washes in PBS, the samples were processed for scanning electron microscopy as described for whole cells (see previous paragraph).
Cell fractionation
5 × 108 cells were harvested by centrifugation and washed once in PBS. The pellet was resuspended in 100 mM Pipes, 2 mM EGTA, and 1 mM MgSO4 containing 1% Nonidet P40 and incubated for 2 min at room temperature to provide a cytoskeleton preparation (Sherwin and Gull, 1989). After centrifugation (at 1,000 g for 5 min), the pellet was resuspended in either 60 mM CaCl2 or 1 M NaCl to depolymerize the subpellicular microtubules, which were incubated for 15 min at room temperature and centrifuged (Kohl et al., 1999). The pellet was resuspended in PBS. Equal cell equivalents of each fraction were resolved by SDS-PAGE and subjected to Western blot analysis.
Western blotting
1–2 ×107 cells were loaded per well on 10% SDS-PAGE gels. Proteins were transferred to nitrocellulose membranes (Hybond ECL Plus; GE Healthcare) and after blocking with 1% TBS-BSA, were incubated with anti-KIF9B antibody (1:500), anti-KIF9A antibody (1:500), L8C4 (1:50), or MAb25 (1:50). The secondary antibody used was an anti–mouse HRP (1:5,000; GE Healthcare).
Multiple alignment and tree inference of kinesin 9 family sequences
The kinesin multiple alignment made by Wickstead and Gull (2006) was updated with new sequences of kinesin 9 family members: Trypanosoma cruzi (NCBI Protein Database accession no. XP_810642.1 and XP_808244.1), Micromonas pusilla (NCBI Protein Database accession no. ACO63266), Mus musculus (NCBI Protein Database accession nos. AAI15766, NP_796026.2, and NP_032472), Leishmania major (NCBI Protein Database accession no. XP_001682276), and Homo sapiens (NCBI Protein Database accession no NP_659464).
All new sequences were incorporated and aligned using the ED program of the package MUST (Management Utilities for Sequences and Trees; Philippe, 1993). Ambiguously aligned regions and gaps were excluded in phylogenetic analyses. Maximum likelihood phylogenetic analyses were performed with the program Treefinder (Jobb et al., 2004), and bootstrap values were calculated from 1,000 replicates. The Whelan and Golding (2001) substitution matrix was used with a γ-distributed variation in substitution rate approximated to four discrete categories.
Online supplemental material
Fig. S1 shows the characterization of the kinesin 9 proteins, Fig. S2 shows the efficiency of RNAi in the KIF9RNAi mutants, and Fig. S3 shows the analysis of the KIF9BRNAi mutant. Video 1 shows T. brucei expressing GFP∷KIF9B, and Videos 2 and 3 show the effect on motility in T. brucei of knockdown of KIF9A and KIF9B, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903139/DC1.
Supplementary Material
Acknowledgments
We acknowledge the electron microscopy department of the Muséum National d’Histoire Naturelle for providing access to their equipment. We wish to thank Derrick Robinson (Bordeaux) for critical reading of the manuscript and anonymous referees for constructive suggestions.
R. Demonchy was funded by the French Ministry of Research. This work was funded by the Pasteur Institut, Centre National de la Recherche Scientifique, and Fonds Dédié Sanofi-Aventis/Ministère de la Recherche.
Footnotes
Abbreviations used in this paper:
- BB
- basal body
- IFA
- immunofluorescence assay
- IFT
- intraflagellar transport
- PFR
- paraflagellar rod
- WT
- wild type
References
- Absalon S., Kohl L., Branche C., Blisnick T., Toutirais G., Rusconi F., Cosson J., Bonhivers M., Robinson D., Bastin P.. 2007. Basal body positioning is controlled by flagellum formation in Trypanosoma brucei. PLoS One. 2:e437. 10.1371/journal.pone.0000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Absalon S., Blisnick T., Kohl L., Toutirais G., Doré G., Julkowska D., Tavenet A., Bastin P.. 2008a. Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes. Mol. Biol. Cell. 19:929–944. 10.1091/mbc.E07-08-0749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Absalon S., Blisnick T., Bonhivers M., Kohl L., Cayet N., Toutirais G., Buisson J., Robinson D., Bastin P.. 2008b. Flagellum elongation is required for correct structure, orientation and function of the flagellar pocket in Trypanosoma brucei. J. Cell Sci. 121:3704–3716. 10.1242/jcs.035626 [DOI] [PubMed] [Google Scholar]
- Bastin P., Sherwin T., Gull K.. 1998. Paraflagellar rod is vital for trypanosome motility. Nature. 391:548. 10.1038/35300 [DOI] [PubMed] [Google Scholar]
- Bastin P., Pullen T.J., Sherwin T., Gull K.. 1999a. Protein transport and flagellum assembly dynamics revealed by analysis of the paralysed trypanosome mutant snl-1. J. Cell Sci. 112:3769–3777. [DOI] [PubMed] [Google Scholar]
- Bastin P., MacRae T.H., Francis S.B., Matthews K.R., Gull K.. 1999b. Flagellar morphogenesis: protein targeting and assembly in the paraflagellar rod of trypanosomes. Mol. Cell. Biol. 19:8191–8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein M., Beech P.L., Katz S.G., Rosenbaum J.L.. 1994. A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J. Cell Biol. 125:1313–1326. 10.1083/jcb.125.6.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhivers M., Landrein N., Decossas M., Robinson D.R.. 2008. A monoclonal antibody marker for the exclusion-zone filaments of Trypanosoma brucei. Parasit Vectors. 1:21. 10.1186/1756-3305-1-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branche C., Kohl L., Toutirais G., Buisson J., Cosson J., Bastin P.. 2006. Conserved and specific functions of axoneme components in trypanosome motility. J. Cell Sci. 119:3443–3455. 10.1242/jcs.03078 [DOI] [PubMed] [Google Scholar]
- Broadhead R., Dawe H.R., Farr H., Griffiths S., Hart S.R., Portman N., Shaw M.K., Ginger M.L., Gaskell S.J., McKean P.G., Gull K.. 2006. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 440:224–227. 10.1038/nature04541 [DOI] [PubMed] [Google Scholar]
- Cole D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., Rosenbaum J.L.. 1998. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141:993–1008. 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe H.R., Shaw M.K., Farr H., Gull K.. 2007. The hydrocephalus inducing gene product, Hydin, positions axonemal central pair microtubules. BMC Biol. 5:33. 10.1186/1741-7007-5-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalier D. 2003. New insights into the assembly of the periaxonemal structures in mammalian spermatozoa. Biol. Reprod. 69:373–378. 10.1095/biolreprod.103.015719 [DOI] [PubMed] [Google Scholar]
- Gadelha C., Wickstead B., McKean P.G., Gull K.. 2006. Basal body and flagellum mutants reveal a rotational constraint of the central pair microtubules in the axonemes of trypanosomes. J. Cell Sci. 119:2405–2413. [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Noda Y.. 2008. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol. Rev. 88:1089–1118. 10.1152/physrev.00023.2007 [DOI] [PubMed] [Google Scholar]
- Ismach R., Cianci C.M., Caulfield J.P., Langer P.J., Hein A., McMahon-Pratt D.. 1989. Flagellar membrane and paraxial rod proteins of Leishmania: characterization employing monoclonal antibodies. J. Protozool. 36:617–624. [DOI] [PubMed] [Google Scholar]
- Jobb G., von Haeseler A., Strimmer K.. 2004. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 4:18. 10.1186/1471-2148-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kohl L., Sherwin T., Gull K.. 1999. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 46:105–109. 10.1111/j.1550-7408.1999.tb04592.x [DOI] [PubMed] [Google Scholar]
- Kohl L., Robinson D., Bastin P.. 2003. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 22:5336–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.F., Witman G.B.. 2007. Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J. Cell Biol. 176:473–482. 10.1083/jcb.200611115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.F., Delmotte P., Robinson M.L., Sanderson M.J., Witman G.B.. 2008. Mutations in Hydin impair ciliary motility in mice. J. Cell Biol. 180:633–643. 10.1083/jcb.200710162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wang C.C.. 2008. KMP-11, a basal body and flagellar protein, is required for cell division in Trypanosoma brucei. Eukaryot. Cell. 7:1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H., Okada Y., Hirokawa N.. 2005. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 15:467–476. 10.1016/j.tcb.2005.07.006 [DOI] [PubMed] [Google Scholar]
- Philippe H. 1993. MUST, a computer package of management utilities for sequences and trees. Nucleic Acids Res. 21:5264–5272. 10.1093/nar/21.22.5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman N., Lacomble S., Thomas B., McKean P.G., Gull K.. 2009. Combining RNA interference mutants and comparative proteomics to identify protein components and dependences in a eukaryotic flagellum. J. Biol. Chem. 284:5610–5619. 10.1074/jbc.M808859200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel L.C., Bonhivers M., Landrein N., Robinson D.R.. 2006. NIMA-related kinase TbNRKC is involved in basal body separation in Trypanosoma brucei. J. Cell Sci. 119:1852–1863. 10.1242/jcs.02900 [DOI] [PubMed] [Google Scholar]
- Qin H., Diener D.R., Geimer S., Cole D.G., Rosenbaum J.L.. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164:255–266. 10.1083/jcb.200308132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston K.S., Hill K.L.. 2008. The flagellum of Trypanosoma brucei: new tricks from an old dog. Int. J. Parasitol. 38:869–884. 10.1016/j.ijpara.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston K.S., Lerner A.G., Diener D.R., Hill K.L.. 2006. Flagellar motility contributes to cytokinesis in Trypanosoma brucei and is modulated by an evolutionarily conserved dynein regulatory system. Eukaryot. Cell. 5:696–711. 10.1128/EC.5.4.696-711.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J.M. 2008. Intraflagellar transport motors in cilia: moving along the cell’s antenna. J. Cell Biol. 180:23–29. 10.1083/jcb.200709133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.J., Rogers G.C., Scholey J.M.. 2000. Microtubule motors in mitosis. Nature. 407:41–47. 10.1038/35024000 [DOI] [PubMed] [Google Scholar]
- Sherwin T., Gull K.. 1989. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 323:573–588. 10.1098/rstb.1989.0037 [DOI] [PubMed] [Google Scholar]
- Snow J.J., Ou G., Gunnarson A.L., Walker M.R., Zhou H.M., Brust-Mascher I., Scholey J.M.. 2004. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat. Cell Biol. 6:1109–1113. 10.1038/ncb1186 [DOI] [PubMed] [Google Scholar]
- Wang Z., Morris J.C., Drew M.E., Englund P.T.. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174–40179. 10.1074/jbc.M008405200 [DOI] [PubMed] [Google Scholar]
- Whelan S., Goldman N.. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18:691–699. [DOI] [PubMed] [Google Scholar]
- Wickstead B., Gull K.. 2006. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol. Biol. Cell. 17:1734–1743. 10.1091/mbc.E05-11-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E., Leal S., Ochatt C., Cross G.A.. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101. 10.1016/S0166-6851(99)00002-X [DOI] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., MacRae T.H., Baines A.J., Gull K.. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491–500. [DOI] [PubMed] [Google Scholar]
- Yokoyama R., O’toole E., Ghosh S., Mitchell D.R.. 2004. Regulation of flagellar dynein activity by a central pair kinesin. Proc. Natl. Acad. Sci. USA. 101:17398–17403. 10.1073/pnas.0406817101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.