Abstract
The PII protein is a signal integrator involved in the regulation of nitrogen metabolism in bacteria and plants. Upon sensing of cellular carbon and energy availability, PII conveys the signal by interacting with target proteins, thereby modulating their biological activity. Plant PII is located to plastids; therefore, to identify new PII target proteins, PII-affinity chromatography of soluble extracts from Arabidopsis leaf chloroplasts was performed. Several proteins were retained only when Mg-ATP was present in the binding medium and they were specifically released from the resin by application of a 2-oxoglutarate-containing elution buffer. Mass spectroscopy of SDS/PAGE-resolved protein bands identified the biotin carboxyl carrier protein subunits of the plastidial acetyl-CoA carboxylase (ACCase) and three other proteins containing a similar biotin/lipoyl-binding motif as putative PII targets. ACCase is a key enzyme initiating the synthesis of fatty acids in plastids. In in vitro reconstituted assays supplemented with exogenous ATP, recombinant Arabidopsis PII inhibited chloroplastic ACCase activity, and this was completely reversed in the presence of 2-oxoglutarate, pyruvate, or oxaloacetate. The inhibitory effect was PII-dose-dependent and appeared to be PII-specific because ACCase activity was not altered in the presence of other tested proteins. PII decreased the Vmax of the ACCase reaction without altering the Km for acetyl-CoA. These data show that PII function has evolved between bacterial and plant systems to control the carbon metabolism pathway of fatty acid synthesis in plastids.
Keywords: Arabidopsis thaliana, biotin carboxyl carrier protein, PII protein, organic acids, fatty acid metabolism
Carbon (C) and nitrogen (N) metabolism pathways are coordinated in plants by a complex cross-talk between different signals. The mechanisms involved in this regulatory network are a fundamental aspect of primary plant metabolism, but to date they are poorly understood. The PII protein is one of the most ancient and conserved actors of signal transduction in archaebacteria, bacteria, and plants. PII signaling proteins are homotrimers composed of 12- to 15-kDa subunits that interpret the metabolic status of the cell. PII binds ATP and 2-oxoglutarate (2-OG), and depending on the degree of ligand binding, it interacts with enzymes, transcription factors, and transporters, and in doing so the activity of these proteins is modified to attain C/N homeostasis (1–6). Although PII proteins are well conserved at the amino acid level, their biological functions, and therefore their interacting protein partners, are diverse. In nonphotosynthetic bacteria, the first studied PII protein was GlnB of Escherichia coli, which regulates glutamine synthetase (GS) activity at both the transcriptional and posttranslational levels by interacting with NtrB (the kinase/phosphatase of the transcription factor NtrA) and GlnE (the GS adenylase) (1). E. coli also contains a second PII protein, GlnK, which regulates ammonium uptake by interacting with AmtB (an ammonium transporter), a function that is common throughout most prokaryotes (7, 8). In Bacillus subtilis, GlnK also interacts with TnrA, a major transcription regulator of N metabolism (9). Meanwhile, PII proteins of N2-fixing bacteria interact with proteins specific to the N2-fixation process. In diazotrophs like Azospirillum brasilense, DraT (interacting with GlnB) and DraG (interacting with GlnZ) are involved in the regulation by ribosylation of the dinitrogenase reductase (10), NifH, while PII directly interacts with NifD and NifK (the subunits of the nitrogenase complex) in N2-fixing Archaea like Methanococcus mariplaudis (11). Cyanobacteria contain a GlnB-type PII, the functions of which differ from the above-mentioned PII proteins. In cyanobacteria, PII signaling is involved in the regulation of N-utilization, nitrate/nitrite and bicarbonate uptake, and gene expression by the global N-control factor, NtcA (3–5). The yeast double-hybrid technique led to the identification of several cyanobacterial PII-interacting proteins: PamA (a putative membrane protein of unknown function) (12), PipX (a small protein uniquely found in cyanobacteria that interacts with both PII and NtcA) (13), and the N-acetyl-L-glutamate kinase (NAGK), which is the controlling enzyme of arginine synthesis (14–16). A plant PII protein was first reported in 1998 (17). Although nuclear-encoded, the protein is located to the chloroplasts of both Arabidopsis thaliana (17) and rice leaves (18). PII knockout mutants exhibit altered levels of C and N metabolites like starch and glutamine when grown under certain N-regimes, thus suggesting that plant PII is involved in regulating C/N homeostasis (19). To date, NAGK is the only PII-interacting protein identified in plants. This was initially shown by yeast double hybrid experiments (14, 18), followed by PII-affinity chromatography in which the NAGK was the unique plant protein identified as specifically interacting with PII (20). The use of recombinant proteins showed that Arabidopsis PII modified NAGK kinetics (20, 21) in a similar way to that described for the Synechococcus PII–NAGK interaction (16). The structure of a PII–NAGK complex has been solved for Arabidopsis (22) and Synechococcus elongatus proteins (23). It is possible that the function of plant PII has further similarities to those found in cyanobacteria because it has been proposed that plant PII regulates chloroplast nitrite uptake via an interaction with a nitrite transporter (24).
In this work, plant PII-interacting proteins have been identified using PII affinity chromatography, soluble chloroplast protein extracts from Arabidopsis rosette leaves, and mass spectrometry. Proteins retained by PII, in the presence of ATP and eluted with 2-OG, contained a common biotin/lipoyl attachment domain motif, and included a subunit of the plastidial ACCase. This heteromeric enzyme is composed of four different proteins including the biotin carboxyl carrier protein (BCCP), and it carries out the first committed step of the lipid biosynthetic pathway by converting acetyl-CoA to malonyl-CoA (25). The Arabidopsis genome also encodes two homomeric ACCases, ACC1 and ACC2, which are dimeric forms located in the cytosol (26) with the exception of the grass family in which they are also found in plastids (27). We report that ATP-activated PII inhibits in vitro chloroplastic ACCase activity via binding to its BCCP subunit, and that this is relieved by 2-OG. This finding indicates that PII function has significantly evolved between bacteria and plants and that PII signaling is not uniquely involved in the regulation of N-metabolism. We propose that plant PII contributes to the fine-tuning of chloroplastic C flow to several metabolic pathways that consume C-skeletons (and ATP).
Results
Identification of PII-Interacting Proteins in Arabidopsis Leaf Chloroplast Extracts.
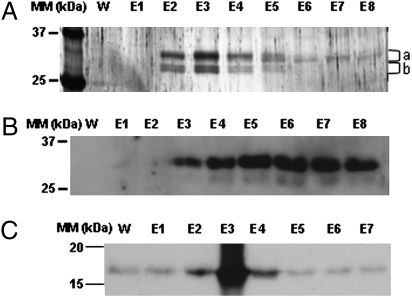
To date, a single PII-interacting protein, the NAGK, has been identified and characterized in plants. This was achieved by using the yeast double hybrid technique (14, 18) and affinity chromatography of protein extracts from Arabidopsis cell cultures (20). To explore further the catalog of PII-interacting proteins in plants, purified recombinant Arabidopsis His-tagged-PII was bound to a nickel affinity resin and associated with soluble proteins from Percoll-purified, intact Arabidopsis rosette leaf chloroplasts. This choice of plant material was made because of the chloroplast localization of PII (17, 18). Previous works have shown that PII–protein interactions depend on Mg-ATP, for binding, and 2-OG for elution (16, 28–30). Therefore, these metabolites were used in our affinity chromatography approach. In this way, several proteins in the molecular mass range of 25–37 kDa were retained by PII in the presence of Mg-ATP and eluted by 2-OG, as judged by SDS/PAGE (Fig. 1A, and see Fig. S1 for the complete SDS/PAGE gel). Gels were sliced and trypsin-digested proteins were analyzed by LC-MS/MS mass spectrometry and bioinformatics using protein sequences in the Arabidopsis TAIR database. This led to the identification of the NAGK (thus validating our protocol) and a group of plastid-localized proteins annotated as “biotin/lipoyl attachment domain-containing” (Table 1). Because this latter protein family exhibited an ATP-dependent affinity for PII that was reversed by 2-OG, their members are good candidates to be PII-regulated targets in vivo. Two identified members of this family, BCCP1 and BCCP2, are the biotin carboxyl carrier subunits of the plastidial heteromeric ACCase, a carboxylating enzyme using biotin as CO2 carrier and initiating the fatty acid biosynthetic pathway. Because the other family members have unknown functions, it was decided to investigate whether PII could modify the carboxylase activity of plastidial ACCase in vitro. PII-affinity chromatography was also repeated, but protein elution was achieved by using 0.5 mM 2-OG; a similar protein pattern was obtained, and Western blotting using BCCP antibodies showed the presence of BCCP in the eluted fractions (Fig. 1B). To confirm this interaction, a reverse approach was carried out where chloroplastic soluble biotin-containing proteins [including BCCP (Fig. S2)] were fixed to immobilized monomeric avidin and their interaction with recombinant Arabidopsis PII was tested in the presence of Mg-ATP. PII was retained and eluted by 5 mM 2-OG as determined by Western blotting with PII antibodies (Fig. 1C).
Fig. 1.
Identification of PII-interacting proteins in leaf chloroplast extracts. Soluble proteins from purified chloroplasts were loaded onto a PII-affinity resin. Unbound proteins were removed by washing with 50 mL of buffer A. Lane W shows the absence of protein at the end of the wash process. Bound proteins were eluted with 5 mM 2-OG (8 × 0.5 mL fractions corresponding to lanes E1–E8). In control experiments omitting Mg-ATP or with NAGK fixed to the affinity resin, these proteins were not bound. (A) Eluted proteins were TCA-precipitated and subjected to SDS/PAGE (12% acrylamide) and silver staining. (B) Bound proteins were eluted with 0.5 mM 2-OG and, after SDS/PAGE, submitted to Western blotting with BCCP antibodies. (C) In a reverse approach, soluble biotin-containing proteins from intact chloroplasts were fixed to immobilized avidin and their interaction with recombinant PII was tested in the presence of Mg-ATP. PII was eluted with 5 mM 2-OG and fractions were evaluated for PII content by Western blotting using PII antibodies. For explanation of a and b, see Table 1 and Fig. S1.
Table 1.
Identified proteins eluted from PII-affinity chromatography by 2-oxoglutarate
| Arabidopsis gene no. | Description | No. of peptides | Percent coverage of protein | Position on Fig. 1 |
| At3g56130 | Biotin/lipoyl attachment domain protein | 8 | 64 | a + b |
| At1g52670 | Biotin/lipoyl attachment domain protein | 5 | 46 | a + b |
| At3g15690 | Biotin carboxyl carrier protein-related | 5 | 40 | b |
| At5g15530 | BCCP2 | 4 | 25 | b |
| At5g16390 | BCCP1 | 4 | 21 | a |
| At3g57560 | NAGK | 4 | 14 | a |
Chloroplastic ACCase Activity Is Regulated by PII.
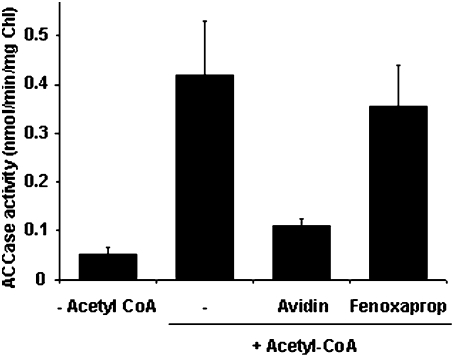
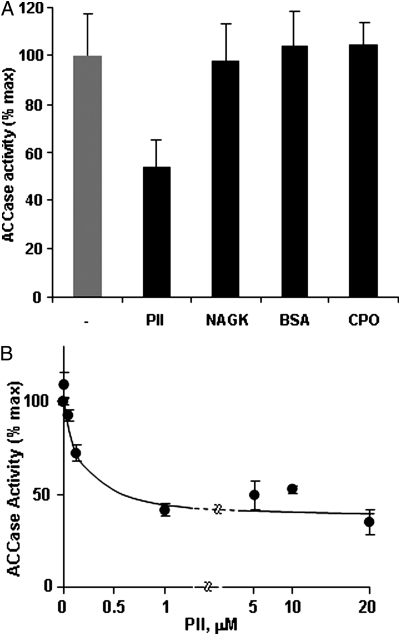
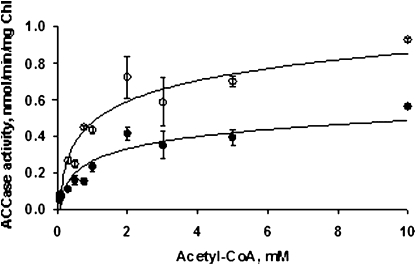
In Arabidopsis, heteromeric ACCase is strictly located to plastids, as is PII. Therefore, intact chloroplasts were isolated from illuminated rosette leaves of a previously described PII null mutant (19) to avoid interaction of endogenous PII with the ACCase in the corresponding extracts. Furthermore, osmotic-shocked, total intact chloroplast extracts were used because plastidial ACCase has been shown to be linked to the chloroplast envelope (31) and DTT was added because ACCase activity is light-activated by a thioredoxin-mediated process (32). First, it was shown that 14C-incorporation (from 14C labeled HCO3−) into stable reaction products strictly depended on the presence of acetyl-CoA (Fig. 2). Avidin, a general inhibitor of ACCase activity, led to a strong reduction of the measured activity, whereas the herbicide fenoxaprop, a specific inhibitor of the cytosolic ACCase (33), did not affect the activity (Fig. 2). Taken together, these data validate the plant material and the method chosen to measure the in vitro activity of plastidial ACCase. This assay was used to investigate the effect of PII on plastidial ACCase activity in the presence of Mg-ATP. The addition of exogenous, recombinant Arabidopsis PII reduced ACCase activity by ≈50% when compared to the control (minus PII) value (Figs. 3 A and B). This effect was PII-dose-dependent, with an apparent IC50 of 150 nM (Fig. 3B). It should be noted that the low PII concentration would bind negligible ATP amounts that should not impact on the ACCase reaction. In marked contrast, the addition of either chloroplastic NAGK or coproporphyrinogen III oxidase (CPO), as recombinant plastid-localized proteins, or bovine serum albumin (BSA) as a nonspecific nonplant protein to the assay medium did not produce any effect on the activity, thereby suggesting that PII specifically modulated the ACCase target (Fig. 3A). PII inhibition of chloroplastic ACCase activity was further characterized by analyzing the effect as a function of acetyl-CoA concentration (Fig. 4). This showed that the 50% reduction in Vmax attributed to the interaction with PII was not associated with a change in the Km for acetyl-CoA, since Lineweaver-Burke analyses of the curves gave Km values of 1.08 mM (in the absence of PII) and 1.23 mM (in the presence of PII). Thus, PII behaved as a typical noncompetitive inhibitor of chloroplastic ACCase.
Fig. 2.
Characterization of chloroplastic ACCase activity. The ACCase activity was assayed by using a total chloroplast extract from the Arabidopsis PIIV1 mutant in the presence, or absence, of 0.5 mM acetyl-CoA. Avidin (0.2 units), a general ACCase inhibitor, and fenoxaprop (100 mM), a specific inhibitor of the cytosolic ACCase, were tested to validate the measured chloroplastic ACCase activity. The data represent the average value (±SE) from 23 (+/−acetyl-CoA), 5 (avidin), and 4 (fenoxaprop) experiments carried out on different chloroplast preparations.
Fig. 3.
PII inhibits chloroplastic ACCase activity. (A) ACCase activity was assayed in the absence (−PII) and presence of 0.36 μM PII (+PII). The specificity of PII inhibition was tested by addition of recombinant NAGK (1.51 μM) or CPO (1.35 μM) (plastidial proteins), or BSA (0.72 μM), a nonplant protein. (B) The response curve of ACCase activity to increasing concentrations of trimeric PII showed that the inhibition was PII-dose-dependent. The data represent the average value (±SE) from 5 (PII, NAGK, CPO, BSA) and 4 (PII-dose-dependence) experiments carried out on different chloroplast preparations.
Fig. 4.
Effect of PII on the kinetic parameters of the ACCase reaction as determined from acetyl-CoA saturation curves. ACCase activity was assayed in the absence (open circles) and the presence (filled circles) of 0.36 μM PII. Kinetic parameters were calculated from Lineweaver–Burke analyses. PII did not alter the Km for acetyl-CoA when the Vmax was reduced by 50%. The data represent the average values (±SEs) from three experiments carried out on different chloroplast preparations.
PII Inhibition of Chloroplastic ACCase Is Released by Metabolites.
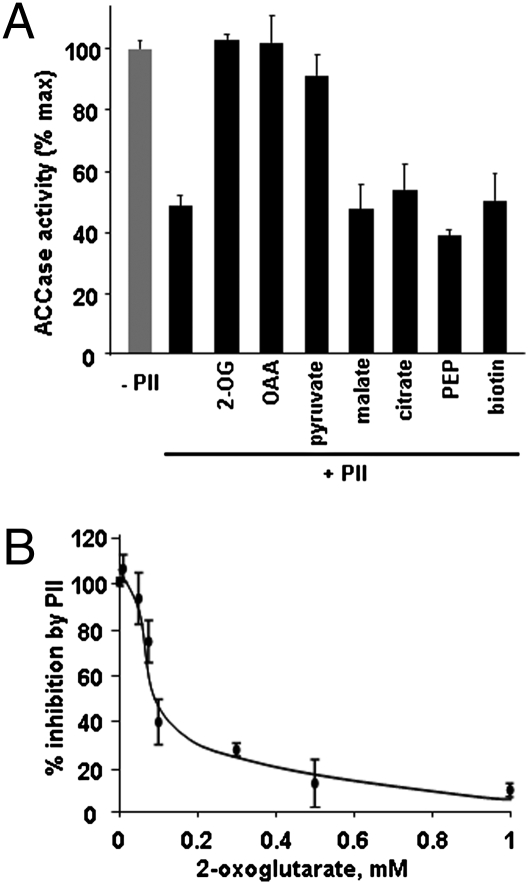
PII–protein interactions are modified by the binding of adenylates and 2-OG to the PII protein, in all cases studied so far the fixing of 2-OG disrupts the PII–protein complex. With respect to plant PII, it has recently been shown that the PII–NAGK interaction is affected by the addition of 2-OG (in the mM range) in the presence of Mg-ATP (29, 30). Because isothermal titration calorimetry showed that PII did not bind various amino acids (34), it was decided to characterize the effect of 2-OG and other organic acids on the PII-induced inhibition of ACCase activity. The presence of 5 mM 2-OG in the assay medium containing PII restored the ACCase activity to a level equivalent to that measured in the absence of PII (Fig. 5A). On the other hand, the addition of L-malate, citrate, or PEP did not relieve the observed PII-induced inhibition (Fig. 5A). However, OAA or pyruvate had the same effect as 2-OG, in that the ACCase activity was restored to that observed in the absence of PII. This was presumably mediated by altered PII–protein interactions because addition of the metabolites to the assay in the absence of PII did not alter the enzymatic activity. Finally, we studied the effect of 2-OG concentration on the maximally inhibited in vitro chloroplastic ACCase activity. At a concentration of 1 mM, this metabolite released almost all of the PII-inhibitory effect with 50% recovery corresponding to ≈0.08 mM 2-OG (Fig. 5B)—the kd for the interaction of 2-OG with PII as determined in ref. 34 and a concentration that is in the physiological range (35). Such data are in good agreement with the current working hypothesis that 2-OG is the central metabolite antagonizing PII–protein interactions. Interestingly, two organic acids (OAA and pyruvate) involved in acetyl-CoA synthesis, appear to be potential regulatory metabolites that can relieve PII-related changes in ACCase activity. It has been shown that OAA can bind to PII (34) whereas it inhibited PII dephosphorylation in Synechocystis (as did 2-OG) probably via an interaction with PII (36). On the other hand, pyruvate did not alter PII dephosphorylation (36) and it was not tested in (34). Finally, the addition of biotin to the assay did not affect the PII inhibition of ACCase activity (Fig. 5A).
Fig. 5.
Effect of metabolites on PII-inhibited chloroplastic ACCase activity. (A) ACCase activity was assayed in the presence of 0.36 μM PII and selected metabolites (5 mM). In control assays omitting PII, the metabolites did not alter the ACCase activity. (B) Recovery of the ACCase activity by increasing concentrations of 2-OG. The data in A represent the average values (±SEs) from nine (+/−PII), eight (2-OG), and three (other metabolites) experiments carried out on different chloroplast preparations. The data in B represent the average values (±SEs) from three experiments carried out on different chloroplast preparations.
Discussion
This study identifies a family of “biotin-binding proteins” as target proteins of the PII protein in chloroplasts from Arabidopsis rosette leaves. Based on previous work involving bacterial and plant systems, the behavior of these proteins with respect to the ATP-dependent interaction with PII and their subsequent elution by 2-OG is expected for PII target proteins. Indeed, ATP binding to PII brings about the conformational changes required for PII–protein interactions and allows the formation of the 2-OG binding site (37). Two of the identified proteins are BCCP subunits of the plastidial ACCase, but the physiological function(s) of the other “biotin-containing” proteins remain to be discovered. In subsequent reconstituted, in vitro assays it was found that addition of exogenous PII to chloroplast extracts down-regulated the measured ACCase activity by 50% (in the presence of Mg-ATP). These observations are interpreted by the negative modulation of ACCase activity when PII interacts with the BCCP subunit and that ATP (which is required for ACCase activity) can control this activity indirectly via PII. However, to have a coherent physiological sense and to conform to the known regulatory mechanism of PII–protein interactions, this process must show a reversible metabolite control. To date, the major metabolite disrupting PII–protein interactions is 2-OG, as recently documented for the plant PII–NAGK complex (29, 30). In our assay, 2-OG fully reversed the PII-induced inhibition of ACCase activity (Fig. 5), and based on our different affinity chromatography experiments (Fig. 1 and Table 1), PII no longer interacts with BCCP in the presence of this metabolite. The low concentration dependence of the 2-OG effect (Fig. 5B) is in agreement with measured 2-OG levels in spinach leaves (35) and the kd for 2-OG binding to PII (34). Interestingly, OAA and pyruvate had similar effects on the PII-specific inhibition (Fig. 5A) and this could be physiologically relevant because they are implicated in metabolic pathways leading to acetyl-CoA production. We propose that PII binds to the BCCP subunit of the plastidial ACCase in the presence of Mg-ATP. This interaction reduces the catalytic activity of the enzyme without altering the Km for acetyl-CoA (Fig. 4) by a mechanism yet to be determined. PII is released from the ACCase complex when 2-OG (OAA or perhaps pyruvate) binds to PII, thus relieving the inhibition. In this way, lipid biosynthesis in the plastids will be controlled by energy status and C-skeleton availability. This new control mechanism must be incorporated into a complex regulatory network because the carboxyltransferase subunit of plastid ACCase is phosphorylated—although the physiological significance of this modification is not yet known (38)—and subject to a thioredoxin-dependent redox control (32) that up-regulates the enzyme, with both processes taking place in the light. Thus, the chloroplastic ACCase in illuminated leaves is expected to display a potentially active state; however, its activity should be negatively modulated via PII by the high ATP concentration unless sufficient C-skeletons are available in the plastid stroma. Interestingly, we showed that PII behaved as a typical noncompetitive inhibitor of ACCase, with an unchanged Km for acetyl-CoA and a decreased Vmax. This means that changes in acetyl-CoA levels will not affect the extent of PII-induced inhibition. ATP only is expected to affect the activity via PII binding to the BCCP subunit. Organic acid concentrations are cardinal in reversing the effect by releasing the PII. 2-OG is the carbon skeleton required to fuel the GS/glutamate synthase (GOGAT) pathway involved in ammonium assimilation and amino acid synthesis in chloroplasts (39). The actual 2-OG content in this organelle will depend on both transport and GS/GOGAT activity. Pyruvate is the precursor of acetyl-CoA synthesis; its accumulation in the plastids would be a sign that lipid metabolism is limiting. The ensuing metabolic scenario implies a balance between C (fatty acid)/N (amino acid) pathways as modulated by the complex interplay between posttranslational modifications and environmental biochemical factors in relation to the physiological context.
Because we identified different members of a “biotin-containing” protein family as potential PII-interacting targets, from a structural point of view there should be a common PII interaction domain. It is possible that PII interacts with the region involved in biotin fixation, thus affecting the efficiency of this cofactor in the enzymatic reaction. However, free biotin did not act as a competitive inhibitor with respect to the observed PII-induced effect on ACCase activity (Fig. 5A). This could suggest that the PII-interaction domain and the inhibitory mechanism do not involve the biotin-binding site. Solving the PII-ACCase structure would be of great interest to clarify these points.
These data show that plant PII has gained new functions during the evolutionary process. To date, bacterial PII is involved uniquely in different aspects of N-metabolism, and plant and cyanobacterial PII share the NAGK as a common target (again an enzyme involved in N-metabolism). The modulation of plastidial ACCase activity by PII is an example of a role in lipid metabolism for this regulatory protein. Although, we have characterized the chloroplastic ACCase activity in plant leaves, the regulation of C and N metabolisms by PII could be of major importance in other plant organs like the seed where amino acid and fatty acid synthesis must be coordinated during the filling process (40). Much effort has been spent to manipulate seed ACCase activity with the goal of increasing oil content. Success depends on the precise knowledge of regulatory mechanisms acting on the enzyme in vivo, thus guiding the development of pertinent biotechnological strategies. Our results may provide clues to achieve this important goal and to improve lipid filling in oleaginous seeds.
Materials and Methods
Plant Growth Conditions.
Wild-type and PII mutant (19) plants were grown in a greenhouse at 70% relative humidity, with a light intensity of 200 μmol·m−2·s−1 and a day–night regime of 8 h at 22°C and 16 h at 20°C, respectively. The total rosette of 6-week-old plants was used for intact chloroplast isolation.
Chloroplast Preparation.
Intact rosette leaf chloroplasts were Percoll-purified as described in ref. 24. The plants were maintained in darkness during 24 h to reduce starch levels and minimize chloroplast damage, and were subsequently transferred to light (150 μE·m−2·s−1) for 20 min to activate the plastidial ACCase. Before use, the intact chloroplasts were broken by an osmotic shock (pelleted and resuspended in buffer without sorbitol) and a freeze–thaw cycle.
Expression and Purification of Recombinant Proteins.
The fusion constructs used to produce C-terminal 6-His-tagged Arabidopsis PII and N-terminal 6-His-tagged NAGK proteins (minus their transit peptides) are described in refs. 21 and 41. The coding sequence of CPO was reverse-transcribed from Arabidopsis total RNA, and the resulting PCR-amplified cDNA was inserted into a modified pSBETA expression vector as described for NAGK (21). PII, NAGK, and CPO recombinant proteins were produced in E. coli and purified by His-Select Nickel Affinity gel (Sigma) chromatography as in ref. 21.
PII-Affinity Chromatography of Soluble Chloroplast Proteins.
His6-PII (25 mg) was immobilized onto 2 mL of His-Select Nickel Affinity gel in buffer A [50 mM Tris·HCl (pH 7.5), 150 mM NaCl, 3 mM ATP, 6 mM MgCl2, and a protease inhibitor mixture] by gentle shaking for 30 min at room temperature. Soluble chloroplast proteins from 30–40 g of leaves (10.5 mg of chlorophyll in 15 mL of buffer A) were loaded onto the resin at a flow rate of 0.2 mL·min−1. The flow-through was collected and then reloaded onto the resin. After washing with 50 mL of buffer A, bound proteins were eluted with 5 mL of buffer A containing either 0.5 or 5 mM 2-OG. The eluted proteins were precipitated with 10% (wt/vol) trichloroacetic acid and stored for 12 h at −20°C. After centrifugation (12,000 × g, 30 min, 4°C), the pellet was solubilized in 20 μL of Laemmli denaturation buffer and subjected to SDS/PAGE (12% acrylamide) as described in ref. 42. Proteins were detected in gel by silver nitrate staining compatible with further analysis by mass spectrometry (43). BCCP was detected by Western blotting (19) using antibodies raised against recombinant Arabidopsis BCCP2 (44).
Immobilized Monomeric Avidin Affinity Chromatography.
The Pierce monomeric avidin resin was prepared as per the manufacturer’s instructions (except that phosphate was replaced by Tris). Soluble chloroplast proteins (in buffer A) from 35 g of leaves were incubated for 1 h at room temperature with the resin to fix biotin-containing proteins. The column was washed with 20 mL of buffer A before adding 5 μg of recombinant PII. After washing with 20 mL of buffer A, PII was eluted with 10 mL of buffer A containing 5 mM 2-OG, and fractions were analyzed as described above using PII-antibodies (19).
Identification of PII Target Proteins by Mass Spectrometry.
In-gel protein digestion was performed with a Progest system (Genomic Solution) as in ref. 45. Resulting peptide extracts were suspended in 25 μL of 0.08% trifluoroacetic acid and 2% acetonitrile (ACN). An Ultimate 3000 LC system (Dionex) connected to an LTQ Orbitrap mass spectrometer (Thermo Fisher) was used for LC-MS/MS analyses. Peptide mixtures (4 μL) were loaded onto a PepMap C18 precolumn (0.3 × 5 mm, 100 Å, 5 μm; Dionex) at a 20 μL·min−1 flow rate. After 4 min, the precolumn was connected to a PepMap C18 nanocolumn (0.075 × 15 cm, 100 Å, 3 μm) and subjected to a 2–36% linear gradient of buffer B (0.1% formic acid, 80% ACN) in buffer A (0.1% formic acid, 2% ACN) at 300 nL·min−1 for 50 min. Ionization was performed with a 1.3-kV spray voltage applied to an uncoated capillary probe (PicoTip EMITER 10-μm tip inner diameter; New Objective). Peptide ions were automatically analyzed by the data-dependent method as follows: full MS scan (m/z 300–1,600) on the Orbitrap analyzer and MS/MS on the four most abundant precursors on the LTQ linear ion trap. Only +1, +2, and +3 charged peptides were subjected to MS/MS, with an exclusion window of 1.5 min, and peptide fragmentation parameters of Qz = 0.22, activation time = 50 ms, and collision energy = 35%. Protein identification was performed with Bioworks 3.3.1 (Thermo Fisher) using an A. thaliana protein database (Tair version 7). The search parameters were trypsin-specificity with 1 missed cleavage, variable methionine oxidation, fixed cysteine alkylation, and mass tolerance of 1.4 Da for precursor ions and 0.5 Da for fragment ions. A multiple-threshold filter was applied at the peptide level: Xcorr magnitude up to 1.7, 2.2, and 3.5 for mono-, di-, and tricharged peptides, respectively, peptide probability lower than 0.05, ΔCn > 0.1 with a minimum of two different peptides for an identified protein.
Enzyme Assays.
Chloroplastic ACCase activity was assayed as in ref. 46. An aliquot of the purified chloroplast fraction, corresponding to 100 μg of chlorophyll (≈400 μg of protein), was assayed in 200 μL of medium containing 100 mM Tricine (pH 8.2), 50 mM KCl, 3 mM ATP, 6 mM MgCl2, 5 mM DTT, 0.5 mM acetyl-CoA, and 10 mM [14C]NaHCO3 (1 mCi/mM) for 20 min at 30°C. The enzymatic activity was linear during this time. Reactions were stopped by adding 50 μL of 6 M HCl. Half of the reaction mixture was placed on Whatman 3-mm paper, washed, and dried, before the radioactivity was determined by using a scintillation counter (Ready Gel Mixture; Beckman Coulter). Assays omitting acetyl-CoA or chloroplasts served as controls. Recombinant proteins underwent gel-filtration [Sephadex G-25 (GE Healthcare) in 100 mM Tricine (pH 8.2)] to remove imidazole and glycerol before use. To test the effect of proteins (PII, NAGK, CPO, and BSA) and metabolites (inhibitors, organic acids, and substrates), assays were carried out in the presence of defined amounts of each molecule as described in the figure legends.
Supplementary Material
Acknowledgments
BCCP2 antibodies were a kind gift from Dr. J. J. Thelen (University of Missouri, Columbia). This work was supported by Fundación Alfonso Martín Escudero (MAD 1-2-105), Madrid; Centre National de la Recherche Scientifique; and Université Paris-Sud 11.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910097107/DCSupplemental.
References
- 1.Arcondeguy T, Jack R, Merrick M. PII signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol Mol Biol Rev. 2001;65:80–105. doi: 10.1128/MMBR.65.1.80-105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ninfa AJ, Jing P. PII signal transduction proteins: sensors of α-ketoglutarate that regulate nitrogen metabolism. Curr Opin Microbiol. 2005;8:168–173. doi: 10.1016/j.mib.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Forchhammer K. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol Rev. 2004;28:319–333. doi: 10.1016/j.femsre.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Osanai T, Tanika K. Keeping in touch with PII: PII-interacting proteins in unicellular cyanobacteria. Plant Cell Physiol. 2007;48:908–914. doi: 10.1093/pcp/pcm072. [DOI] [PubMed] [Google Scholar]
- 5.Forchhammer K. PII signal transducers: novel functional and structural insights. Trends Microbiol. 2008;16:65–72. doi: 10.1016/j.tim.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Uhrig RG, Ng KKS, Moorhead GBG. PII in higher plants: a modern role for an ancient protein. Trends Plant Sci. 2009;14:505–511. doi: 10.1016/j.tplants.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Coutts G, Thomas G, Blakey D, Merrick M. Membrane sequestration of the signal transduction protein GlnK by the ammonium transporter AmtB. EMBO J. 2002;21:536–545. doi: 10.1093/emboj/21.4.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand A, Merrick M. In vitro analysis of the Escherichia coli AmtB-GlnK complex reveals a stoichiometric interaction and sensitivity to ATP and 2-oxoglutarate. J Biol Chem. 2006;281:29558–29567. doi: 10.1074/jbc.M602477200. [DOI] [PubMed] [Google Scholar]
- 9.Heinrich A, et al. Interaction of the membrane-bound GlnK-AmtB complex with the master regulator of nitrogen metabolism TnrA in Bacillus subtilis. J Biol Chem. 2006;281:34909–34917. doi: 10.1074/jbc.M607582200. [DOI] [PubMed] [Google Scholar]
- 10.Heurgo LF, et al. Interactions between PII proteins and nitrogenase regulatory enzymes DraT and DraG in Azospirillum brasilense. FEBS Lett. 2006;580:5232–5236. doi: 10.1016/j.febslet.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 11.Dodsworth JA, Leigh JA. Regulation of nitrogenase by 2-oxoglutarate-reversible, direct binding of a PII-like nitrogen sensor protein to dinitrogenase. Proc Natl Acad Sci USA. 2006;103:9779–9784. doi: 10.1073/pnas.0602278103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osanai T, Sato S, Tabata S, Tanaka K. Identification of PamA as a PII-binding membrane protein important in nitrogen-related and sugar-catabolic gene expression in Synechocystis sp. PCC 6803. J Biol Chem. 2005;280:34684–34690. doi: 10.1074/jbc.M507489200. [DOI] [PubMed] [Google Scholar]
- 13.Espinosa J, Forchhammer K, Burillo S, Contreras A. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol Microbiol. 2006;61:457–469. doi: 10.1111/j.1365-2958.2006.05231.x. [DOI] [PubMed] [Google Scholar]
- 14.Burillo S, Luque I, Feuntes I, Contreras A. Interactions between the nitrogen signal transduction protein PII and N-acetyl glutamate kinase in organisms that perform oxygenic photosynthesis. J Bacteriol. 2004;186:3346–3354. doi: 10.1128/JB.186.11.3346-3354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinrich A, Maheswaran M, Ruppert U, Forchhammer K. The Synechococcus elongatus PII signal transduction protein controls arginine synthesis by complex formation with N-acetyl-L-glutamate kinase. Mol Microbiol. 2004;52:1303–1314. doi: 10.1111/j.1365-2958.2004.04058.x. [DOI] [PubMed] [Google Scholar]
- 16.Maheswaran M, Urbanke C, Forchhammer K. Complex formation and catalytic activation by the PII signaling protein of N-acetyl-L-glutamate kinase from Synechococcus elongatus strain PCC 7942. J Biol Chem. 2004;279:55202–55210. doi: 10.1074/jbc.M410971200. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh M-H, Lam H-M, van de Loo FJ, Coruzzi G. A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc Natl Acad Sci USA. 1998;95:13965–13970. doi: 10.1073/pnas.95.23.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugiyama K, Hayakawa T, Kudo T, Ito T, Yamaya T. Interaction of N-acetyl glutamate kinase with a PII-like protein in rice. Plant Cell Physiol. 2004;45:1768–1778. doi: 10.1093/pcp/pch199. [DOI] [PubMed] [Google Scholar]
- 19.Ferrario-Méry S, et al. Physiological characterisation of Arabidopsis mutants affected in the expression of the putative regulatory protein PII. Planta. 2005;223:28–39. doi: 10.1007/s00425-005-0063-5. [DOI] [PubMed] [Google Scholar]
- 20.Chen YM, et al. The PII signal transduction protein of Arabidopsis thaliana forms an arginine-regulated complex with plastid N-acetyl glutamate kinase. J Biol Chem. 2006;281:5726–5733. doi: 10.1074/jbc.M510945200. [DOI] [PubMed] [Google Scholar]
- 21.Ferrario-Méry S, Besin E, Pichon O, Meyer C, Hodges M. The regulatory PII protein controls arginine biosynthesis in Arabidopsis. FEBS Lett. 2006;580:2015–2020. doi: 10.1016/j.febslet.2006.02.075. [DOI] [PubMed] [Google Scholar]
- 22.Mizuno Y, Moorhead GB, Ng KK. Structural basis for the regulation of N-acetylglutamate kinase by PII in Arabidopsis thaliana. J Biol Chem. 2007;282:35733–35740. doi: 10.1074/jbc.M707127200. [DOI] [PubMed] [Google Scholar]
- 23.Llácer JL, et al. The crystal structure of the complex of PII and acetylglutamate kinase reveals how PII controls the storage of nitrogen as arginine. Proc Natl Acad Sci USA. 2007;104:17644–17649. doi: 10.1073/pnas.0705987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrario-Méry S, Meyer C, Hodges M. Chloroplast nitrite uptake is enhanced in Arabidopsis PII mutants. FEBS Lett. 2008;582:1061–1066. doi: 10.1016/j.febslet.2008.02.056. [DOI] [PubMed] [Google Scholar]
- 25.Sasaki Y, Nagano Y. Plant acetyl-CoA carboxylase: Structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci Biotechnol Biochem. 2004;68:1175–1184. doi: 10.1271/bbb.68.1175. [DOI] [PubMed] [Google Scholar]
- 26.Mekhedov S, Martinez de Ilarduya O, Ohlrogge JB. Toward a functional catalog of the plant genome. A survey of genes for lipid biosynthesis. Plant Physiol. 2000;122:389–401. doi: 10.1104/pp.122.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konishi T, Shinohara K, Yamada K, Sasaki Y. Acetyl-CoA carboxylase in higher plants; most plants other than Gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol. 1996;37:117–122. doi: 10.1093/oxfordjournals.pcp.a028920. [DOI] [PubMed] [Google Scholar]
- 28.Kamberov ES, Atkinson MR, Ninfa AJ. The Escherichia coli signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J Biol Chem. 1995;270:17797–17807. doi: 10.1074/jbc.270.30.17797. [DOI] [PubMed] [Google Scholar]
- 29.Beez S, Fokina O, Herrmann C, Forchhammer K. N-acetyl-L-glutamate kinase (NAGK) from oxygenic phototrophs: PII signal transduction across domains of life reveals novel insights in NAGK control. J Mol Biol. 2009;389:748–758. doi: 10.1016/j.jmb.2009.04.053. [DOI] [PubMed] [Google Scholar]
- 30.Feria Bourrellier AB, Ferrario-Méry S, Vidal J, Hodges M. Metabolite regulation of the interaction between Arabidopsis thaliana PII and N-acetyl-L-glutamate kinase. Biochem Biophys Res Commun. 2009;387:700–704. doi: 10.1016/j.bbrc.2009.07.088. [DOI] [PubMed] [Google Scholar]
- 31.Thelen JJ, Ohlogge JB. The multisubunit acetyl-CoA carboxylase is strongly associated with the chloroplast envelope through non-ionic interactions to carboxyltransferase subunits. Arch Biochem Biophys. 2002;400:245–257. doi: 10.1016/S0003-9861(02)00025-5. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki Y, Kozaki A, Hatano M. Link between light and fatty acid synthesis: thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proc Natl Acad Sci USA. 1997;94:11096–11101. doi: 10.1073/pnas.94.20.11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konishi T, Sasaki Y. Compartmentalization of two forms of acetyl-CoA carboxylase in plants and the origin of their tolerance toward herbicides. Proc Natl Acad Sci USA. 1994;91:3598–3601. doi: 10.1073/pnas.91.9.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith CS, Weljie AM, Moorhead GBG. Molecular properties of the putative nitrogen sensor PII from Arabidopsis thaliana. Plant J. 2003;33:353–360. doi: 10.1046/j.1365-313x.2003.01634.x. [DOI] [PubMed] [Google Scholar]
- 35.Winter H, Robinson DG, Heldt HW. Subcellular volumes and metabolic concentrations in spinach leaves. Planta. 1994;193:530–535. [Google Scholar]
- 36.Ruppert U, Irmler A, Kloft N, Forchhammer K. The novel protein phosphatase PphA from Synechocystis PCC 6803 controls dephosphorylation of the signalling protein PII. Mol Microbiol. 2002;44:855–864. doi: 10.1046/j.1365-2958.2002.02927.x. [DOI] [PubMed] [Google Scholar]
- 37.Yildiz O, Kalthoff C, Raunser S, Kühlbrandt W. Structure of GlnK1 with bound effectors indicates regulatory mechanism for ammonia uptake. EMBO J. 2007;26:589–599. doi: 10.1038/sj.emboj.7601492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savage LJ, Ohlrogge JB. Phosphorylation of pea chloroplast acetyl-CoA carboxylase. Plant J. 1999;18:521–527. doi: 10.1046/j.1365-313x.1999.00478.x. [DOI] [PubMed] [Google Scholar]
- 39.Gálvez S, Lancien M, Hodges M. Are isocitrate dehhydrogenases and 2-oxoglutarate involved in the regulation of glutamate synthesis? Trends Plant Sci. 1999;4:484–490. doi: 10.1016/s1360-1385(99)01500-9. [DOI] [PubMed] [Google Scholar]
- 40.Baud S, Boutin J-P, Miquel M, Lepiniec L, Rochat C. An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem. 2002;40:151–160. [Google Scholar]
- 41.Smith CS, Zaplachinski ST, Muench D, Moorhead GBG. Expression and purification of the chloroplast putative nitrogen sensor, PII, of Arabidopsis thaliana. Protein Expr Purif. 2002;25:342–347. doi: 10.1016/s1046-5928(02)00018-9. [DOI] [PubMed] [Google Scholar]
- 42.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 43.Sinha P, Poland J, Schnolzer M, Rabilloud T. A new silver staining apparatus and procedure for matrix-assisted laser desorption/ionization-time of flight analysis of proteins after two-dimensional electrophoresis. Proteomics. 2001;7:835–840. doi: 10.1002/1615-9861(200107)1:7<835::AID-PROT835>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 44.Thelen JJ, Mekhedov S, Ohlrogge JB. Brassicaceae express multiple isoforms of biotin carboxyl carrier protein in a tissue-specific manner. Plant Physiol. 2001;125:2016–2028. doi: 10.1104/pp.125.4.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Albertin W, et al. Differential regulation of gene products in newly synthesized Brassica napus allotetraploids is not related to protein function nor subcellular localization. BMC Genomics. 2007;8:56. doi: 10.1186/1471-2164-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nikolau BJ, Hawke JC, Slack CR. Acetyl-Coenzyme A carboxylase in maize leaves. Arch Biochem Biophys. 1981;211:605–612. doi: 10.1016/0003-9861(81)90495-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.