For many decades, cancer research focused on genetic changes that led to tumorigenesis. However, it is now established that epigenetics also plays an important role in cancer initiation and development (1, 2). The cancer epigenome is marked by a global decrease in methylation, leading to genomic instability and increased methylation at promoters, which in turn leads to gene silencing (3). The importance of this abnormal DNA methylation in silencing cancer-related genes has recently gained a great deal of attention. Detecting altered methylation in tumors on a genome-wide scale is not always easy, but its importance has led to the development of many assays including restriction enzyme digestion, ligation mediated PCR, bisulfite sequencing technologies such as methylation sensitive PCR, incubation of DNA with methylated DNA binding proteins or antibodies to methylcytosine among others. These approaches have their strengths and weaknesses and in this issue of PNAS, Diede et al. describe an approach employing physical properties of DNA called denaturation analysis of methylation differences (DAMD) (4). The method relies on the fact that methylated CpG dinucleotides have different melting characteristics compared to unmethylated CpG dinucleotides. Although the specific conditions described in the paper limit the analyses to CpG islands (i.e., areas with a high density of CpG dinucleotides), the method can be modified in such a way as to also examine DNA methylation differences in genomic regions that are less CpG rich.
The authors use the altered melting behavior of methylated CpG dinucleotides to easily and rapidly screen for CpG-rich regions, which have become abnormally methylated relative to control samples and compare the results to those obtained by other approaches. The direct head-to-head comparison with two existing methods, MeDIP and MBD binding, showed that the DAMD method is more sensitive. Importantly, when the authors used the DAMD method to identify promoters methylated in pediatric medulloblastomas relative to normal cerebellum, they identified that several key developmental pathways had become abnormally methylated and hence silenced. These included the sonic hedgehog, the retinoic acid receptor, the bone morphogenetic protein and notch pathways, which were disabled not only in medulloblastoma cell lines but also in uncultured pediatric medulloblastomas, which are a lethal kind of childhood brain cancer. Proper functioning of these signaling pathways are vitally important for the generation of cerebellar granule cells both in vivo and in vitro. Retinoic acid, BMP, Wnt, Shh, and Notch signaling, along with fibroblast growth factor, specify and mediate the proliferation of granule cell precursor cells and their subsequent differentiation into mature cerebellar granule cell neurons (5, 6).
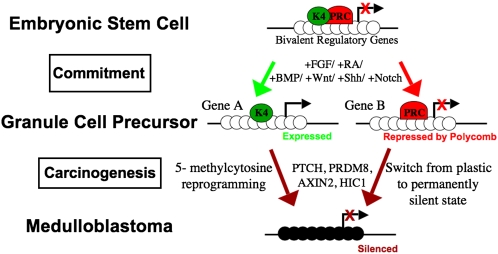
The findings of Diede et al. are particularly timely when coupled with other recent studies which have shown the inappropriate silencing of differentiation related genes in adult tumors (7–9). Genome-wide epigenomic analyses have shown that these kinds of genes have a particular type of chromatin structure in embryonic stem cells, which precludes their expression until the genes are called upon during later stages of development (10, 11). The adoption of the so-called “bivalent chromatin” domains in embryonic stem cells appears to keep these key regulators in a poised state so that they can be rapidly activated or alternatively kept silent at later stages of development. These bivalent regulatory genes have both the histone mark of the polycomb repressive complex (H3-K27me3) and an active histone mark, H3-K4me3 (Fig. 1). As cells commit to one lineage or another, the bivalent chromatin structure is resolved into either an active configuration, maintaining the H3K4me3 mark, or a repressed state maintaining the polycomb complex, depending upon whether expression of the marked gene is necessary for lineage progression.
Fig. 1.
In embryonic stem cells, promoters of developmentally important genes are “bivalent”—having both polycomb repressive complex (PRC) and the active (H3K4me3) histone modification. In response to specific signaling molecules, ES cells become committed to Granule Cell Precursors, losing this bivalency: active promoters lose PRC binding and retain H3K4me3 whereas repressed promoters lose H3K4me3 and retain PRC binding. Many of these polycomb target genes in ES cells become abnormally methylated in cancer, potentially resulting from a failure to differentiate properly. Open circles indicate unmethylated CpG sites, dark circles indicate methylated CpG sites. Arrows indicate transcription start sites. K4 indicates H3K4me3 and PRC indicates polycomb repressive complex.
DNA methylation is not normally used in embryonic stem cells to suppress the activities of genes which have CpG islands within their promoters because the 5-methylcytosine mark is far less plastic than histone modifications which silence genes. Numerous studies have shown that genes, which are subject to this bivalency, are far more likely to undergo inappropriate gene silencing by DNA methylation during the formation of cancer (8, 9, 12, 13). The reason for this propensity to undergo de novo DNA methylation is not understood but clearly the DNA methylation mark is more robust and stable than modifications of histones. Thus once genes have become silenced by DNA methylation, a cell’s differentiation possibilities are restricted. Importantly, the de novo DNA methylation mechanism does not only silence active genes but is also commonly observed in genes that have been kept silenced by the polycomb complex in a process that we have called “epigenetic switching” (14), reducing their regulatory plasticity.
At first sight, it might seem that this epigenetic switch would be of limited relevance to cancer because there is no corresponding change in gene expression. However, acquisition of methylation in a CpG island in a gene such as p16, which is a known polycomb regulated gene (15), might condemn that gene to irreversible epigenetic silence. The results of Diede et al. are particularly interesting because they suggest that exactly this kind of inflexibility with respect to epigenetic marks occurs on key developmental regulators in a childhood tumor such as medulloblastoma. Few studies to date have examined pediatric tumors for DNA methylation changes, even though it might be expected that they have a more developmental and hence epigenetic etiology compared to adult tumors, which arise much later in life after complex developmental decisions have been made. Diede et al. go on to show that the PTCH1 gene, which is a key player in the sonic hedgehog pathway can be resurrected from silence by treatment with the DNA methylation inhibitor 5-aza-2’-deoxycytidine. Genes that have become inappropriately silenced by application of 5-methylcytosine marks to their promoters seem to be particularly vulnerable to pharmacological reactivation. Therefore, the increased biological stability of silencing afforded by DNA methylation, counterintuitively makes these regulatory genes vulnerable to drug-induced reactivation. Restoration of their expression or plasticity and reestablishing proper signaling pathways may have great therapeutic advantages by reducing aberrant proliferative activity and inducing differentiation programs. Furthermore, nucleoside analogs such as 5-aza-cytidine can cross the blood–brain barrier and have been successful in clinical trials of myelodysplastic syndrome (16), thus clinical trials in pediatric tumors might be feasible.
The DAMD approach relying on a simple difference in the physical properties of methylated CpG islands has therefore led to the identification of aberrantly methylated regulatory genes and important developmental pathways that have been altered in medulloblastoma. These findings offer ideas for how these changes may have arisen and how they may be used to target a deadly pediatric disease.
Footnotes
The authors declare no conflict of interest.
See companion article on page 234.
References
- 1.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 4.Diede SJ, et al. DNA methylation of developmental genes in pediatric medullablastomas identified by denaturation analysis of methylation differences. Proc Natl Acad Sci USA. 2009;107:234–239. doi: 10.1073/pnas.0907606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salero E, Hatten ME. Differentiation of ES cells into cerebellar neurons. Proc Natl Acad Sci USA. 2007;104:2997–3002. doi: 10.1073/pnas.0610879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Millen KJ, Gleeson JG. Cerebellar development and disease. Curr Opin Neurobiol. 2008;18:12–19. doi: 10.1016/j.conb.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 9.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 10.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 12.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn MA, et al. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–10289. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gal-Yam EN, et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci USA. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bracken AP, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenaux P, et al. International Vidaza High-Risk MDS Survival Study Group Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–232. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]