Summary
Flagellar motility in Listeria monocytogenes (Lm) is restricted to temperatures below 37°C due to the opposing activities of the MogR transcriptional repressor and the GmaR anti-repressor. Previous studies have suggested that both the DegU response regulator and MogR regulate expression of GmaR. In this report, we further define the role of DegU for GmaR production and flagellar motility. We demonstrate that deletion of the receiver domain of DegU has no effect on flagellar motility in Lm. Using transcriptional reporter fusions, we determined that gmaR is co-transcribed within an operon initiating with fliN. Furthermore, the fliN-gmaR promoter (pfliN-gmaR) is transcriptionally activated by DegU and is also MogR-repressed. DNA affinity purification, gel mobility shift, and footprinting analyses revealed that both DegU and MogR directly bind fliN-gmaR promoter region DNA and that the binding sites do not overlap. Quantitative analysis of gmaR transcripts in ΔmogR bacteria indicated that transcriptional activation of pfliN-gmaR by DegU is not inherently temperature-dependent. However, GmaR protein was not detectable at 37°C in ΔmogR bacteria, indicating that a temperature-dependent, post-transcriptional mechanism limits GmaR production to temperatures below 37°C. Our findings reveal that flagellar motility in Lm is governed by both temperature-dependent transcriptional and post-transcriptional regulation of the GmaR anti-repressor.
Keywords: Listeria monocytogenes, flagellar motility, GmaR, MogR, DegU, post-transcriptional regulation
Introduction
Bacteria can inhabit a diverse range of niches by sensing and adapting to environmental fluctuations through rapid changes in gene transcription and protein expression. Listeria monocytogenes (Lm) is a Gram-positive bacterium that thrives in a vast range of environments and over a wide spectrum of temperatures (3°C–43°C). As a facultative intracellular pathogen, Lm replicates and spreads within the intracellular environment of host cells (Vazquez-Boland et al., 2001). Transition from the extracellular milieu to an intracellular lifestyle requires several environmental signals that lead to the up-regulation of virulence factors and the reciprocal down regulation of other genes, such as those required for flagellar motility.
Flagellar motility is a highly advantageous, but energetically demanding survival mechanism utilized by bacteria in the extracellular environment. Flagellar motility allows the bacterium to move towards, or retreat from specific environmental conditions required for optimal bacterial growth (Armitage, 1999). Since flagellar motility in Lm increases adherence to abiotic and cellular surfaces, it is important for biofilm formation and cellular invasion (Lemon et al., 2007, O’Neil & Marquis, 2006). However, while flagellar motility can enhance cellular invasion (O’Neil & Marquis, 2006), the continual production of flagella during infection can stimulate innate immune responses that are inhibitory for bacterial survival during infection (Hayashi et al., 2001, Molofsky et al., 2006). Therefore, Lm along with several other pathogens down-regulate flagellar motility at physiological temperature (37°C) (Peel et al., 1988, Kapatral & Minnich, 1995, Akerley & Miller, 1993, Ott et al., 1991). Temperature is also a signal that coordinately regulates virulence factor expression in Lm. The master virulence gene activator, PrfA, is regulated by a temperature-dependent secondary structure in the 5′ untranslated region (UTR) of prfA RNA, which inhibits translation of PrfA protein at non-physiological temperatures, and thus limits virulence gene expression to temperatures of 37°C and above (Johansson et al., 2002). Conversely, transcription of flagellar motility genes is repressed at 37°C due to the binding activity of the MogR transcriptional repressor, restricting flagellar motility to low temperatures (30°C and below) (Grundling et al., 2004, Shen & Higgins, 2006).
The bacterial flagellum is a complex substructure requiring the coordinate assembly of multiple proteins; therefore, most bacterial systems have a 3–4 tiered hierarchal regulatory cascade controlling the temporal expression and production of flagella (Aldridge & Hughes, 2001, McCarter, 2006). Regulation of flagellar motility in Lm differs substantially from other bacterial species as Lm lacks conserved master regulators that govern hierarchal regulation of flagella production (Chilcott & Hughes, 2000). In Lm, MogR binds directly to flagellar motility gene promoters and represses transcription of all flagellar motility genes in a non-hierarchal manner (Shen & Higgins, 2006). At temperatures below 37°C, the MogR anti-repressor GmaR antagonizes MogR repression activity by binding directly to MogR (Shen et al., 2006). Temperature-dependent expression of GmaR restricts transcription of flagellar motility genes to low temperatures, however it is not known how GmaR expression is initiated as temperature decreases below 37°C (Shen et al., 2006). Interestingly, transcription of gmaR is MogR-repressed and therefore GmaR-regulated; as GmaR is initially produced, transcription of gmaR and production of GmaR protein are consequently up-regulated (Shen et al., 2006). Epistasis and microarray analysis have shown that in addition to GmaR, the DegU response regulator is also required to relieve MogR repression at low temperatures (Shen et al., 2006, Shen & Higgins, 2006). Constitutive expression of GmaR in a ΔdegU strain restores flagellar motility gene expression (Shen et al., 2006), indicating that GmaR acts downstream of DegU and is likely DegU-regulated.
Bacterial two-component signal transduction systems (TCS) enable bacteria to sense environmental changes such as temperature and transduce them into transcriptional responses. A typical TCS consists of a sensor histidine kinase (HK) that detects environmental signals and a cognate response regulator (RR) that binds to DNA to mediate a cellular response. Communication between the HK and RR is controlled by a phosphotransfer event from HK to RR that changes the conformation of the RR DNA-binding domain, thus altering DNA binding specificity of the RR (Khorchid & Ikura, 2006, Bijlsma & Groisman, 2003). Regulation of flagellar motility in several bacterial species is mediated by the activities of TCS that can function as repressors such as the Bordetella pertussis BvgAS system or function as activators such as the FlgRS system of Campylobacter jejuni (Wosten et al., 2004). Interestingly, the Bacillus subtilis (Bs) DegU RR has the unique ability to either repress or activate transcription of flagellar motility genes depending on the phosphorylation state of the RR receiver domain (Amati et al., 2004, Kobayashi, 2007).
The BsDegS sensor kinase alters the DNA binding specificity of BsDegU by mediating phosphorylation of a conserved phosphoryl acceptor site (Asp56) located in the BsDegU N-terminal receiver domain (Mukai et al., 1990, Dahl et al., 1991). When phosphorylated, BsDegU binds and represses the fla-che flagellar motility gene operon (Amati et al., 2004). However, transcriptional profiling of a BsDegU deletion strain revealed that BsDegU also activates flagellar motility gene transcription (Kobayashi, 2007). The phosphorylation state of BsDegU is therefore critical for its specific regulatory roles since phosphorylation of the receiver domain alters BsDegU DNA-binding activity (Dahl et al., 1992, Kobayashi, 2007). In Bs, DegU plays multiple roles in gene regulation for flagellar motility, but also functions in regulating exoprotease production, competence development, biofilm formation, and swarming motility (Dahl et al., 1992, Hamoen et al., 2000, Kobayashi, 2007, Verhamme et al., 2007) where BsDegU can function as a transcriptional activator, co-activator, or repressor. The LmDegU RR is highly homologous to the BsDegU RR (63% identical, 78% similar). Similar to BsDegU, LmDegU is required for flagellar motility (Knudsen et al., 2004). However, unlike BsDegU, LmDegU is an orphan response regulator since a cognate sensor kinase is absent from the Lm genome. Nonetheless, it was recently reported that a phosphoryl acceptor site mutation (D55N) in LmDegU reduces flagellin transcription and motility in Lm (Mauder et al., 2008, Gueriri et al., 2008).
In this report, we further define the role of the DegU response regulator in GmaR production and temperature-dependent flagellar motility in Lm. We determined that LmDegU can be phosphorylated in vitro, however the receiver domain is dispensable for DegU-dependent regulation of flagellar motility. We further demonstrated that DegU activates gmaR transcription by binding directly to fliN-gmaR promoter region DNA; however, DegU-dependent activation of fliN-gmaR is temperature-independent. We also determined that a post-transcriptional mechanism limits GmaR protein production to low temperatures, as gmaR transcript levels are similar at both 37°C and 30°C in ΔmogR bacteria, yet GmaR protein is differentially expressed. Thus, our findings reveal that flagellar motility in Lm is governed by both temperature-dependent transcriptional and post-transcriptional control of the GmaR anti-repressor.
Results
The receiver domain of DegU is dispensable for flagellar motility
Temperature-dependent expression of the GmaR anti-repressor restricts flagellar motility gene expression to low temperatures in Lm (Shen et al., 2006). Although the DegU response regulator is known to be required for gmaR transcription, the regulatory mechanisms governing temperature-dependent production of GmaR remain elusive. We previously reported that DegU protein levels are temperature-independent (Shen et al., 2006), therefore changes in DegU activity may confer temperature specificity for flagellar motility gene transcription. LmDegU is highly homologous to the Bacillus subtilis (Bs) DegU RR, which contains an N-terminal receiver domain and a C-terminal helix-turn-helix (HTH) DNA binding motif (Fig. 1A). The N-terminal conserved phosphoryl acceptor site Asp56 of BsDegU is phosphorylated by the cognate sensor kinase DegS (Dahl et al., 1991). The phosphorylation state of BsDegU is critical for its specific regulatory roles since phosphorylation of the receiver domain alters BsDegU DNA-binding activity (Dahl et al., 1992). We therefore hypothesized that phosphorylation of the LmDegU receiver domain may modify LmDegU DNA-binding activity and possibly confer temperature specificity to flagellar motility.
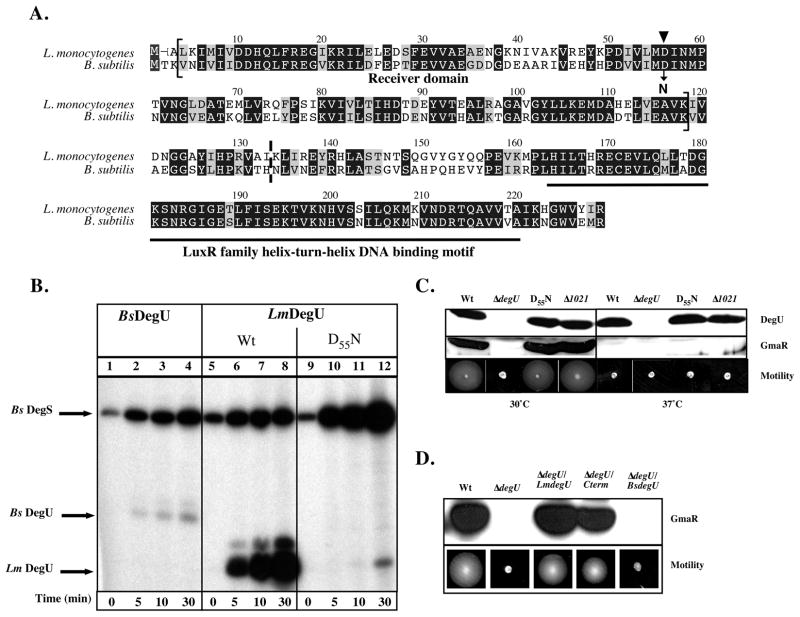
Figure 1. Phosphorylation of DegU.
A. Alignment of amino acid residues of LmDegU and BsDegU proteins. Conserved identical residues are blocked in black and similar residues are blocked in grey. The LmDegU phosphorylation receiver domain (aa 3-117) is indicated by brackets and contains the conserved phosphoryl acceptor site D55 (▼). The LuxR/FixJ family helix-turn-helix motif (aa 162-219) is underlined in black. The N-terminal truncation site in ΔdegU/Cterm is marked by a dashed line between aa 132 and aa 133.
B. in vitro phosphorylation of DegU. Purified BsDegS sensor kinase was incubated with either BsDegU (lanes 1–4), wild-type (Wt) LmDegU (lanes 5–8), or LmDegUD55N (lanes 9–12) in the presence of [γ32P]-ATP. LmDegUD55N has a phosphoryl acceptor site mutation Asp55 to Asn. Reactions were incubated at room temperature (RT) 5, 10, or 30 min. Proteins were resolved by SDS-PAGE and phosphorylation was detected using autoradiography.
C. Western blot and motility analysis of DegUD55N and Δlmo1021 bacteria. DegU and GmaR protein levels were examined using Western blot analysis of whole cell lysates prepared from cultures grown at either 30°C or 37°C for 18 h. A GmaR- or DegU- specific polyclonal antibody was used for detection. Motility was examined in low agar (0.3%) motility plates inoculated with a straight needle and incubated for 48 h.
D. Western blot and motility analysis of ΔdegU/Cterm and ΔdegU/BsDegU bacteria. A DegU-negative strain was complemented with either full-length LmDegU (ΔdegU/LmdegU), a N-terminally truncated DegU (ΔdegU/Cterm), or full-length BsDegU (ΔdegU/BsdegU). Whole cell lysates were prepared from cultures grown at 30°C for 18 h. Western blot analysis was used to detect GmaR using a GmaR-specific polyclonal antibody. Motility was also examined at 30°C in low agar (0.3%) motility plates inoculated with a straight needle and incubated for 48 h.
Genetic mutation of the conserved phosphoryl acceptor site Asp56 to Asn in BsDegU (D56N) inhibits phosphorylation in vitro and alters BsDegU DNA binding specificity (Dahl et al., 1991). It was recently published that the corresponding amino acid change in LmDegU (D55N) results in bacteria that are less motile than wild-type and produce less flagellin transcripts (Mauder et al., 2008, Gueriri et al., 2008). In vitro phosphorylation studies of His6-tagged BsDegS and LmDegU, confirmed that the BsDegS HK can phosphorylate LmDegU and that the D55N alteration abolishes the phosphotransfer event (Fig. 1B and (Gueriri et al., 2008)). The low level of detectable 32P-labeled LmDegUD55N at the 30 min time point (Fig. 1B) is likely not physiologically relevant since a similar observation has been previously reported with CheYD55N in Escherichia coli and is the result of a phosphoester group on a heterologous residue (Lukat et al., 1991, Bourret et al., 1990). To determine if the D55N phosphoryl acceptor site substitution in Lm affects Lm flagellar motility, Western blot and motility assays were performed using a LmDegUD55N substituted strain. Analysis revealed that temperature-dependent expression of GmaR in LmDegUD55N bacteria was similar to wild-type and did not affect flagellar motility in a low agar motility assay (Fig. 1C), unlike what has been previously reported (Mauder et al., 2008, Gueriri et al., 2008). Although LmDegU is an orphan response regulator, a type II histidine kinase similar to BsDegS could potentially trans-phosphorylate LmDegU as seen previously with the NarL response regulator in E. coli (Schroder et al., 1994). Deletion of lmo1021 (Δ1021), the only putative type II sensor kinase in Lm, also did not affect GmaR expression or flagellar motility (Fig. 1C). Furthermore, additional in vitro phosphorylation studies did not detect phosphorylation of LmDegU by Lmo1021 (data not shown). Taken together, these results strongly suggest that phosphorylation of the conserved LmDegU phosphoryl acceptor site does not significantly regulate LmDegU activity. If the D55 phosphoryl acceptor site of LmDegU is dispensable, then it is possible that the entire N-terminal receiver domain may be dispensable for DegU-dependent regulation of GmaR. To examine this hypothesis, LmDegU was N-terminally truncated at amino acid 133 and heterologously expressed in a DegU-negative strain (ΔdegU/Cterm). The ΔdegU/Cterm strain, harboring the LmDegU HTH DNA binding domain and lacking the receiver domain, still produced GmaR and was motile at low temperature (Fig. 1D). Collectively, these results demonstrate that LmDegU is able to function to produce GmaR in the absence of phosphorylation of the conserved phosphoryl acceptor site and that only the C-terminal portion of DegU containing the DNA binding domain is sufficient. In addition, whereas LmDegU and BsDegU are highly homologous, BsDegU does not complement LmΔdegU (Fig. 1D), reinforcing the contrasting roles DegU plays in these bacterial species.
Temperature-dependent transcription of gmaR initiates from the fliN promoter
Whereas hierarchal regulation in most bacterial species ensures ordered production and assembly of flagella, MogR repression of all flagellar motility gene promoters in Lm results in a non-hierarchal regulatory scheme. Derepression of MogR by the GmaR anti-repressor is absolutely necessary for flagellar motility gene transcription to occur at low temperatures. To further understand temperature-dependent expression of GmaR, we sought to determine the mechanism of DegU-dependent regulation of gmaR transcription.
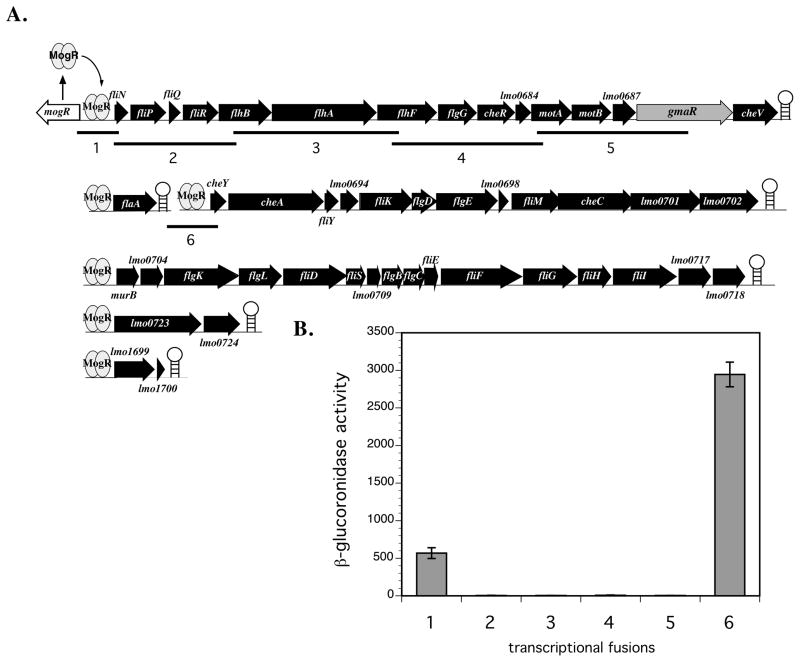
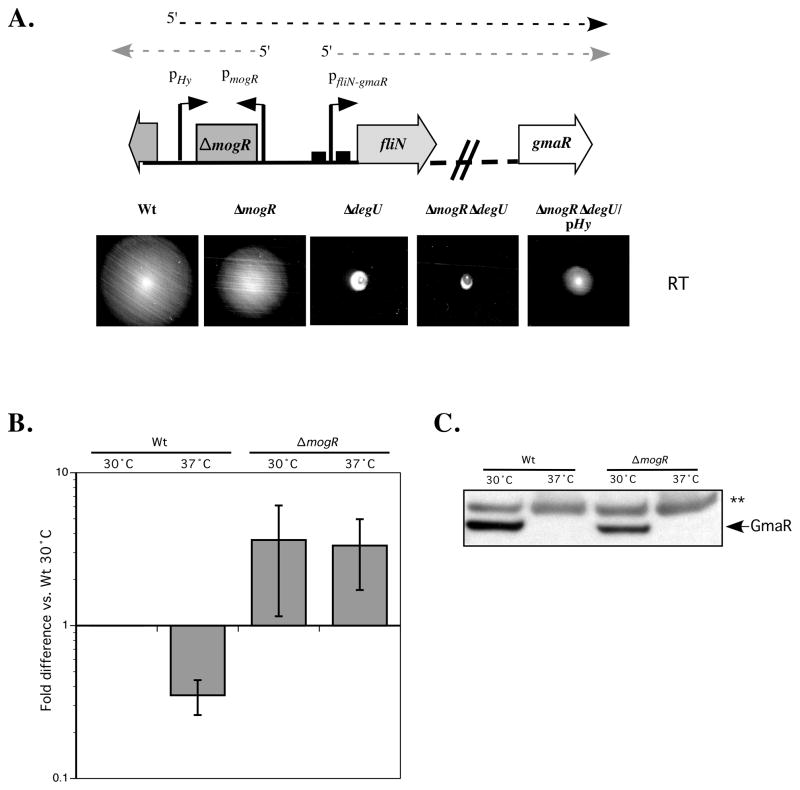
gmaR is located in the first operon of the flagellar motility gene locus ~10 kb downstream of the fliN promoter (pfliN) (Fig. 2A). To determine if pfliN is the only promoter controlling transcription of gmaR, we constructed transcriptional fusions of regions of the fliN-gmaR operon to a gusA reporter and determined β-glucoronidase activity (Fig. 2B). Transcriptional fusions containing the fliN and cheY promoter regions fused to gusA were used as controls and demonstrated β-glucoronidase activity as expected (Fig. 2B; lane 1 and 6, respectively), whereas transcriptional fusions containing DNA regions from within the fliN-gmaR operon yielded no β-glucoronidase activity (Fig. 2B; lanes 2–5). These results suggest that the MogR-regulated promoter directly upstream of fliN (pfliN-gmaR) is the only promoter controlling transcription of gmaR. Since GmaR functions as a MogR anti-repressor and the fliN-gmaR promoter is MogR repressed (Shen et al., 2006), these data indicate that production of gmaR transcripts is positively regulated by GmaR.
Figure 2. gmaR is transcribed from a promoter upstream of fliN.
A. Genetic organization of the Lm flagellar motility gene locus. The gene encoding the MogR anti-repressor, gmaR, is located in the first operon of the flagellar motility gene locus with transcription initiating upstream of fliN and terminating at a predicted transcriptional terminator (open circle on ladder). mogR is divergently transcribed from fliN and MogR functions to repress transcription of all six flagellar motility gene promoters. Numbered underlined fragments correspond to the transcriptional fusions described in Figure 2B.
B. Analysis of fliN-gmaR promoter activity as determined by β-glucuronidase assay. DNA regions from the fliN-gmaR operon (labeled 1–5 in Fig. 2A) were fused to a gusA reporter and integrated in single copy into the tRNAArg locus of wild-type Lm. As a positive control, the cheY promoter region was also fused to a gusA reporter (labeled 6 in Fig. 2A). Bacteria were grown at 30°C in BHI broth for 18–20 h. β-glucuronidase activities represent the means and standard deviations of three independent experiments.
DegU activates gmaR transcription by binding directly to fliN-gmaR promoter region DNA
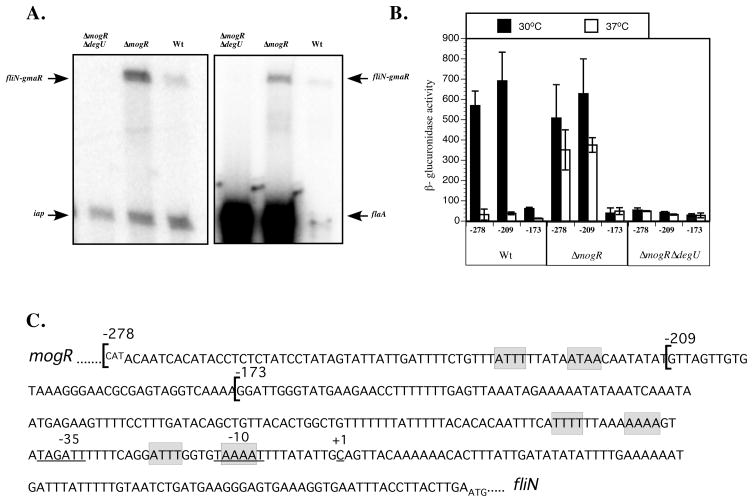
While gmaR transcription is repressed by MogR at 37°C, it is unknown how transcription of gmaR is initiated as temperatures decrease below physiological levels. Given our previous findings that gmaR transcripts are absent in ΔdegU bacteria and that GmaR functions downstream of DegU (Shen et al., 2006), we hypothesized that DegU may activate the pfliN-gmaR promoter to initiate transcription of gmaR. Analysis of pfliN-gmaR activity by both primer extension and transcriptional fusions revealed that DegU is required for transcriptional activation of the fliN-gmaR promoter (Fig. 3A and 3B). DegU-dependent transcriptional activation was specific for the fliN-gmaR promoter and was not required for transcription from other flagellar motility gene promoters (flaA shown in Fig. 3A and unpublished data). Surprisingly, DegU-mediated activation of pfliN-gmaR in a MogR-negative strain (ΔmogR) occurred at both 30°C and 37°C (Fig. 3B). This result indicates that the inherent ability of DegU to activate pfliN-gmaR transcription is temperature-independent.
Figure 3. The fliN-gmaR promoter is DegU-activated.
A. Analysis of fliN-gmaR transcripts by primer extension. RNA was extracted from ΔmogRΔdegU, ΔmogR and wild-type (Wt) strains grown at 30°C in BHI broth for 6 h. Transcript specific primers were end-labeled with [γ32P]-ATP. Primer extension products were separated on a 5% denaturing acrylamide gel and detected by a phosphorimager. Primers for flaA and iap were included in independent experiments as controls.
B. Analysis of fliN-gmaR promoter activity as determined by β-glucuronidase assay. Transcriptional fusions of fliN-gmaR promoter region DNA to gusA were integrated in single copy into the tRNAArg locus of wild-type (Wt), ΔmogR, or ΔmogRΔdegU bacteria. Cultures were grown in BHI broth at 30°C or 37°C for 18–20 h prior to analysis. β-glucuronidase activities represent the means and standard deviations of three independent experiments. Numbers correspond to fusions harboring −278, −209, or −173 through +274 relative to the transcriptional start of fliN-gmaR.
C. DNA sequence of the mogR-fliN intergenic region. The DNA sequence begins with the start codon (negative strand) for mogR and ends with the start codon for fliN. The transcriptional start site of the fliN-gmaR promoter (+1) was identified by primer extension and is underlined along with the predicted −35 and −10 elements. Transcriptional fusions of the fliN-gmaR promoter region DNA (−278, −209, −173 through +274 relative to the transcriptional start) are indicated by brackets and were fused to a gusA reporter and analyzed in Figure 2B. The predicted MogR binding sites are shaded in grey.
To identify the DNA elements within the pfliN-gmaR region that are required for DegU-dependent activation, promoter region truncations spanning −278, −209, and −173 to +274 nucleotides relative to the transcriptional start site were examined using reporter fusions to gusA (Fig. 3C). The pfliN-gmaR truncation fusions revealed that DNA elements located between −209 and −173 nucleotides upstream of the predicted fliN-gmaR transcriptional start site are required for DegU-dependent activation (Fig. 3B). To further support the transcriptional fusion studies, DNA affinity purification was used to determine if DegU binds directly to the fliN-gmaR promoter region. Biotin-labeled fliN-gmaR promoter region DNA coupled to Strepavidin-beads was incubated with lysates isolated from ΔdegU, wild-type, or ΔmogR bacteria. Bound proteins were eluted and then separated on an SDS-PAGE gel and analyzed by Western blot (Fig. 4A). In support of the requirement for DegU in transcriptional activation of pfliN-gmaR (Fig. 3A and 3B), DegU present in both wild-type and ΔmogR lysates was shown to bind to the −278 to +100 fliN-gmaR promoter region DNA fragment (Fig. 4A). Since DegU did not bind the −173 to +100 fliN-gmaR promoter region DNA, this result strengthens the hypothesis that DegU activates the fliN-gmaR promoter region by binding to DNA encompassing sequences −173 to −209 nucleotides upstream of the transcriptional start site (Fig. 3B and Fig. 4A). As expected, MogR bound both the −278 and −173 to +100 fliN-gmaR promoter region DNA fragments (Fig. 4A) since predicted MogR binding sites are contained within both of these DNA fragments (Fig. 3C).
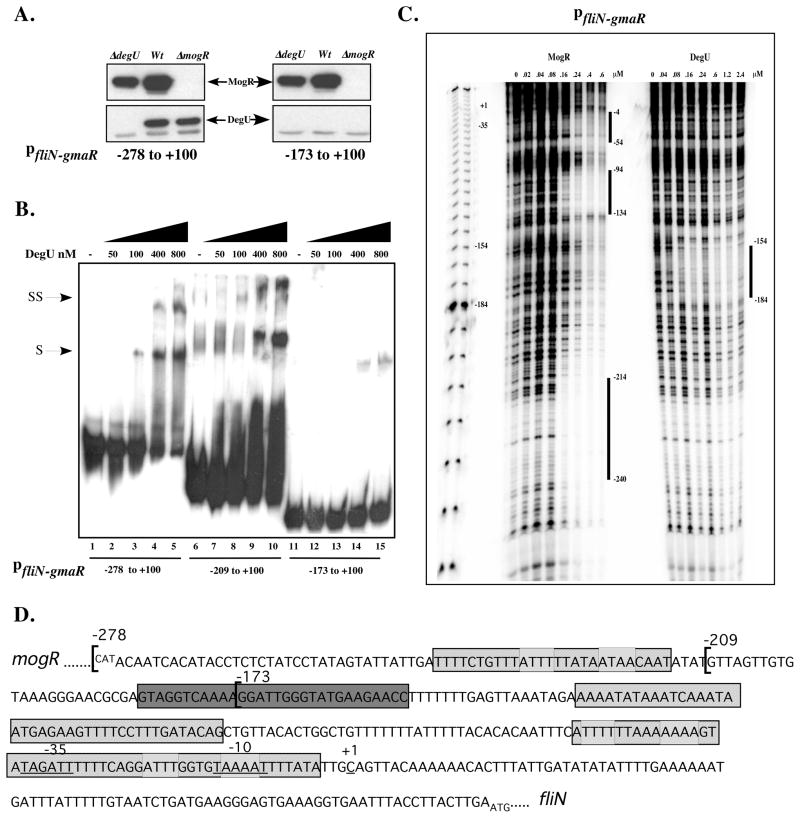
Figure 4. DegU binds directly to fliN-gmaR promoter region DNA.
A. Affinity purification of proteins that specifically bind fliN-gmaR promoter region DNA. Whole cell lysates of wild-type (Wt), ΔdegU, or ΔmogR bacteria were incubated at RT with magnetic Dynabeads coupled to fliN-gmaR specific promoter region fragments (−278 to +100 or −173 to +100 relative to the transcriptional start site). DNA-Bead complexes were washed, proteins were eluted and then separated by SDS-PAGE. DegU- or MogR- specific polyclonal antibody was used for Western blot detection. Arrows indicate DegU and MogR.
B. Gel shift analysis of DegU binding to fliN-gmaR promoter region DNA. Radiolabeled fliN-gmaR promoter region DNA fragments spanning −278, −209, or −173 to +100 relative to the transcriptional start site were incubated at RT with increasing amounts of purified His6-tagged LmDegU. Binding reactions were separated by non-denaturing PAGE and detected by autoradiography. Shifted (S) and Super-shifted (SS) complexes were detected.
C. DNase I footprint analysis of MogR and DegU binding to fliN-gmaR promoter region DNA. A radiolabeled DNA probe spanning the fliN-gmaR promoter region from −278 to +100 relative to the transcriptional start site was incubated at RT with increasing amounts of purified His6-tagged LmDegU or MogR and subsequently treated with DNaseI. Samples were run on a denaturing 6% acrylamide gel and the footprint was detected by phosphoimager. Binding sites are labeled with negative numbers relative to the transcriptional start site.
D. DNA sequence of the mogR-fliN intergenic region showing DegU and MogR binding sites. The DNA sequences corresponding to the three regions of MogR binding as identified in Figure 4C by DNaseI footprint analysis are shaded with cross-hatches. The predicted MogR binding sites are marked with light grey boxes. The DegU footprint as identified in Figure 4C is shaded with dark grey.
Gel mobility shift analysis was subsequently used to determine if His-tagged purified LmDegU can bind pfliN-gmaR region DNA in vitro in the absence of cell lysates. Increasing concentrations of DegU incubated with radiolabeled fliN-gmaR promoter region DNA fragments, spanning −278 and −209 to +100 relative to the transcriptional start site, resulted in the formation of shifted (S) and super-shifted (SS) DNA complexes (Fig. 4B, lanes 1–10), whereas incubation of DegU with the −173 to +100 DNA fragment resulted in only partially shifted DNA complexes (S) at high DegU concentrations (Fig. 4B, lanes 11–15). These results indicated that purified LmDegU was able to bind to fliN-gmaR promoter region DNA spanning −278 and −209 to +100, but was unable to efficiently bind to fliN-gmaR promoter region DNA spanning −173 to +100 nucleotides relative to the start of transcription. DegU did not require any additional bacterial factors to bind fliN-gmaR promoter region DNA since purified DegU alone was sufficient for binding and shifting the DNA fragments. Identical results were obtained when the DNA affinity purification and gel mobility shift analyses were performed at 37°C, suggesting that the in vitro DNA binding ability of DegU is not affected by temperature (data not shown). Taken together, the DNA affinity purification, gel mobility shift analysis, and transcriptional activation data indicate that DegU binding to fliN-gmaR promoter region DNA requires sequences within −173 and −209 nucleotides upstream of the transcriptional start site and that DegU binding mediates transcriptional activation of the fliN-gmaR promoter.
To further localize the specific DNA binding site(s) for DegU and MogR within the mogR-fliN intergenic region, DNase I footprinting analysis was performed using fliN-gmaR promoter region DNA and purified His-tagged LmDegU and His-tagged MogR. At low concentrations, MogR bound to three distinct regions of the fliN-gmaR promoter region DNA; −4 to −54, −94 to −134, and −214 to −240 relative to the transcriptional start site (Fig. 4C and 4D), which is similar to MogR binding to flaA promoter region DNA (Shen & Higgins, 2006). The DegU footprint spanned nucleotides −184 to −154 relative to the transcriptional start site (Fig. 4C and 4D). The DegU footprint corresponds to the fliN-gmaR promoter region sequences required for DegU binding that were identified by DNA affinity purification (Fig. 4A), gel mobility shift analysis (Fig. 4B), as well as DegU-dependent transcriptional activation using gusA reporter fusions (Fig. 3B). Interestingly, the MogR and DegU binding sites do not overlap as determined by DNase I footprinting analysis (Fig. 4C and 4D).
A post-transcriptional mechanism controls temperature-dependent GmaR production
We have previously shown that both ΔmogRΔdegU and ΔdegU constitutively expressing GmaR (ΔdegU/cgmaR) are non-motile at low temperatures (<37°C) despite evidence that flagellin is produced (Shen & Higgins, 2006, Shen et al., 2006). Since DegU is required for transcriptional activation of the first operon in the Lm flagellar motility gene cluster (Fig. 3A and 3B), we hypothesized that the DegU-dependent motility defect of ΔmogRΔdegU bacteria is due to the absence of essential flagellar biosynthetic components encoded in the fliN-gmaR operon that are required for proper flagellar assembly and motility. Effectively, DegU-dependent transcriptional activation of the entire fliN-gmaR operon and not just gmaR is required for flagellar motility in Lm at low temperatures. To confirm this hypothesis, a Lm strain was constructed in which a promoter (pHy) was inserted upstream of the fliN-gmaR promoter in a ΔmogRΔdegU strain to provide constitutive transcription of the fliN-gmaR operon in a MogR-negative, DegU-negative background (Fig. 5A, ΔmogRΔdegU/pHy). Motility assay analysis revealed that the ΔmogRΔdegU/pHy strain was slightly motile at low temperature. While the ΔmogRΔdegU/pHy strain was not as motile as MogR-negative bacteria, the lack of increased motility is likely due to placement of the pHy promoter and promoter strength relative to the native DegU-activated pfliN-gmaR promoter. However, the fact that constitutive expression of the fliN-gmaR operon overcomes the DegU-dependent motility defect, strongly suggests that the requirement for DegU in Lm flagellar motility is strictly for transcriptional activation of the fliN-gmaR operon. Since we have established that DegU is capable of activating transcription of the fliN-gmaR promoter and that this activation does not appear to be temperature-dependent (Fig. 3B), it remained to be determined how GmaR production is restricted to low temperatures (Fig. 1C).
Figure 5. A post-transcriptional mechanism controls temperature-dependent production of GmaR.
A. Constitutive expression of the fliN-gmaR operon in ΔmogRΔdegU. Native promoters for mogR and fliN-gmaR are represented by bent arrows (pmogR and pfliN-gmaR). MogR binding sites are marked as boxes overlapping the fliN-gmaR promoter. Divergent transcripts initiating from the native promoters are drawn as light dashed arrows. The pHy promoter was inserted upstream of the native fliN-gmaR promoter in a ΔmogRΔdegU strain to constitutively drive expression of the fliN-gmaR operon. Transcription initiating from this promoter is drawn as a dark dashed line. For the motility assay analysis, strains were inoculated into low agar (0.3%) motility plates with a straight needle and incubated at RT for 48 h.
B. Real-Time quantitative PCR analysis of gmaR transcripts. RNA was extracted from wild-type (Wt) and ΔmogR strains grown at either 30°C or 37°C for 24 h. Samples were DNaseI treated and reverse-transcribed with random hexamers to generate cDNA. Relative gene expression was quantified by using Real-Time PCR and the Pfaffl method (2−ΔΔCT). Results represent the average and standard deviation of three independent experiments. The iap gene was used as an internal standard and the Wt 30°C sample was set as the calibrator.
C. GmaR protein analysis by Western blot. Total protein samples from cultures used in B were processed for SDS-PAGE and Western blot analysis. A GmaR-specific polyclonal antibody was used for detection. ** indicates a non-specific band that is shown as a loading control.
To determine if a post-transcriptional mechanism controls temperature-dependent GmaR production, we analyzed gmaR transcript and GmaR protein levels in a ΔmogR strain, which allows temperature-independent transcription of gmaR to occur (Fig. 3B). Quantitative Real-Time PCR analysis revealed that gmaR transcript levels in ΔmogR bacteria do not change in response to temperature (Fig. 5B). This result correlated with transcriptional fusion studies indicating that in ΔmogR bacteria, DegU can activate transcription of pfliN-gmaR at low temperatures (30°C) and at 37°C (Fig. 3B). Collectively, these results reveal that although DegU is required for activation of the fliN-gmaR promoter, transcriptional activation is temperature-independent in the absence of MogR. However, since temperature-dependent expression of GmaR protein is observed in ΔmogR bacteria (Fig. 5C), a temperature-dependent, post-transcriptional mechanism must control GmaR production. The data in Figure 5 indicates that two levels of regulation (transcriptional and post-transcriptional) govern production of GmaR and thus the temperature specificity of Lm flagellar motility.
Discussion
Temperature-dependent regulation of flagellar motility in Lm is mediated by the activities of the MogR transcriptional repressor, the MogR anti-repressor GmaR, and the DegU response regulator. In this study, we determined that DegU-dependent transcriptional activation of the first operon in the flagellar motility gene cluster, which contains gmaR, is required for Lm flagellar motility at low temperatures. DegU activates transcription of the fliN-gmaR operon by binding directly to fliN-gmaR promoter region DNA. DegU-dependent activation does not depend on the phosphorylation of the conserved DegU phosphoryl acceptor site (Asp55) and is temperature-independent. Furthermore, in ΔmogR bacteria, DegU-dependent activation of fliN-gmaR transcription is constitutive. Our data indicate that temperature specificity of flagellar motility in Lm occurs through a temperature-dependent, post-transcriptional mechanism that limits GmaR expression to temperatures below 37°C. At permissive temperatures, we hypothesize that GmaR production leads to positive auto-regulation of gmaR transcription and subsequent transcriptional up-regulation of all flagellar motility genes. Therefore, our results demonstrate that flagellar motility in Lm is restricted to low temperatures by both MogR transcriptional repression and post-transcriptional regulation of the GmaR anti-repressor.
While MogR repression and GmaR anti-repression regulate transcription of all flagellar motility gene promoters, DegU is required to activate transcription of pfliN-gmaR for production of GmaR (Fig. 3A and data not shown). Furthermore, constitutive expression of the fliN-gmaR operon in ΔmogRΔdegU bacteria revealed that DegU-dependent transcription of additional flagellar biosynthetic components within the fliN-gmaR operon is necessary for flagellar motility (Fig. 5A). Since the fliN-gmaR operon encodes several proteins comprising the flagella export apparatus, the complete lack of flagella in ΔmogRΔdegU bacteria, despite production of the flagellin subunit (Shen & Higgins, 2006), is likely explained by the absence of proteins necessary for flagellin secretion and flagellum assembly. Therefore, DegU-dependent activation of pfliN-gmaR is necessary for production of GmaR, but is also required for the proper assembly of flagella.
DegU-dependent activation of pfliN-gmaR
Our results indicated that LmDegU binds directly to fliN-gmaR promoter region DNA at −154 to −184 nucleotides upstream of the fliN-gmaR transcriptional start site and that binding of DegU to this region is essential for transcriptional activation of the fliN-gmaR promoter (Fig. 3 and 4). Gel mobility shift and DNaseI footprinting analyses using purified protein confirmed that DegU binds fliN-gmaR promoter region DNA independent of additional cellular factors (Fig. 4B and 4C). Since the gel mobility shift analysis showed a super-shifted complex at higher concentrations of DegU (Fig. 4B), but footprinting analysis only indicated one DNA binding region (Fig. 4C and 4D), it is possible that DegU binds either as a multimer or that DegU binds multiple sites within the −154 to −184 region. Although the DegU binding site is located 94 nucleotides upstream of the translational start site of MogR (Fig. 4D), MogR expression was found to be constitutive under all conditions examined and unaffected by the deletion of DegU (data not shown). While many transcriptional activators bind close to the −35 region of their regulated promoters, it is not unprecedented for activators to bind promoter region DNA further upstream of the transcriptional start site and still directly activate transcription. DegU binding to the −184 to −154 region upstream of the fliN-gmaR promoter may facilitate bending of the DNA to allow contact of DegU with RNA polymerase or may change the conformation of the promoter region DNA to enhance RNA polymerase binding (Rhodius & Busby, 1998). It is also possible that DegU may be an essential co-activator that recruits another protein factor that then makes direct contact with RNA polymerase to activate transcription of fliN-gmaR (Rhodius & Busby, 1998). In fact, BsDegU functions as a priming protein in Bs competence development. Binding of BsDegU to the comK promoter facilitates ComK binding and auto-regulation of comK transcription. Therefore, BsDegU is important for initiation of the competence auto-regulatory loop when ComK levels are insufficient to support comK transcription (Hamoen et al., 2000, Maamar & Dubnau, 2005). Similarly, it is unknown if LmDegU is acting alone at the fliN-gmaR promoter or if a co-activator is involved in transcription initiation. However, if LmDegU is interacting with another factor, the interaction domain is presumably contained within the C-terminal portion of DegU, since the N-terminal domain is dispensable for transcriptional activation of fliN-gmaR (Fig. 1D).
MogR binding to the pfliN-gmaR promoter region
We previously demonstrated that MogR binds specifically to TTTT-N5-AAAA operator sites found within Lm flagellar motility gene promoter regions (Shen & Higgins, 2006). MogR binding to a minimum of two operator sites overlapping the −10 or −35 regions of flagellar promoters results in occlusion of RNA polymerase binding and represses transcription of flagellar motility genes (Shen & Higgins, 2006). Sequence analysis of the fliN-gmaR promoter region revealed that a nearly consensus (one mismatch) MogR binding site overlaps the −10 region of the fliN-gmaR promoter and a second consensus binding site is located just upstream of the −35 region (Fig. 3C). DNaseI footprinting analysis confirmed that MogR binds the −10 and −35 regions of the fliN-gmaR promoter as well as two additional sites further upstream (Fig. 4C and 4D). A similar MogR footprinting pattern was observed previously with the flaA promoter region (Shen & Higgins, 2006); however it remains to be determined if the additional upstream binding sites aid in MogR repression. It is possible that MogR binding to multiple sites and/or MogR multimerization between sites may produce a higher order DNA-protein structure. The crystal structure of MogR bound to a DNA binding site indicates that MogR functions as a dimer (Shen et al., 2009). Therefore, it is possible that multimers of MogR dimers are required for MogR-dependent repression of flagellar motility. Footprinting analysis of the fliN-gmaR promoter region indicated that although both MogR and DegU bind within the region, the MogR binding sites do not overlap the DegU binding site and therefore it is possible that both factors could be bound to the promoter region simultaneously (Fig. 4C and 4D). Gel shift analysis of simultaneous DegU and MogR binding of pfliN-gmaR promoter region DNA provides additional support that neither factor occludes the binding of the other and that both factors could potentially bind pfliN-gmaR DNA concurrently (Fig. S1). Depending on the relative on/off rates for the binding of MogR and DegU to their respective DNA sites, it is therefore possible that DegU-dependent transcriptional activation could occur as a rare event during a MogR-repressed state (i.e. 37°C).
Phosphorylation of LmDegU
In this study, we demonstrated that although the LmDegU RR is capable of being phosphorylated (Fig. 1B), DegU-dependent activation of flagellar motility does not require phosphorylation of the conserved phosphoryl acceptor site (Fig. 1C and 1D). In fact, DegU-dependent activation of fliN-gmaR could be obtained using just the C-terminal DNA-binding domain (Fig. 1D). Since DegU can function without the N-terminal receiver domain that is typically required for signal recognition (Fig. 1A), it is likely that LmDegU is constitutively active. Nevertheless we cannot rule out the possibility that the C-terminal fragment used in our studies is a constitutively active form of DegU. While there exist several examples of response regulators that do not require phosphorylation for activity (Rotter et al., 2006, Hong et al., 2007), it is surprising that DegU activity for Lm flagellar motility appears to be phosphorylation-independent due to the high homology between LmDegU and BsDegU (Fig. 1A). The phosphorylation state of the BsDegU receiver domain alters BsDegU function as either an activator or repressor of flagellar motility (Kobayashi, 2007, Tsukahara & Ogura, 2008). However, despite the similarities between BsDegU and LmDegU, important amino acid differences must exist to confer DNA-binding specificity in Lm, as a LmΔdegU strain can not be complemented with BsDegU (Fig. 1D). Since phosphorylation appears not to have a significant role in DegU-dependent activation of flagellar motility in Lm, it is not surprising that LmDegU is an orphan response regulator that lacks a cognate sensor kinase and deletion of the only related type II sensor kinase in Lm (lmo1021) had no affect on flagellar motility (Fig. 1C).
Temperature-dependent control of flagellar motility
Although DegU is required for flagellar motility, we have shown that DegU does not regulate flagellar motility in a temperature-dependent manner as previously hypothesized. While DegU-dependent transcription of the fliN-gmaR operon is absolutely required for flagellar motility (Fig. 5A), DegU is able to activate transcription of the fliN-gmaR operon at both 30°C and 37°C (Fig. 3B), indicating that DegU activity is temperature-independent. In support of this finding, DNA affinity purification and gel mobility shift analyses at 37°C revealed that DegU binding to the fliN-gmaR promoter region is unaffected by temperature (data not shown). Whereas analysis of the fliN-gmaR promoter in ΔmogR bacteria showed that gmaR transcription is initiated at 37°C (Fig. 3B), it was unknown if gmaR transcripts were stable at 37°C. Quantitative Real-Time PCR analysis demonstrated that gmaR transcripts were present in equivalent levels at both 30°C and 37°C (Fig. 5B). Despite the abundance of gmaR transcripts in ΔmogR bacteria, GmaR protein was not detectable at 37°C (Fig. 5C). Therefore, a post-transcriptional mechanism regulates temperature-dependent GmaR expression. This temperature-dependent, post-transcriptional mechanism functions in addition to the transcriptional repression of gmaR by MogR; therefore at 37°C both transcriptional and post-transcriptional mechanisms prevent GmaR production. It is likely that the post-transcriptional mechanism involves either translational control or protein stability, since we have already demonstrated that gmaR transcripts are stable at elevated temperatures (Fig. 5B). Studies are currently underway to determine the exact mechanism(s) responsible for the temperature-dependent, post-transcriptional control of GmaR, and to identify potential cis- and/or transacting factors involved.
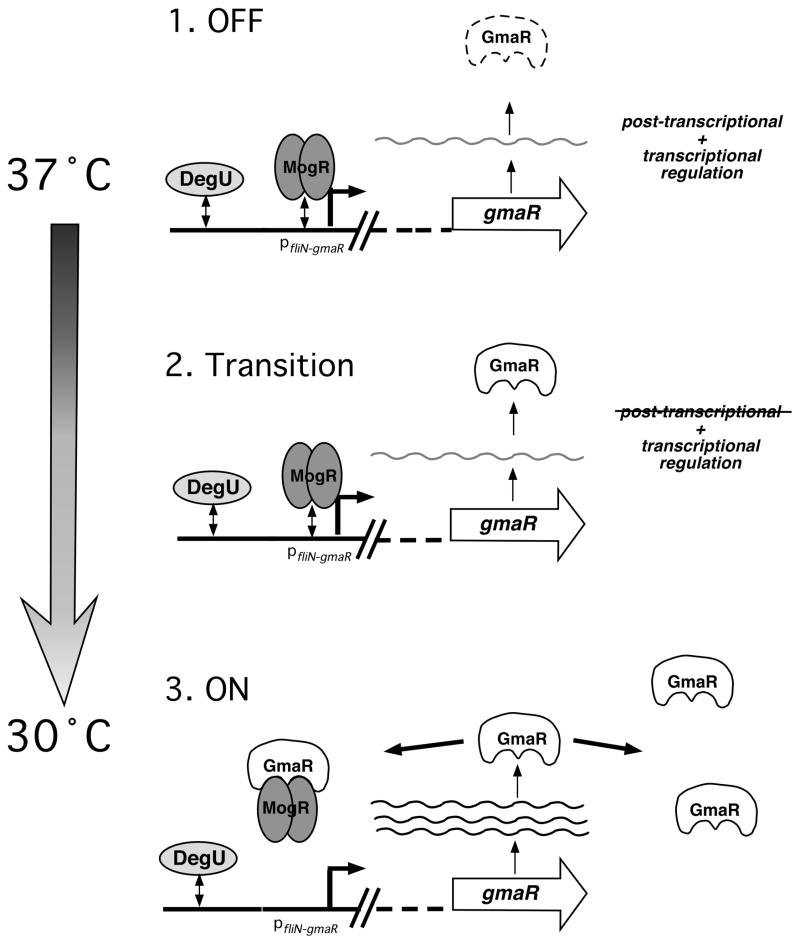
We favor the following model for temperature-dependent regulation of GmaR and flagellar motility in Lm (Fig. 6). When flagellar motility is “OFF” (1), MogR binds and represses all flagellar motility gene promoters at elevated temperatures (37°C and above). DegU can also bind DNA upstream of the fliN-gmaR promoter and activate transcription at 37°C, but due to stringent MogR repression, transcription of fliN-gmaR is minimized at elevated temperatures. In addition to MogR transcriptional repression of fliN-gmaR, a post-transcriptional mechanism prevents GmaR protein production from gmaR transcripts at 37°C (Fig. 6, OFF). As temperatures decrease below 37°C (2), the inherent on/off rates of MogR and DegU to their respective binding sites in the pfliN-gmaR promoter region will generate gmaR transcripts that in the absence of the temperature-dependent post-transcriptional regulatory mechanism, will now yield GmaR protein (Fig. 6, Transition). Once GmaR protein is initially produced (3), GmaR can up-regulate gmaR transcript levels by alleviating MogR repression from the fliN-gmaR promoter (Fig. 6, ON). Elevated levels of GmaR protein can remove MogR from all flagellar motility gene promoters permitting flagellar motility gene transcription at low temperatures. If the post-transcriptional mechanism involves GmaR protein instability at higher temperatures, reduced levels of GmaR at 37°C would release MogR protein to reinitiate repression of gmaR and other flagellar motility genes, initiating a transition from an ON to an OFF state.
Figure 6. Model of temperature-dependent regulation of GmaR expression.
1. OFF: At 37°C, when flagellar motility is OFF, the opposing activities of the MogR repressor and the DegU activator at pfliN-gmaR results in minimal fliN-gmaR transcripts. However, a temperature-dependent, post-transcriptional mechanism inhibits GmaR production.
2. Transition: As the temperature decreases below 37°C, the post-transcriptional mechanism is no longer active, therefore fliN-gmaR transcripts result in GmaR protein production.
3. ON: Once GmaR is expressed at low temperatures, GmaR can remove MogR from the fliN-gmaR promoter, up-regulating transcription of gmaR. Elevated levels of GmaR protein results in the relief of MogR repression from all flagellar motility gene promoters, allowing flagellar motility gene transcription and flagellar motility.
In this report, we have defined the requirement of DegU for flagellar motility in Lm, but the question still lingers as to why a response regulator has evolved to activate a complex bacterial system constitutively. Moreover, we have uncovered a post-transcriptional layer of regulation through which temperature-dependent control of flagellar motility occurs. Since flagellar motility is an energetically demanding process, it is not surprising that several layers of regulation would be needed to ensure a committed “OFF” regulatory state at physiological temperatures (37°C). Similarly, as temperatures fluctuate, RNA and protein levels within the bacterium also fluctuate until a steady-state commitment (ON or OFF) for flagellar motility can be reached (Fig. 6). Inherent in this model, auto-regulation of gmaR transcription by GmaR protein allows for an adaptive temporal response to temperature fluctuations. Therefore, by incorporating both transcriptional and post-transcriptional responses, Lm has the ability to stringently regulate expression of flagellar motility in response to environmental temperatures.
Experimental Procedures
Bacterial strains and growth media
Listeria monocytogenes (Lm) and Escherichia coli (Ec) strains used in this study are listed in Supplemental Table 1 and Supplemental Table 2, respectively. Specific details for construction of bacterial strains are located in Supplemental Materials. Primers used in this study are listed in Supplemental Table 3. Ec strains were grown in Luria-Bertani (LB) media for plasmid isolation and protein purification. All Lm strains are in the EGDe background and were grown in Brain Heart Infusion (BHI) broth. Antibiotics were used at the following concentrations: chloramphenicol at 20 μg/ml for selection of pPL3 derivatives in Ec, 5 μg/ml for selection of integrated pPL3 derivatives in Lm, and 7.5 μg/ml for pCON1 derivatives in Lm; 100 μg/ml carbenicillin for pCON1 derivatives in Ec; 30 μg/ml kanamycin for pET28a vectors in Ec. All plasmid constructs were confirmed by automated DNA sequencing. Plasmids were isolated from XL1-Blue or CLG190 strains and transformed into SM10 for conjugative transfer into Lm or electroporated directly into Lm.
Purification of His6-tagged proteins
Ec strains DH-E1446, DH-E1451, DH-E1521, and DH-E1523 expressing LmDegU-His6, LmDegUD55N-His6, BsDegU-His6 and BsDegS-His6 were grown in LB medium at 37°C to OD600=0.5, and protein expression was induced with 100 μM IPTG. The induced culture was grown at 30°C for 4 h. Bacteria were pelleted, and the His-tagged proteins were purified using Ni-NTA spin columns (Qiagen) according to the manufacturer’s instructions. Purified His-tagged proteins used for in vitro phosphorylation were dialyzed in buffer E [100mM Tris HCL (pH 8.0), 200 mM KCl, 4 mM MgCl2, 4 mM CaCl2, 0.5 mM DTT, 0.1 mM EDTA, 5% glycerol] (Dahl et al., 1991).
in vitro phosphorylation
in vitro phosphorylation assays were performed by mixing DegU and DegS proteins together in a final volume of 40 μL. The phosphorylation reaction was initiated by the addition of 5 μL of ATP containing 20 μCi [γ32P]-ATP diluted in cold ATP. The final concentration of reaction components were 0.5 μM DegS, 2.7 μM DegU, 1 μM cold ATP, and 0.15 μM [γ32P]-ATP. Reactions were stopped at the time points indicated by removing 10 μL of the reaction and placing it into a tube containing 10 μL of 2X FSB [0.0625M Tris (pH 6.8), 2% SDS, 10% glycerol, 0.01% bromophenol blue]. The in vitro phosphorylation reactions were boiled for 2 min at 95°C and resolved by SDS-PAGE. The resulting gel was dried and then analyzed by autoradiography.
Western blot analysis
Bacterial cultures were grown standing at either 30°C or 37°C for 18–20 h. A culture volume equivalent of 1 ml of OD600 = 1.5 was pelleted and resuspended in 75 μl of TE/lysozyme (10 mM Tris-HCL [pH 8.0], 1 mM EDTA, 3 mg/ml lysozyme) and incubated at 37°C for 1 h. After the 1 h incubation at 37°C, an equal volume of 2X FSB was added. Samples were boiled for 5 min at 95°C and then centrifuged for 1 min at 16,000 × g. Sixty microliters of the boiled sample was loaded onto a 6% SDS-PAGE gel for analysis of GmaR or a 12% SDS-PAGE gel for analysis of DegU. Western blot analysis was performed as previously described (Shen et al., 2006) using a polyclonal antibody specific for GmaR or DegU.
Motility assay analysis
A single colony was inoculated with a straight needle into BHI agar (0.3%). Plates were incubated at 30°C or 37°C for 24 h.
β-glucuronidase assay
β-glucuronidase assays were performed as previously described for β-galactosidase assays except that 4-methylumbelliferyl-β-D-glucuronide was used in the place of 4-methylumbelliferyl-β-D-galactoside (Grundling et al., 2004).
Primer extension
Oligonucleotide primers #375, #133, #326 were used for primer extension analysis of fliN, iap, and flaA transcripts, respectively. Primer extension was performed as previously described (Grundling et al., 2004).
Affinity purification
To generate bacterial cell lysates, 500 ml of a 16–18 h culture of ΔdegU, ΔmogR, or wild-type Lm were pelleted and resuspended in 20 ml of lysis buffer [10 mM Tris-HCl (pH 7.5), 50mM NaCl] supplemented with Complete protease inhibitor mixture (Roche). Cultures were passed 3X through a French press at 4°C and stored at −80 °C. The 5′ primers #715 or #718 and the 3′ biotinylated primer #731 were used to amplify the pfliN-gmaR DNA region from EGDe genomic DNA. Affinity purification was performed as previously described (Grundling et al., 2004) with the following exceptions. A low salt binding buffer (LSBB) was used [10% glycerol, 2 mM MgCl2, 10 mM Tris-HCL (pH 7.5), 100 mM NaCl, 0.5 mM DTT, 4 mM EDTA]. Binding reactions consisted of 100 μl of Dynabead-DNA complex (1 mg of beads, 40 ng of DNA), 0.9 ml of 5X BB, and 3.5 ml of bacterial cell lysate. Binding reactions were incubated 3 h at RT (18–25°C) on a rotisserie. Proteins were visualized by Western blot analysis using a DegU or MogR specific antibody.
DNaseI footprinting analysis
DNaseI footprinting analysis was performed as previously reported (Shen & Higgins, 2006) with the following exceptions. The fliN-gmaR promoter region DNA was amplified with primer pair #715 and #736 to generate a DNA fragment spanning −278 to +100 relative to the transcriptional start site of pfliN-gmaR. The resulting PCR product was digested with EagI and radiolabeled using Klenow enzyme as previously described (Shen & Higgins, 2006). Reactions were terminated by adding 200 μl of cold Stop solution [6.45ml of 200% ETOH, 18 μg/ml of Bovine tRNA (R4752 Sigma), 500 μl of NH4OAc] to each sample, and placed immediately in an ETOH dry ice bath for 20 min to precipitate the DNA. Samples were centrifuged at 16,000 × g for 15 min at 4°C, washed with 70% ETOH, and then resuspended in formamide loading buffer. Samples were loaded onto a denaturing 6% acrylamide gel and analyzed with a phosphoimager.
Real-Time quantitative PCR
Wild-type Lm EGDe and ΔmogR strains were grown 16–18 h at 30°C or 37°C and RNA was extracted as previously reported (Grundling et al., 2004). A total of 3 RNA samples were collected for each strain from three independent experiments. Approximately 10 μg of each RNA sample was digested with 8 units of DNaseI (NEB) in a final volume of 100 μl for 70 min at 37°C to eliminate any trace genomic DNA contamination. cDNA were synthesized from 8 μl (~800ng) of DNaseI-treated RNA using random hexamers with a SuperScript III First-Strand Synthesis System Kit (Invitrogen) following manufacturer’s instructions. No Reverse Transcriptase samples were included and treated identically with the exception that the reverse transcriptase was eliminated from the protocol. The cDNA samples were used in qRT-PCR to quantify mRNA levels of gmaR and iap by using iQ SYBR Green Supermix (BioRad) and a MiQ cycler (BioRad). Gene specific primers #732 and #733 were used to amplify a 151 nucleotide (nt) fragment of gmaR spanning nt 1517 to nt 1667. Gene specific primers #734 and #735 were used to amplify a 121 nt fragment of iap spanning nt 757 to nt 877. The specificity of each primer pair was monitored through melting curves and the primer pair efficiencies were calculated using a standard curve (102% for gmaR and 110% for iap). Relative gene expression was quantified by using the Pfaffl method of data analysis. The amount of gmaR mRNA was normalized using iap as an internal standard (iap is known to be transcribed equivalently at both 30°C and 37°C) and then compared to the calibrator condition (gmaR levels in the wild-type 30°C sample). Therefore, the relative level of gmaR mRNA at 30°C in a wild-type strain thus corresponded to 1. mRNA quantification was performed in duplicate from two independent cDNA pools generated from total RNA extracted from three independent experiments.
Supplementary Material
Acknowledgments
We would like to especially thank Aimee Shen for construction of the pET28a expression vectors, the Δlmo1021 strain, and for providing helpful discussions and guidance at the onset of this project. We would like to thank Laura Burrack and Ann Hochschild for critical review of the manuscript and providing helpful discussions. We would also like to thank Pricilla Yang and members of her laboratory for assistance with the qRT-PCR. This work was supported by U.S. Public Health Service grant AI-053669 from the National Institutes of Health and grant MCB-0718559 from the National Science Foundation (DEH).
References
- Akerley BJ, Miller JF. Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. J Bacteriol. 1993:175, 3468–3479. doi: 10.1128/jb.175.11.3468-3479.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge P, Hughes KT. How and when are substrates selected for type III secretion? Trends Microbiol. 2001:9, 209–214. doi: 10.1016/s0966-842x(01)02014-5. [DOI] [PubMed] [Google Scholar]
- Amati G, Bisicchia P, Galizzi A. DegU-P represses expression of the motility fla-che operon in Bacillus subtilis. J Bacteriol. 2004;186:6003–6014. doi: 10.1128/JB.186.18.6003-6014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage JP. Bacterial tactic responses. Adv Microb Physiol. 1999;41:229–289. doi: 10.1016/s0065-2911(08)60168-x. [DOI] [PubMed] [Google Scholar]
- Bijlsma JJ, Groisman EA. Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol. 2003;11:359–366. doi: 10.1016/s0966-842x(03)00176-8. [DOI] [PubMed] [Google Scholar]
- Bourret RB, Hess JF, Simon MI. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl MK, Msadek T, Kunst F, Rapoport G. Mutational analysis of the Bacillus subtilis DegU regulator and its phosphorylation by the DegS protein kinase. J Bacteriol v. 1991;173:2539–2547. doi: 10.1128/jb.173.8.2539-2547.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl MK, Msadek T, Kunst F, Rapoport G. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J Biol Chem. 1992;267:14509–14514. [PubMed] [Google Scholar]
- Gründling A, Burrack LS, Bouwer HG, Higgins DE. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc Natl Acad Sci USA. 2004;101:12318–12323. doi: 10.1073/pnas.0404924101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueriri I, Bay S, Dubrac S, Cyncynatus C, Msadek T. The Pta-AckA pathway controlling acetyl phosphate levels and the phosphorylation state of the DegU orphan response regulator both play a role in regulating Listeria monocytogenes motility and chemotaxis. Mol Microbiol. 2008 doi: 10.1111/j.1365-2958.2008.06496.x. [DOI] [PubMed] [Google Scholar]
- Hamoen LW, Van Werkhoven AF, Venema G, Dubnau D. The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis. Proc Natl Acad Sci U S A. 2000;97:9246–9251. doi: 10.1073/pnas.160010597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- Hong E, Lee HM, Ko H, Kim DU, Jeon BY, Jung J, Shin J, Lee SA, Kim Y, Jeon YH, Cheong C, Cho HS, Lee W. Structure of an atypical orphan response regulator protein supports a new phosphorylation-independent regulatory mechanism. J Biol Chem. 2007;282:20667–20675. doi: 10.1074/jbc.M609104200. [DOI] [PubMed] [Google Scholar]
- Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- Kapatral V, Minnich SA. Co-ordinate, temperature-sensitive regulation of the three Yersinia enterocolitica flagellin genes. Mol Microbiol. 1995;17:49–56. doi: 10.1111/j.1365-2958.1995.mmi_17010049.x. [DOI] [PubMed] [Google Scholar]
- Khorchid A, Ikura M. Bacterial histidine kinase as signal sensor and transducer. Int J Biochem Cell Biol. 2006;38:307–312. doi: 10.1016/j.biocel.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Knudsen GM, Olsen JE, Dons L. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol Lett. 2004;240:171–179. doi: 10.1016/j.femsle.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Kobayashi K. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol. 2007;66:395–409. doi: 10.1111/j.1365-2958.2007.05923.x. [DOI] [PubMed] [Google Scholar]
- Lemon KP, Higgins DE, Kolter R. Flagellar motility is critical for Listeria monocytogenes biofilm formation. J Bacteriol. 2007;189:4418–4424. doi: 10.1128/JB.01967-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukat GS, Lee BH, Mottonen JM, Stock AM, Stock JB. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem. 1991;266:8348–8354. [PubMed] [Google Scholar]
- Maamar H, Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauder N, Williams T, Fritsch F, Kuhn M, Beier D. Response regulator DegU of Listeria monocytogenes controls temperature-responsive flagellar gene expression in its unphosphorylated state. J Bacteriol. 2008 doi: 10.1128/JB.00258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter LL. Regulation of flagella. Curr Opin Microbiol. 2006;9:180–186. doi: 10.1016/j.mib.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Byrne BG, Whitfield NN, Madigan CA, Fuse ET, Tateda K, Swanson MS. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai K, Kawata M, Tanaka T. Isolation and phosphorylation of the Bacillus subtilis degS and degU gene products. J Biol Chem. 1990;265:20000–20006. [PubMed] [Google Scholar]
- O’Neil HS, Marquis H. Listeria monocytogenes flagella are used for motility, not as adhesins, to increase host cell invasion. Infect Immun. 2006;74:6675–6681. doi: 10.1128/IAI.00886-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Messner P, Heesemann J, Marre R, Hacker J. Temperature-dependent expression of flagella in Legionella. J Gen Microbiol. 1991;137:1955–1961. doi: 10.1099/00221287-137-8-1955. [DOI] [PubMed] [Google Scholar]
- Peel M, Donachie W, Shaw A. Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE and western blotting. J Gen Microbiol. 1988;134:2171–2178. doi: 10.1099/00221287-134-8-2171. [DOI] [PubMed] [Google Scholar]
- Rhodius VA, Busby SJ. Positive activation of gene expression. Curr Opin Microbiol. 1998;1:152–159. doi: 10.1016/s1369-5274(98)80005-2. [DOI] [PubMed] [Google Scholar]
- Rotter C, Muhlbacher S, Salamon D, Schmitt R, Scharf B. Rem, a new transcriptional activator of motility and chemotaxis in Sinorhizobium meliloti. J Bacteriol. 2006;188:6932–6942. doi: 10.1128/JB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder I, Wolin CD, Cavicchioli R, Gunsalus RP. Phosphorylation and dephosphorylation of the NarQ, NarX, and NarL proteins of the nitrate-dependent two-component regulatory system of Escherichia coli. J Bacteriol. 1994;176:4985–4992. doi: 10.1128/jb.176.16.4985-4992.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A, Higgins DE. The MogR transcriptional repressor regulates non-hierarchal expression of flagellar motility genes and virulence in Listeria monocytogenes. PLoS Pathog. 2006;2:e30. doi: 10.1371/journal.ppat.0020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A, Higgins DE, Panne D. Recognition of AT-rich DNA binding sites by the MogR repressor. Structure. 2009;17:769–777. doi: 10.1016/j.str.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A, Kamp HD, Grundling A, Higgins DE. A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Dev. 2006;20:3283–3295. doi: 10.1101/gad.1492606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara K, Ogura M. Promoter selectivity of the Bacillus subtilis response regulator DegU, a positive regulator of the fla/che operon and sacB. BMC Microbiol. 2008;8:8. doi: 10.1186/1471-2180-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhamme DT, Kiley TB, Stanley-Wall NR. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol Microbiol. 2007;65:554–568. doi: 10.1111/j.1365-2958.2007.05810.x. [DOI] [PubMed] [Google Scholar]
- Wosten MM, Wagenaar JA, van Putten JP. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J Biol Chem. 2004;279:16214–16222. doi: 10.1074/jbc.M400357200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.