Abstract
Vitamin D signaling through its nuclear vitamin D receptor has emerged as a key regulator of innate immunity in humans. Here we show that hormonal vitamin D, 1,25-dihydroxyvitamin D3, robustly stimulates expression of pattern recognition receptor NOD2/CARD15/IBD1 gene and protein in primary human monocytic and epithelial cells. The vitamin D receptor signals through distal enhancers in the NOD2 gene, whose function was validated by chromatin immunoprecipitation and chromatin conformation capture assays. A key downstream signaling consequence of NOD2 activation by agonist muramyl dipeptide is stimulation of NF-κB transcription factor function, which induces expression of the gene encoding antimicrobial peptide defensin β2 (DEFB2/HBD2). Pretreatment with 1,25-dihydroxyvitamin D3 synergistically induced NF-κB function and expression of genes encoding DEFB2/HBD2 and antimicrobial peptide cathelicidin in the presence of muramyl dipeptide. Importantly, this synergistic response was also seen in macrophages from a donor wild type for NOD2 but was absent in macrophages from patients with Crohn disease homozygous for non-functional NOD2 variants. These studies provide strong molecular links between vitamin D deficiency and the genetics of Crohn disease, a chronic incurable inflammatory bowel condition, as Crohn's pathogenesis is associated with attenuated NOD2 or DEFB2/HBD2 function.
Keywords: Diseases/Genetic, DNA/Structure, DNA/Transcription, Gene/Regulation, Gene/Transcription, Immunology/Defensins, Immunology/Cellular Response, Immunology/Innate Immunity
Introduction
Vitamin D has a range of physiological activities (1) and has emerged recently as a direct regulator of immune system function in humans (2, 3). It is obtained from limited dietary sources and ultraviolet B-induced conversion of 7-dehydrocholesterol to vitamin D3 in skin (1). Hormonal 1,25-dihydroxyvitamin D3 (1,25D)2 is produced by hepatic generation of 25-hydroxvitamin D3 (25D), the major circulating metabolite, followed by CYP27B1-catalyzed 1α-hydroxylation in kidney and peripheral tissues (1, 4, 5). Notably, activated macrophages and dendritic cells express CYP27B1 (2, 3), which, unlike the renal enzyme, is regulated primarily by immune inputs. 1,25D signals through its cognate VDR nuclear receptor (1, 6), which binds DNA as a heterodimer with retinoid X receptors to vitamin D-response elements (VDREs), direct repeats of PuG(G/T)TCA motifs separated by 3 bp (DR3) (1).
A molecular basis for regulation by vitamin D of innate immunity emerged with observations by ourselves (7) and others (8, 9) that 1,25D induces expression of genes encoding antimicrobial peptides (AMPs), vanguards of responses against microbial pathogens (3, 10). 1,25D modestly induced defensin β2 (DEFB2/HBD2) and strongly stimulated cathelicidin antimicrobial peptide (CAMP) gene expression through consensus VDREs (7). Neither VDRE is conserved in mice, and the element in the CAMP gene is imbedded in a human/primate-specific Alu repeat (8). Notably, human macrophages stimulated through toll-like receptors (TLRs) strongly induced CYP27B1 and VDR expression, rendering cells responsive to circulating levels of 25D (11). This study showed that the magnitude of the downstream induction of CAMP by 25D in TLR-stimulated macrophages was strongly dependent on variations in 25D levels within the physiological range (11).
At higher latitudes, solar ultraviolet B is insufficient for cutaneous vitamin D3 synthesis for periods of several months around winter. This, coupled with vitamin D-poor diets, leads to seasonal variations in circulating 25D levels and widespread vitamin D deficiency (3, 5), which has been linked to certain cancers and to infectious and autoimmune diseases. Clinical and preclinical studies (e.g. Refs. 12 and 13) have suggested that 1,25D deficiency may contribute to the pathogenesis of inflammatory bowel diseases (IBD), including Crohn disease (CD), a chronic incurable inflammatory condition, which is believed to arise from defective innate immune regulation of intestinal bacterial load (12, 14). North-South gradients in rates of CD have been described in Europe and North America (12, 15, 16), although data concerning seasonal variations in CD relapse rates are conflicting (17–19). In addition, VDR gene polymorphisms correlate with susceptibility to CD and to ulcerative colitis (20, 21). Associations between VDR signaling and IBD are also supported by animal models, where vitamin D sufficiency is associated with reduced frequency of onset and improved IBD symptoms in animals with established disease (22, 23).
Here we provide further evidence for the key role of vitamin D signaling in regulating innate immunity in humans and its links to inflammatory bowel disease. We find that 1,25D is a direct and robust inducer of expression of the gene encoding pattern recognition receptor NOD2/CARD15/IBD1 in cells of monocytic and epithelial origin. NOD2/CARD15 detects muramyl dipeptide (MDP), a lysosomal breakdown product of bacterial peptidoglycan common to Gram-minus and Gram-positive bacteria (24, 25). We also find that pretreatment of cells with 1,25D leads to a synergistic induction of genes encoding HBD2 and CAMP upon introduction of MDP in cells expressing functional NOD2 but not in cells from patients with Crohn disease homozygous for an inactivating mutation of the NOD2 gene. These studies provide a key link between vitamin D signaling and the genetics of Crohn disease as attenuated NOD2 or HBD2 function has been linked to the CD pathogenesis (12, 26–28), and show that NOD2 regulation by 1,25D markedly affects the amplitude of downstream AMP gene regulation induced by MDP.
RESULTS AND DISCUSSION
Induction of NOD2 by 1,25D in Human Monocytic and Epithelial Cells
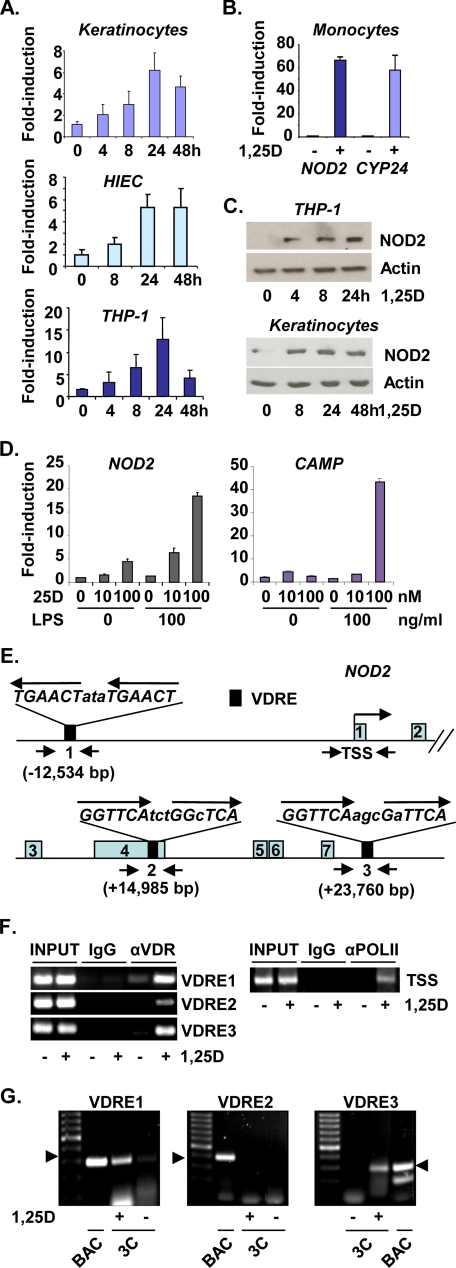
Initially, NOD2 was identified as a potential 1,25D target gene during Affymetrix Hu133A oligonucleotide microarray analyses of 1,25D-treated poorly differentiated human SCC4 head and neck squamous carcinoma head and neck squamous carcinoma cells, where its expression was induced 5.4- and 8.5-fold after 8 and 24 h of treatment, respectively. Regulation of NOD2 was further investigated in primary cultures and lines of human epithelial and monocytic cells. 1,25D robustly induced NOD2 mRNAs in primary human keratinocytes and in immortalized human crypt intestinal epithelial cells (HIEC (29)) (Fig. 1A), as well as in well differentiated human SCC25 head and neck squamous carcinoma cells and in HT29 colon carcinoma cells (supplemental Fig. 1, A and B). 1,25D also strongly stimulated NOD2 expression in differentiated human THP-1 macrophage-like cells (Fig. 1A) and primary human monocytes (Fig. 1B), where fold induction was similar to that of 1,25D target gene CYP24, used as a positive control for 1,25D signaling. Western blotting confirmed that 1,25D treatment induced a corresponding increase in NOD2 protein in THP-1 cells and keratinocytes (Fig. 1C).
FIGURE 1.
Hormonal vitamin D induces expression of CD susceptibility gene NOD2/CARD15/IBD1. A, quantitative PCR analyses are shown of reverse transcribed RNA from primary human keratinocytes, immortalized HIEC cells, and differentiated human THP-1 cells treated with 100 nm 1,25D, as indicated. B, 1,25D induces NOD2 expression in cultures of human monocytes treated with 1,25D (100 nm) for 24 h prior to RNA isolation for quantitative PCR analysis. CYP24 mRNA levels were also monitored as a positive control for 1,25D action. C, Western analysis of NOD2 protein expression or β-actin internal control in primary cultures of human keratinocytes or differentiated THP-1 monocytic cells treated with 100 nm 1,25D, as indicated. D, induction of 1,25D target genes in LPS-primed differentiated THP-1 cells incubated with 25D analyzed by qPCR. Cells were incubated with LPS as indicated for 24 h prior to treatment with 25D for 12 h at the concentrations indicated. Expression of 1,25D target gene CAMP was monitored as a positive control, along with expression of NOD2. Error bars in A, B, and D indicate S.E. E, schematic representation of the human NOD2 gene, with VDREs indicated by black boxes, and exons (gray) are numbered. Putative VDREs were identified as described (38) using the NCBI Build 35 Human May 2004 (hg17) genome assembly (University of California Santa Cruz (UCSC) Genome Browser data base). Regions encompassing putative VDREs or the TSS amplified by PCR in ChIP assays are indicated by inverted arrows. F, ChIP analysis using primary human keratinocytes of 1,25D-dependent binding of the VDR to putative VDREs, along with ChIP analysis of pol II binding to the TSS. Cells were treated with 100 nm 1,25D for 2 h prior to cross-linking for ChIP assay. G, 3C analysis using primary human keratinocytes of 1,25D-dependent association of genomic regions encompassing VDREs 1, 2 and 3 with the NOD2 TSS. No chromatin looping between VDRE2 and the TSS was detected under any conditions. Controls (BAC) are provided, showing that expected PCR products (arrowheads) were generated when primer sets for 3C detection were used for amplification digestion and religation of BAC clones encompassing the NOD2 gene.
Similar to signaling through TLR2/1 (11), exposure of differentiated THP-1 cells to TLR4 ligand LPS strongly induced expression of CYP27B1 (supplemental Fig. 2), thus rendering cells responsive to prohormonal 25D. Incubation with 100 nm 25D, a concentration that corresponds to a vitamin D-replete state (3, 5), strongly stimulated NOD2 expression (Fig. 1D) in LPS-primed THP-1 cells, along with 1,25D target genes CAMP (Fig. 1D) and CYP24 (supplemental Fig. 3), whereas 10 nm 25D, which corresponds to vitamin D deficiency, was substantially less effective (Fig. 1D and supplemental Fig. 3). Note that the fold induction of NOD2 expression seen in the presence of 100 ng/ml LPS and 100 nm 25D (Fig. 1D) was essentially identical to that seen in THP-1 cells treated with 1,25D alone (Fig. 1A), suggesting that it is due essentially entirely to the production of 1,25D. In other experiments, we saw no evidence for a combined effect of 1,25D and LPS on the expression of NOD2 or other 1,25D target genes (data not shown).
Function of Distal VDREs in the NOD2 Gene
To address the mechanism of 1,25D-regulated NOD2 expression, we scanned the human gene for putative VDREs and found three distal sequences: a consensus element at −12.5kb relative to the transcription start site (TSS) and two near consensus sequences at approximately +15 kb in exon 4 and at +24 kb downstream of the gene (Fig. 1E). ChIP assays revealed strong 1,25D-dependent VDR binding to VDREs 1 and 3 and weaker binding to VDRE2, along with strongly 1,25D-dependent association of pol II with the TSS (Fig. 1F), consistent with transcriptional regulation. Function of putative VDREs was further tested by 3C assay (30) (supplemental Fig. 4), which detects loops between distal chromatin sites via formation of diagnostic, ligation-dependent PCR products. 3C assays detected strongly 1,25D-dependent PCR product formation consistent with loops between VDREs 1 or 3 and the TSS (Fig. 1G; 3C), whereas no evidence for looping between VDRE2 and the TSS was found, consistent with its weaker binding of the VDR observed in ChIP assays. In control experiments, all primer sets generated expected PCR products after digestion and religation of BAC clones containing the NOD2 gene (Fig. 1G; BAC). Collectively, these studies reveal that the ligand-bound VDR strongly and directly induces NOD2 gene transcription through VDREs lying at −12.5 and +24 kb.
Synergistic Induction of AMP Gene Expression by 1,25D and MDP
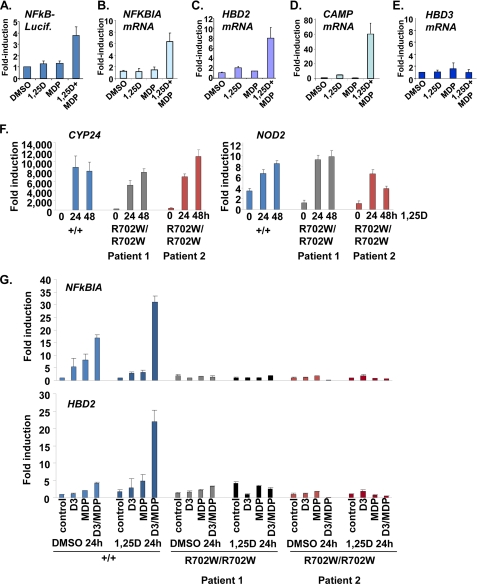
A key downstream consequence of NOD2 activation by MDP is stimulation of NF-κB and induction of HBD2 transcription through promoter-proximal NF-κB-binding sites (33). This signaling pathway was defective in cells expressing the major variant of NOD2 present in a subset of patients with CD (33). We tested the individual and combined effects of 1,25D and MDP on NF-κB function via analysis of NF-κB-sensitive expression of a reporter gene and regulation of endogenous target gene NFKBIA, which encodes NF-κB inhibitor IκBα. Initial experiments were performed in epithelial cells as a number of studies have shown, remarkably, that epithelial-specific NF-κB activity is important for prevention of chronic inflammatory conditions (34).
MDP was introduced by transfection, as described previously (33), into primary cultures of human keratinocytes. Although 1,25D and MDP alone had no or modest effects on NF-κB-driven reporter activity or NFKBIA transcripts, pretreatment with 1,25D followed by MDP markedly enhanced expression of luciferase and NFKBIA (Fig. 2, A and B). Similarly, MDP alone had little effect on HBD2 (Fig. 2C), whereas, as expected (7), 1,25D alone modestly augmented HBD2 mRNA levels. In contrast, 1,25D pretreatment followed by exposure to MDP synergistically induced HBD2 mRNAs (Fig. 2C). We also observed that induction of CAMP by 1,25D was markedly augmented in the presence of MDP (Fig. 2D). In contrast, although HBD3 is part of the same locus as HBD2 (32), its expression was not regulated by 1,25D alone or in combination with MDP (Fig. 2E), consistent with a lack of apparent NF-κB-binding sites within 2.9 kb of the 5′end of the HBD3 gene (32).
FIGURE 2.
Synergistic induction of AMP gene expression by 1,25D and NOD2 ligand MDP. A, synergistic induction of NF-κB-responsive reporter gene expression in keratinocytes by 1,25D and MDP. Cells were transfected with pNF-κB-luciferase (NFkB-Lucif.) and pSG5-β-GAL internal control plasmid, treated with vehicle (−) or 1,25D (100 nm) for 24 h to induce NOD2, and then transfected with ProteoJuice reagent alone or in the presence of MDP prior to analysis of luciferase expression. DMSO, dimethyl sulfoxide. B, RT/qPCR analysis of regulation of endogenous NF-κB target gene NFKBIA by 1,25D and MDP alone or in combination. Cells were treated as in A. C–E, analysis of induction of endogenous HBD2 (C), CAMP (D), or HBD3 (E) transcripts by 1,25D and MDP in primary human keratinocytes. Cells were treated with vehicle (−) or 1,25D (100 nm) for 24 h to induce NOD2. Cells were then transfected with ProteoJuice reagent alone or in the presence of MDP (0.1 μg/ml) prior to RNA extraction, RT, and qPCR. F, similar induction of CYP24 and NOD2 expression by 1,25D in NOD2 wild-type (+/+) macrophages and macrophages homozygous for Crohn's-associated variant (R702W/R702W). Gene expression was analyzed by RT/qPCR in cells treated with 100 nm 1,25D as indicated. G, RT/qPCR analysis of regulation of NFKBIA or HBD2 expression by 1,25D and MDP alone or in combination in wild-type (+/+) or R702W/R702W human macrophages. Cells were treated with 1,25D (100 nm) or MDP alone or in combination for 24 h (DMSO 24h) or pretreated with 1,25D for 24 h (1,25D 24 h) to induce NOD2 expression followed by 24 h of further incubation with 1,25D or MDP, as indicated. Error bars indicate S.E.
Loss of 1,25D-MDP Synergism in Macrophages Lacking Functional NOD2
To confirm the role of NOD2 function in the combined effects of 1,25D and MDP, we compared the regulation of NFKBIA and HBD2 in wild-type human macrophages and those derived from two patients with Crohn disease homozygous for a variant (R702W/R702W) adjacent to the leucine-rich repeats in NOD2 that disrupts MDP signaling (35). Comparable levels of induction by 1,25D of the CYP24 and NOD2 genes were seen in wild-type and R702W/R702W monocytes (Fig. 2F), indicating that vitamin D signaling was functioning similarly in all preparations of cells. Although expression of NOD2 was induced by 1,25D as expected (Fig. 2F), basal expression of the gene was higher in all preparations of cells than in previous isolates of monocytes (e.g. Fig. 1B). Incubation of wild-type monocytes with MDP led to an induction of NFKBIA expression that was augmented by co-treatment with 1,25D, consistent with basal expression of NOD2 (Fig. 2G, top). The combined effects of 1,25D and MDP were enhanced by a 24-h pretreatment with 1,25D followed by further incubation with the two compounds for 24 h, consistent with the induction by 1,25D of NOD2 expression. Similar results were obtained when regulation of HBD2 was analyzed in wild-type macrophages, where incubation with 1,25D and MDP produced a dramatic increase in expression after a 24-h pretreatment (Fig. 2G, bottom). These data are in agreement with the regulation of the NFKBIA and HBD2 genes in keratinocytes seen above. In contrast, combined effects of 1,25D and MDP on HBD2 and NFKB1A were completely absent in R702W/R702W macrophages (Fig. 2G), demonstrating that MDP signaling requires expression of functional NOD2.
Although our present results and those of others (11) show that induction of HBD2 by 1,25D alone is weak or absent in myelomonocytic cells, the strong combined actions of 1,25D and MDP on HBD2 expression suggest that the VDR and NF-κB may cooperate to induce transcription of the gene. In addition, we cannot rule other effects of prolonged 1,25D treatment on signaling through NF-κB that may contribute to the observed synergism.
Physiological and Clinical Implications of 1,25D-regulated NOD2 Expression
The above results (see supplemental Fig. 5 for summary) substantially extend the scope of known effects of vitamin D signaling on innate immunity in humans. They reveal that 1,25D signaling through the VDR is a primary regulator of NOD2 expression and that induction of NOD2 by 1,25D markedly affects the amplitude of downstream AMP gene regulation induced by NOD2 agonist MDP. The results also suggest that vitamin D insufficiency/deficiency would doubly affect HBD2 and CAMP expression by reducing direct induction by 1,25D and by attenuating any synergism with MDP-NOD2 signaling because of reduced NOD2 expression.
There has been some debate as to whether vitamin D deficiency plays a causative role in CD or is merely a consequence of intestinal malabsorption (12, 35). Our observation that 1,25D signaling is a direct inducer of NOD2 expression argues strongly that vitamin D insufficiency/deficiency does play a causative role in the prevalence of CD. The genetics of CD demonstrate that NOD2 insufficiency contributes to development of the disease (26, 27). Importantly, our findings also imply that heterozygotes with inactivating NOD2 mutations might be particularly susceptible to development of CD if concurrently vitamin D-deficient. The data suggest that homozygotes carrying such mutations would be refractory to vitamin D treatment. However, NOD2 is only one of several loci implicated in susceptibility to CD, and NOD2 mutations are absent in some ethnic groups with CD (14), suggesting that vitamin D supplementation may be beneficial to most patients at risk of disease development.
Variations in the HBD2 locus, which is highly polymorphic in the human population, also contribute to CD susceptibility (28). HBD2 copy numbers range from 2 to 10, and the gene is preferentially expressed in colonic epithelia. Notably, a median copy number of four was found in normal individuals or patients with ileal CD, whereas the median copy number in patients with colonic CD was three, and individuals with three or fewer copies had a significantly higher risk of developing colonic CD (28).
The physiological and clinical implications of 1,25D regulation of NOD2 extend beyond CD. Disruption of NOD2 expression in mice led to reduced levels of expression in intestinal Paneth cells of the AMP α-defensin (cryptdin) during infection with Listeria monocytogenes (36). These observations, coupled with our findings of synergistic induction of AMP expression by 1,25D/MDP, suggest that 1,25D-stimulated NOD2 function may contribute to host resistance to a variety of infectious challenges through the generation of AMPs. CAMP protein is expressed in 1,25D-stimulated macrophages and co-localizes with phagosomal mycobacteria (11). Moreover, knockdown of CAMP diminished suppression of phagosomal Mycobacterium tuberculosis replication by 1,25D (37). In conclusion, our findings suggest that regulation of NOD2 expression by 1,25D and its marked effects on the amplitude of downstream AMP production contribute substantially to 1,25D-mediated innate antibacterial responses, as well as providing a strong molecular basis for the contribution of vitamin D deficiency to the pathogenesis of CD.
Supplementary Material
Acknowledgments
We are grateful to Dr. J. F. Beaulieu, Université de Sherbrooke, for kindly providing HIEC cells, to Dr. Josée Dostie (Department of Biochemistry, McGill University) for advice with 3C assays, and to Marianna Orlova (McGill University) for assistance with NOD2 genotyping.
This work was supported by a nutrition grant from McGill University (to J. H. W. and M. B.).

The on-line version of this article (available at http://www.jbc.org) contains “Experimental Procedures” and supplemental Figs. 1–5.
- 1,25D
- 1,25-dihydroxyvitamin D3
- 25D
- 25-hydroxyvitamin D3
- AMP
- antimicrobial peptides
- CAMP
- cathelicidin antimicrobial peptide
- CARD15
- caspase-recruitment domain-containing protein 15
- CD
- Crohn disease
- HBD2
- human defensin β2
- HBD3
- human defensin β3
- HIEC
- human crypt intestinal epithelial cells
- IBD
- inflammatory bowel disease
- NOD2
- nucleotide-binding oligomerization domain 2
- MDP
- muramyl dipeptide
- pol II
- RNA polymerase II
- TLR
- toll-like receptor
- TSS
- transcription start site
- VDR
- vitamin D receptor
- VDRE
- vitamin D-response element
- ChIP
- chromatin immunoprecipitation
- 3C
- chromosome conformation capture
- BAC
- bacterial artificial chromosome
- qPCR
- quantitative PCR
- RT
- reverse transcription
- LPS
- lipopolysaccharide.
REFERENCES
- 1.Lin R., White J. H. (2004) BioEssays 26, 21–28 [DOI] [PubMed] [Google Scholar]
- 2.Liu P. T., Modlin R. L. (2008) Curr. Opin. Immunol. 20, 371–376 [DOI] [PubMed] [Google Scholar]
- 3.White J. H. (2008) Infect. Immun. 76, 3837–3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prosser D. E., Jones G. (2004) Trends. Biochem. Sci. 29, 664–673 [DOI] [PubMed] [Google Scholar]
- 5.Holick M. F. (2007) N. Engl. J. Med. 357, 266–281 [DOI] [PubMed] [Google Scholar]
- 6.Rochel N., Wurtz J. M., Mitschler A., Klaholz B., Moras D. (2000) Mol. Cell. 5, 173–179 [DOI] [PubMed] [Google Scholar]
- 7.Wang T. T., Nestel F. P., Bourdeau V., Nagai Y., Wang Q., Liao J., Tavera-Mendoza L., Lin R., Hanrahan J. W., Mader S., White J. H. (2004) J. Immunol. 173, 2909–2912 [DOI] [PubMed] [Google Scholar]
- 8.Gombart A. F., Borregaard N., Koeffler H. P. (2005) FASEB J. 19, 1067–1077 [DOI] [PubMed] [Google Scholar]
- 9.Weber G., Heilborn J. D., Chamorro Jimenez C. I., Hammarsjo A., Törmä H., Stahle M. (2005) J. Invest. Dermatol. 124, 1080–1082 [DOI] [PubMed] [Google Scholar]
- 10.Jenssen H., Hamill P., Hancock R. E. (2006) Clin. Microbiol. Rev. 19, 491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu P. T., Stenger S., Li H., Wenzel L., Tan B. H., Krutzik S. R., Ochoa M. T., Schauber J., Wu K., Meinken C., Kamen D. L., Wagner M., Bals R., Steinmeyer A., Zügel U., Gallo R. L., Eisenberg D., Hewison M., Hollis B. W., Adams J. S., Bloom B. R., Modlin R. L. (2006) Science. 311, 1770–1773 [DOI] [PubMed] [Google Scholar]
- 12.Lim W. C., Hanauer S. B., Li Y. C. (2005) Nat. Clin. Pract. Gastroenterol. Hepatol. 2, 308–315 [DOI] [PubMed] [Google Scholar]
- 13.Liu N., Nguyen L., Chun R. F., Lagishetty V., Ren S., Wu S., Hollis B., DeLuca H. F., Adams J. S., Hewison M. (2008) Endocrinology 149, 4799–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho J. H. (2008) Nat. Rev. Immunol. 8, 458–466 [DOI] [PubMed] [Google Scholar]
- 15.Loftus E. V., Jr., Sandborn W. J. (2002) Gastroenterol. Clin. North Am. 31, 1–20 [DOI] [PubMed] [Google Scholar]
- 16.Loftus E. V., Jr. (2004) Gastroenterology 126, 1504–1517 [DOI] [PubMed] [Google Scholar]
- 17.Zeng L., Anderson F. H. (1996) Scand. J. Gastroenterol. 31, 79–82 [DOI] [PubMed] [Google Scholar]
- 18.Lewis J. D., Aberra F. N., Lichtenstein G. R., Bilker W. B., Brensinger C., Strom B. L. (2004) Gastroenterology 126, 665–673 [DOI] [PubMed] [Google Scholar]
- 19.Aratari A., Papi C., Galletti B., Angelucci E., Viscido A., D'Ovidio V., Ciaco A., Abdullahi M., Caprilli R. (2006). Dig. Liver Dis. 38, 319–323 [DOI] [PubMed] [Google Scholar]
- 20.Simmons J. D., Mullighan C., Welsh K. I., Jewell D. P. (2000) Gut 47, 211–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin K., Radlmayr M., Borchers R., Heinzlmann M., Folwaczny C. (2002) Digestion. 66, 121–126 [DOI] [PubMed] [Google Scholar]
- 22.Cantorna M. T., Munsick C., Bemiss C., Mahon B. D. (2000) J. Nutr. 130, 2648–2652 [DOI] [PubMed] [Google Scholar]
- 23.Froicu M., Weaver V., Wynn T. A., McDowell M. A., Welsh J. E., Cantorna M. T. (2003) Mol. Endocrinol. 17, 2386–2392 [DOI] [PubMed] [Google Scholar]
- 24.Girardin S. E., Boneca I. G., Viala J., Chamaillard M., Labigne A., Thomas G., Philpott D. J., Sansonetti P. J. (2003) J. Biol. Chem. 278, 8869–8872 [DOI] [PubMed] [Google Scholar]
- 25.Inohara N., Ogura Y., Fontalba A., Gutierrez O., Pons F., Crespo J., Fukase K., Inamura S., Kusumoto S., Hashimoto M., Foster S. J., Moran A. P., Fernandez-Luna J. L., Nuñez G. (2003) J. Biol. Chem. 278, 5509–5512 [DOI] [PubMed] [Google Scholar]
- 26.Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cézard J. P., Belaiche J., Almer S., Tysk C., O'Morain C. A., Gassull M., Binder V., Finkel Y., Cortot A., Modigliani R., Laurent-Puig P., Gower-Rousseau C., Macry J., Colombel J. F., Sahbatou M., Thomas G. (2001) Nature. 411, 599–603 [DOI] [PubMed] [Google Scholar]
- 27.Ogura Y., Bonen D. K., Inohara N., Nicolae D. L., Chen F. F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R. H., Achkar J. P., Brant S. R., Bayless T. M., Kirschner B. S., Hanauer S. B., Nuñez G., Cho J. H. (2001) Nature 411, 603–606 [DOI] [PubMed] [Google Scholar]
- 28.Fellermann K., Stange D. E., Schaeffeler E., Schmalzl H., Wehkamp J., Bevins C. L., Reinisch W., Teml A., Schwab M., Lichter P., Radlwimmer B., Stange E. F. (2006) Am. J. Hum. Genet. 79, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy E., Beaulieu J. F., Delvin E., Seidman E., Yotov W., Basque J. R., Ménard D. (2000). J. Lipid Res. 41, 12–22 [PubMed] [Google Scholar]
- 30.Dostie J., Dekker J. (2007) Nat. Protoc. 2, 988–1002 [DOI] [PubMed] [Google Scholar]
- 31.Cantorna M. T., Mahon B. D. (2004) Exp. Biol. Med. (Maywood) 229, 1136–1142 [DOI] [PubMed] [Google Scholar]
- 32.Jia H. P., Schutte B. C., Schudy A., Linzmeier R., Guthmiller J. M., Johnson G. K., Tack B. F., Mitros J. P., Rosenthal A., Ganz T., McCray P. B., Jr. (2001). Gene 263, 211–218 [DOI] [PubMed] [Google Scholar]
- 33.Voss E., Wehkamp J., Wehkamp K., Stange E. F., Schröder J. M., Harder J. (2006) J. Biol. Chem. 281, 2005–2011 [DOI] [PubMed] [Google Scholar]
- 34.Pasparakis M. (2009) Nat. Rev. Immunol. 9, 778–788 [DOI] [PubMed] [Google Scholar]
- 35.van Heel D. A., Ghosh S., Butler M., Hunt K. A., Lundberg A. M., Ahmad T., McGovern D. P., Onnie C., Negoro K., Goldthorpe S., Foxwell B. M., Mathew C. G., Forbes A., Jewell D. P., Playford R. J. (2005) Lancet 365, 1794–1796 [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi K. S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nuñez G., Flavell R. A. (2005) Science 307, 731–734 [DOI] [PubMed] [Google Scholar]
- 37.Liu P. T., Stenger S., Tang D. H., Modlin R. L. (2007) J. Immunol. 179, 2060–2063 [DOI] [PubMed] [Google Scholar]
- 38.Wang T. T., Tavera-Mendoza L. E., Laperriere D., Libby E., MacLeod N. B., Nagai Y., Bourdeau V., Konstorum A., Lallemant B., Zhang R., Mader S., White J. H. (2005) Mol. Endocrinol. 19, 2685–2695 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.