Abstract
We describe a novel approach to genetic association analyses with proteins sub-divided into biologically relevant smaller sequence features (SFs), and their variant types (VTs). SFVT analyses are particularly informative for study of highly polymorphic proteins such as the human leukocyte antigen (HLA), given the nature of its genetic variation: the high level of polymorphism, the pattern of amino acid variability, and that most HLA variation occurs at functionally important sites, as well as its known role in organ transplant rejection, autoimmune disease development and response to infection. Further, combinations of variable amino acid sites shared by several HLA alleles (shared epitopes) are most likely better descriptors of the actual causative genetic variants. In a cohort of systemic sclerosis patients/controls, SFVT analysis shows that a combination of SFs implicating specific amino acid residues in peptide binding pockets 4 and 7 of HLA-DRB1 explains much of the molecular determinant of risk.
INTRODUCTION
Proteins of the major histocompatibility complex (MHC; HLA in humans) participate in wide-ranging immunological processes. The class I (HLA-A, -B and -C) and class II (HLA-DR, -DQ and -DP) molecules are widely expressed on hematopoietic and non-hematopoietic cell surfaces, and function to bind short peptides in such a way that the combination of peptide and MHC are recognized by clonotypic T-cell receptors, resulting in T-cell activation. Class I molecules also function as the ligands for natural killer (NK) receptors.
MHC molecules are extremely polymorphic. The extensive polymorphism of HLA sequences reflects the need for the presentation of diverse repertoires of peptides for effective immune surveillance. The IMGT/HLA Database (1) (www.ebi.ac.uk/imgt/hla/) describes over 2000 class I and 1000 class II alleles. The majority of the protein sequence variation occurs within discrete areas that are involved in peptide binding and T-cell receptor interaction. Other variations may affect interactions with the accessory proteins CD4 and CD8, or NK receptors.
This allelic variation in the ability of different MHC molecules to bind peptides and activate effector cells in the immune system underlies their association with infection, autoimmune disease, drug sensitivity and tissue transplantation success. In model systems, the peptide epitopes derived from infectious agents such as human immunodeficiency virus, Epstein-Barr virus and others have been elucidated, as have the specific MHC residues involved in their binding/presentation (2–5). In the case of autoimmune diseases, while many allelic associations are clear—e.g. HLA-B*2705/2/4/7 and ankylosing spondylitis (6,7), HLA-DRB1*0401/*0404/*0405 and rheumatoid arthritis (8,9), HLA-DRB1*0301 HLA-DQA1*0501 DQB1*0201 and DRB1*0401/2/4/5 DQA1*0301 HLA-DQB1*0302 and type 1 diabetes (10,11)—the actual antigenic peptides responsible for these associations are not. However, knowing the nature of the critical MHC amino acid residues involved can allow reasonable predictions about peptide epitopes. Such predictions are important for the design of novel vaccines and the understanding of autoimmunity.
Typically, the significant association of a given normal or pathologic immune response with one or more HLA alleles or haplotypes is based on statistical analysis followed by a manual inspection of linear sequence alignments with the goal of identification of those amino acid residues that occur more commonly in individuals with the given response. More specific analytic and computational approaches have been developed to efficiently identify combinations of amino acids that may be causative in differential disease risk (12–14). These approaches, however, do not explicitly take into account biological information about the MHC molecule under study.
Here we describe a novel method for the analysis of MHC/disease associations that additionally incorporates structural and functional information about the HLA molecules (antigenic peptide binding, TcR binding, etc.) to help illuminate the biological nature of disease associations based on variations in these functional ‘sequence features’ to augment allele-based association analyses. Sequence features may be defined based on purely structural, e.g. ‘α-helical segment 1’ or functional characteristics, e.g. ‘peptide binding’, or a combination of both. Sequence features can be large (e.g. the entire HLA-DRB1 polypeptide) or small (e.g. the loop between beta-strands 1 and 2 of HLA-DRB1), overlapping and non-contiguous (e.g. the peptide antigen binding pocket 7 of HLA-DRB1). There are no restrictions on what protein sub-region can be labeled as a sequence feature. Variation of each sequence feature is then based on the known primary sequences of all alleles of a given HLA molecule. Since this sequence variation can be seen in multiple alleles, we have termed this the ‘variant type’ for a given sequence feature. In genetic terms, the sequence feature variant type (SFVT) can be thought of as an ‘allele’ comprising a haplotype of particular amino acids within a single protein. A single allele of a particular HLA molecule can then be represented as an SFVT feature vector in which the number of dimensions corresponds to the number of sequence features defined for the locus. The resulting association studies can be automated to produce information that is (i) based on experimentally determined structure–function relationships as well as allele and individual amino acid level variation and (ii) statistically informative due to the opportunity to combine groups of individuals rather than separate them by HLA allele.
We have applied this SFVT analysis, to a cohort of patients with systemic sclerosis (scleroderma, SSc). SSc is an autoimmune disorder characterized by organ fibrosis (skin, lungs, heart, kidneys), vasculopathy and the production of autoantibodies to nuclear antigens such as centromeric proteins, topoisomerase I, RNA polymerase III and others. Over the past 20 years, several groups have shown associations between particular HLA class II alleles and the presence of disease (15–23). These include HLA-DR3, -DR11 and -DR7. Strong associations have also been seen when subsets of SSc patients are analyzed. For example, expression of anti-centromere antibody is associated with DQB1*0501 and other DQB1 alleles that lack leucine at position 26 (24,25). Anti-topoisomerase is associated with the DR-DQ haplotype DRB1*1104 DQB1*0301 (24,26,27), and anti-U3-RNP with the DRB1*1302 DQB1*0604 haplotype (28).
Using the SFVT approach, we have been able to confirm and extend the previous association between SSc and HLA-DRB1 at the allele level. Moreover, sequence features corresponding to antigenic peptide binding pockets that predispose to disease risk and protection were delineated. These data provide evidence that this novel method of analyzing MHC association data can identify the most likely molecular determinants of disease association for future functional study.
RESULTS
Definition of human leukocyte antigen protein sequence features and variant types
Sequence features for all classical HLA class I and II proteins were defined at the amino acid level. The procedure used to define sequence features is described in detail in the Materials and Methods; representative sequence features for HLA-DRB1 are shown in Table 1.
Table 1.
Representative sequence feature types for HLA-DRB1
| Sequence feature ID | Sequence feature name | Feature type | Sequence feature definition (amino acid position) | Sequence feature length (amino acid) | Number of variant typesa |
|---|---|---|---|---|---|
| Hsa_HLA-DRB1_SF1 | Hsa_HLA-DRB1_allele | Standard allele designation | NA | NA | 497 |
| Hsa_HLA-DRB1_SF4 | Hsa_HLA-DRB1_mature protein | Structural—Complete protein | 1 … 237 | 237 | 52 |
| Hsa_HLA-DRB1_SF5 | Hsa_HLA-DRB1_beta 1 domain | Structural—Domain | 1 … 95 | 95 | 69 |
| Hsa_HLA-DRB1_SF12 | Hsa_HLA-DRB1_loop between beta-strands 1 and 2 | Structural—Secondary structure motif | 19, 20, 21, 22 | 4 | 5 |
| Hsa_HLA-DRB1_SF13 | Hsa_HLA-DRB1_beta-strand 2 | Structural—Secondary structure motif | 23 … 32 | 10 | 28 |
| Hsa_HLA-DRB1_SF21 | Hsa_HLA-DRB1_alpha-helix 2 | Structural—Secondary structure motif | 65 … 72 | 8 | 29 |
| Hsa_HLA-DRB1_SF128 | Hsa_HLA-DRB1_T-cell receptor binding | Functional | 60, 64, 65, 66, 67, 69, 70, 71, 73, 76, 77, 78, 80, 81, 82, 84, 85 | 17 | 81 |
| Hsa_HLA-DRB1_SF127 | Hsa_HLA-DRB1_peptide antigen binding | Functional | 9, 11, 13, 26, 28, 30, 37, 47, 56, 57, 60, 61, 67, 70, 71, 74, 77, 78, 81, 82, 85, 86, 89, 90 | 24 | 351 |
| Hsa_HLA-DRB1_SF137 | Hsa_HLA-DRB1_peptide antigen binding pocket 7 | Functional | 28, 30, 47, 61, 67, 71 | 6 | 53 |
| Hsa_HLA-DRB1_SF163 | Hsa_HLA-DRB1_alpha-helix 2_peptide binding | Structural_Functional combination | 67, 70, 71 | 3 | 21 |
| Hsa_HLA-DRB1_SF164 | Hsa_HLA-DRB1_alpha-helix 2_T-cell receptor binding | Structural_Functional combination | 65, 66, 67, 69, 70, 71 | 6 | 24 |
| Hsa_HLA-DRB1_SF152 | Hsa_HLA-DRB1_beta-strand 2_peptide antigen binding pocket | Structural_Functional combination | 28, 30 | 2 | 9 |
| Hsa_HLA-DRB1_SF156 | Hsa_HLA-DRB1_alpha-helix 1_peptide antigen binding pocket | Structural_Functional combination | 61 | 1 | 1 |
| Hsa_HLA-DRB1_SF98 | Hsa_HLA-DRB1_variant position 67b | Sequence alteration—single amino acid variation | 67 | 1 | 3 |
aIn some cases the number of variant types can be larger for smaller sequence features due to incomplete information. For example, for Hsa_HLA-DRB1_SF4 only 52 complete sequences for the mature protein are available, whereas virtually all alleles have complete sequence for the peptide antigen binding region (Hsa_HLA_DRB1_SF127).
bSynonymous with many other Sequence Features, e.g. Hsa_HLA-DRB1_alpha-helix 2_variant position 67; Hsa_HLA-DRB1_beta 1 domain_variant position 67; Hsa_HLA-DRB1_peptide antigen binding pocket 7_variant position 67.
Four general categories of sequence features were identified—structural, functional, sequence alteration and combinational. Structural SFs include protein domains (e.g. Hsa_HLA-DRB1_beta 1 domain) and secondary structure motifs (e.g. Hsa_HLA-DRB1_beta-strand 2). Figure 1A shows functional SFs that include protein regions known to provide some important biological function (e.g. Hsa_HLA-DRB1_peptide binding; Hsa_HLA-DRB1_TCR binding). Sequence alteration SFs are simply those single amino acid positions in which sequence variation has been observed (e.g. Hsa_HLA-DRB1_variant position 67). Combinational SFs were constructed by identifying the intersection between SFs of the other types, for example those residues in alpha helix 2 that are involved in peptide binding (Hsa_HLA-DRB1_alpha-helix 2_peptide binding).
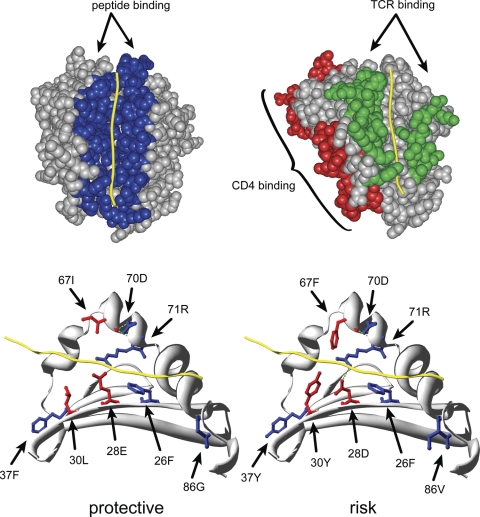
Figure 1.
Three-dimensional models of HLA sequence features. (A) Space-filled representations of HLA-DR1 highlighting functional sequence features. Yellow—peptide antigen; blue—peptide antigen binding (Hsa_HLA-DRA_SF43 and Hsa_HLA-DRB1_SF127); green—T-cell receptor binding (Hsa_HLA-DRA_SF44 and Hsa_HLA-DRB1_SF128); red—putative CD4 receptor binding (Hsa_HLA-DRB1_SF131). See Supplementary Material, Table S1 for a listing of the amino acids comprising TCR, CD4 and CD8 contact sequence features highlighted here. (B) Ribbon diagram of the HLA-DR1 β-chain highlighting the composite sequence feature identified in Table 5. The DR1 α-chain is not depicted in order to more clearly visualize the residues of the peptide-binding groove. Yellow—peptide antigen; red—residues of peptide binding pocket 7 that differ between the protective and risk composite sequence feature; green—predicted H-bonds.
Over 2000 unique protein sequence features for the 10 classical HLA class I and II loci were defined (Table 2). A complete list of all currently defined sequence features can be found in Supplementary Material, Table S1. This list is not intended to be static. As new functions are discovered for HLA proteins, new sequence features can be defined and appended to the list without affecting the identity of those previously defined.
Table 2.
Summary of sequence features of HLA class I and II
| HLA locus | Structural | Functional | Single amino acid polymorphism | Unique structural_ functional | Total unique SFs |
|---|---|---|---|---|---|
| HLA-A | 56 | 17 | 163 | 75 | 311 |
| HLA-B | 57 | 12 | 169 | 44 | 282 |
| HLA-C | 58 | 15 | 128 | 53 | 254 |
| HLA-DRB1 | 43 | 19 | 83 | 36 | 181 |
| HLA-DRB3 | 42 | 15 | 25 | 45 | 127 |
| HLA-DRB4 | 4 | 1 | 6 | 0 | 11 |
| HLA-DRB5 | 41 | 18 | 17 | 42 | 118 |
| HLA-DRA | 41 | 19 | 1 | 46 | 107 |
| HLA-DQA1 | 40 | 22 | 52 | 34 | 148 |
| HLA-DQB1 | 45 | 22 | 64 | 33 | 164 |
| HLA-DPA1 | 9 | nd | 19 | nd | 28 |
| HLA-DPB1 | 9 | nd | 35 | nd | 44 |
nd, not done.
For each HLA locus, one common and well-documented allele was selected as a reference, and the specific amino acid residues found at each position in a given sequence feature captured as the definition of variant type #1 (VT1). For example, for HLA-DRB1 the DRB1*0101 allele was chosen as the reference. The amino acid sequence for the ‘Hsa_HLA-DRB1_beta-strand 2_peptide antigen binding’ sequence feature in HLA-DRB1*0101 allele is 26L_28E_30C, defining Hsa_HLA-DRB1_SF153_VT1 (Table 3). Since HLA-DRB1*0102 and HLA-DRB1*0103 have the same amino acid sequences at these positions, they are also VT1 for SF153. However, HLA-DRB1*0113 has different amino acids at positions 26 and 30 and thus defined the second variant type Hsa_HLA-DRB1_SF153_VT2 as 26F_28E_30L and so on. Using this strategy, detailed comparisons of sequence similarity can be described that are not apparent using the standard allele nomenclature. For example, while DRB1*0301 and *0304 share the same amino acid sequence, and thus are of the same variant type (VT3), for SF153, the *0302 and *0303 alleles are distinct (VT4); *0307 defined yet another variant type (VT5) for SF153; *0701 and *0703 are related to *0113 through SF153_VT2 sequence identity. The sequence relationships between all known HLA alleles are now explicitly described for each sub-region of the encoded proteins. Since each SF is treated individually, a comprehensive assessment of sequence similarity is captured when the variant types are compared. Finally, the approach also reduces the complexity seen when each allele is treated as an independent entity. For example, in the case of HLA-DRB1, the ∼700 known alleles are collapsed into just 11 distinct variant types for SF153, in theory leading to increased statistical power in disease association studies.
Table 3.
Variant types for sequence feature 153, ‘Hsa_HLA-DRB1_beta-strand 2_peptide antigen binding’ sequence feature for selected HLA-DRB1 alleles
| Allele | 26a | 28 | 30 | Motif | Variant typeb | Variant type definition |
|---|---|---|---|---|---|---|
| DRB1*0101 | L | E | C | LEC | Type 1 | 26L_28E_30C |
| DRB1*0102 | LEC | Type 1 | 26L_28E_30C | |||
| DRB1*0103 | LEC | Type 1 | 26L_28E_30C | |||
| DRB1*0113 | F | L | FEL | Type 2 | 26F_28E_30L | |
| DRB1*0301 | Y | D | Y | YDY | Type 3 | 26Y_28D_30Y |
| DRB1*0302 | F | Y | FEY | Type 4 | 26F_28E_30Y | |
| DRB1*0303 | F | Y | FEY | Type 4 | 26F_28E_30Y | |
| DRB1*0304 | Y | D | Y | YDY | Type 3 | 26Y_28D_30Y |
| DRB1*0307 | F | D | Y | FDY | Type 5 | 26F_28D_30Y |
| DRB1*0401 | F | D | Y | FDY | Type 5 | 26F_28D_30Y |
| DRB1*0701 | F | L | FEL | Type 2 | 26F_28E_30L | |
| DRB1*0703 | F | L | FEL | Type 2 | 26F_28E_30L | |
| DRB1*0709 | F | F | FEF | Type 6 | 26F_28E_30F | |
| DRB1*0801 | F | D | Y | FDY | Type 5 | 26F_28D_30Y |
| DRB1*0832 | H | LEH | Type 7 | 26L_28E_30H | ||
| DRB1*1101 | F | D | Y | FDY | Type 5 | 26F_28D_30Y |
| DRB1*1104 | F | D | Y | FDY | Type 5 | 26F_28D_30Y |
| DRB1*1159 | Y | D | Y | YDY | Type 3 | 26Y_28D_30Y |
| DRB1*1201 | H | LEH | Type 7 | 26L_28E_30H | ||
| DRB1*1202 | H | LEH | Type 7 | 26L_28E_30H | ||
| DRB1*1301 | F | D | Y | FDY | Type 5 | 26F_28D_30Y |
| DRB1*1315 | F | Y | FEY | Type 4 | 26F_28E_30Y | |
| DRB1*1327 | Y | D | Y | YDY | Type 3 | 26Y_28D_30Y |
| DRB1*1402 | F | Y | FEY | Type 4 | 26F_28E_30Y | |
| DRB1*1403 | F | Y | FEY | Type 4 | 26F_28E_30Y | |
| DRB1*1501 | F | D | Y | FDY | Type 5 | 26F_28D_30Y |
| DRB1*1601 | F | D | Y | FDY | Type 5 | 26F_28D_30Y |
aPolymorphic amino acid residues 26, 28 and 30 in the peptide binding region of beta strand 2.
bSeven of the 11 variant types for this sequence feature are shown. The full name for each variant type is: Hsa_HLA-DRB1_SF153_VTn, where n is the type number assigned.
The complete set of SF and SFVT definitions is being made available through three public database resources that are committed to their maintenance and consistency (www.immport.org, www.ebi.ac.uk/imgt/hla/, www.ncbi.nlm.nih.gov/gv/mhc/). One of the authors (S.G.E.M.) maintains the IMGT/MHC database of new HLA alleles as they are defined and will create the relevant variant type vectors during their regular database version release process for distribution to other database resources, including ImmPort and dbMHC, and use by other investigators.
SFVT analysis of systemic sclerosis
In order to assess the utility of the SFVT approach for HLA disease associations, we analyzed a data set of HLA typing information for a cohort of 1300 subjects with systemic sclerosis together with 1000 healthy controls. Four-digit typing data for HLA-DRB1 was used to prepare the complete SFVT feature vector for all 181 DRB1 sequence features. Chi-square analysis was used to determine which sequence features and corresponding variant types demonstrated significant association with the presence or absence of disease (see Materials and Methods for details).
Table 4 shows the 21 SFVTs with the most significant association with the disease. [A complete list of SFVTs with significant association (corrected P < 0.05) can be found in Supplementary Material, Table S2.] In some cases, the amino acid sequences from significantly associated SFVTs were over-represented in the control cohort suggesting that these sequences might have a protective effect (e.g. Hsa_HLA-DRB1_SF155_VT2), with odds ratios <1. In other cases, the specific SFVT was over-represented in the disease cohort suggesting that these sequences increased the risk of developing disease (e.g. Hsa_HLA-DRB1_SF98_VT3), with odds ratios >1. Variant types from sequence features of a wide range of sizes were identified, ranging from the entire polypeptide sequence to single amino acids. The comprehensive SFVT analysis necessarily produces some overlapping results. For example, Hsa_HLA-DRB1_SF163, SF164, SF165 and SF21 all describe different sequence features within the same amino acid stretch (65–72). In this case, the SFs share the same set of variant positions among the alleles typed in the cohort, resulting in identical odds ratios and P-values. The sequence features highlighted are those with the smallest number of common amino acids and can thus be thought of as ‘tagging’ this family of features. Similarly, Hsa_HLA-DRB1_SF161, SF74 and SF15 all include residue 37, which appears to be the sole contributor of disease association for this family of features. Sequence feature Hsa_HLA-DRB1_SF91 describes the single residue 58, which is biallellic (A/E) in this sample, leading to variant types with the same P-value and reciprocal odds ratios.
Table 4.
HLA-DRB1 SFVTs associated with systemic sclerosis
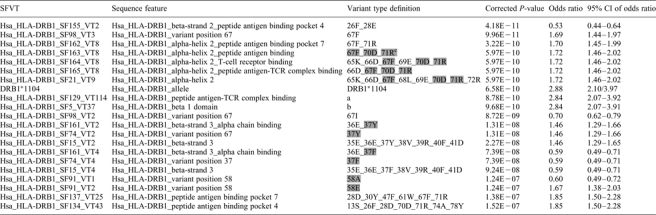 |
a11S_13S_26F_28D_30Y_47F_60Y_61W_64Q_66D_67F_70D_71R_74A_77T_78Y_80R_81H_82N_84G_85V_86V_89F_90T.
b1G_2D_3T_4R_5P_6R_7F_8L_9E_10Y_11S_12T_13S_14E_15C_16H_17F_18F_19N_20G_21T_22E_23R_24V_25R_26F_27L_28D_29R_30Y_31F_32Y_33N_34Q_35E_36E_37Y_38V_39R_40F_41D_42S_43D_44V_45G_46E_47F_48R_49A_50V_51T_52E_53L_54G_55R_56P_57D_58E_59E_60Y_61W_62N_63S_64Q_65K_66D_67F_68L_69E_70D_71R_72R_73A_74A_75V_76D_77T_78Y_79C_80R_81H_82N_83Y_84G_85V_86V_87E_88S_89F_90T_91V_92Q_93R_94R_95V.
cSFVTs with identical odds ratios and P-values are highlighted in gray within groups. The highlighted residues are common to all sequence features in the group.
There is evidence for dependencies in association between amino acid positions that would not be apparent in strategies that investigate single polymorphic amino acid associations. For example, the most significant SFVT found is Hsa_HLA-DRB1_SF155_VT2 consisting of residues 26 and 28 (P-value 4.18 × 10−11). Interestingly, position 26 was found to be a phenylalanine (F) residue both in risk and protective alleles, whereas position 28 was preferentially glutamic acid (E) in protective alleles and aspartic acid in risk alleles. However, SFVTs for position 28 by itself were not as strongly associated with disease, with a more than five log difference in P-value (8.15 × 10−6). This indicates a dependency on position 26 for the strong association found with differences at position 28 (see in what follows).
The entire protein sequence from each HLA-DRB1 allele can also be considered as an SFVT, in essence replicating the traditional method of association testing. Thus, included in the list of significantly associated SFVTs is the allele HLA-DRB1*1104, which was found to be over-represented in the disease cohort with an odds ratio of 2.88 and a P-value of 6.58 × 10−10. This confirms previous studies on this (29) and other SSc cohorts (15,17,18,30).
Fourteen amino acid positions were found repeatedly in significant SFVTs. We generated a set of temporary sequence features (tSF) corresponding to all combinations of these amino acid positions, and performed the SFVT statistical analysis. Two protective tSFVTs (tSF155_VT2, tSF517_VT4) and five risk-associated tSFVT's (tSF1501_VT9, tSF1557_VT10, tSF1592_VT9, tSF1612_VT10, tSF1498_VT12) showed extremely low corrected P-values (<10−11), and were composed of overlapping amino acid groups. For example, tSF1557_VT10 is composed of amino acids 28, 70, 71 and 86 and SF1498_VT12 is composed of 26, 67, 71 and 86. We assembled another tSF composed of the union of seven amino acid residues (26, 28, 30, 37, 67, 70, 71, 86) in the most highly significant sequence features. We examined the amino acid sequence of this tSF (SFVT 13036_14) for all alleles with either statistically significant association with SSc or extreme odds ratios. We found that risk alleles with odds ratios substantially >1 tended to have specific amino acids at each of these positions, which were different from those found in protective alleles with odds ratios substantially <1 (Table 5). For example, protective alleles tended to have an asparagine or phenylalanine at residue 37, whereas the risk alleles tended to have a tyrosine at position 37. Thus, protective alleles are characterized by the sequence 26F_28E_30Y/L_37N/F_67L/I_70Q/D_71K/R_86G, whereas risk alleles have the sequence 26F_28D_30Y_37Y_67F/I_70D_71R_86V.
Table 5.
Correlation between disease risk and composite sequence feature
| Allele/SFVTa | Adjusted P-value | Odds ratio | Number of cases | Number of controls | 26 | 28 | 30c | 37 | 67 | 70 | 71 | 86 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DRB1*1104 | 6.58E−10 | 2.88 | 179 | 50 | 26F | 28D | 30Y | 37Y | 67F | 70D | 71R | 86V |
| DRB1*0402 | 1.00E+00 | 2.13 | 33 | 12 | 26F | 28D | 30Y | 37Y | 67I | 70D | 71E | 86V |
| DRB1*0801 | 3.73E−01 | 1.83 | 66 | 28 | 26F | 28D | 30Y | 37Y | 67F | 70D | 71R | 86G |
| DRB1*0804 | 1.00E+00 | 1.61 | 54 | 26 | 26F | 28D | 30Y | 37Y | 67F | 70D | 71R | 86V |
| 13036_14b | 1.70E−10 | 2.48 | 235 | 76 | 26F | 28D | 30Y | 37Y | 67F | 70D | 71R | 88V |
| Consensus amino acids associated with risk of SSc | 26F | 28D | 30Y | 37Y | 67F | 70D | 71R | 86V | ||||
| DRB1*0701 | 2.68E−05 | 0.59 | 183 | 226 | 26F | 28E | 30L | 37F | 67I | 70D | 71R | 86G |
| DRB1*1402 | 4.26E−01 | 0.40 | 12 | 23 | 26F | 28E | 30Y | 37N | 67L | 70Q | 71R | 86G |
| DRB1*0302 | 6.40E−03 | 0.34 | 17 | 38 | 26F | 28E | 30Y | 37N | 67L | 70Q | 71K | 86G |
| Consensus amino acids associated with protection from SSc | 26F | 28E | 86G | |||||||||
aAlleles with an adjusted SFVT P-value >0.05; or odds ratio >1.5 or <0.67 and frequency >1%.
bComposite SFVT 13036_14 is found in HLA_DRB1 alleles 1104, 0804 and 0806.
cResidue 30 is not independently associated with disease after conditional analysis (see text).
Finally, the SFVT results can be used to identify amino acid residues for a conditional analysis to quantify the risk contributed by each residue. A full conditional analysis is beyond the scope of this paper, but a study of amino acids 26, 28 and 30 has been done (Table 6) as an illustration. The combination of 26 and 28 has a stronger association with disease than either residue individually (26F_28E is the highest ranked SFVT effect); in fact residue 26 by itself is not associated with disease (uncorrected overall P-values: 26 and 28 P < 1.30 × 10−9; 26 alone P = 0.11; 28 alone P < 2.22 × 10−5, Table 6). Residue 30 is individually very strongly associated with disease (uncorrected overall P < 3.58 × 10−5) and found in a number of the highest ranked SFs. Residue 26 has moderate LD with 28 and high LD with 28 and 30, while 28 and 30 have very high LD (31). A series of conditional haplotype method (CHM) analyses show that both 26 and 28 together influence disease risk differentiation compared with each amino acid individually. The association of these two residues with protection from disease was highly significant (26 and 28 condition on 26F, P < 6.47 × 10−11; condition on 28E, P < 6.64 × 10−7). The combination 26F_28E is very significantly protective compared with the remaining homogenous risk set of all other observed haplotypes. CHM analyses also show that residue 30 has little effect on this protective effect (26F_28E_30L versus 26_F28_E_30Y, P = 0.12), nor does it alter the risk effect of any other haplotypes of 26 and 28 (Table 6).
Table 6.
Conditional haplotype method analyses of HLA-DRB1 residues 26, 28 and 30
| Amino acid residue(s)/condition | χ2 | P-valuea |
|---|---|---|
| Single amino acid analysis | ||
| 26 overall | 4.43 | 0.11 |
| 28 overall | 21.43 | 2.22E−05 |
| 30 overall | 28.76 | 3.58E−05 |
| Pairwise amino acid analysis | ||
| 26 and 28 overall | 47.33 | 1.30E−09 |
| 26F_28Eb | 40.87 | 1.63E−09 |
| 26 and 28 overall, minus 26F_28E | 1.10 | 0.78 |
| 28 and 30 overall | 51.68 | 6.74E−09 |
| 28E_30L | 24.50 | 7.42E−07 |
| 28E_30Y | 18.74 | 1.50E−05 |
| 28 and 30 overall, minus 28E_30L/Y | 3.10 | 0.68 |
| 28E_30Y | 21.25 | 4.03E−06 |
| 26 and 30 overall | 31.25 | 5.60E−05 |
| 26F_30L | 24.50 | 7.42E−07 |
| 26 and 30 overall, minus 26F_30L | 4.32 | 0.63 |
| Triple amino acid analysis | ||
| 26, 28 and 30 overall | 52.34 | 1.45E−09 |
| 26F_28E_30L | 24.50 | 7.42E−07 |
| 26F_28E_30Y | 18.74 | 1.50E−05 |
| 26, 28 and 30, minus 26F_28E_30L/Y | 3.77 | 0.71 |
| 26, 28 and 30, condition on 26F_28E | 2.37 | 0.12 |
aThe P-values are uncorrected to allow direct comparisons of effects.
bThe individual contributions to the overall chi-square heterogeneity test considering all amino acid haplotypes are listed, these differ from the corrected P-values in Table 4 which are based on presence versus absence of this variant in patients and controls.
DISCUSSION
The SFVT approach is a novel method for analyzing the association of highly polymorphic proteins with any phenotypic characteristic. It was specifically designed to facilitate the analysis of disease association with HLA gene products in the human MHC, but could be applied to any analogous situation, such as the association of viral pathogen sequence variation and measures of virulence. In developing the approach, we have created a catalog of relevant sequence features and their variant types for all HLA class I and class II proteins, enabling the efficient mapping of information obtained in traditional high-resolution HLA typing studies to known structural and functional elements of the protein. The current compendium is completely extensible, as new sequence features can easily be added without changing the current list, and has the potential to be easily automated so that existing data sets can be re-analyzed quickly.
There are two major advantages of this approach to association analysis. First, it focuses on molecular variants that are likely to have biological importance—structural features of the three-dimensional protein, functional domains and combinations of the two. This information is not explicitly utilized when information is only analyzed at the level of the whole allele. Analysis of individual polymorphic amino acids is similar to the SFVT in the extreme case where each amino acid is considered as an SF. However, this does not take into account any information that puts each residue into structural and functional context. In this regard, Salamon et al., used set covering computation to describe all the possible unique combinations of amino acids in HLA DQA1-DQB1 proteins distinguishing predisposition to Type 1 diabetes (12). While this method is unbiased to the extreme, it is far more computationally intensive than the SFVT method and does not take advantage of biological knowledge relating to protein structure and function.
Second, the SFVT method can increase the statistical power of smaller data sets. In a given case–control cohort, rare disease-associated alleles may not show statistical significance. However, these alleles may harbor causative SVFTs in common with other associated alleles, thus allowing appropriate allele grouping and thereby increasing the sample size and decreasing degrees of freedom, leading to increased statistical power when such cohorts are analyzed using this approach.
The analysis of the SSc data set illustrates the applicability of this method. By including the entire polypeptide sequence as one of the sequence features, this analysis confirms the positive association seen with HLA-DRB1*1104 (previously DR5) in Caucasian and Hispanic SSc patients. This allele contains the amino acid sequence 26F, 28D, 70D and 78Y in the peptide-binding pocket 4. These residues are also seen in HLA-DRB1*0804 which is associated with SSc in African Americans (29). Sequence variation in peptide antigen binding pockets 4 and 7 has been shown to greatly influence the type of peptides presented by a particular DRB1 allele, e.g. myelin basic protein bound by HLA-DRB1*1501 versus DRB1*0401 (32). Intriguingly, there is a significant difference in risk between these two alleles in our analysis of SSc as well.
Only three of the seven alleles in Table 5 showed significant disease associations at the P < 0.05 level, and yet many of them carry an SFVT strongly associated with disease. For example, DRB1*0804 (primarily an African allele) carries the same sequence at this composite sequence feature as DRB1*1104 (a Caucasoid allele) and yet does not demonstrate a significant P-value on its own due to the relatively small sample size. However, when tested as an allele in the African-American SSc patients versus race-matched controls, it was strongly associated with disease, similar to DRB1*1104 in European-derived Caucasians and Hispanics (29). When the association is tested on the basis of the composite sequence feature, then DRB1*0804 and DRB1*1104 individuals are combined, yielding a highly significant result. Thus the SFVT approach identified stronger associations with sub-regions of the protein than with entire alleles although ethnicity also needs to be considered.
Distinguishing between the effects on disease of specific amino acids and SFs is not simple, due to the well-known complex LD patterns of the polymorphic amino acids in the HLA classical genes (31, and G. Thomson et al., in preparation). Conditional analyses can help to disentangle causative effects from those due to LD with a causative agent, but these are complex, as many different comparisons must be made. In this data set, a preliminary conditional analysis shows that while residue 30 appears in many sequence features associated with the disease, its contribution appears to be by virtue of LD with residues 26 and 28, not due to an independent effect. For amino acids with very high LD it may be impossible to distinguish the causative amino acid(s). Cross ethnic studies can aid in such cases if there is a sufficiently different LD pattern, but for some amino acids this is not the case.
These results suggest that not only is HLA-DRB1*1104 associated with disease risk for systemic sclerosis, but also known structural and functional elements, especially beta strands 2 and 3, alpha helix 2 and peptide binding pockets 4 and 7. An inspection of the predicted structure of the high-risk allele shows several bulky aromatic amino acids protruding into the peptide-binding groove of risk alleles that could dramatically influence the repertoire of peptides bound by HLA-DRB1 (Fig. 1B). These data are consistent with the hypothesis that the ability of particular HLA class II alleles to bind and present auto-antigenic peptides determines the extent of T-cell help stimulated, which in turn determines the degree of auto-antibody production. This was suggested by the analysis of 28 Japanese SSc patients who were tested for the presence of anti-topoisomerase antibodies (19). The authors noted that both the presence and amount of anti-topoisomerase was associated with those HLA-DRB1 alleles containing the linear sequence 67FLEDR71. This sequence includes residues that make up peptide binding pockets 4 and 7. In our analysis, three of the four risk alleles contain this sequence. But in addition, our analysis finds a more extensive and select group of localized amino acids that appear to be responsible for the underlying association. The analysis to determine if the same HLA-DRB1 SFVT association is found with auto-antibody presence in our cohort is ongoing. Moreover, there is strong linkage disequilibrium (LD) within HLA, and associations between HLA-DQB1*0301 and related alleles have been seen, particularly in patients who have anti-topoisomerase antibodies (24). We have completed the SFVT assignments for HLA-DQB1 and -DQA1 and are now analyzing them for association with SSc.
In conclusion, we have developed a novel approach for the analysis of HLA genetic associations in which the proteins are broken down into smaller sequence features. These sequence features can be large (e.g. complete protein domains) or small (e.g. single amino acids), they can be based on structural features (e.g. a particular beta strand) or functional features (e.g. a peptide binding region), they can be continuous or discontinuous with respect to the linear sequence, and they can overlap with each other. Once the sequence features have been defined, the corresponding variant types found in the population of HLA alleles are identified. This approach then allows for the independent analysis of disease association with any SFVT regardless of which HLA alleles carry the variation. In order to test the hypothesis that this approach will both provide stronger statistical correlations with disease and highlight the critical functional parts of the HLA molecule, we have used this approach to analyze the correlations of HLA-DRB1 sequence features in a cohort of study participants with systemic sclerosis and show that a sequence feature composed of specific amino acid residues in peptide binding pockets 4 and 7 of HLA-DRB1 (residues 26, 28, 30, 37, 67, 70, 71, 86) best explains the molecular determinant of HLA-DRB1*1104 associated disease risk for systemic sclerosis. Although this study is focused on the analysis of HLA, this approach can be applied in any circumstance in which associations between sequence polymorphisms and phenotypic characteristics are being investigated.
MATERIALS AND METHODS
Definition of human leukocyte antigen protein sequence features
A protein sequence feature is any part of a protein that is expected to have some biological or experimental relevance and is defined by a specified combination of amino acid positions encoded relative to a consensus allele. There are virtually no restrictions to what constitutes a sequence feature. Sequence features may be defined based on purely structural characteristics (e.g. ‘α-helical segment 1’), functional characteristics (e.g. ‘peptide binding’) or a combination of both, and can vary in size from the entire protein to a single amino acid residue. Sequence features can be contiguous or discontinuous in the primary amino acid sequence. Each sequence feature is defined to be unique, though they can overlap with each other either partially or completely. The list of sequence features is expected to evolve; new sequence features can be added to the list as needed at any point in the future without affecting previously defined sequence features.
To define parts of each HLA protein that might correspond to the molecular determinants of disease association, four general categories of sequence features were proposed—structural, functional, sequence alterations and combinational. Structural sequence features are defined purely based on secondary and tertiary structural elements of the folded protein (e.g. beta strand 1). Functional sequence features are defined based on experimental evidence that specific amino acid positions play a defined role in a specific functional property of the protein (e.g. antigenic peptide binding). Sequence alteration sequence features are defined based on variations detected in the human population (e.g. amino acid position 67 in HLA-DRB1). Combinational sequence features combine structural and functional characteristics through their intersection as detailed below (e.g. peptide binding positions in beta strand 1). By convention, protein positions are numbered starting at the N-terminal amino acid of the mature HLA protein; leader sequences are numbered in the opposite direction with negative integers. Each sequence feature is given a descriptive name that reflects the categorical types, but is actually defined as the string of the specific amino acid positions that constitutes the sequence feature. In some cases, sequence features have been generated that correspond to the same amino acid position string (e.g. when a combinational sequence feature generates a single amino acid variant position). In those cases, the descriptive names are considered to be synonyms of the same sequence feature as defined by the amino acid string.
HLA protein structural features were defined based on two sources of information. Secondary structure sequence features were derived from annotations in relevant records of HLA 3D protein structures in the Protein Data Bank (www.pdb.org). Domain definitions, including transmembrane and cytoplasmic regions, were derived from motif information in the relevant UniProt record (www.uniprot.org). The specific HLA reference proteins and database records used are shown in Supplementary Material, Table S3.
HLA protein functional sequence features were defined based on specific functional annotation in relevant IMGT/3Dstructure-DB database records (33) (http://imgt.cines.fr/3Dstructure-DB). These functional features are focused on amino acid contact residues known to be involved in mediating specific non-covalent interactions with antigenic peptides, T-cell receptors, the CD4 and CD8 co-receptors, KIR proteins, beta 2-microglobulin (for class I), class II beta chains (for class II alpha chains) and class II alpha chains (for class II beta chains). For some HLA molecules, the interacting residue positions were verified and augmented based on experimental evidence from the literature. For each classical HLA locus, the interacting residue positions for several HLA proteins are identified from the IMGT/3Dstructure-DB database records and the literature. These functionally relevant residue positions were then mapped to the co-ordinates of the reference HLA allele sequence for the locus (Supplementary Material, Table S3) using the allele alignment data from the IMGT/HLA database. Thus, for each locus the residue positions known to be involved in a specific interaction were identified from several allele sequences and thus would constitute a functional sequence feature. For such functional sequence features, specific Gene Ontology terms were incorporated into the name in order to facilitate interoperability with other data sources utilizing related GO annotations, e.g. ‘peptide antigen binding’: GO:0042605 (22,34).
HLA protein sequence alteration sequence features were defined from the multiple sequence alignments for each HLA locus using all allele sequence contained in Release 2.24.0 (16 January 2009) of the IMGT/HLA database (1). Every position in which more than one amino acid was found in at least one other allele was defined as a single amino acid variation subtype.
HLA combinational sequence features were defined by determining those amino acids that correspond to the overlap between all structural sequence features with all functional sequence features, i.e. the intersection of the positional definitions.
In addition to the sequence feature descriptive name, which reflects the structural and functional characterization, and the definitions based on the amino acid position string, each sequence feature is annotated with a unique identifier that comprises the following information delimited by an underscore: the species name three-letter abbreviation, the locus name, sequence feature label in the form of ‘SF’ followed by a number that uniquely refers to the sequence feature for the given HLA locus. An example of a sequence feature ID would be Hsa_HLA-DRB1_SF155 for the beta-strand 2 positions involved in peptide binding at pocket 4 (the sequence feature descriptive name is Hsa_HLA-DRB1_beta-strand 2_peptide antigen binding pocket 4) defined by amino acid positions 26, 28 of HLA-DRB1. A complete list of all sequence features defined for all classical HLA class I and II proteins is provided in Supplementary Material, Table S1, and can be found at www.immport.org, www.ebi.ac.uk/imgt/hla/ and www.ncbi.nlm.nih.gov/gv/mhc/.
Definition of human leukocyte antigen protein SFVTs
Once the sequence features were defined for each HLA protein, all of the SFVTs were determined. To define the different variant types, multiple sequence alignment data were obtained from the IMGT/HLA Database for each locus. For each sequence feature, the sequences were determined for each allele by combining the amino acid residues from the allele sequence at the amino acid positions defined by each sequence feature. The unique sequences are then identified and are assigned a variant type (VT) ID, which comprises the following information delimited by an underscore: species name three-letter abbreviation, the locus name, sequence feature label and the variant type label in the form of ‘VT’ followed by a number that uniquely refers to the variant type for the sequence feature. VT1 refers to the motif sequence contained in the selected reference allele. An example of a unique SFVT would be Hsa_HLA-DRB1_SF155_VT2 to refer the unique motif sequence type Phe, Glu at positions 26 and 28. Any allele that includes the same motif sequence is assigned the same variant type designation for that sequence feature. As with the sequence features themselves, the numbering order of variant types does not imply any biologically relevant relationships, but rather reflects the historical nature of the HLA nomenclature at a given locus. Alleles whose sequence information is unknown at any of sequence feature amino acid positions are designated as carrying the ‘unknown’ variant type for the sequence feature.
Once all of the variant types for each of the sequence features have been defined for a given allele, the traditional HLA allele can be represented as a SFVT feature vector in n-dimensional space in which n corresponds to the number of sequence features defined for the HLA locus in question (e.g. 181 dimensions for HLA-DRB1). The SFVT feature vector can then be used to test for significant associations with human populations segregated based on interesting phenotypic characteristics using χ2 statistical analysis.
Systemic sclerosis data set
To test the utility of the SFVT approach as a way to rapidly isolate the molecular determinants of disease associations, we utilized molecular HLA typing data from a large cohort of 1300 systemic sclerosis patients and 1000 healthy controls that was assembled at the University of Texas Health Science Center at Houston. The characteristics of this study group (ethnicities, SSc clinical sub-phenotypes, autoantibody subgroups and standard HLA case–control analyses, including χ2 and exact logistic regression with appropriate corrections for multiple testing) have been fully described (29). The ethnic makeup of the population is shown in Supplementary Material, Table S4. For this analysis, all ethnic groups were analyzed together, and no multivariate analysis for clinical sub-types (e.g. ‘diffuse’ versus ‘limited’ systemic sclerosis or presence of particular autoantibodies) was undertaken in this first analysis. The individuals in the cohort were typed for HLA-DRB1, HLA-DQA1 and HLA-DQB1 at the 4-digit level of resolution. The quality control of the data ensured that the 4-digit HLA types of the individuals were defined in congruence with current nomenclature standards (35). All subjects gave informed consent to be in the cohort and UT Houston and UT Southwestern Institutional Review Boards have approved studies with their data.
For each subject in the data set, the allele designation for HLA locus typed is transformed into the SFVT feature vector that was determined for that allele. The resulting data set matrix of subjects and the SFVT feature vectors was used in the association analysis.
SFVT statistical association analysis
The general goal of the SFVT approach is to analyze the SFVT transformed data set so that sequence features and SFVTs exhibiting significant association with disease are identified in a robust, unbiased manner. The association test can be carried out using a number of standard test approaches, e.g. χ2 heterogeneity testing, or a resampling procedure. The approach involves the assessment of a relatively large number of SFVTs. In this study, several strategies were employed to avoid false positive associations, including the generation of pseudo-replicate data sets and initial filtering based on significant sequence features common to both data sets and P-value adjustments to control for multiple hypothesis testing. The following steps have been used for the analysis:
Partition data into two half data sets for parallel analysis
Perform χ2 analysis on 2 × n contingency table for each SF in each half data set
Adjust P-values to control for differences in degrees of freedom between SFs
Select those SF considered to be significant in both half data sets
Perform χ2 analysis on 2 × 2 contingency table of SFVT for all selected SFs in the full data set
Adjust P-values to control for multiple hypothesis testing
Select those SFVT significantly different between cases and controls
Segregate based on odds ratio (OR)
Determine impact of all amino acid residues in all significant SFVTs
Assemble all contributing amino acid residues into a new composite sequence feature
Measure P-value and OR of the new composite SFVTs.
Random sampling of the data set
The systemic sclerosis data set matrix was sampled randomly and partitioned into two subsets that contained equal number of cases and controls (650 SSc patients and 500 controls in each pseudo-replicate data set). These two data sets were then analyzed separately to identify which SFs and SFVTs showed a skewed distribution separately in order to select those SFVTs to be used for a combined final analysis.
Statistical analysis
For every sequence feature, a 2 × n contingency table was constructed in which the occurrence of n SFVTs in systemic sclerosis cases and controls were compared. The chi-square statistic χ2 was calculated with k = n−1 degrees of freedom. The P-value from the distribution then gives the probability of observing the distribution of the variant types of the sequence feature among the cases and controls, or more extreme values. The χ2 test was performed on each sequence feature using SAS®9 (SAS Institute, Inc., Cary, NC). The number of pairwise comparisons possible for n SFVTs is n(n−1)/2, (which equals k*(k + 1)/2). The corrected P-value was obtained by multiplying the P-value from the 2 × n χ2 test by the number of pairwise comparisons for that SFVT. The sequence features whose corrected P-values were less than or equal to 0.01 in both half data sets were selected for further analysis.
For each selected sequence feature, the χ2 analysis was used again in a series of 2 × 2 tables to test if the distribution of a particular variant type in comparison with all other variant types (i.e. Type ‘X’ versus non-Type ‘X’) of the sequence feature is associated with disease. The χ2 statistic was thus calculated for the variant type of the sequence feature on the original complete data set. The odds ratios for each variant type were also calculated. The P-value of each of the variant types of the sequence feature was adjusted to correct for multiple comparisons by multiplying the P-value with the number of multiple comparisons for that SF, which in this case was the number of variant types of the sequence feature minus 1. A complete list of SF, SFVT, P-values and odds ratios from this analysis is provided in Supplementary Material, Table S2.
Determination of amino acid positions contributing to disease association for all SFVTs
From the list of significant SFVTs obtained, all variant positions from alleles in the cohort in two or more significant SFVTs were selected. In addition, those variant positions with unknown sequence information in any subset of alleles in the cohort are filtered out, giving rise to the following set of potentially relevant amino acid positions: 11, 13, 14, 25, 26, 28, 30, 37, 58, 67, 70, 71, 74, 86. tSFs of length 2–14 positions were defined for all possible combinations derived from the selected 14 positions. The SFVTs for the temporary SFs were identified, and χ2 tests were performed on the tSFs and tSFVTs as described earlier. The χ2 P-values of the tSFs were adjusted to control for differences in degrees of freedom between tSFs. The χ2 P-values of the tSFVTs were adjusted to control for multiple hypothesis testing. Among the tSFVTs with odds ratios greater than equal to 1.5 and lesser than equal to 0.67, those that do not completely overlap with any other tSFVTs of similar odds ratios are provided in Supplementary Material, Table S5, along with their P-values and odds ratios.
Determining the actual disease associated amino acids
The CHM (14,36) was applied to a subset of amino acids of interest to illustrate the next step in analysis of the data, i.e. determining which amino acids are most likely causative in disease risk, as distinct from associations due to LD with causative amino acids. In CHM analysis, heterogeneity testing is performed to determine if stratification by an additional amino acid of specific haplotype combinations at one or more other amino acid sites affects the disease risk. Significant effects imply that the additional amino acid either itself directly affects disease risk, or is in LD with another amino acid that does.
Crystal structure modelling
The structure of HLA-DR1 in complex with a peptide antigen derived from influenza hemagglutinin (PDB ID: 1fyt) (37) was used in the analyses in Figure 1. Space-filled models were produced using MBT Protein Workshop (38) (http://mbt.sdsc.edu/software/applications). Visualization of the amino acid residues involved in the composite sequence feature associated with risk to and protection from SSc was performed using SwissPdb Viewer DeepView v4.0.1 (39) (http://spdbv.vital-it.ch/). Residue changes were performed using the DeepView mutate function, and for each amino acid the rotamer with the lowest clash score and highest probability was chosen. (See http://spdbv.vital-it.ch/mutation_guide.html for a description of how these parameters are calculated.) Images from DeepView were rendered using POV-Ray raytracer (http://www.povray.org/).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the National Institutes of Health [contracts N01-AI40076 and N01-AR02251; grants P50-AR054144, UL1-RR024148 and UL1-RR024982].
Supplementary Material
REFERENCES
- 1.Robinson J., Malik A., Parham P., Bodmer J.G., Marsh S.G. IMGT/HLA database–a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55:280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- 2.Frahm N., Baker B., Brander C. Identification and Optimal Definition of HIV-derived cytotoxic T-lymphocyte (CTL) epitopes for the study of CTL escape, functional avidity, and viral evolution. In: Korber B.T., Haynes B.C., Koup R., Moore J.P., Walker B.D., Watkins D.I., editors. HIV Molecular Immunology 2008. Los Alamos, NM: Los Alamos National Laboratory, Theoretical Biology and Biophysics; 2008. pp. 3–24. [Google Scholar]
- 3.Lautscham G., Rickinson A., Blake N. TAP-independent antigen presentation on MHC class I molecules: lessons from Epstein-Barr virus. Microbes Infect. 2003;5:291–299. doi: 10.1016/s1286-4579(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 4.Miles J.J., Elhassen D., Borg N.A., Silins S.L., Tynan F.E., Burrows J.M., Purcell A.W., Kjer-Nielsen L., Rossjohn J., Burrows S.R., et al. CTL recognition of a bulged viral peptide involves biased TCR selection. J. Immunol. 2005;175:3826–3834. doi: 10.4049/jimmunol.175.6.3826. [DOI] [PubMed] [Google Scholar]
- 5.Tynan F.E., Elhassen D., Purcell A.W., Burrows J.M., Borg N.A., Miles J.J., Williamson N.A., Green K.J., Tellam J., Kjer-Nielsen L., et al. The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation. J. Exp. Med. 2005;202:1249–1260. doi: 10.1084/jem.20050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan M.A., Mathieu A., Sorrentino R., Akkoc N. The pathogenetic role of HLA-B27 and its subtypes. Autoimmun. Rev. 2007;6:183–189. doi: 10.1016/j.autrev.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Reveille J.D. Major histocompatibility genes and ankylosing spondylitis. Best Pract. Res. Clin. Rheumatol. 2006;20:601–609. doi: 10.1016/j.berh.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Imboden J.B. The immunopathogenesis of rheumatoid arthritis. Annu. Rev. Pathol. 2009;4:417–434. doi: 10.1146/annurev.pathol.4.110807.092254. [DOI] [PubMed] [Google Scholar]
- 9.Newton J.L., Harney S.M., Wordsworth B.P., Brown M.A. A review of the MHC genetics of rheumatoid arthritis. Genes Immun. 2004;5:151–157. doi: 10.1038/sj.gene.6364045. [DOI] [PubMed] [Google Scholar]
- 10.Erlich H., Valdes A.M., Noble J., Carlson J.A., Varney M., Concannon P., Mychaleckyj J.C., Todd J.A., Bonella P., Fear A.L., et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57:1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson G., Valdes A.M., Noble J.A., Kockum I., Grote M.N., Najman J., Erlich H.A., Cucca F., Pugliese A., Steenkiste A., et al. Relative predispositional effects of HLA class II DRB1-DQB1 haplotypes and genotypes on type 1 diabetes: a meta-analysis. Tissue Antigens. 2007;70:110–127. doi: 10.1111/j.1399-0039.2007.00867.x. [DOI] [PubMed] [Google Scholar]
- 12.Salamon H., Tarhio J., Ronningen K., Thomson G. On distinguishing unique combinations in biological sequences. J. Comput. Biol. 1996;3:407–423. doi: 10.1089/cmb.1996.3.407. [DOI] [PubMed] [Google Scholar]
- 13.Valdes A.M., McWeeney S., Thomson G. HLA class II DR-DQ amino acids and insulin-dependent diabetes mellitus: application of the haplotype method. Am. J. Hum. Genet. 1997;60:717–728. [PMC free article] [PubMed] [Google Scholar]
- 14.Valdes A.M., Thomson G. Detecting disease-predisposing variants: the haplotype method. Am. J. Hum. Genet. 1997;60:703–716. [PMC free article] [PubMed] [Google Scholar]
- 15.Gladman D.D., Keystone E.C., Baron M., Lee P., Cane D., Mervert H. Increased frequency of HLA-DR5 in scleroderma. Arthritis Rheum. 1981;24:854–856. doi: 10.1002/art.1780240614. [DOI] [PubMed] [Google Scholar]
- 16.Arnett F.C., Bias W.B., McLean R.H., Engel M., Duvic M., Goldstein R., Freni-Titulaer L., McKinley T.W., Hochberg M.C. Connective tissue disease in southeast Georgia. A community based study of immunogenetic markers and autoantibodies. J. Rheumatol. 1990;17:1029–1035. [PubMed] [Google Scholar]
- 17.Reveille J.D., Durban E., MacLeod-St Clair M.J., Goldstein R., Moreda R., Altman R.D., Arnett F.C. Association of amino acid sequences in the HLA-DQB1 first domain with antitopoisomerase I autoantibody response in scleroderma (progressive systemic sclerosis) J. Clin. Invest. 1992;90:973–980. doi: 10.1172/JCI115974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briggs D., Stephens C., Vaughan R., Welsh K., Black C. A molecular and serologic analysis of the major histocompatibility complex and complement component C4 in systemic sclerosis. Arthritis Rheum. 1993;36:943–954. doi: 10.1002/art.1780360711. [DOI] [PubMed] [Google Scholar]
- 19.Kuwana M., Kaburaki J., Okano Y., Inoko H., Tsuji K. The HLA-DR and DQ genes control the autoimmune response to DNA topoisomerase I in systemic sclerosis (scleroderma) J. Clin. Invest. 1993;92:1296–1301. doi: 10.1172/JCI116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuwana M., Kaburaki J., Mimori T., Tojo T., Homma M. Autoantigenic epitopes on DNA topoisomerase I. Clinical and immunogenetic associations in systemic sclerosis. Arthritis Rheum. 1993;36:1406–1413. doi: 10.1002/art.1780361013. [DOI] [PubMed] [Google Scholar]
- 21.Morel P.A., Chang H.J., Wilson J.W., Conte C., Falkner D., Tweardy D.J., Medsger T.A., Jr HLA and ethnic associations among systemic sclerosis patients with anticentromere antibodies. Hum. Immunol. 1995;42:35–42. doi: 10.1016/0198-8859(94)00078-5. [DOI] [PubMed] [Google Scholar]
- 22.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agarwal S.K., Tan F.K., Arnett F.C. Genetics and genomic studies in scleroderma (systemic sclerosis) Rheum. Dis. Clin. North. Am. 2008;34:17–40. doi: 10.1016/j.rdc.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Arnett F.C. HLA and autoimmunity in scleroderma (systemic sclerosis) Int. Rev. Immunol. 1995;12:107–128. doi: 10.3109/08830189509056707. [DOI] [PubMed] [Google Scholar]
- 25.Reveille J.D., Owerbach D., Goldstein R., Moreda R., Isern R.A., Arnett F.C. Association of polar amino acids at position 26 of the HLA-DQB1 first domain with the anticentromere autoantibody response in systemic sclerosis (scleroderma) J. Clin. Invest. 1992;89:1208–1213. doi: 10.1172/JCI115704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morel P.A., Chang H.J., Wilson J.W., Conte C., Saidman S.L., Bray J.D., Tweardy D.J., Medsger T.A., Jr Severe systemic sclerosis with anti-topoisomerase I antibodies is associated with an HLA-DRw11 allele. Hum. Immunol. 1994;40:101–110. doi: 10.1016/0198-8859(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 27.Whyte J., Artlett C., Harvey G., Stephens C.O., Welsh K., Black C., Maddison P.J., McHugh N.J. HLA-DQB1 associations with anti-topoisomerase-1 antibodies in patients with systemic sclerosis and their first degree relatives. United Kingdom Systemic Sclerosis Study Group. J. Autoimmun. 1994;7:509–520. doi: 10.1006/jaut.1994.1037. [DOI] [PubMed] [Google Scholar]
- 28.Arnett F.C., Reveille J.D., Goldstein R., Pollard K.M., Leaird K., Smith E.A., Leroy E.C., Fritzler M.J. Autoantibodies to fibrillarin in systemic sclerosis (scleroderma). An immunogenetic, serologic, and clinical analysis. Arthritis Rheum. 1996;39:1151–1160. doi: 10.1002/art.1780390712. [DOI] [PubMed] [Google Scholar]
- 29.Arnett F.C., Gourh P., Shete S., Ahn C.W., Honey R., Agarwal S.K., Tan F.K., McNearney T., Fischbach M., Fritzler M.J., et al. Major histocompatibility complex (MHC) class II alleles, haplotypes, and epitopes which confer susceptibility or protection in the fibrosing autoimmune disease systemic sclerosis: analyses in 1300 Caucasian, African-American and Hispanic cases and 1000 controls. Ann. Rheum. Dis. 2009 doi: 10.1136/ard.2009.111906. ard.2009.111906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunckley H., Jazwinska E.C., Gatenby P.A., Serjeantson S.W. DNA-DR typing shows HLA-DRw11 RFLPs are increased in frequency in both progressive systemic sclerosis and CREST variants of scleroderma. Tissue Antigens. 1989;33:418–420. doi: 10.1111/j.1399-0039.1989.tb01686.x. [DOI] [PubMed] [Google Scholar]
- 31.Lancaster A. Berkley, CA: University of California at Berkley; 2006. Interplay of selection and molecular function in HLA genes. PhD thesis, Integrative Biology. [Google Scholar]
- 32.Smith K.J., Pyrdol J., Gauthier L., Wiley D.C., Wucherpfennig K.W. Crystal structure of HLA-DR2 (DRA*0101, DRB1*1501) complexed with a peptide from human myelin basic protein. J. Exp. Med. 1998;188:1511–1520. doi: 10.1084/jem.188.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaas Q., Ruiz M., Lefranc M.P. IMGT/3Dstructure-DB and IMGT/StructuralQuery, a database and a tool for immunoglobulin, T cell receptor and MHC structural data. Nucleic Acids Res. 2004;32:D208–D210. doi: 10.1093/nar/gkh042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diehl A.D., Lee J.A., Scheuermann R.H., Blake J.A. Ontology development for biological systems: immunology. Bioinformatics. 2007;23:913–915. doi: 10.1093/bioinformatics/btm029. [DOI] [PubMed] [Google Scholar]
- 35.Marsh S.G., Albert E.D., Bodmer W.F., Bontrop R.E., Dupont B., Erlich H.A., Geraghty D.E., Hansen J.A., Hurley C.K., Mach B., et al. Nomenclature for factors of the HLA system, 2004. Tissue Antigens. 2005;65:301–369. doi: 10.1111/j.1399-0039.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- 36.Thomson G., Barcellos L.F., Valdes A.M. Searching for additional disease loci in a genomic region. Adv. Genet. 2008;60:253–292. doi: 10.1016/S0065-2660(07)00411-7. [DOI] [PubMed] [Google Scholar]
- 37.Hennecke J., Carfi A., Wiley D.C. Structure of a covalently stabilized complex of a human alphabeta T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. EMBO J. 2000;19:5611–5624. doi: 10.1093/emboj/19.21.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreland J.L., Gramada A., Buzko O.V., Zhang Q., Bourne P.E. The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics. 2005;6:21. doi: 10.1186/1471-2105-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.