Abstract
Transcription from the mouse mammary tumor virus (MMTV) promoter can be induced by progestins. The progesterone receptor (PR) binds to a cluster of five hormone responsive elements (HREs) and activates the promoter by synergistic interactions with the ubiquitous transcription factor, nuclear factor 1 (NF1). Progesterone treatment of cells in culture leads to activation of the Src/Ras/Erk/Msk1 cascade. Selective inhibition of Erk, or its target kinase Msk1, interferes with chromatin remodeling and blocks MMTV activation. A complex of activated PR, Erk and Msk1 is recruited to promoter after 5 min of hormone treatment and phosphorylates histone H3 at serine 10. This modification promotes the displacement of HP1γ and subsequent chromatin remodeling. Progestin treatment leads to the recruitment of the BAF complex, which selectively displaces histones H2A and H2B from the nucleosome containing the HREs. The acetyltransferase PCAF is also required for induction of progesterone target genes and acetylates histone H3 at K14, an epigenetic mark, which interacts with Brg1 and Brm, anchoring the BAF complex to chromatin. In nucleosomes assembled on either MMTV or mouse rDNA promoter sequences, SWI/SNF displaces histones H2A and H2B from MMTV, but not from the rDNA nucleosome. Thus, the outcome of nucleosome remodeling by purified SWI/SNF depends on DNA sequence. The resultant H3/H4 tetramer particle is then the substrate for subsequent events in induction. Thus, initial activation of the MMTV promoter requires activation of several kinases and PCAF leading to phosphoacetylation of H3, and recruitment of BAF with subsequent removal of H2A/H2B.
Introduction
The promoter of the mouse mammary tumor virus (MMTV) provirus is a well-characterized example of transcriptional control by steroid hormones in which the chromatin organization plays an important role [Richard-Foy and Hager, 1987]. The provirus integrated in the host cell chromatin is virtually silent in the absence of hormones, but responds with rapid transcriptional activation to the addition of either glucocorticoids or progestins. The receptors for these hormones bind to a cluster of HREs in the MMTV promoter and facilitate the interaction of ubiquitous transcription factors including Nuclear Factor 1 (NF1) [Di Croce et al., 1999] and the octamer transcription factor, Oct1/OTF1 [Bruggemeier et al., 1991] with their target sites located between the HREs and the TATA box. This results in a synergistic activation of transcription by the hormone receptors and NF1 (for a review see [Beato et al., 1995]). How synergism between PR and NF1 occurs is a question that has attracted considerable attention, but the mechanism is not simply cooperative DNA binding of the various proteins to the MMTV promoter DNA [Bruggemeier et al., 1990].
Chromatin organization and factor binding
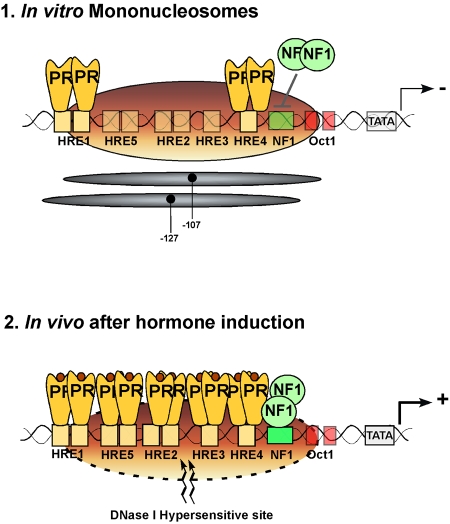
The LTR region of MMTV is organized into positioned nucleosomes [Richard-Foy and Hager, 1987] and hormone induction leads to the appearance of a DNase I-hypersensitive region over the promoter chromatin [Zaret and Yamamoto, 1984], suggesting an influence of hormone induction on the chromatin organization of the promoter (Figure 1). A role for nucleosome phasing in MMTV regulation has been postulated based on studies with breast cancer cell lines carrying a single copy of MMTV reporter stably integrated and on in vitro nucleosome assembly studies [Truss et al., 1995]. Though the exact positioning of nucleosome over the MMTV promoter has been debated [Fragoso et al., 1995], a dominant nucleosome phase in breast cancer cells precludes binding of NF1, but permits steroid hormone receptors (SHRs) to recognize one properly oriented HRE within the HRE cluster [Truss et al., 1995] (Figure 1). The different affinities of SHRs and NF1 for nucleosomally-organized target sites can be reproduced in vitro [Eisfeld et al., 1997; Pina et al., 1990a] and reflect the different ways in which the two proteins recognize their cognate DNA sequences [Beato and Eisfeld, 1997]. SHRs only contact a narrow region of the HRE DNA double helix and can therefore bind if this section is exposed, while NF1 embraces the complete circumference of the helix and thus cannot interact with target sites within nucleosomes. When both SHRs and NF1 are added simultaneously to isolated MMTV mononucleosomes, the receptors bind to the accessible HREs, but NF1 is unable to recognize its target sites (Figure 1) [Pina et al., 1990a], suggesting that additional components are required for simultaneous factor binding as detected in intact cells by genomic footprinting analysis following hormone treatment [Truss et al., 1995].
Figure 1. Schematic representation of the main cis elements in the MMTV promoter and their occupancy in nucleosomes assembled in vitro (upper panel) and in intact cells after hormone induction (lower panel).
The positions covered by the main population of histone octamers are indicated by the grey ovals. The HREs, the NF1 binding site and the TATA box are indicated. The numbers refer to the distance in nucleotides from the transcription start site. The hormone receptor (PR) dimers are depicted in yellow and the NF1 dimer by green circles.
When introduced in Saccharomyces cerevisiae engineered to express GR or PR, the MMTV promoter is organized into positioned nucleosomes, is silent in the absence of hormone, and responds poorly to expression of NFI or to a NFI-VP16 fusion, but can be induced by hormone treatment [Chavez et al., 1995]. Deletion of the HREs disrupts nucleosome positioning and makes the promoter responsive to NFI-VP16, suggesting that the HRE region would repress access to the NFI site by positioning a nucleosome [Candau et al., 1996]. The nucleosomal organization of the MMTV promoter in vitro is similar in yeast and animal cells, implying that the DNA sequence contains conformational or topological information which determines nucleosome positioning [Pina et al., 1990b] and thus modulates the accessibility of transcription factors to cis-acting elements.
Hormone induction was believed to result in a displacement of the nucleosome over the promoter, thus allowing free access of NF1 to its binding site and transcriptional activation [Richard-Foy and Hager, 1987]. However, genomic footprinting of the chromosomal MMTV promoter shows that hormone induction does not lead to displacement, but rather to a rearrangement of nucleosomes, which is necessary for simultaneous binding of receptors, NFI and OTF1 [Truss et al., 1995]. Thus, contrary to the situation in the test tube, the presence of a positioned nucleosome in vivo is not an obstacle, but rather a prerequisite for simultaneous factor binding and optimal induction of the MMTV promoter (Figure 1) [Truss et al., 1995]. This is strongly supported by the observation that nucleosome depletion in yeast, while enhancing the accessibility of the MMTV promoter for hormone receptors and NF1, eliminates the functional synergism between these two factors [Chavez and Beato, 1997]. Moreover, in a cell-free system, synergistic binding of receptors and NF1 to the MMTV promoters depends on its previous assembly into minichromosomes with positioned nucleosomes and on preincubation in the presence of ATP [Di Croce et al., 1999].
FRET experiments and footprints
Fluorescence photobleaching experiments in living cells containing a cluster of 200 MMTV promoters in tandem have provided direct evidence that the hormone-occupied GR undergoes rapid exchange between chromatin and the nucleoplasmic compartment with a half-life of less than a minute [McNally et al., 2000]. This rapid exchange has also been reported for the ERα [Stenoien et al., 2001a; Stenoien et al., 2001b]. In agreement with the difficulties in visualizing a footprint of GR on the MMTV promoter in vivo, this has led to the formulation of a “hit-and-run” model of receptor action [Rigaud et al., 1991]. However, following dexamethasone treatment of hepatic cells, such footprints have been reported on the GREs of the tyrosine aminotransferase enhancer [Becker et al., 1986; Espinas et al., 1994]. Similarly, after progestin addition to breast cancer cells, DNase I and DMS footprints are observed on a single copy chromosomal MMTV promoter [Truss et al., 1995]. Moreover, following incubation of MMTV minichromosomes with purified PR in Drosophila embryo extracts, clear DNase I footprints are detected over the HREs [Di Croce et al., 1999]. These results demonstrate that a significant fraction of target promoters must be occupied by the receptors during the time of treatment with the footprinting agents, DMS or DNase I. How these findings can be reconciled with the very rapid exchange of GR-GFP observed in vivo remains to be elucidated. One possibility is that the majority of the GR binding events observed in the MMTV cluster in vivo are non-productive and do not lead to stable complexes containing receptors and NF1. Non-productive does not imply irrelevant. Possibly, many of these weak interactions are needed to form a stable complex of PR and NF1. Indeed, in the absence of NF1, it is exceedingly difficult to generate a receptor footprint on MMTV chromatin in vitro. This can only be achieved after addition of very high concentrations of receptor that push the equilibrium to the DNA bound state [Di Croce et al., 1999]. Nevertheless, even in the absence of NF1, receptor concentrations which do not generate a footprint can activate chromatin transcription to some extent, indicating that an interaction is taking place. That this short-lived interaction could correspond to the rapid exchange observed in vivo is suggested by the observation that after hormone induction only a small proportion of the MMTV clusters exhibit the phenomenon of chromatin decondensation, indicative of active transcription [Muller et al., 2001].
Hormone-dependent chromatin remodeling
Role of histone H1
The nature of the hormone-induced nucleosomal change that permits simultaneous binding of transcription factors to the MMTV promoter remains obscure. Depletion of histone H1 from the MMTV promoter could be a possibility [Bresnick et al., 1992] and a role for histone H1 phosphorylation in modulating MMTV activation has been postulated [Lee and Archer, 1998]. However, the MMTV promoter is regulated in budding yeast, which lacks linker histones [Chavez et al., 1995], and also in minichromosomes assembled in the absence of histone H1 [Di Croce et al., 1999]. Incorporation of H1 into MMTV minichromosomes increases nucleosome spacing and reduces access of general transcription factors to the promoter, thus inhibiting basal transcription [Koop et al., 2003]. However, transcription activation by PR and NF1 is enhanced in H1 containing minichromosomes [Koop et al., 2003]. This unexpected effect is due to a more homogeneous positioning of nucleosomes in the presence of H1 [Vicent et al., 2002] and, as a consequence, a better binding of PR [Koop et al., 2003; Vicent et al., 2002] and a higher proportion of promoters participating in transcription [Koop et al., 2003]. Moreover, in the presence of bound PR in an extract from Drosophila embryos, H1 is phosphorylated and subsequently removed from the promoter upon transcription initiation [Koop et al., 2003].
Histone acetylation
Additional possibilities for chromatin remodeling are recruitment by the receptors of chromatin remodeling activities, either ATP-dependent, such as the SWI/SNF complex [Cote et al., 1994], or ATP-independent, such as histone acetyltransferases (HATs) [Lee et al., 1993]. This latter possibility has received considerable attention following the discovery that several steroid receptor coactivators exhibit HAT activity, including members of the SRC-1 family, P/CAF and p300/CBP [Brown et al., 2000].
The role of histone acetylation in hormone induction of the MMTV promoter in not clear. High doses of histone deacetylase inhibitors, butyrate or TSA, lead to intense hyperacetylation of core histones and inhibit hormone induction [Bartsch et al., 1996; Bresnick et al., 1990] without altering nucleosome positioning [Bresnick et al., 1991]. However, low doses of the inhibitors activate the MMTV promoter in the absence of hormone and generate a DNAse I-hypersensitive site similar to that observed following hormone treatment [Bartsch et al., 1996]. Derepressing concentrations of the inhibitors of histone deacetylases enhance hormone-activated transcription, and thus the extent of hormonal induction is not affected [Bartsch et al., 1996]. Similar results are obtained with these inhibitors in the cell-free transcription assay with MMTV minichromosomes, suggesting that inhibitor-sensitive acetylation is not involved in mediating the synergism between receptors and NF1. More recently, a role for histone deacetylase 1 (HDAC1) has been proposed for MMTV activation [Qiu et al., 2006]. HDAC1, generally considered as a corepressor, acts as a coactivator of the glucocorticoid receptor (GR) and its histone deacetylase activity is regulated by a p300-dependent acetylation [Qiu et al., 2006]. The mechanism of this effect remains unclear.
ATP-dependent remodeling
The SWI/SNF complex, or other ATP-dependent chromatin remodeling factors, could destabilize the interaction between the DNA double helix and the histone octamer and thus facilitate access to otherwise masked cis elements. Hormone-induced chromatin remodeling might also facilitate the assembly of the transcription initiation complex (TIC) on the TATA box, and/or contribute to efficient transcription elongation through downstream nucleosomes.
There is evidence for a requirement of the SWI/SNF complex in glucocorticoid gene regulation in yeast [Yoshinaga et al., 1992] and in animal cells [Muchardt and Yaniv, 1993], and hSWI/SNF seems to be required for chromatin remodeling initiated by glucocorticoids [Fryer and Archer, 1998]. The situation appears to be different for progestins, which act through the same HREs of the MMTV promoter [Fryer and Archer, 1998]. We have reproduced the synergism between SHR and NF1 in a cell-free system driven by MMTV minichromosomes assembled in extracts from Drosophila embryos [Di Croce et al., 1999]. These minichromosomes exhibit nucleosomes positioned in the same way as in cultured cells and the NF1 site in the promoter is not accessible [Venditti et al., 1998]. Addition of hormone-activated GR or PR leads to ATP-dependent remodeling of these minichromosomes, as demonstrated by topoisomer analysis [Di Croce et al., 1999], and to simultaneous binding of receptors and NF1. This activity can be partly reproduced with purified MMTV minichromosomes with recombinant dISWI, but not with purified dCHRAC or ySWI/SNF complex [Di Croce et al., 1999]. Moreover, incubation of the minichromosomes in the Drosophila extract in the presence of receptors and NF1 leads to recruitment of dISWI, and dNURF301, two subunits of the NURF complex [Gdula et al., 1998]. Thus, in the Drosophila embryo extract, PR recruits NURF to the MMTV promoter chromatin, and NURF remodels the minichromosomes in an ATP-dependent fashion.
In mammalian cells with a single MMTV promoter integrated in chromatin we have recently found the hormone-induced recruitment of both Brg1- and ISWI-containing complexes to the promoter [Vicent et al., 2006b; Vicent et al., 2004]. Moreover, in experiments with MMTV mononucleosomes assembled in vitro, PR is able to recruit purified yeast Swi/Snf to the promoter, where it catalyzes the ATP-dependent displacement of H2A/H2B dimers [Vicent et al., 2004]. Though we cannot formally exclude the possibility of a replacement of the H2A/H2B dimers by dimers containing histone variants, our results strongly suggest that the activated MMTV nucleosome with bound SHRs and NF1 is a H3/H4 tetramer particle. This assumption is further supported by our previous findings showing that MMTV promoter sequences assembled in vitro on H3/H4 tetramers, adopt similar positions as on a histone octamer, but in contrast with sequences assembled on the octamer, are able to bind SHRs and NF1 [Spangenberg et al., 1998].
There are two human SWI/SNF-like complexes and both contain ATPase subunits similar to yeast Swi2/Snf2, hBRM (human Brahma) or BRG1 (Brahma-Related Gene 1), as well as a series of other subunits, some of which differ in various cell types [Wang, 2003]. The hSWI/SNF-α complex, also called BAF (BRG1/hBRM-associated factor), contains either hBRM or BRG1 as the central ATPase subunit and is an orthologue of the yeast SWI/SNF. The hSWI/SNF-β complex, also called PBAF (Polybromo-associated BAF), contains only hBRM and is an orthologue to the yeast RSC complex [Mohrmann and Verrijzer, 2005]. The BAF and PBAF complexes share many subunits, but also contain subtype-specific subunits: BAF250 and hBRM are only found in BAF, whereas BAF180 and BAF 200 are only found in PBAF [Mohrmann and Verrijzer, 2005; Yan et al., 2005].
In breast cancer cells that contain BAF and PBAF, we have recently found that BAF is essential during the activation of MMTV and other endogenous progesterone-induced genes. BAF is recruited via an interaction between PR and the BAF57 subunit of the complex and is anchored by H3K14ac, generated by PCAF, suggesting a mechanism for the cooperation between two types of chromatin remodeling activities. The recruited BAF catalyzes the ATP-dependent displacement of histones H2A/H2B required for NF1 to gain access to the promoter site. Thus, synergism between the two transcription factors PR and NF1 is mediated by the cooperation between two chromatin-remodeling machines, BAF and PCAF [Vicent et al., 2009].
Link between protein kinases and transcriptional control
In addition to their transcriptional effect, steroid hormones also crosstalk to kinase signaling pathways, and this crosstalk is essential for the proliferative response of breast cancer cells to estrogens [Migliaccio et al., 1996] and progesterone [Migliaccio et al., 1998].
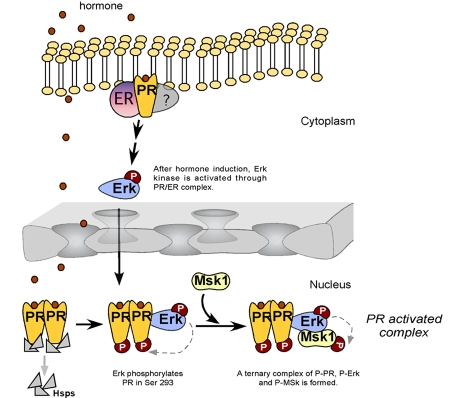
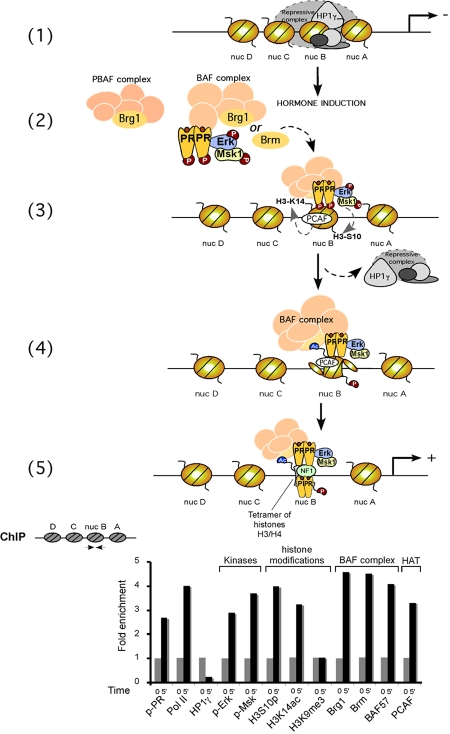
What could be the connection between the rapid hormonal activation of cytoplasmic signaling cascades and the events taking place at the MMTV promoter? We have investigated the function of Erk activation by progesterone, which is mediated via an interaction between PR and ERα (Figure 2 and [Ballare et al., 2003]). We observed that progestin activation of the MMTV promoter can be blocked by inhibiting Erk activation either with antiestrogens, with small molecular inhibitors (PD 98059), with dominant negative kinase mutant, or with RNAi against Erk [Vicent et al., 2006a]. As Erk phosphorylated PR at ser294 in response to progesterone, it was possible that the transcriptional inhibition of MMTV induction was due to a lack of PR phosphorylation. However, a similar inhibition of MMTV induction was observed when the activation of Msk1 was prevented by small molecular inhibitors (H89), a dominant negative mutant of Msk, or down-regulation of Msk1 with an specific RNAi [Vicent et al., 2006a]. Unexpectedly, after 5 minutes of hormone treatment, we detected a ternary complex of phosphorylated PR and activated Erk and Msk1 (Figure 2), which is selectively recruited to the MMTV promoter nucleosome containing the HREs (Figure 3) [Vicent et al., 2006a]. Concomitant with the recruitment of the ternary complex, histone H3 becomes phosphorylated at serine 10 and acetylated at lysine 14, only on the nucleosome containing the HREs and not on adjacent nucleosomes (Figure 3) [Vicent et al., 2006a]. Blocking H3 phosphoacetylation precludes displacement of a repressive complex containing HP1γ and recruitment of BAF, preventing H2A/H2B displacement and promoter activation.
Figure 2. Hypothetical model for the initial steps of MMTV promoter induction.
Progestins bind to cytoplasmic PR/ER complexes, and activate the Src/Ras/Erk pathway, leading to nuclear accumulation of activated Erk. Most of PR is nuclear and complexed to chaperones (Hsps). Upon binding of progestins, PR homodimers dissociate from chaperones and a fraction of PR is phosphorylated by pErk, which also phosphorylates Msk1. A “PR -activated complex” composed of pPR/pErk/pMsk1 is formed.
Figure 3. Hypothetical model for the role of “PR activated complex” in chromatin.
Upper Panel: (1) An HP1γ-containing complex is bound to the promoter in the absence of hormone; (2) PR-activated complex binds BAF, but not PBAF; (3) PR-activated complex is recruited to the nucleosome B upon hormone induction, followed by H3 phosphoacetylation. The repressive complex is displaced from the promoter; (4) Recruitment of the BAF complex to the nucleosome B is stabilized by PCAF-dependent H3K14 acetylation, enabling ATP-dependent H2A/H2B displacement (and/or histone variants incorporation); (5) This metastable opening facilitates NF1 binding, generating a stable platform for the assembly of further PR molecules, BAF, coactivators and the basal transcriptional machinery, including RNA polymerase II. Lower Panel: T47D-MTVL cells were untreated (0) or treated for 5 min with 10 nM R-5020 and subjected to ChIP assays with the indicated specific antibodies. A schematic representation of the ChIPs described in (Vicent et al., 2006a) and (Vicent et al., 2009) is shown.
Microarray analysis of the progesterone response in breast cancer cells (Ballare et al., manuscript in preparation), showed that about 25% of the hormonally-regulated genes are sensitive to the inhibition of Erk activation. This shows that the behavior of the MMTV promoter does not represent a rare exception, but also suggests that other signaling pathways are involved in mediating progesterone action. We know that the PI3K/Akt pathway is involved in the regulation of stromal cell proliferation in the endometrium [Vallejo et al., 2005], and also in the proliferative response of breast cancer cells to estrogens [Castoria et al., 2001]. There is also evidence that the cAMP/PKA pathway is implicated in estrogen action ([Aronica et al., 1994; Cho and Katzenellenbogen, 1993]), and that the JAK/STAT5 plays an important role in glucocorticoid and progesterone effects in mammary gland ([Buser et al., 2007; Stocklin et al., 1996; Stoecklin et al., 1997]). It remains to be established whether the crosstalk of the SHR with these different pathways acts merely by regulating the phosphorylation of key transcription factors and coregulators, or whether, as in the case of the Erk pathway, modifications of structural components of the chromatin also play an important role.
Conclusions
The results summarized in this review contribute to a better understanding of the molecular mechanisms of promoter activation by progesterone. During activation of the MMTV promoter, the chromatin constitutes a key component essential for regulation of transcription by allowing synergism between the nuclear receptors and NF-1, causing full activation of the promoter.
The linker histone H1, known as a repressive component of chromatin needed for condensation, increases MMTV transcription by improving the positioning of nucleosomes. PR interacts with an exposed HRE on the surface of a nucleosome and initiates chromatin remodeling as a prerequisite for the subsequent steps of gene activation.
The nucleosomal DNA sequence, as part of chromatin, contains the information required to direct the outcome of the subsequent remodeling event. The SWI/SNF complex, depending on the DNA sequence, can displace histones H2A and H2B or slide the nucleosome toward one end of the DNA fragment. Thus, the “final outcome” in each case is completely different, depending on structural features of the underlying DNA, which confer the differential response to SWI/SNF action and should be further developed.
The results with the MMTV promoter underline the significance of incorporating a precise knowledge of the structure of native DNA sequences in chromatin. Evidence for the positive contributions of chromatin structure and dynamics to the process of gene regulation continues to accumulate at an increasing pace. A classical example is a situation in which a positioned nucleosome brings in close proximity cis-regulatory elements located some 200 bp apart and in this way facilitates the interaction between DNA-bound factors. There are several examples of this type of regulation (for a recent report see [Zhao et al., 2001]), and one case involves an interaction of the estrogen receptor with the transcription factor NF1 [Schild et al., 1993].
A particularly illustrative example of the significance of chromatin is the regulation of the Human Immunodeficiency Virus type 1 (HIV-1), the etiologic agent of AIDS. Recent advances have indicated that HIV-1 encoded proteins interact with chromatin-remodeling complexes and histone-modifying enzymes, implying that chromatin remodeling plays an important role in the HIV-1 life cycle [Henderson et al., 2004; Mahmoudi et al., 2006]. Nucleosomes are positioned on the HIV-1 LTR and act as barriers to transcription. Following cellular activation, these nucleosomes are modified and repositioned, allowing for activation of viral gene expression [el Kharroubi and Martin, 1996; Verdin et al., 1993].
Another example where chromatin has been shown to be essential is the gene regulation of the rat albumin gene enhancer. In its active state, in liver nuclei, the enhancer is organized into a nuclease-accessible array of three positioned nucleosome-like particles N1, N2, and N3, with bound transcription factors [McPherson et al., 1993]. The N1 nucleosome-like particle at the albumin enhancer is positioned over the FoxA binding sites and is DNase I-hypersensitive in hepatic cells. In contrast, in non-liver tissues, where the FoxA sites are not occupied, nucleosome-like particles are not precisely positioned over the enhancer, and the enhancer is not hypersensitive [McPherson et al., 1993]. It has been observed that FoxA and GATA-4 bound their sites in compacted chromatin and opened the local nucleosomal domain in the absence of ATP-dependent enzymes [Cirillo et al., 2002].
These are just a few examples of the intricacies that the chromatin organization of promoters and regulatory regions impose on gene regulation. This complexity is partly due to the fact that one of the key events during cell differentiation in complex organisms is the progressive chromatin-mediated silencing of genes that are not required in a particular cell lineage and the proper chromatin organization of those genes that will be needed at some later stage of differentiation or in response to external signals. Understanding this differential chromatin organization of particular genome regions is the main challenge for epigenetic research in the coming years.
Acknowledgments
G.P.V. was a recipient of a fellowship of the Ramón y Cajal Programme. The experimental work was supported by grants from the European Union (HEROIC integrated project), the Departament d´Innovacio Universitat I Empresas (DIUE), Ministerio de Educación y Ciencia (MEC) BMC 2003-02902 and Fondo de Investigación Sanitaria (FIS) PI0411605 and CP04/00087.
Abbreviations
- BAF
BRG1/hBRM-associated factor
- BRG1
Brahma-related gene 1
- CBP
CREB binding protein
- CHRAC
chromatin accessibility complex
- Erk
extracellular signal-regulated kinase
- ERα
estrogen receptor α
- GR
glucocorticoid receptor
- HATs
histone acetyltransferases
- HDAC1
histone deacetylase 1
- HIV-1
human immunodeficiency virus type 1
- HP1γ
heterochromatin protein 1γ
- HREs
hormone responsive elements
- ISWI
imitation SWI remodeling factor
- MMTV
mouse mammary tumor virus
- Msk1
mitogen and stress response kinase-1
- NF1
nuclear factor 1
- NURF
nucleosome remodeling factor
- PBAF
polybromo-associated BAF
- PCAF
P300/CBP-associated factor
- PR
progesterone receptor
- SHRs
steroid hormone receptors
- SRC-1
steroid receptor coactivator-1
- SWI/SNF
Switch/Sucrose nonfermentable nucleosome remodeling complex
- TIC
transcription initiation complex
References
- Aronica S. M., Kraus W. L., Katzenellenbogen B. S. Estrogen action via the cAMP signaling pathway: stimulation of adenylate cyclase and cAMP-regulated gene transcription. Proc Natl Acad Sci U S A. 1994;91:8517–21. doi: 10.1073/pnas.91.18.8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballare C., Uhrig M., Bechtold T., Sancho E., Di Domenico M., Migliaccio A., Auricchio F., Beato M. Two domains of the progesterone receptor interact with the estrogen receptor and are required for progesterone activation of the c-Src/Erk pathway in mammalian cells. Mol Cell Biol. 2003;23:1994–2008. doi: 10.1128/MCB.23.6.1994-2008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch J., Truss M., Bode J., Beato M. Moderate increase in histone acetylation activates the mouse mammary tumor virus promoter and remodels its nucleosome structure. Proc Natl Acad Sci U S A. 1996;93:10741–6. doi: 10.1073/pnas.93.20.10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M., Herrlich P., Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–7. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- Beato M., Eisfeld K. Transcription factor access to chromatin. Nucleic Acids Res. 1997;25:3559–63. doi: 10.1093/nar/25.18.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P. B., Gloss B., Schmid W., Strahle U., Schutz G. In vivo protein-DNA interactions in a glucocorticoid response element require the presence of the hormone. Nature. 1986;324:686–8. doi: 10.1038/324686a0. [DOI] [PubMed] [Google Scholar]
- Bresnick E. H., John S., Berard D. S., LeFebvre P., Hager G. L. Glucocorticoid receptor-dependent disruption of a specific nucleosome on the mouse mammary tumor virus promoter is prevented by sodium butyrate. Proc Natl Acad Sci U S A. 1990;87:3977–81. doi: 10.1073/pnas.87.10.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick E. H., John S., Hager G. L. Histone hyperacetylation does not alter the positioning or stability of phased nucleosomes on the mouse mammary tumor virus long terminal repeat. Biochemistry. 1991;30:3490–7. doi: 10.1021/bi00228a020. [DOI] [PubMed] [Google Scholar]
- Bresnick E. H., Bustin M., Marsaud V., Richard-Foy H., Hager G. L. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 1992;20:273–8. doi: 10.1093/nar/20.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. E., Lechner T., Howe L., Workman J. L. The many HATs of transcription coactivators. Trends Biochem Sci. 2000;25:15–9. doi: 10.1016/s0968-0004(99)01516-9. [DOI] [PubMed] [Google Scholar]
- Bruggemeier U., Rogge L., Winnacker E. L., Beato M. Nuclear factor I acts as a transcription factor on the MMTV promoter but competes with steroid hormone receptors for DNA binding. Embo J. 1990;9:2233–9. doi: 10.1002/j.1460-2075.1990.tb07393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemeier U., Kalff M., Franke S., Scheidereit C., Beato M. Ubiquitous transcription factor OTF-1 mediates induction of the MMTV promoter through synergistic interaction with hormone receptors. Cell. 1991;64:565–72. doi: 10.1016/0092-8674(91)90240-y. [DOI] [PubMed] [Google Scholar]
- Buser A. C., Gass-Handel E. K., Wyszomierski S. L., Doppler W., Leonhardt S. A., Schaack J., Rosen J. M., Watkin H., Anderson S. M., Edwards D. P. Progesterone receptor repression of prolactin/signal transducer and activator of transcription 5-mediated transcription of the β-casein gene in mammary epithelial cells. Mol Endocrinol. 2007;21:106–25. doi: 10.1210/me.2006-0297. [DOI] [PubMed] [Google Scholar]
- Candau R., Chavez S., Beato M. The hormone responsive region of mouse mammary tumor virus positions a nucleosome and precludes access of nuclear factor I to the promoter. J Steroid Biochem Mol Biol. 1996;57:19–31. doi: 10.1016/0960-0760(96)00262-2. [DOI] [PubMed] [Google Scholar]
- Castoria G., Migliaccio A., Bilancio A., Di Domenico M., de Falco A., Lombardi M., Fiorentino R., Varricchio L., Barone M. V., Auricchio F. PI3-kinase in concert with Src promotes the S-phase entry of oestradiol-stimulated MCF-7 cells. Embo J. 2001;20:6050–9. doi: 10.1093/emboj/20.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez S., Candau R., Truss M., Beato M. Constitutive repression and nuclear factor I-dependent hormone activation of the mouse mammary tumor virus promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6987–98. doi: 10.1128/mcb.15.12.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez S., Beato M. Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter. Proc Natl Acad Sci U S A. 1997;94:2885–90. doi: 10.1073/pnas.94.7.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Katzenellenbogen B. S. Synergistic activation of estrogen receptor-mediated transcription by estradiol and protein kinase activators. Mol Endocrinol. 1993;7:441–52. doi: 10.1210/mend.7.3.7683375. [DOI] [PubMed] [Google Scholar]
- Cirillo L. A., Lin F. R., Cuesta I., Friedman D., Jarnik M., Zaret K. S. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–89. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cote J., Quinn J., Workman J. L., Peterson C. L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Di Croce L., Koop R., Venditti P., Westphal H. M., Nightingale K. P., Corona D. F., Becker P. B., Beato M. Two-step synergism between the progesterone receptor and the DNA-binding domain of nuclear factor 1 on MMTV minichromosomes. Mol Cell. 1999;4:45–54. doi: 10.1016/s1097-2765(00)80186-0. [DOI] [PubMed] [Google Scholar]
- Eisfeld K., Candau R., Truss M., Beato M. Binding of NF1 to the MMTV promoter in nucleosomes: influence of rotational phasing, translational positioning and histone H1. Nucleic Acids Res. 1997;25:3733–42. doi: 10.1093/nar/25.18.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Kharroubi A., Martin M. A. cis-acting sequences located downstream of the human immunodeficiency virus type 1 promoter affect its chromatin structure and transcriptional activity. Mol Cell Biol. 1996;16:2958–66. doi: 10.1128/mcb.16.6.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinas M. L., Roux J., Ghysdael J., Pictet R., Grange T. Participation of Ets transcription factors in the glucocorticoid response of the rat tyrosine aminotransferase gene. Mol Cell Biol. 1994;14:4116–25. doi: 10.1128/mcb.14.6.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso G., John S., Roberts M. S., Hager G. L. Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev. 1995;9:1933–47. doi: 10.1101/gad.9.15.1933. [DOI] [PubMed] [Google Scholar]
- Fryer C. J., Archer T. K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- Gdula D. A., Sandaltzopoulos R., Tsukiyama T., Ossipow V., Wu C. Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes Dev. 1998;12:3206–16. doi: 10.1101/gad.12.20.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A., Holloway A., Reeves R., Tremethick D. J. Recruitment of SWI/SNF to the human immunodeficiency virus type 1 promoter. Mol Cell Biol. 2004;24:389–97. doi: 10.1128/MCB.24.1.389-397.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop R., Di Croce L., Beato M. Histone H1 enhances synergistic activation of the MMTV promoter in chromatin. Embo J. 2003;22:588–99. doi: 10.1093/emboj/cdg052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. Y., Hayes J. J., Pruss D., Wolffe A. P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- Lee H. L., Archer T. K. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. Embo J. 1998;17:1454–66. doi: 10.1093/emboj/17.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T., Parra M., Vries R. G., Kauder S. E., Verrijzer C. P., Ott M., Verdin E. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J Biol Chem. 2006;281:19960–8. doi: 10.1074/jbc.M603336200. [DOI] [PubMed] [Google Scholar]
- McNally J. G., Muller W. G., Walker D., Wolford R., Hager G. L. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–5. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- McPherson C. E., Shim E. Y., Friedman D. S., Zaret K. S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. 1993;75:387–98. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- Migliaccio A., Piccolo D., Castoria G., Di Domenico M., Bilancio A., Lombardi M., Gong W., Beato M., Auricchio F. Activation of the Src/p21ras/Erk pathway by progesterone receptor via cross-talk with estrogen receptor. Embo J. 1998;17:2008–18. doi: 10.1093/emboj/17.7.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio A., Di Domenico M., Castoria G., de Falco A., Bontempo P., Nola E., Auricchio F. Tyrosine kinase/p21ras/MAP-kinase pathway activation by estradiol-receptor complex in MCF-7 cells. Embo J. 1996;15:1292–300. [PMC free article] [PubMed] [Google Scholar]
- Mohrmann L., Verrijzer C. P. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim Biophys Acta. 2005;1681:59–73. doi: 10.1016/j.bbaexp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Muchardt C., Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. Embo J. 1993;12:4279–90. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W. G., Walker D., Hager G. L., McNally J. G. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J Cell Biol. 2001;154:33–48. doi: 10.1083/jcb.200011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina B., Truss M., Ohlenbusch H., Postma J., Beato M. DNA rotational positioning in a regulatory nucleosome is determined by base sequence. An algorithm to model the preferred superhelix. Nucleic Acids Res. 1990b;18:6981–7. doi: 10.1093/nar/18.23.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina B., Bruggemeier U., Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990a;60:719–31. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Zhao Y., Becker M., John S., Parekh B. S., Huang S., Hendarwanto A., Martinez E. D., Chen Y., Lu H., Adkins N. L., Stavreva D. A., Wiench M., Georgel P. T., Schiltz R. L., Hager G. L. HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol Cell. 2006;22:669–79. doi: 10.1016/j.molcel.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Richard-Foy H., Hager G. L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. Embo J. 1987;6:2321–8. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud G., Roux J., Pictet R., Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991;67:977–86. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- Schild C., Claret F. X., Wahli W., Wolffe A. P. A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. Embo J. 1993;12:423–33. doi: 10.1002/j.1460-2075.1993.tb05674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg C., Eisfeld K., Stunkel W., Luger K., Flaus A., Richmond T. J., Truss M., Beato M. The mouse mammary tumour virus promoter positioned on a tetramer of histones H3 and H4 binds nuclear factor 1 and OTF1. J Mol Biol. 1998;278:725–39. doi: 10.1006/jmbi.1998.1718. [DOI] [PubMed] [Google Scholar]
- Stenoien D. L., Patel K., Mancini M. G., Dutertre M., Smith C. L., O'Malley B. W., Mancini M. A. FRAP reveals that mobility of oestrogen receptor-α is ligand- and proteasome-dependent. Nat Cell Biol. 2001b;3:15–23. doi: 10.1038/35050515. [DOI] [PubMed] [Google Scholar]
- Stenoien D. L., Nye A. C., Mancini M. G., Patel K., Dutertre M., O'Malley B. W., Smith C. L., Belmont A. S., Mancini M. A. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor α-coactivator complexes in living cells. Mol Cell Biol. 2001a;21:4404–12. doi: 10.1128/MCB.21.13.4404-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocklin E., Wissler M., Gouilleux F., Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–8. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- Stoecklin E., Wissler M., Moriggl R., Groner B. Specific DNA binding of Stat5, but not of glucocorticoid receptor, is required for their functional cooperation in the regulation of gene transcription. Mol Cell Biol. 1997;17:6708–16. doi: 10.1128/mcb.17.11.6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truss M., Bartsch J., Schelbert A., Hache R. J., Beato M. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. Embo J. 1995;14:1737–51. doi: 10.1002/j.1460-2075.1995.tb07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo G., Ballare C., Baranao J. L., Beato M., Saragueta P. Progestin activation of nongenomic pathways via cross talk of progesterone receptor with estrogen receptor β induces proliferation of endometrial stromal cells. Mol Endocrinol. 2005;19:3023–37. doi: 10.1210/me.2005-0016. [DOI] [PubMed] [Google Scholar]
- Venditti P., Di Croce L., Kauer M., Blank T., Becker P. B., Beato M. Assembly of MMTV promoter minichromosomes with positioned nucleosomes precludes NF1 access but not restriction enzyme cleavage. Nucleic Acids Res. 1998;26:3657–66. doi: 10.1093/nar/26.16.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E., Paras P., Jr., Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. Embo J. 1993;12:3249–59. doi: 10.1002/j.1460-2075.1993.tb05994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent G. P., Melia M. J., Beato M. Asymmetric binding of histone H1 stabilizes MMTV nucleosomes and the interaction of progesterone receptor with the exposed HRE. J Mol Biol. 2002;324:501–17. doi: 10.1016/s0022-2836(02)01101-4. [DOI] [PubMed] [Google Scholar]
- Vicent G. P., Ballare C., Zaurin R., Saragueta P., Beato M. Chromatin remodeling and control of cell proliferation by progestins via cross talk of progesterone receptor with the estrogen receptors and kinase signaling pathways. Ann N Y Acad Sci. 2006a;1089:59–72. doi: 10.1196/annals.1386.025. [DOI] [PubMed] [Google Scholar]
- Vicent G. P., Nacht A. S., Smith C. L., Peterson C. L., Dimitrov S., Beato M. DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol Cell. 2004;16:439–52. doi: 10.1016/j.molcel.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Vicent G. P., Ballare C., Nacht A. S., Clausell J., Subtil-Rodriguez A., Quiles I., Jordan A., Beato M. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol Cell. 2006b;24:367–81. doi: 10.1016/j.molcel.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Vicent G. P., Zaurin R., Nacht A. S., Li A., Font-Mateu J., Le Dily F., Vermeulen M., Mann M., Beato M. Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLoS Genet. 2009;5:e1000567. doi: 10.1371/journal.pgen.1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr Top Microbiol Immunol. 2003;274:143–69. doi: 10.1007/978-3-642-55747-7_6. [DOI] [PubMed] [Google Scholar]
- Yan Z., Cui K., Murray D. M., Ling C., Xue Y., Gerstein A., Parsons R., Zhao K., Wang W. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–7. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga S. K., Peterson C. L., Herskowitz I., Yamamoto K. R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Yamamoto K. R. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984;38:29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]
- Zhao X., Pendergrast P. S., Hernandez N. A positioned nucleosome on the human U6 promoter allows recruitment of SNAPc by the Oct-1 POU domain. Mol Cell. 2001;7:539–49. doi: 10.1016/s1097-2765(01)00201-5. [DOI] [PubMed] [Google Scholar]