Abstract
[PIN+] is the prion form of Rnq1 in Saccharomyces cerevisiae and is necessary for the de novo induction of a second prion, [PSI+]. The function of Rnq1, however, is little understood. The limited availability of defective rnq1 alleles impedes the study of its structure-function relationship by genetic analysis. In this study, we isolated rnq1 mutants that are defective in the stable maintenance of the [PIN+] prion. Since there is no rnq1 phenotype available that is applicable to a direct selection or screening for loss-of-function rnq1 mutants, we took advantage of a prion inhibitory agent, Rnq1Δ100, to develop a color-based genetic screen. Rnq1Δ100 eliminates the [PSI+] prion in the [PIN+] state but not in the [pin−] state. This allows us to find loss-of-[PIN+] rnq1 mutants as white [PSI+] colonies. Nine rnq1 mutants with single-amino-acid substitutions were defined. These mutations impaired the stable maintenance of [PIN+] and, as a consequence, were also partially defective in the de novo induction of [PSI+]. Interestingly, eight of the nine alleles were mapped to the N-terminal region of Rnq1, which is known as the non-prion domain preceding the asparagine and glutamine rich prion domain of Rnq1. Notably, overexpression of these rnq1 mutant proteins restored [PIN+] prion activity, suggesting that each of the rnq1 mutants was not completely inactive. These findings indicate that the N-terminal non-prion domain of Rnq1 harbors a potent activity to regulate the maintenance of the [PIN+] prion.
Key words: Rnq1, [PIN+], Sup35, [PSI+], yeast prion
Introduction
A prion is known as a “protein only” genetic element,1 capable of transmitting its self-propagating conformation to an otherwise non-infectious, non-amyloidogenic protein in an infectious manner.2 The mammalian prion protein, PrP, is known to cause numerous infectious diseases including scrapie (sheep), bovine spongiform encephalopathy (BSE, cow), chronic wasting (deer and elk), kuru and Creutzfeld-Jacob disease (humans). Prions also exist in Saccharomyces cerevisiae as non-Mendelian inheritable elements. [PSI+] is one such yeast prion that has been well characterized by genetic and biochemical studies. [PSI+] is the prion form of Sup35, which is also known as the polypeptide release factor eRF3 and is essential for terminating protein synthesis at stop codons3,4 (reviewed in ref. 5). When Sup35 is in the [PSI+] state, ribosomes often read through stop codons due to impaired release factor function, causing a non-Mendelian trait easily detected by nonsense suppression.6–8 The molecular mechanisms of both maintenance and transmission of [PSI+] are fascinating subjects in prion biology.
There are some reports that the maintenance or de novo appearance of one prion is affected by heterologous prion variants.9,10 The prion [PIN+], named for its [PSI+] inducing properties,11–13 is required for efficient [PSI+] appearance,14 but not for [PSI+] propagation.15 [PIN+] is the prion form of Rnq1, a protein of unknown function. Rnq1 is one of several known yeast proteins containing an asparagine (N) and glutamine (Q) rich prion domain, hence named so for rich in N and Q.13 Two models, “seeding” and “titration,” have been proposed to explain how heterologous prions (e.g., [PIN+]) facilitate the de novo appearance of [PSI+]. According to the seeding model, a heterologous pre-existing protein in the prion conformation templates the conversion of Sup35 into its prion form, which then proceeds to seed its own rapid and separate aggregation. Supporting data for the model was reported in vivo16 and in vitro.17 The alternative titration model postulates that pre-existing heterologous prions or prion-like aggregates capture and inactivate an inhibitor that prevents conversion of Sup35 into a prion.11,12 As of yet, neither model has been proved or disproved.
To date, it is widely accepted that Rnq1 plays a positive role in facilitating the conversion of other prions in [PIN+] cells.11,12 Previously, the minimum domains of Rnq1 and their roles in [PIN+] prion maintenance have been determined by deletion analyses.13,18,19 However, these deletion analyses do not define the specific residues required for the maintenance of [PIN+] or the ability of [PIN+] to promote [PSI+] induction. Thus, the molecular roles of Rnq1, including its structure-function relationship to prion inducing activity, remain largely unknown. This is in part due to the limited availability of defective rnq1 alleles. Although some rnq1 derivatives are known, an extensive or systematic isolation of loss-of-function alleles in rnq1 has not been reported. The biological activity of Rnq1 to facilitate the de novo appearance of [PSI+] in [PIN+] cells is not easily, or directly, applicable to a genetic selection for loss-of-[PIN+] mutations in rnq1. To conduct an unbiased comprehensive search for rnq1 alleles that disable [PIN+] activity, we took advantage of a prion inhibitor, Rnq1Δ100.19,20 Rnq1Δ100 lacks the N-terminal non-Q/N rich domain and eliminates [PSI+] when overproduced in the [PIN+] state but not in the [pin-] state, allowing us to find loss-of-[PIN+] rnq1 mutations as [PSI+] colonies. Using this genetic system, we isolated nine rnq1 mutants with single amino acid substitutions. Interestingly, eight of these are localized to the N-terminal non-prion domain of Rnq1.
Results
Isolation of rnq1 mutants defective in the maintenance of [PIN+].
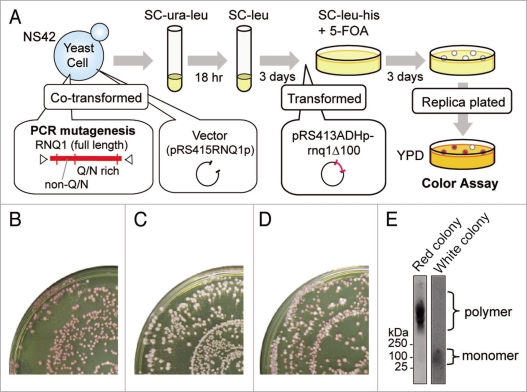
Overexpression of Rnq1Δ100 is inhibitory to the maintenance of [PSI+] in the [PIN+] state, not in the [pin−] state, causing a white-to-red color change of [PSI+] ade1-14 (nonsense) colonies.19 Therefore, any rnq1 mutation that impaired [PIN+] would still yield a white colony in the presence of overexpressed Rnq1Δ100. We took advantage of this color-based assay to select for rnq1 mutants. The rationale of mutant selection, summarized below, is illustrated in Figure 1A. The parental strain NS42 was [PSI+] [PIN+] ade1-14, its chromosomal RNQ1 gene was deleted by rnq1::KanMX and Rnq1 was instead expressed from pRS416RNQ1p-RNQ1 (URA3 marker) using the authentic promoter of RNQ1. Strain NS42 was transformed with a PCR-mutated rnq1 plasmid (pRS415RNQ1p-rnq1: see Materials and Methods, marked with LEU2), and Ura+ Leu+ transformants were first selected in SC-ura-leu medium. Then, Ura− (i.e., pRS416RNQ1p-RNQ1 segregant) Leu+ (i.e., pRS415RNQ1p-rnq1 transformant) colonies were grown in SC+ura-leu medium. These plasmid-shuffled cells were transformed with pRS413ADHp-rnq1Δ100 (marked with HIS3)20 and grown on SC-leu-his+5-FOA plates. Leu+ His+ 5-FOA resistant colonies were replica-plated onto YPD plates for color assay.
Figure 1.
Procedures to screen for mutant alleles. (A) Schematic of the screen for rnq1 mutants. A wild-type RNQ1 plasmid in strain NS42 ([PSI+] [PIN+] rnq1::KanMX) was replaced with mutant rnq1 plasmids by plasmid shuffling. These colonies were white, representing the [PSI+] state, and were further transformed with a Rnq1Δ100 plasmid that is able to eliminate [PSI+] prion when cells are [PIN+]. These doubly transformed cells were examined for colony color by replica plating on YPD. (B) [PIN+] control. Cells shuffled with wild-type RNQ1 turned red upon expression of Rnq1Δ100. (C) [pin−] control. Cells shuffled with an empty plasmid no longer synthesized Rnq1, became [pin−], and remained white as these were resistant to Rnq1Δ100's action. (D) Loss-of-function rnq1 colonies appeared upon mutagenesis. Cells shuffled with mutagenized rnq1 generated white colonies among the majority of red colonies upon expression of Rnq1Δ100. (E) Representative tests for the [PIN+] or [pin−] status of red and white colonies by SDD-AGE. Left, a red colony with a SDS-stable polymer ([PIN+]). Right, a white colony without a SDS-stable polymer ([pin−]).
In control experiments, colonies turned red in the presence of wild type Rnq1, and remained white in the absence of Rnq1 (Fig. 1B and C), showing that Rnq1Δ100 eliminates [PSI+] in the [PIN+] state, not in the [pin−] state, as expected. Under these conditions, there appeared a significant number of white colonies amongst a majority of red colonies (Fig. 1D). Here, we confirmed that white colonies were truly [pin−] by monitoring SDS-stable polymers of Rnq1, a [PIN+] prion signature, detectable by SDD-AGE (Fig. 1E). In white colonies, Rnq1 exists as a monomer. Approximately 40,000 mutant clones were screened and 150 mutants were isolated. The resulting Rnq1-aggregation defective mutants were characterized by DNA sequencing. Of 150 mutant clones examined (including those carrying multiple mutations, frameshift or nonsense mutations in rnq1), nine distinct single-amino-acid substitutions were finally defined to impair (or weaken) Rnq1Δ100-mediated [PSI+] elimination.
Sequence and protein stability analysis of rnq1 mutants.
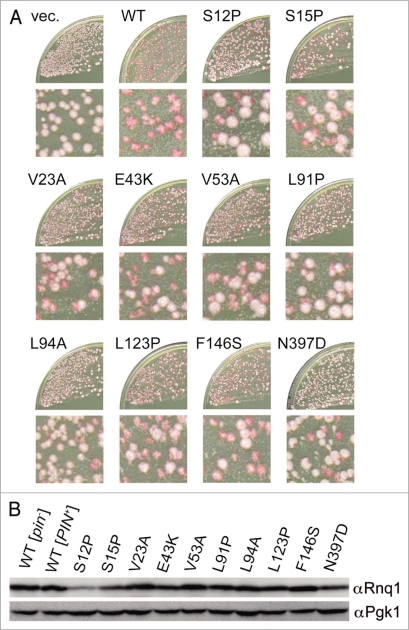
There seemed to be phenotypic diversity in the defined rnq1 mutants, as some mutants gave rise to mostly white colonies while others gave rise to a mixture of white and red colonies, upon substitution of wild-type RNQ1 with rnq1 alleles by plasmid shuffling in the NS42 strain (Fig. 2A). We included a rnq1 mutant having a L94A amino acid change in the mutant library since it has been reported that the L94A protein is defective in binding to a member of the Hsp40 chaperone family, Sis1, and its overexpression is toxic in [PIN+] cells.26 The quantitative analysis of loss-of-[PIN+] frequency by rnq1 alleles will be described shortly.
Figure 2.
Rnq1 mutants defective in stable propagation of [PIN+]. The defined rnq1 mutants contain the indicated single amino acid substitutions. (A) The appearance of white colonies upon substitution of rnq1 alleles for wild-type RNQ1 in NS42 strain after plasmid shuffling. NS42 ([PSI+] [PIN+] rnq1::KanMX) cells containing plasmid pRS415 (empty vector, vec.) or its derivatives carrying wild-type (WT) or mutant rnq1 alleles were transformed with pRS413ADHp-rnqΔ100, and the double transformants were plated on YPD. The frequency of red-to-white colony conversion varied depending on the individual mutations. Enlargements are also shown. (B) Protein levels of mutant Rnq1s in [PIN+] cells. The [PIN+] or [pin−] status of colonies of strain NS43 ([psi−] [PIN+]) carrying pRS415RNQ1p derivatives expressing wild-type or mutant Rnq1s was assigned by fluorescence microscopy using a Rnq1-GFP reporter plasmid, pRS413CUP1p-RNQ1-GFP. After segregation of plasmid pRS413CUP1p-RNQ1-GFP, [PIN+] cells from each rnq1 mutant were lysed by boiling in SDS sample buffer and analyzed by western blotting using anti-Rnq1 (upper) and anti-Pgk1 (lower, loading control) antibodies.
Protein levels of mutant Rnq1s in these transformant cells were examined by western blotting. Since expression of rnq1 alleles after plasmid shuffling essentially produced a mixture of white and red colonies, we first classified the [PIN+] or [pin−] status of individual colonies by monitoring Rnq1 aggregation with a Rnq1 and green fluorescent protein (Rnq1-GFP) fusion protein. [PIN+] and [pin−] colonies were determined by the presence or absence of Rnq1-GFP foci, respectively, and were then individually examined for Rnq1 levels after segregation of a Rnq1-GFP-reporter plasmid by western blotting. It is known that Rnq1 protein is relatively unstable in [pin−] cells.20 To avoid degradation of mutant Rnq1 proteins during manipulation, harvested yeast cells were directly boiled with SDS-sample buffer to prepare Rnq1 proteins for SDS-PAGE. The abundance of mutant Rnq1 proteins in these transformants was either unaffected (V23A, V53A, L91P, L94A, L123P, F146S and N397D) or reduced partially (S12P, S15P and E43K) relative to wild-type (Fig. 2B). Three immunoblotting experiments using independent cell cultures confirmed the reproducibility. Similar propensity appeared in [pin−] cells (data not shown). Therefore, we assume that, for the seven unaffected mutants, the loss of [PSI+] elimination ability of Rnq1Δ100 was mainly caused by the loss of protein activity, but not stability, of Rnq1; in addition, for the other three mutants, it might be also caused by the instability of Rnq1 mutant protein.
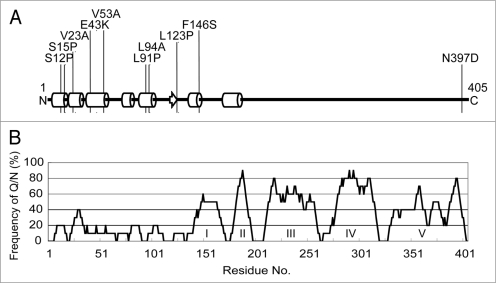
DNA sequence analysis revealed that eight out of the nine selected rnq1 mutations, as well as the L94A mutation, are mapped within the N-terminal non-Q/N rich (i.e., non-prion) domain, mostly localized to the α-helix sub-domains (Fig. 3). It is intriguing that the truncated N-terminal non-prion domain of Rnq1, Rnq1Δ100, inhibits the maintenance of [PSI+] when rnq1Δ100 was overexpressed, while multiple single mutations in the same domain hampers the maintenance of [PIN+], suggesting that the N-terminal non-prion domain is not completely dispensable and is rather important for proper functioning of [PIN+] as well as proper, direct or indirect, interaction with a second prion, [PSI+].
Figure 3.
Defective rnq1 mutations are localized to the N-terminal non-prion forming domain of Rnq1. (A) Secondary protein structure prediction of Rnq1 generated from the PSIPRED Protein Structure Prediction Server [http://bioinf.cs.ucl.ac.uk/psipred].35 Circular cylinders, an arrow and bold lines indicate α-helices, β-sheet and coils, respectively. Numbers represent the amino acid positions from the first Met codon. Positions of amino acid substitutions are shown. (B) Schematic diagram of non-Q/N rich and Q/N rich regions in the Rnq1 protein. The frequency of Q/N residues was calculated from every ten amino acid interval. Q/N-rich sub-regions are indicated in roman numerals.19 Note that amino acid positions are shown in the same scale in (A and B) for comparison.
Impaired [PIN+] phenotype of rnq1 mutants.
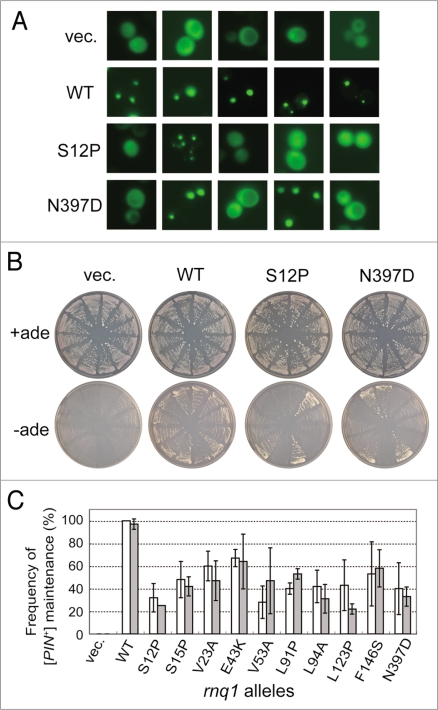
We assumed that the defined rnq1 mutants and the L94A mutant might not be complete loss-of-function mutants but rather retain a residual activity because substitution of rnq1 alleles for RNQ1 in NS42 strain generated a mixture of white ([pin−]) and red ([PIN+]) colonies at different ratios (as shown in Fig. 2A). The frequency of prion curing was examined quantitatively by measuring [PIN+] or [pin−] status of plasmid-shuffled NS43 colonies. Two different methods were used to monitor prion status. First, Rnq1 aggregates were monitored with a Rnq1-GFP probe, and second, the Pin+ phenotype, the de novo induction of [PSI+] upon overexpression of Sup35 NM polypeptide was examined. First, when Rnq1-GFP was expressed from the CUP1 promoter in the presence of CuSO4, Rnq1-GFP formed punctate foci in [PIN+] cells, while it showed mostly cytoplasmic, dispersed fluorescence in [pin−] cells, as expected (Fig. 4A). Under these conditions, three sets of 20 randomly chosen rnq1 colonies were examined for Rnq1-GFP fluorescence (as shown in Fig. 4A); the [PIN+] or [pin−] status was scored and summarized in Figure 4C. SDD-AGE analysis also reinforced the results (data not shown). Secondly, the Pin+ phenotype was examined using the same sets of 12 randomly chosen rnq1 colonies for their ability to induce [PSI+]. Accordingly, these colonies were transformed with plasmid pCUP1p-SUP35NM, a pRS413 (HIS3)-based plasmid overexpressing Sup35's NM prion domain from the CUP1 promoter.19 Transformants were grown on SC-his-leu medium plates supplemented with 5-FOA and 50 µM CuSO4 for 3 days (about 72 hr), subsequently streaked on SC+ade and SC-ade plates, and then grown for 7 days. As shown in Figure 4B, colonies appeared on SC-ade plates in the [PIN+] state but not in the [pin−] state, such that rnq1 mutants were able to produce Ade+ colonies due to their prion status. The frequency of Pin+ colonies estimated by this means is shown in Figure 4C. The quantitative analysis of [PIN+] colonies based on these two different assays is more or less consistent, and clearly demonstrates that the selected rnq1 alleles are defective in the stable maintenance of [PIN+], although they may retain some residual capacity.
Figure 4.
Quantitative analysis of defects of rnq1 mutants in [PIN+] propagation. (A) Fluorescence microscopy was performed to determine the [PIN+] or [pin−] status of each colony using a Rnq1-GFP reporter. Upon substitution of mutant Rnq1s for the wild-type Rnq1 by plasmid shuffling in strain NS43 ([psi−] [PIN+]), three sets of 20 independent colonies were randomly chosen and again transformed with pRS413CUP1p-Rnq1-GFP to visualize the [PIN+] or [pin−] state with Rnq1-GFP fusion protein. Rnq1-GFP expression under the control of the CUP1 promoter was induced by 50 µM CuSO4 for 3 days on SC plates. Panels show five randomly chosen fluorescent images of each Rnq1 sample. Samples: vec., an empty plasmid; WT, wild-type Rnq1; S12P and N397D, mutant Rnq1s. (B) Pin+ activity monitored by de novo appearance of [PSI+] colonies. Three sets of 12 independent plasmid-shuffled colonies were transformed with plasmid pRS413CUP1p-SUP35NM, and upon overexpression of the Sup35NM domain, these cells were grown on SC+ade (control) and SC-ade plates for seven days. (C) The frequency of appearance of Rnq1-GFP foci (open boxes) and de novo induction of [PSI+] (gray boxes) is expressed as the mean of three independent experiments with standard deviations.
Transmission of [PIN+] from rnq1 mutants to wild-type Rnq1.
To confirm [PIN+] prion maintained with the rnq1 mutants (S12P, V23A and L94A), we examined transmission of [PIN+] from plasmid-shuffled rnq1 mutants to wild-type Rnq1 strain by mating. The rnq1 shuffled mutants, in which mutant Rnq1s were expressed from either the weak RNQ1 promoter or the strong ADH promoter (see below), were mated with [psi−] [pin−] wild-type RNQ1 cells (NPK175 or NPK569), and plasmids carrying the rnq1 alleles were segregated from these diploids. The [PIN+] status of the resulting diploids that express only wild-type Rnq1 was analyzed by the Rnq1-GFP fluorescence assay and the [PSI+]-inducibility (Pin+) test. As shown in Table 1, [PIN+] phenotype was transmitted from S12P, V23A and L94A mutants with both RNQ1 and ADH promoters to wild-type Rnq1 although the transmission frequency is seemingly lower in the rnq1 mutants expressed from the RNQ1 promoter than that from the ADH promoter. In these experiments, the imperfect transmission frequency was due to prion loss during growth and, unexpectedly, the transmission frequency to wild-type Rnq1 is seemingly greater than the frequency of [PIN+] maintenance (as shown in Fig. 4C). We assume that mating with wild-type Rnq1 might prevent prion loss during a long incubation time. Importantly, [PIN+] phenotype was transmitted in all or some cells, indicating that the rnq1 alleles maintained a [PIN+] prion.
Table 1.
Transmission of [PIN+] from Rnq1 mutants to wild-type Rnq1
| Frequency of [PIN+] transmission from rnq1 alleles expressed from: | ||||
| rnq1 alleles | RNQ1 promoter | ADH promoter | ||
| GFP foci | [PSI+] induction | GFP foci | [PSI+] induction | |
| empty vector | 0/10 | 0/10 | 0/5 | 0/5 |
| wild-type | 10/10 | 10/10 | 5/5 | 4/5 |
| S12P | 3/20 | 5/20 | 5/5 | 5/5 |
| V23A | 7/10 | 7/10 | 5/5 | 5/5 |
| L94A | 7/10 | 8/10 | 5/5 | 5/5 |
The plasmid-shuffled rnq1 strains were mated with [psi−] [pin−] wild-type RNQ1 cells; After confirming the presence of SDS-stable Rnq1 polymers in the selected diploids by SDD-AGE, the rnq1 mutant plasmids were segregated from these diploids; The [PIN+] state of these diploid colonies was examined upon transformation with Rnq1-GFP expressing plasmid or Sup35-NM domain overexpressing plasmid; Colonies showing Rnq1-GFP foci and [PSI+] inducibility were counted.
Interaction of Sis1 with mutant Rnq1 proteins.
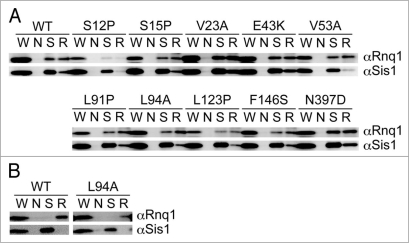
Sis1, an essential Hsp40 chaperone, is required for the propagation of the [PIN+] prion state, probably through catalyzing generation of [PIN+] seeds.27 Sis1 not only promotes [PIN+] prion formation, but remains stably bound to the prion in a 1:1 complex.28 We examined whether mutant Rnq1 proteins are unable to interact with Sis1 by immunoprecipitation in [PIN+] cells since it is known that Sis1 does not bind to Rnq1 when cells are [pin−].27 Derivatives of NS43 ([psi−] [PIN+]) strain in which RNQ1 was replaced by rnq1 alleles after plasmid shuffling were transformed with a Rnq1-GFP-expression plasmid, and colonies that formed punctate foci were assigned as [PIN+]. The resulting [PIN+] cells were used to prepare lysates after segregation of the Rnq1-GFP-expression plasmid. The [PIN+] state of these cells was confirmed by SDD-AGE analysis (data not shown). Proteins that were immunoprecipitated by anti-Sis1 antibody or anti-Rnq1 antibody were analyzed by immunoblotting with anti-Rnq1 or anti-Sis1 antibodies, respectively. As shown in Figure 5A, Sis1 remained bound to most mutant Rnq1 proteins, with the exception of S12P. Three immunoblotting experiments using independent cell cultures reproduced the weak recovery of Rnq1-S12P protein by immunoprecipitation. We assume that the S12P mutant protein lost or weakened the ability to interact with the antibody, not with Sis1. Nevertheless, it could not be excluded at present that the S12P mutant protein might be degraded during growth or manipulation as suggested by the immunoblotting experiment (Fig. 2B). Under these conditions, Rnq1 did not interact with Sis1 in [pin−] (Fig. 5B) as reported previously.
Figure 5.
Immunoprecipitation of Sis1 complexes. (A) [PIN+] (NS43) cells transformed with pRS415RNQ1p plasmids expressing wild-type (WT) or the indicated mutant Rnq1 proteins were harvested, and subjected to immunoprecipitation with anti-Sis1 (S), anti-Rnq1 (R), or no (N) antibodies. Immunoprecipitated proteins were analyzed by western blotting using anti-Rnq1 (αRnq1) and anti-Sis1 (αSis1) antibodies. Lanes: W, whole cell lysate; N, no immunoprecipitating antibody; S, anti-Sis1 antibody immunoprecipitation; R, anti-Rnq1 antibody immunoprecipitation. Immunoblotting was repeatedly performed using three to five independent shuffled transformants. (B) [pin−] cells transformed with pRS415RNQ1p plasmids expressing wild-type (WT) or Rnq1-L94A proteins were harvested, and subjected to the immunoprecipitation analysis as shown in (A).
A classical, hydrophobic chaperone-binding motif, LGKLALL,29 has been proposed to be the Sis1-binding site in Rnq1 at positions 91–97.26 Two relevant substitutions, L91P and L94A, less efficiently bound to Sis1 in [PIN+] cells (Fig. 5A), roughly consistent with a previous report.26 The binding efficiency will be discussed shortly. L94A, as well as wild-type Rnq1 protein, did not interact with Sis1 in [pin−] (Fig. 5B).
Suppression of impaired [PIN+] phenotype by hyperexpression of mutant Rnq1 proteins.
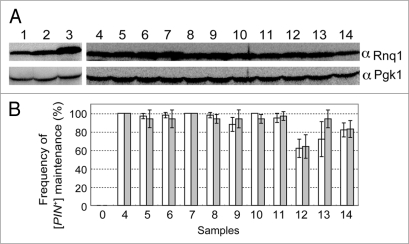
As mentioned above, rnq1 mutants had residual activity to propagate [PIN+] when expressed from pRS415RNQ1p-rnq1 plasmids using the authentic promoter of RNQ1. This, in turn, suggests that the observed impairment of rnq1 could be compensated for by overexpression of mutant Rnq1s. To test this possibility, rnq1 alleles were cloned into pRS413ADHp carrying the strong ADH promoter. These pRS413ADHp plasmids carrying wild-type and mutant rnq1 alleles were transformed into the NS43 (rnq1::KanMX) strain, and the protein abundance of each mutant Rnq1 was examined by western blotting (Fig. 6A). As expected, Rnq1 levels were several fold higher when driven by the ADH promoter (Fig. 6A, lane 3) as opposed to the RNQ1 promoter (Fig. 6A and compare lane 3 with lane 2). It is also noteworthy that expression levels of Rnq1 from plasmid pRS415RNQ1p-RNQ1 were similar to those expressed from the chromosome in the wild-type strain (Fig. 6A and compare lane 2 with lane 1). Under these conditions, the abundance of the mutant Rnq1 proteins expressed from the ADH promoter was equivalent to that of the wild-type Rnq1 synthesized with plasmid pRS413ADHp-RNQ1 (Fig. 6A, lanes 4 through 14).
Figure 6.
Suppression of defective rnq1 mutants by overexpression of mutant Rnq1 proteins. (A) Overexpression of Rnq1 proteins examined by western blotting. Experimental conditions and procedures are the same as described in Figure 2 except that rnq1 alleles are expressed from the strong ADH promoter substituted for the authentic RNQ1 promoter. Yeast strains used were NS43 ([psi−] [PIN+]) derivatives whose chromosomal RNQ1 allele is either wild type (lane 1) or nullified with rnq1::KanMX (lanes 2 through 14). Plasmids: 1, plasmid free control; 2, pRS413RNQ1p-RNQ1; 3 and 4, pRS413ADHp-RNQ1; 5, pRS413ADHp-S12P; 6, pRS413ADHp-S15P; 7, pRS413ADHp-V23A; 8, pRS413ADHp-E43K; 9, pRS413ADHp-V53A; 10, pRS413ADHp-L91P; 11, pRS413ADHp-L94A, 12, pRS413ADHp-L123P; 13, pRS413ADHp-F146S; 14, pRS413ADHp-N397D. (B) Frequency of appearance of Rnq1-GFP foci (open boxes) and de novo induction of [PSI+] (gray boxes). Experimental conditions and procedures are the same as described in Figure 5. Three sets of 20 independent colonies were chosen for observation of Rnq1-GFP fluorescence and 12 for monitoring Pin+ activity generated by plasmid shuffling for each rnq1 allele were examined and scored. Results are expressed as the mean of three independent experiments with standard deviations.
The compensatory effect of mutant Rnq1 overexpression on [PIN+] propagation was then investigated via the same procedures as described above (see Fig. 4). NS43 ([psi−] [PIN+]) cells were transformed with pRS413ADHp-rnq1 (mutant) plasmids and three sets of 20 or 12 randomly chosen transformant colonies were examined for the formation of Rnq1-GFP fluorescent foci or the de novo induction of [PSI+] upon overexpression of Sup35 NM polypeptide, respectively. The data are summarized in Figure 6B. The impaired activity of rnq1 mutants in stable maintenance of [PIN+] was mostly compensated for by overexpression of mutant Rnq1 proteins (although the suppression level of L123P remained partial). Therefore, all the defined rnq1 mutants are markedly, but not completely, defective in [PIN+] propagation. It is rather surprising that no single mutant resulting in complete loss-of-function was obtained through this screen given that the genetic screen itself worked properly.
Discussion
We used a [PIN+]-dependent [PSI+] inhibitor, Rnq1Δ100, as a proxy for the [PIN+] status of the cell to identify rnq1 mutants with defects in [PIN+] propagation. Based on the colony color assay, we identified nine rnq1 alleles that are defective in stable [PIN+] propagation. All these rnq1 mutants showed remarkable reductions in Rnq1 aggregate formation (as monitored with an Rnq1-GFP fluorescent probe) and in de novo [PSI+] prion induction (by Sup35 NM overexpression). Furthermore, we found that Rnq1-L94A was also defective in stable [PIN+] propagation. Interestingly, nine of these rnq1 mutations are mapped within the N-terminal, so-called non-prion domain of Rnq1, and only one allele, N397D, is mapped at the C-terminus (see Fig. 3). This was surprising since the C-terminus (amino acids 153–405) of Rnq1 was shown to be sufficient for joining or forming a prion aggregate.13 It is widely accepted that the prion forming ability of prion proteins resides within the Q/N-rich regions.13,30–32 The C-terminus of Rnq1 is Q/N rich, composed of several Q/N tracts separated by hydrophobic regions (see Fig. 3).
This discrepancy could be explained by assuming that while the C-terminus of Rnq1 is sufficient for forming a prion aggregate, the N-terminus of Rnq1 plays a regulatory role in [PIN+] propagation. Consistent with this prediction, deletions within the N-terminus of Rnq1 cause [PIN+] to be transmitted inefficiently,18,20 and expression of Rnq1Δ100, which lacks the N-terminal 100 amino acids, inhibits propagation of [PSI+] in a [PIN+] background.19 Moreover, the N-terminus of Rnq1 is known to be responsible for binding to the chaperone Sis1,26 which is essential for [PIN+] propagation.26,27,33 During the course of writing this manuscript, we became aware of a very recent article that warrants mention. Bardill and True34 have tried to isolate mutants in Rnq1 that are unable to propagate the [PIN+] prion using a chimeric reporter, called RRP, that is composed of the C-terminus prion-forming domain of Rnq1 (amino acids 153–405) and the M and C domains of Sup35 (amino acids 124–685). Using this reporter, they defined five specific mutations in the C-terminus of Rnq1 (F184S, S223P, Q239R, N297S and Q298R) that decreased the ability of [PIN+] to induce [PSI+]. However, these mutants do not appear to significantly affect [PIN+] propagation within the context of the full-length form of Rnq1. They speculated that one possible reason for this discrepancy is the addition of the N-terminus. Considering this speculation, our present findings are consistent with theirs, and it is rather surprising that the results shown by two separate approaches indicate, albeit unexpectedly, the importance of the N-terminus non-prion domain for the propagation of [PIN+] prion.
The nine rnq1 mutants isolated in our screen and L94A exhibit interesting features. First, most of them are mapped within the predicted α-helical regions of the N-terminus of Rnq1. Although the function of these α-helixes is not known, it is reasonable to speculate that these are involved in modulating proper function of the C-terminal prion domain by intra- or inter-molecular interactions. The C-terminus of Rnq1 is Q/N rich, and therefore readily makes protein-protein interactions, leading to self-aggregation of Rnq1 in the absence of the N-terminal non-Q/N rich domain.19 Second, despite extensive screening for loss-of-function rnq1 alleles, all the defined rnq1 mutants are not completely null and possess residual activities to some extent. In fact, the observed defects in [PIN+] propagation could be compensated for by overexpression of the mutant Rnq1 proteins. Therefore, it seems likely that the propagation of the [PIN+] prion is difficult to disrupt by single amino acid substitutions. This, in turn, suggests that Rnq1's ability to promote prion seeding with homologous (Rnq1) or heterologous (Sup35) proteins could not be attributed to a specific unique residue but to several residues or regions within Rnq1. Probably the mode of protein-protein interaction for prion seeding requires multiple interactions between [PIN+] prion seeds and the recipient proteins.
It is generally believed that [PIN+] is required for efficient [PSI+] appearance,14 but not for [PSI+] propagation.15 However, the existence of Rnq1Δ100, which inhibits the maintenance of [PSI+], might suggest a possible involvement of Rnq1 prion in the maintenance of heterologous prion [PSI+],19 and that Rnq1Δ100 is defective in catalyzing the maintenance of [PSI+]. Assuming this possibility, we could speculate that the given rnq1 mutants were isolated primarily as those that regained the function to catalyze the maintenance of [PSI+], not as loss-of-[PIN+] mutants. Given this scenario, perhaps, the putative [PSI+]-maintaining activity might be located in soluble N-terminal region of Rnq1, and Rnq1Δ100 lost this activity and hindered the activity of the full-length Rnq1 by forming the Rnq1Δ100·Rnq1 tight aggregates in [PIN+] cells. Therefore, one might speculate that the rnq1 mutants recovered allow for suppression based on the ability of soluble N-terminal domain to perform some function. Perhaps, the mutations change the folding such that the N-terminus is available for that function. Alternatively, a small soluble pool of Rnq1 created by these mutations may be enough. Since this is one of many explanations, further experiments are required to uncover the mechanism.
By in vitro peptide array analysis, the Sis1-binding site in Rnq1 was determined to be amino acids 91-97, LGKLALL, and these residues are also a classical, hydrophobic chaperone-binding motif.26,29 It has been reported that the capacity of Rnq1-GFP to interact with Sis1 is strongly, but not completely, reduced by mutations in this motif, L91A, L94A and L97A.26 In accordance with this indication, L91P and L94A Rnq1 proteins were immunoprecipitated with Sis1 less efficiently compared with wild-type Rnq1 (Fig. 5A). The degree of reduction, however, was not striking in our study, which was presumably owing to differences in experimental conditions from the previous report. This could suggest that there is a somewhat significant change in the interaction with Sis1, leading to unstable [PIN+] propagation. The Sis1-binding site remains to be investigated rigorously.
In sum, these ten rnq1 alleles specifically impair [PIN+] propagation. Additionally, these findings clearly suggest an importance for the N-terminal non-prion domain of Rnq1 to regulate [PIN+] propagation. These rnq1 mutants will prove useful for elucidating intra- and inter-molecular interactions required for the [PIN+] propagation and heterologous prion induction.
Materials and Methods
Strains and manipulations.
S. cerevisiae strains used in this study are: NS42 ([PSI+] [PIN+] MATa ade1-14 leu2-3, 112 ura3-52 his3Δ200 trp1-289 rnq1::KanMX [pRS416RNQ1p-RNQ1 (ARS/CEN, URA3 marker)]), NS43 ([psi−] derivative of NS42 constructed by transient overexpression of HSP104), NPK175 ([psi−] [pin−] MATα ade1-14 leu2 ura3 trp1)21 and NPK569 ([psi−] [pin−] MATa ade1-14 leu2 ura3 his3).20 The RNQ1 deletion strain was generated by transformation with a PCR product of the rnq1::KanMX sequence (American Type Culture Collection catalog no. 4013435) essentially as described previously.19 The yeast media used were YPD (1% yeast extract, 2% polypeptone, 2% dextrose) and synthetic complete glucose (SC) (0.67% yeast nitrogen base without amino acid [DIFCO] and 2% dextrose) supplemented with adenine, leucine, uracil, histidine, tryptophan, or 5-fluoroorotic acid (5-FOA) as required. Yeast cells were grown at 30°C in YPD or in SC media with appropriate supplements. To monitor colony color based on the ade1-14 nonsense suppression, yeast cells were grown on YPD plates for 4 days at 30°C. Transformation was performed using Frozen-EZ Yeast Transformation II (ZYMO Research, Orange, CA) according to the manufacturer's instructions.
Plasmids.
Plasmids used are pRS400 series vectors (Stratagene). pRS416RNQ1p-RNQ1 (ARS/CEN, URA3 marker) or pRS415RNQ1p-RNQ1 (ARS/CEN, LEU2 marker) carrying RNQ1 from nucleotide positions −228 (counted from the transcript start site) to 1653 were constructed by cloning a PCR fragment amplified with P1 (5′-TTT GAG CTC GGT ATT TCA AAC GCA TAC-3′) and P2 (5′-ATA CTC GAG TTT GAT TTT CAG AAA CTT G-3′) primers into the BamHI-XhoI site of pRS416 or pRS415. Expression vectors contain the ADH, CYC1 or CUP1 promoter and the CYC1 terminator for expression of exogenous sequences cloned into the BamHI-XhoI site as described previously.19 Other expression plasmids used are: Rnq1, pRS413CYC1p-RNQ1 (ARS/CEN, HIS3 marker)20 and pRS413ADHp-RNQ1 (ARS/CEN, HIS3 marker);20 Rnq1Δ100, pRS413ADHp-rnq1Δ100 (HIS3 marker);20 Rnq1-GFP, pRS413CUP1p-RNQ1-GFP (ARS/CEN, HIS3 marker)21 and pRS414CUP1p-RNQ1-GFP (ARS/CEN, TRP1 marker);19 Sup35NM, pRS413CUP1p-SUP35NM (ARS/CEN, HIS3 marker)19 and pRS414CUP1p-SUP35NM (ARS/CEN, TRP1 marker).19 pRS415RNQ1p-rnq1(L94A) was constructed by site directed mutagenesis with P3 primer (5′-GCT AAC AAA GCT GCT TTT CCT AGT TGC-3′) using pRS415RNQ1p-RNQ1 as a template.
Isolation of rnq1 mutants.
The 1.5-kb fragment containing the RNQ1 coding sequence, 5′-flanking (RNQ1 promoter) and 3′-flanking (RNQ1 terminator) sequences was amplified by error-prone PCR using P4 (5′-ATG CCT GTC TCG TCC AAA CG-3′) and P5 (5′-CCT GCA GAG ATA CAA CTC TG-3′) primers in vitro, and co-transformed into the NS42 strain with a linearized, BglII-SnaBI, plasmid pRS415RNQ1p (ARS/CEN, LEU2 marker) carrying partial sequences for the RNQ1 promoter and terminator. The mutagenized rnq1 sequences were inserted in vivo into the vector by homologous recombination with the RNQ1 promoter and the RNQ1 terminator sequences. The resulting Ura+ Leu+ transformants were selected in SC-ura-leu liquid and incubated for 18 hr, and then cultured in SC+ura-leu liquid for 3 days. These cells were transformed with pRS413ADHp-rnq1Δ100 (ARS/CEN, HIS3 marker)20 and grown on SC-leu-his+5-FOA plates. Leu+ His+ 5-FOA resistant colonies were replica plated onto YPD plates to screen for white (i.e., [PSI+]) colonies. Plasmids were confirmed for reproducible phenotypes and characterized by DNA sequence analysis.
Induction of [PSI+] element.
[PSI+] was induced in [psi−] cells upon transformation with pCUP1p-SUP35NM (HIS3 or TRP1 marker). Transformants were grown in SC medium lacking selective nutrients and supplemented with 5-FOA and 50 µM CuSO4 for 3 days and were subsequently streaked on SC-ade and SC+ade plates (for control), and then grown for 7 days on SC-ade or for 4 days on SC+ade.
Protein analysis.
Yeast cells were lysed with glass beads as described previously,22 or lysed by boiling in SDS-sample buffer.23 SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and semi-denaturing detergent-agarose gel electrophoresis in the presence of 1% SDS (SDD-AGE24,25) were carried out as described previously.21 The immunoblot experiments were performed using anti-Rnq1 antibody,21 anti-Sis1 antibody19 and anti-Pgk1 antibody (catalog no. A-6457; Molecular Probes). Immunoprecipitations were performed as described previously.19
Acknowledgements
We thank Colin Crist and Keita Oishi for critical reading of the manuscript and valuable comments. This work was supported in part by grants from The Ministry of Education, Sports, Culture, Science and Technology of Japan (MEXT).
Abbreviations
- S. cerevisiae
Saccharomyces cerevisiae
- Q/N rich
asparagine and glutamine rich
- SC
synthetic complete
- 5-FOA
5-fluoroorotic acid
- SDS-PAGE
SDS-polyacrylamide gel electrophoresis
- SDD-AGE
semi-denaturing detergent-agarose gel electrophoresis
Footnotes
Previously published online: www.landesbioscience.com/journals/prion/article/10388
References
- 1.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB. Early evidence that a protease-resistant protein is an active component of the infectious prion. Cell. 2004;116:109. doi: 10.1016/s0092-8674(03)01032-8. [DOI] [PubMed] [Google Scholar]
- 3.Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, et al. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrenberg M, Hauryliuk V, Crist CG, Nakamura Y. Translation termination, prion [PSI+], and ribosome recycling. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control in biology and medicine. New York: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 6.Liebman SW, Sherman F. Extrachromosomal ψ+ determinant suppresses nonsense mutations in yeast. J Bacteriol. 1979;139:1068–1071. doi: 10.1128/jb.139.3.1068-1071.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 8.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 9.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA. 2002;99:16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol Cell Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN+] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 12.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 13.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 14.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI+] and [PIN+]: a two-prion system in yeast? EMBO J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci USA. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vitrenko YA, Gracheva EO, Richmond JE, Liebman SW. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J Biol Chem. 2007;282:1779–1787. doi: 10.1074/jbc.M609269200. [DOI] [PubMed] [Google Scholar]
- 18.Vitrenko YA, Pavon ME, Stone SI, Liebman SW. Propagation of the [PIN+] prion by fragments of Rnq1 fused to GFP. Curr Genet. 2007;51:309–319. doi: 10.1007/s00294-007-0127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurahashi H, Ishiwata M, Shibata S, Nakamura Y. A regulatory role of the Rnq1 nonprion domain for prion propagation and polyglutamine aggregates. Mol Cell Biol. 2008;28:3313–3323. doi: 10.1128/MCB.01900-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurahashi H, Shibata S, Ishiwata M, Nakamura Y. Selfish prion of Rnq1 mutant in yeast. Genes Cells. 2009;14:659–668. doi: 10.1111/j.1365-2443.2009.01297.x. [DOI] [PubMed] [Google Scholar]
- 21.Kurahashi H, Nakamura Y. Channel mutations in Hsp104 hexamer distinctively affect thermotolerance and prion-specific propagation. Mol Microbiol. 2007;63:1669–1683. doi: 10.1111/j.1365-2958.2007.05629.x. [DOI] [PubMed] [Google Scholar]
- 22.Crist CG, Nakayashiki T, Kurahashi H, Nakamura Y. [PHI+], a novel Sup35-prion variant propagated with non-Gln/Asn oligopeptide repeats in the absence of the chaperone protein Hsp104. Genes Cells. 2003;8:603–618. doi: 10.1046/j.1365-2443.2003.00661.x. [DOI] [PubMed] [Google Scholar]
- 23.Horvath A, Riezman H. Rapid protein extraction from Saccharomyces cerevisiae. Yeast. 1994;10:1305–1310. doi: 10.1002/yea.320101007. [DOI] [PubMed] [Google Scholar]
- 24.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- 25.Liebman SW, Bagriantsev SN, Derkatch IL. Biochemical and genetic methods for characterization of [PIN+] prions in yeast. Methods. 2006;39:23–34. doi: 10.1016/j.ymeth.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 26.Douglas PM, Treusch S, Ren HY, Halfmann R, Duennwald ML, Lindquist S, Cyr DM. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc Natl Acad Sci USA. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+] Mol Biol Cell. 2003;14:1172–1181. doi: 10.1091/mbc.E02-09-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DePace AH, Santoso A, Hillner P, Weissman JS. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell. 1998;93:1241–1252. doi: 10.1016/s0092-8674(00)81467-1. [DOI] [PubMed] [Google Scholar]
- 31.Patel BK, Liebman SW. “Prion-proof” for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN+] J Mol Biol. 2007;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masison DC, Maddelein ML, Wickner RB. The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation. Proc Natl Acad Sci USA. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007;26:3794–3803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bardill JP, True HL. Heterologous Prion Interactions Are Altered by Mutations in the Prion Protein Rnq1p. J Mol Biol. 2009;388:583–596. doi: 10.1016/j.jmb.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]