Abstract
Auxin homeostasis is pivotal for normal plant growth and development. The superroot2 (sur2) mutant was initially isolated in a forward genetic screen for auxin overproducers, and SUR2 was suggested to control auxin conjugation and thereby regulate auxin homeostasis. However, the phenotype was not uniform and could not be described as a pure high auxin phenotype, indicating that knockout of CYP83B1 has multiple effects. Subsequently, SUR2 was identified as CYP83B1, a cytochrome P450 positioned at the metabolic branch point between auxin and indole glucosinolate metabolism. To investigate concomitant global alterations triggered by knockout of CYP83B1 and the countermeasures chosen by the mutant to cope with hormonal and metabolic imbalances, 10-day-old mutant seedlings were characterized with respect to their transcriptome and metabolome profiles. Here, we report a global analysis of the sur2 mutant by the use of a combined transcriptomic and metabolomic approach revealing pronounced effects on several metabolic grids including the intersection between secondary metabolism, cell wall turnover, hormone metabolism, and stress responses. Metabolic and transcriptional cross-talks in sur2 were found to be regulated by complex interactions between both positively and negatively acting transcription factors. The complex phenotype of sur2 may thus not only be assigned to elevated levels of auxin, but also to ethylene and abscisic acid responses as well as drought responses in the absence of a water deficiency. The delicate balance between these signals explains why minute changes in growth conditions may result in the non-uniform phenotype. The large phenotypic variation observed between and within the different surveys may be reconciled by the complex and intricate hormonal balances in sur2 seedlings decoded in this study.
Keywords: Abiotic/environmental stress, hormonal regulation, metabolomics, secondary metabolism/natural products, transcriptome analysis, development
INTRODUCTION
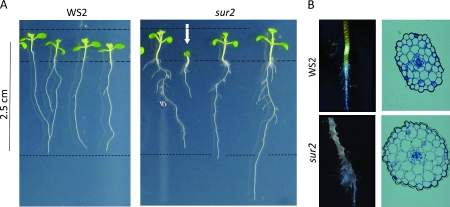
The superroot2 (sur2) mutant in Arabidopsis thaliana was initially identified in a forward genetics screen for auxin overproducers (Delarue et al., 1998). As for the sur1 (superroot1) mutant (Boerjan et al., 1995), the name sur2 was assigned to recognize the increased number of lateral roots as the main phenotypic characteristic of high auxin levels (Figure 1A). SUR2 was suggested to control auxin conjugation and thereby regulate auxin homeostasis. Indication of a high auxin phenotype is also the observed cellular expansion of the hypocotyls and lateral and adventitious root formation (Figure 1B). That the phenotype cannot be exclusively ascribed as an auxin phenotype is apparent by the epinastic coteledons and excessive root hair formation—a phenotypic effect resembling high ethylene levels (Figure 1B). Later, SUR2 was identified as the cytochrome P450 enzyme CYP83B1 (Bak et al., 2001; Barlier et al., 2000), which catalyzes the conversion of indole-3-acetaldoxime to an S-alkyl-thiohydroximate in the presence of a thiol donor in the indole glucosinolate biosynthetic pathway (Bak et al., 2001). The identification of the SUR2 locus as encoding an enzyme involved in biosynthesis of a secondary metabolite (Bak and Feyereisen, 2001; Bak et al., 2001; Hansen et al., 2001) refuted the hypothesis of SUR2 as a modulator of auxin homeostasis, and demonstrated an unexpected coupling of auxin and indole glucosinolate synthesis (Bak et al., 2001).
Figure 1.
Phenotype of 10-Day-Old Seedlings Grown on Vertical Agar Plates Showing Phenotypic Differences between sur2 Knockout Mutant and Wild-Type Seedlings.
(A) The white arrow underlines the heterogeneity observed among the sur2 mutants.
(B) Transverse section of plastic embedded hypocotyls of wild-type and sur2 showing radial expansion of cortical cells and close-up of adventitious root on the hypocotyls.
Other forward and reverse genetic screens, which were not focused on auxin selection criteria, resulted in the identification of the same locus as RTN1 from the runt size of the rnt1 mutant plants (Bak et al., 2001; Winkler et al., 1998) as ATR4 (altered tryptophan regulation4) based on altered feedback regulation of tryptophan biosynthesis in the presence of the toxic tryptophan analog 5-methyl-tryptophan (Smolen and Bender, 2002) and as RED1 (red light elongated1) based on a screen for perturbed phytochrome B signaling (Hoecker et al., 2004). The large diversity of names assigned to the same locus indicates that SUR2/CYP83B1 is involved in a complex metabolic grid and regulatory network. Furthermore, the irregular physiological phenotype observed between and within the different surveys has, until now, not been understood.
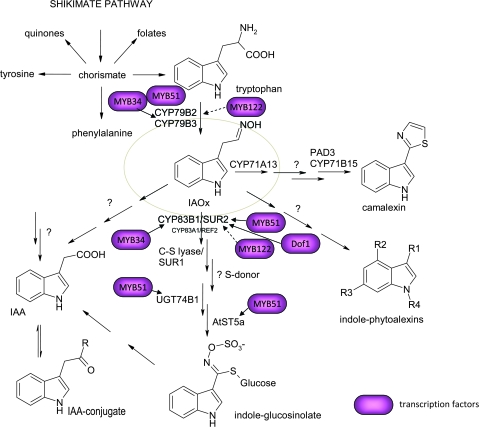
Glucosinolates are secondary metabolites characteristic to the Capparales order (Rodman et al., 1996) and serve as important determinants in plant–insect interactions. Upon cell disruption, glucosinolates are hydrolyzed by myrosinases and the unstable aglycone moieties rearrange into isothiocyanates and nitriles that may deter herbivores (Wittstock and Halkier, 2002). Indole glucosinolates are derived from tryptophan, aromatic from tyrosine and phenylalanine, and aliphatic from methionine. In A. thaliana, only indole and aliphatic glucosinolates are present. Identification of a residual low level of indole glucosinolates in the sur2 knockout mutant, while the levels of aliphatic glucosinolates were unaffected (Bak et al., 2001; Naur et al., 2003), suggested the presence of an additional enzyme catalyzing the same conversion. This enzyme was later identified as CYP83A1 (Bak and Feyereisen, 2001). CYP83A1 preferentially catalyzes the conversion of methionine derived oximes to aliphatic glucosinolates (Bak and Feyereisen, 2001), and exhibits a 50-fold reduced affinity towards indole-3-acetaldoxime in comparison to CYP83B1. Accordingly, indole-3-acetaldoxime is not considered a physiological substrate of CYP83A1 in planta (Bak and Feyereisen, 2001). Knockout lines of CYP83A1 results in plants designated to ref2. These plants have no visual phenotype in comparison to wild-type but were shown to have reduced levels of sinapoyl malate and absence of aliphatic glucosinolates in the leaves, indicating a link between aliphatic glucosinolates and phenylpropanoids in A. thaliana (Hemm et al., 2003). The absence of a high auxin phenotype in ref2/CYP83A1 mutants underpins that indole-3-acetaldoxime is not a physiological substrate for CYP83A1. The SUR2/CYP83B1 catalyzed conversion of indole-3-acetaldoxime results in the formation of a reactive aci-nitro product that is conjugated to an unknown thiol-donor as part of the enzyme reaction (Bak and Feyereisen, 2001; Bak et al., 2001; Halkier and Gershenzon, 2006). The final steps of the indole glucosinolates pathway are catalyzed by SUR1, which is a C–S lyase (Mikkelsen et al., 2004), followed by an S-glucosylation catalyzed by UGT74B1 (Grubb et al., 2004) and finally by a sulfurylation reaction catalyzed by AtSP5a (Piotrowski et al., 2004) (Figure 2).
Figure 2.
Schematic Representation of Indole Glucosinolate Pathway Showing the Importance of Indole-3-Acetaldoxime as a Central Metabolite for Indole Secondary Metabolism in Arabidopsis thaliana.
R can either be a glucose moiety or an amino acid. Known positive regulators of transcription involved in this grid are indicated in magenta.
In A. thaliana, the fundamental role of SUR2/CYP83B1 reflects the importance of indole-3-acetaldoxime (Figure 2). In A. thaliana, indole-3-acetaldoxime is not only a direct precursor for IGs, but may also be converted into a large number of other pathogen-induced metabolites. These include camalexin, the major phytoalexin in A. thaliana (Glawischnig et al., 2004; Nafisi et al., 2007, Böttcher et al., 2009), and several other indole-metabolites induced as a result of microbial infection (Bednarek et al., 2005; Hagemeier et al., 2001). The carbon skeleton for biosynthesis of indole compounds is derived from the shikimate pathway, which channels up to 20% of the total carbon flux and thus offers high capacity for metabolic re-configuration (Herrmann, 1995). Despite the pivotal role of auxin in plants, its biosynthesis has not been fully elucidated. Several independent pathways that are not mutually exclusive have been reported (see review by Woodward and Bartel, 2005). In A. thaliana, and most likely in all glucosinolate-producing plants, indole-3-acetaldoxime is an intermediate in an auxin biosynthetic pathway (Figure 2). The central position of indole-3-acetaldoxime at a crossroad of several pathways explains why the SUR2 locus was first related to auxin homeostasis in the sur2 knockout mutant.
The complex phenotype associated with perturbation of the metabolic indole-3-acetaldoxime grid implies tight regulation at multiple levels and sites to control the flux of metabolites into the different pathways in response to external stimuli. Several transcription factors have recently been shown to be involved in the regulation of this indole-metabolic grid. R2R3 MYB transcription factors of subgroup 12 (Kranz et al., 1998; Stracke et al., 2001) play a major role (Figure 2), and MYB28 and MYB29, belonging to the subgroup 12, have been shown to be involved in regulation of aliphatic glucosinolate biosynthesis (Hirai et al., 2007). Previously, ATR1/AtMYB34 has been demonstrated to control auxin homeostasis by regulating the balance between auxin and indole glucosinolates synthesis (Celenza et al., 2005) and AtMYB51 has been identified as a positive regulator of indole glucosinolates biosynthesis, also under stress conditions with no impact on auxin homeostasis (Gigolashvili et al., 2007). AtMYB122 is the closest homolog of AtMYB51 in A. thaliana, and an overexpression experiment suggested that AtMYB122 also controls auxin homeostasis partly based on regulation of indole glucosinolates biosynthesis (Gigolashvili et al., 2007). Finally, the transcription factor AtDof1, which belongs to the DOF family (DNA binding with One Finger), has also been shown to be involved in the regulation of the indole glucosinolates pathway at stress conditions (Skirycz et al., 2006).
Previous analyses of the sur2 mutant have mainly focused on its high auxin and indole glucosinolate phenotype based on the overflow of indole-3-acetaldoxime into indole-3-acetic acid (IAA) biosynthesis as a result of interruption of the biosynthesis of IGs.
Here, we report a global analysis of the sur2 mutant by the use of a combined transcriptomic and metabolomic approach revealing pronounced effects on metabolites outside the indole metabolic grid. Metabolic and transcriptional cross-talk between glucosinolates and phenylpropanoids in sur2 was found to be regulated by complex interactions between both positively and negatively acting transcription factors. The complex phenotype of sur2 may thus not only be assigned to elevated levels of auxin, but also to ethylene and abscisic acid responses as well as drought responses in the absence of a water deficiency. The delicate balance between these signals explains why minute changes in growth conditions may result in non-uniform phenotype.
RESULTS
Phenotypic Characteristics
This study is based on the comparison of wild-type and SUR2/CYP83B1 knockout plants 10 d after germination and growth on agar in vertically positioned square Petri dishes under a 16-h light regime. Under these conditions, the previously reported mutant phenotype (Bak and Feyereisen, 2001; Delarue et al., 1998) ascribed to overproduction of auxin was observed (Figure 1). The mutant attributes are a more developed secondary root system often initiated from the hypocotyls, extended hypocotyls, and epinastic cotyledons. As previously reported (Bak and Feyereisen, 2001; Delarue et al., 1998), the phenotype was not uniform. Some seedlings developed slowly and remained relatively smaller than others (Figure 1, white arrow)—a phenomenon that has not previously been explained (Delarue et al., 1998). In this study, we observed a number of additional phenotypic traits: longer primary roots indicative of a response to drought stress and thickened hypocotyls and hyponastic petioles, two features that are characteristic ethylene responses. The thickened hypocotyls were the result of radially expanded cortical cells and not additional cell layers (Figure 1B).
Knockout of SUR2 Has a Profound Impact on the Transcriptome
A global analysis was carried out to provide a general overview of the biological processes perturbed in sur2 to resolve how sur2 copes with loss of a functional CYP83B1.
To reveal alterations in the global transcriptome, the gene expression pattern of the sur2 mutant was compared to that of wild-type seedlings and monitored using the Agilent Arabidopsis 2 Oligo Microarray containing 21 500 different 60-mer oligonucleotide probes (Supplemental Data 1 and Supplemental Figure 1). According to The Arabidopsis Information Resource (TAIR7 release), this micro array covers ∼80% of the coding genome of A. thaliana and thus offers a good global view of the transcriptome. Based on a cut-off p-value of 0.001 (Supplemental Data 1), 301 genes were selected as being differentially expressed in sur2. These 301 genes represent approximately 1.1% of the predicted proteins encoded by the A. thaliana genome. As expected, the top-ranked gene was SUR2 because its mRNA is lacking in the mutant. The majority (66%) of the top-ranking differentially expressed genes were up-regulated in the sur2 mutant. The relative fold changes observed using the micro array data and by real-time PCR were very similar (Supplemental Table 1).
To elucidate which biological processes are mostly affected, the 301 top-ranked genes with altered expression levels were sorted according to their putative involvement in different biological processes using the gene ontology program BiNGO (Biological Network Gene Ontology) (Maere et al., 2005). BiNGO provides an unbiased approach to identify groups of genes involved in related biological processes or responses that are significantly over- or under-represented. BiNGO analysis is based on the information available in Gene Ontology Annotations (GO) and computes significant alterations in the representation of those groups. As the gene annotation is far from complete, only a subset of the 301 high-ranked genes was annotated in GO and thus retained in the BiNGO analysis performed. Of the 175 genes that were retained in the BiNGO analysis (Supplemental Data 2 and Supplemental Figure 1), 121 genes were up-regulated and 54 were down-regulated. The results of the BiNGO analysis clearly demonstrate that genes in a number of specific processes and responses are significantly over-represented in the sur2 mutant, while only a few are significantly under-represented. Based on the BiNGO analysis, five subgroups of up-regulated processes in the sur2 transcriptome were formed and graphically represented (Figure 3). The most significantly over-represented GO group in the sur2 mutant was general stimuli responses, which could be further divided into chemical, biotic, abiotic, hormone, and specifically abscisic acid responses (Figure 3, dashed oval). This demonstrates extensive re-programming of signaling processes in sur2. Secondary metabolism was also notably altered in sur2 mutant, with up-regulation of genes involved in synthesis of indole and other aromatic compounds and down-regulation of genes involved in phenylpropanoid synthesis (Supplemental Data 2 and Supplemental Figure 1; Figure 3). A group of eight genes involved in the GO group carbohydrate metabolism and related to cell wall elongation and was found to be significantly over-represented in the sur2 mutant (Supplemental Data 2). Finally, a small subset of genes involved in the GO group hormone-mediated signaling was identified as being over-represented in the mutant, essentially related to abscisic acid (Supplemental Data 2). The 54 down-regulated genes were related to the GO groups lignin biosynthesis and metabolism, amino acid-derived metabolism, and phenylpropanoid biosynthesis and metabolism.
Figure 3.
Biological Processes Affected in the sur2 Knockout Mutant Based on Expression Analysis using the Agilent 22K Array.
The 301 genes most significantly affected in their expression were analyzed in BiNGO in order to identify the biological processes significantly affected. The figure is based on the data extracted from the BiNGO analysis (Supplemental Data 2). The representation includes sorting by p-value (color) and size of each group. When GO-groups only referred to a small number of genes, those genes were preferred.
Multiple Transcription Factors Are Affected
Transcription factors (TFs) are important for the coordination of gene expression in response to endogenous and external stimuli and enable regulation of key enzymes in different metabolic and signaling pathways to optimize growth and development, and to counteract environmental challenges. TFs are particularly important in regulation of secondary metabolism to optimize the profile and levels of secondary metabolites in response to abiotic and biotic stress (Endt et al., 2002). Accordingly, the altered expression levels of TFs signify and may facilitate elucidation of alterations in secondary metabolism. In the sur2 mutant seedlings, 23 TFs belonging to 11 different classes were found among the 301 high-ranked genes with significantly altered expression p-value < 0.001 (Table 1). In A. thaliana, TFs constitute ∼6% of the coding genome (Riechmann and Ratcliffe, 2000). Accordingly, the total number of TFs is neither over- nor under-represented among the 301 top-ranked genes with altered expression in the sur2 mutant. The affected TFs were not uniformly distributed between the different classes of TFs. A strong prevalence of R2R3 MYB (five genes), WRKY (four genes), NAC (four genes), and HDZip (three genes) families (Table 1) was identified.
Table 1.
Transcription Factors Significantly Affected in their Expression in sur2 Knockout Mutant.
| Name | Gene ID | p-value | Fold |
| MYB family (133 members according to http://Arabidopsis.med.ohio-state.edu/AtTFDB) | |||
| MYB7 | AT2G16720 | 7.62E–05 | +2.0 |
| MYB45 | AT3G48920 | 7.84E–05 | +1.9 |
| MYB34 | AT5G60890 | 1.57E–04 | +2.1 |
| MYB36 | AT5G57620 | 5.34E–04 | +1.7 |
| MYB4 | AT4G38620 | 1.00E–03 | +1.7 |
| WRKY family (74 members according to Kalde et al. (2003)) | |||
| WRKY54 | AT2G40750 | 9.24E–05 | −1.7 |
| WRKY23 | AT2G47260 | 1.64E–04 | +1.7 |
| WRKY3 | AT2G03340 | 3.65E–04 | −2.2 |
| NAC family (96 members according to http://Arabidopsis.med.ohio-state.edu/AtTFDB) | |||
| ANAC034/ANAC035 | AT2G02450 | 1.78E–04 | −1.8 |
| ANAC087 | AT5G18270 | 2.65E–04 | +1.6 |
| ANAC029 | AT1G69490 | 4.18E–04 | −1.7 |
| NAC/ATAF1 | AT1G01720 | 9.25E–04 | +1.4 |
| HDZip family (47 members according to Henriksson et al. (2005)) | |||
| HB12 | AT3G61890 | 2.00E–04 | +2.0 |
| HB7 | AT2G46680 | 2.65E–04 | +2.0 |
| HB53 | AT5G66700 | 8.66E–04 | +1.9 |
| bZip family (73 members according to http://Arabidopsis.med.ohio-state.edu/AtTFDB) | |||
| bZIP37/ABF3 | AT4G34000 | 1.10E–04 | +1.8 |
| bZIP55/GBF3 | AT2G46270 | 8.23E–04 | +1.8 |
| ZF-HD family (15 members according to http://Arabidopsis.med.ohio-state.edu/AtTFDB) | |||
| HB29 | AT1G69600 | 1.51E–04 | +1.8 |
| ARR family (15 members according to http://Arabidopsis.med.ohio-state.edu/AtTFDB) | |||
| ARR17 | AT3G56380 | 3.70E–04 | −2.0 |
| RAV family (11 members according to http://Arabidopsis.med.ohio-state.edu/AtTFDB) | |||
| RAV1 | AT1G13260 | 4.06E–04 | −1.7 |
| SBP family (15 members according to http://Arabidopsis.med.ohio-state.edu/AtTFDB) | |||
| SPL3 | AT2G33810 prevent early flowering | 5.60E–04 | −1.6 |
| GeBP family (16 members according to http://Arabidopsis.med.ohio-state.edu/AtTFDB) | |||
| AT4G00250 | 8.07E–04 | −2.1 | |
| bHLH family (161 members according to http://Arabidopsis.med.ohio-state.edu/AtTFDB) | |||
| bHLH135 | AT1G74500 | 8.49E–04 | +1.6 |
Transcription factors with a known function are shown in bold.
The R2R3 MYB TFs are of particular interest because their main function is to regulate secondary metabolism in plants (Stracke et al., 2001). Five R2R3 MYB TFs in the sur2 mutant were identified as up-regulated (P < 0.001) (Table 1), of which four have been ascribed a function. AtMYB34/ATR1 was up-regulated 2.1 times. This is in agreement with that MYB34 has previously been shown to function as an activator of the indole glucosinolates pathway by regulating the coordinated expression of CYP79B2, CYP79B3, and CYP83B1 (Celenza et al., 2005). Similarly, AtMYB4 was up-regulated 1.7-fold and has previously been characterized as a negative regulator of the phenylpropanoid pathway repressing the expression of C4H/CYP73A5 (Jin et al., 2000). As a consequence, knockout of AtMYB4 results in plants that are more resistant to UV light because they produce and accumulate more of the sun-protectants sinapoyl malate and sinapoyl glucose (Jin et al., 2000). AtMYB7 belongs to the same subgroup as AtMYB4 (Stracke et al., 2001) and was up-regulated 2.0-fold. AtMYB4, as well as AtMYB7, possess the motif ‘PDLNL’, a motif that is associated with a negative regulator (Kazan, 2006; Preston et al., 2004). AtMYB45 belongs to a different subfamily and was up-regulated 2.0-fold. Phylogenetic analyses show that MYB34 (up-regulated 2.1-fold) clusters in the same subgroup as AtMYB18 (Ballesteros et al., 2001; Stracke et al., 2001). Finally, AtMYB36 was identified as up-regulated. The function of this TF is currently unknown, but based on phylogenetic analyses, AtMYB36 is a close paralog to AtMYB37 and AtMYB38. The latter two MYB transcription factors are known to be involved in shoot axillary meristem initiation (Muller et al., 2006). AtMYB36 is expressed in roots (Muller et al., 2006) and thus may be involved in axillary meristem initiation in roots.
Cytokinins are regarded as negative regulators of axillary meristem initiation in roots. This effect is mediated by TFs belonging to the type-A ARR class. These are negative regulators of cytokinin signaling (To et al., 2004). Ten type-A ARR TFs are encoded by the A. thaliana genome. In sur2, ARR17 was 2.0-fold down-regulated. Although the function of ARR17 is currently unknown, down-regulation of an ARR TF involved in cytokinin signaling is in agreement with the elongated root phenotype of sur2.
The WRKY TFs constitute a large class of plant-specific TFs with 74 members in A. thaliana (Kalde et al., 2003). Three of them were identified in our analysis of alterations in the expression patterns between wild-type and the sur2 mutant (Table 1). WRKY23 was up-regulated 1.7-fold, whereas WRKY54 and WRKY3 were down-regulated 1.7 and 2.2-fold, respectively. Of these three WRKY TFs, only WRKY54 has been studied previously and shown to control reactions associated with stress and pathogen attack (Kalde et al., 2003).
Among the large class of NAC TFs, ATAF1 was up-regulated 1.4-fold. ATAF1 was one of the first NAC TFs identified in A. thaliana and is known as a drought-responsive gene suggested to negatively regulate the expression of stress-responsive genes under drought stress (Lu et al., 1996). The functions of the three other NAC TFs that are up- or down-regulated in the sur2 mutant are currently unknown.
RAV1 was up-regulated 1.7-fold in the sur2 mutant. RAV1 belongs to a small family of TFs, composed of only 11 members in A. thaliana (Table 1). CARAV1 is the ortholog of RAV1, a TF in pepper that triggers resistance to bacterial infections and tolerance to osmotic stresses (Sohn et al., 2006).
HBZip genes are plant-specific TFs that regulate developmental processes and several have been shown to mediate responses to external stimuli (Henriksson et al., 2005). Three TFs belonging to this family, HBZip7, -12 and 53 showed 2.0, 2.0, and 1.9-fold up-regulation in the sur2 mutant, respectively, and have all been described as regulators of growth under conditions of water deficiency (Henriksson et al., 2005; Olsson et al., 2004) and probably respond to ABA. Two TFs of the bZip family, bZip55 and bZip37, were both up-regulated 1.8-fold. They belong to the class of basic leucine zipper TFs and have been described as involved in abscisic acid responses (Kang et al., 2002; Lu et al., 1996). The classes of TF that are differentially expressed concur well with the phenotype as well as with the up-regulation of gene products belonging to the LEA protein (late embryogenesis abundant) family (AT2G35300, AT2G42560, AT3G17520), GDSL-motif lipase/hydrolase protein, oleosin (At2g25890), LTP (Lipid Transfer Proteins), and other proteins (AT4G11650, AT4G15910) that are all characterized as being induced by drought—all of which were found among the 301 ranked genes (Supplementary Data, drought and ABA). However, ABA-biosynthetic genes are not differentially expressed in sur2, even when the top 1000 genes are considered. This suggests that ABA responses are elicited without stimulation of ABA-biosynthesis.
Auxin and Ethylene
The sur2 knockout mutant was initially isolated based on its high auxin phenotype. Transcriptomic analysis of 10-day-old seedlings identified five up-regulated genes belonging to the known early-auxin-responsive genes (Abel and Theologis, 1996) within the 301 genes with a p-value below 0.001 (Supplemental Data 1; light-yellow table fill indicates top 301 genes). Three GH3genes: GH3.1 (AT2G14960, 4.7-fold), GH3.3 (AT2G23170, 6.5-fold), GH3.5 (AT4G27260, 3.4-fold), and one Aux/IAA gene: IAA17 (AT1G04250, 1.7-fold) were identified (note the unfortunate ambiguity in naming: the GH3-genes mentioned here are not members of the GH3 family of glycosyl hydrolases). To further the analysis of the dataset, the p-value cutoff was adjusted to 0.008. This identified two additional GH3genes: GH3.6 (AT5G54510, 1.8-fold) and GH3.17 (AT1G28130, 1.5-fold), and four Aux/IAA genes: IAA1 (1.8-fold), IAA19 (AT3G15540, 1.7-fold), IAA20 (AT2G46990, 1.6-fold), and IAA29 (AT4G32280 1.6-fold), indicative of major changes in expression of early-auxin-responsive genes in agreement with the observed visual phenotype.
In 5–8-day-old sur2 seedlings, the level of reversible IAA conjugates is decreased and the level of free IAA is increased (Delarue et al., 1998)—an observation that formed the original hypothesis of SUR2 as a regulator of free IAA by controlling the amount of hydrolysable IAA conjugates (Delarue et al., 1998). Later, the levels of irreversible IAA catabolites, such as oxindole-3-acetic acid conjugates and IAA-Asp amide conjugates, were shown to be increased in 3-week-old seedlings (Barlier et al., 2000), indicating that the sur2 mutant modulated its metabolism towards irreversible catabolism of IAA to cope with the high levels of free IAA (Ostin et al., 1998). Whereas IAA-Asp cannot be hydrolyzed to free IAA in vivo, IAA–Leu is considered a hydrolysable conjugate (Bartel and Fink, 1995). In agreement with the observed shift in sur2 towards formation of irreversible IAA catabolites, we identified repressed expression of ILL1 (At5g56650, 1.6-fold) and ILL2 (At5g56660, 1.8-fold, respectively), encoding an IAA amino acid hydrolases with very high activity toward several reversible IAA conjugates (Bartel and Fink, 1995; LeClere et al., 2002). Both ILL1 and ILL2 were found beyond the top 301 differentially expressed genes (p-value 0.005), but their combined suppression suggests that sur2 attempts to inhibit the release of free auxin from the pool of reversible amino acid conjugated IAA by reducing the levels of the IAA-conjugate hydrolases.
Enzymes involved in conjugating amino acids to auxin and therefore involved in controlling auxin homeostasis are encoded by the GH3 genes listed above as early auxin-response genes (Staswick et al., 2005). Free IAA can also be conjugated to glucose and based on in vitro studies, several UGTs have been implied to be involved in this process (Jackson et al., 2002). The most prominent candidate found was UGT84B1, whereas UGT75D1 was not found to glucosylate IAA. UGT75D1 has previously been annotated as an IAA UGT (Graham and Thornburg, 1997) (www.cazy.org/fam/GT1.html). We have evaluated the ability of UGT75D1, as well as others of the differentially expressed UGTs, to glucosylate IAA in in vitro assay using heterologous expressed UGTs, and observed that UGT75B1 possesses a high activity with IAA as acceptor substrate (Table 2). In agreement with this, UGT75B1 was up-regulated whereas UGT75D1 was not (Table 3).
Table 2.
In vitro screening of heterologously expressed UGTs that showed the most significant changes in expression level between sur2 knock-out mutant and wild-type towards a set of six commercially available indole and phenylpropanoid compounds.
| Name | Group | S1 | S2 | S3 | S4 | S5 | S6 |
| UGT73B2* | D | +++ | (+) | (+) | ++ | + | (+) |
| UGT73B3 | D | (+) | − | − | +++ | (+) | (+) |
| UGT73B5 | D | (+) | − | − | − | − | (+) |
| UGT72B1 | E | (+) | − | (+) | − | +++ | − |
| UGT72E2 | E | − | − | − | − | (+) | − |
| UGT71C3* | E | +++ | − | − | + | ++ | (+) |
| UGT74F1 | L | +++ | (+) | (+) | ++ | (+) | − |
| UGT75B1 | L | +++ | +++ | − | (+) | − | +++ |
| UGT84A2 | L | +++ | (+) | − | (+) | ++ | +++ |
| UGT84A3 | L | ++ | (+) | (+) | − | − | +++ |
S1: indole-3-carboxylic acid; S2: indole-3-acetic acid; S3: indole-3-carboxaldehyde; S4: coniferyl alcohol; S5: sinapoyl-alcohol; S6: sinapic acid.
* UGTs that were not identified based on their direct up-regulation, but were selected because of their homology to other up-regulated UGTs (Table 3). Based on the intensity of the signal corresponding to the formation of a glucosylated product, a relative scale was established: +++: high conversion; ++: medium conversion; +: low conversion; (+): minute conversion; -: no conversion.
Table 3.
List of UDP Glucosyl-Transferases Identified as Up- or Down-Regulated in sur2 Knockout Mutant by a Microarray Experiment (Sorted by Group).
| AGI entry | Name | Group | Rank | p-value |
| Up-regulated | ||||
| At2g22930 | UGT79B8 | A | 13 | 8.48E–02 |
| At2g16890 | UGT90A1 | C | 8 | 6.65E–02 |
| At4g34131 | UGT73B3 | D | 1 | 2.25E–04 |
| At2g15490 | UGT73B4 | D | 11 | 7.88E–02 |
| At2g15480 | UGT73B5 | D | 3 | 1.60E–02 |
| At4g01070 | UGT72B1 | E | 9 | 7.03E–02 |
| At1g07250 | UGT71C4 | E | 7 | 4.91E–02 |
| At5g66690 | UGT72E2 | E | 2 | 9.17E–03 |
| At2g43840 | UGT74F1 | L | 12 | 7.93E–02 |
| At1g05560 | UGT75B1 | L | 10 | 7.78 E–02 |
| At4g14090 | UGT75C1 | L | 5 | 2.91 E–02 |
| At3g21560 | UGT84A2 | L | 4 | 2.71 E–02 |
| At4g15490 | UGT84A3 | L | 6 | 4.24 E–02 |
| Down-regulated | ||||
| At1g50580 | UGT79B5 | A | 11 | 5.07E–02 |
| At5g03490 | UGT89A2 | B | 5 | 1.31E–02 |
| At1g10400 | UGT90A2 | C | 10 | 4.51E–02 |
| At2g36760 | UGT73C2 | D | 1 | 6.55E–05 |
| At2g36800 | UGT73C5 | D | 3 | 2.17E–03 |
| At2g29750 | UGT71C1 | E | 8 | 4.23E–02 |
| At2g29740 | UGT71C2 | E | 9 | 4.46E–02 |
| At1g30530 | UGT78D1 | F | 2 | 9.13E–04 |
| At5g17030 | UGT78D3 | F | 4 | 8.92E–03 |
| At3g55700 | UGT76F2 | H | 7 | 2.32E–02 |
| At2g28080 | UGT86A2 | K | 12 | 6.23E–02 |
| At4g15500 | UGT84A4 | L | 6 | 1.91E–02 |
Ranks are given for UGTs according to the microarray experiment after statistical analysis.
The thickened hypocotyl phenotype of sur2 is indicative of increased exposure to ethylene (Figure 1B). This feature of sur2 has not previously been reported, but is not entirely unexpected because auxin concentrations beyond the normal physiological range stimulate ethylene evolution, as auxin is known to induce ACC-oxidase. In agreement with this, ACC-oxidase (At2g19590) catalyzing the last step in ethylene biosynthesis is up-regulated by a factor of 1.6 (p-value 0.002) in the mutant, while none of the ACC-synthases is (the ACC-synthases considered are listed in Supplemental File 1). De Paepe et al. (2004) and Millenaar et al. (2006) have identified a set of ethylene-induced genes. Thirty-three of the top 301 differentially regulated genes were found to be influenced by ethylene by these authors. An additional 65 ethylene-responsive genes are identified in sur2 if the list is extended to the 1000 most significantly differentially expressed genes (p-value < 0.008; see Supplemental Data 1, ‘ethylene differentially’). The radially expanded stem phenotype is known to be associated with increased expression of cell wall hydrolases (Maclachlan et al., 1968). To further the analysis of genes differentially expressed in sur2, a comparison to the list of carbohydrate-active enzymes compiled at www.cazy.org was done. Several members of glycosyl hydrolase (GH) families GH16, GH28, GH35, and GH36 as well as polysaccharide lyase (PL1) and carbohydrate esterase (CE8) were indeed up-regulated and found among the top 301 differentially regulated genes (Supplemental Data 1, 1000 genes versus CAZy), and may be responsible for the observed radial expansion. However, none of the genes encoding these enzymes is known to respond uniquely to ethylene. Increased levels of enzymes belonging to the GH16 family may as well be indicative of auxin action.
Cell Wall Biosynthesis and Turnover
It has previously been reported that in sur2, the hypocotyl cortical and epidermal cells disintegrate and peel off (Bak et al., 2001). The elongated and thickened hypocotyls and the beginning disintegration of the hypocotyl epidermal and cortical cells suggest changes in cell wall turnover. This may be ascribed to an auxin effect, as seedlings engaged in auxin-stimulated growth may be expected to display an increased rate of cell wall turnover (Labavitch and Ray, 1974). There are no indications of up-regulated cell wall biosynthesis among the top 301 differentially expressed genes in sur2 (Supplementary Data/1000 genes versus CAZy) with the probable exception of the up-regulated arabinogalactan proteins (AGP), hydroxyproline-rich glycoproteins (HPRG), and proline-rich glycoproteins (PRG) (Supplemental Data 1, Other cell wall proteins).
Several glycosyl hydrolases (GHs) are up-regulated in sur2. GH-catalyzed turnover of cell wall polysaccharides is also an intrinsic property of expanding cell walls. Two β-galactosidase members of the GH35 family have been implicated in cell expansion (Valero and Labrador, 1993) and are up-regulated in the mutant as are several xyloglucan endo-transglycosylases belonging to the GH16 family (Rose et al., 2002). Members of the GH16 family are involved in auxin-promoted insertion of new polysaccharides and in stress relaxation of cell walls during expansion.
Indole and Phenylpropanoid Content Is Perturbed in sur2
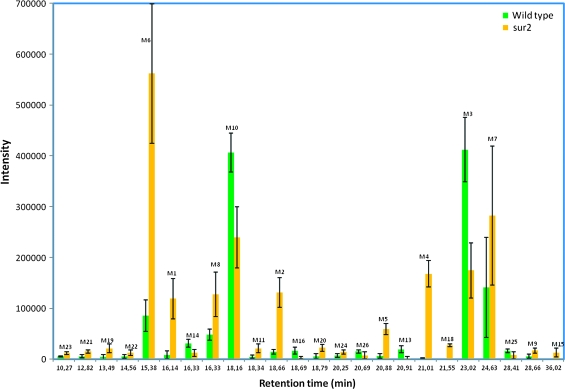
To gain an overall picture of metabolic re-routing and perturbation of the metabolome, 10-day-old sur2 and wild-type seedlings were extracted with 85% MeOH and the MeOH extracts were analyzed by LC–MS. Such LC–MS-based protocols enable the detection of several hundred metabolites (e.g. De Vos et al., 2007; Kristensen et al., 2005). To detect significant changes, 10 independent replicates of the LC–MS profiles of sur2 and wild-type seedlings were generated and analyzed using MetAlign. MetAlign allows background subtraction, alignment, and normalization to reveal statistically significant accumulating metabolites in LC–MS datasets (Kuzina et al., 2009; De Vos et al., 2007; Lommen, 2009; Tikunov et al., 2005). To allow direct comparison and to be able to directly link changes in the metabolome with changes in the transcriptome, analysis of the metabolite content of sur2 mutant and wild-type seedlings was performed using seedlings grown under the very same conditions as those used for the transcriptome profiling. After background subtraction, normalization, and alignment of the LC–MS profiles, 228 masses were detected that could be identified as being differentially accumulated in at least nine of the 10 replicates of either sur2 or wild-type (Supplemental File 2). Subsequently, significant differences in the 228 masses identified between the two plant lines were identified using a t-test (Supplemental Data 3). Out of the initially 228 masses detected by MetAlign, 23 masses, each corresponding to a specific metabolite, were selected for further analysis based on compliance with the following criteria: (1) a more than two-fold difference in intensity between mutant and wild-type, and (2) obtained signal sizes significantly distinct from the background (Table 4). Seventeen out of 23 secondary metabolites were present in higher amounts in the sur2 mutant compared to wild-type (Figure 4), underpinning the activation of secondary metabolite synthesis in the mutant, as predicted from the gene expression analyses. The metabolites M16 and M13 in wild-type were not abundant in sur2 and the metabolites M4, M18, and M15 in sur2 were not abundant in wild-type, suggesting that they may represent metabolites that are only present in either wild-type or sur2.
Table 4.
Metabolites Significantly Altered in the sur2 Knockout Mutant.
| Name | Observed ion | Calculated ion | Mol. formula | Fold | p-value | Tissues (R vs S) |
| Up-regulated | ||||||
| M1* | 365.1204 | 365.1207 | C16H22NaO8 | 13.9 | 6.90E–06 | R>>>S |
| M2* | 346.0880 | 346.0897 | C15H17NNaO7 | 9.0 | 4.40E–09 | R>S |
| M4* | 347.1665 | 347.1667 | C14H28NaO8 | 90.6 | 8.20E–09 | R=S |
| M5* | 359.1213 | 359.1214 | C16H20N2NaO6 | 9.7 | 8.50E–09 | S>R |
| M6 | 365 | 6.6 | 8.77E–07 | R>>>S | ||
| M7 | 425 | 3.2 | 8.57E–03 | R>S | ||
| M8* | 395.1298 | 395.1313 | C17H24NaO9 | 2.7 | 2.20E–04 | R>>>S |
| M11 | 377 | 4.6 | 1.38E–04 | S>>>R | ||
| M15 | 489 | 14.2 | 1.28E–03 | − | ||
| M18 | 492 | 26.6 | 5.00E–10 | R>>S | ||
| M19 | 277 | 4.7 | 9.27E–05 | S>>R | ||
| M20* | 360.1048 | 360.1054 | C16H19N1NaO7 | 4.0 | 9.41E–06 | R>S |
| M9 | 427 | 3.0 | 5.30E–05 | − | ||
| M21 | 298 | 2.6 | 9.05E–06 | R=S | ||
| M22 | 294 | 2.4 | 1.81E–03 | − | ||
| M23 | 138 | 2.2 | 9.56E–06 | R>>S | ||
| M24 | 211 | 2.0 | 3.54E–04 | − | ||
| Down-regulated | ||||||
| M3* | 363.0687 | 363.0687 | C15H16NaO9 | 2.5 | 5.18E–08 | S>>>R |
| M10* | 409.1111 | 409.1105 | C17H22NaO10 | 1.7 | 2.07E–06 | S>>>R |
| M13 | 186 | 10.9 | 5.74E–06 | R=S | ||
| M14 | 349 | 2.5 | 1.31E–05 | R=S | ||
| M16 | 323 | 6.1 | 1.06E–04 | − | ||
| M25 | 465 | 2.0 | 3.46E–03 | − | ||
| M26 | 348 | 2.0 | 7.17E–03 | − | ||
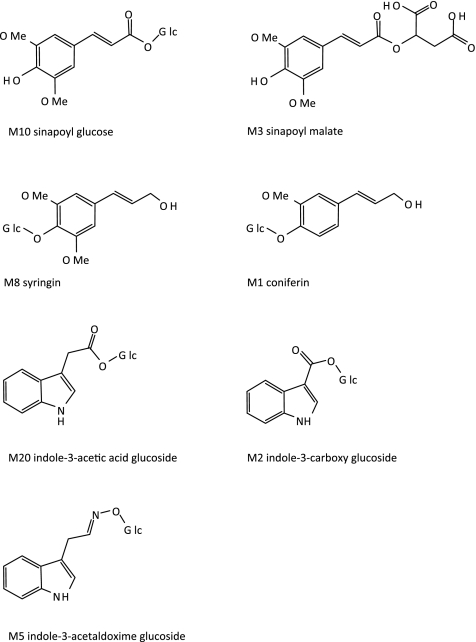
Accurate mass were measured for metabolites marked with * and the deduced molecular formulas. Relative occurrence in roots (R) and shoots (S). Metabolites of which the structure could be elucidated are presented in Figure 5.
Figure 4.
Metabolites Identified as Accumulating at Significantly Different Levels in the sur2 Mutant and Wild-Type Plants Based on Average Values.
Columns represent the metabolites that were significantly detected as up- or down-regulated with a cut-off of two-fold and with a p-value of 0.0001 (see Supplemental File 2). Major metabolites subsequently identified are represented with a *. Sinapoyl glucose (M10) had less than a two-fold variation, but was kept because of its high intensity and because it is known to be co-regulated with sinapoyl malate (M3).
To obtain further information on the 23 masses identified (Table 4 and Figure 4), their distribution in roots and above-ground tissues was determined. Of the 23 masses, eight accumulated preferentially in the roots of the sur2 mutant. M3 and M10 accumulated preferentially in the above-ground parts of wild-type plants reflecting down-regulation of the biosynthesis of these two metabolites in the sur2 mutant shoots (Figure 1 and Table 4). These results show that the major differences in the metabolite profiles of mutant and wild-type seedlings may be ascribed to differences in secondary metabolite content in mutant and wild-type roots.
Currently, only a fraction of the total number of different secondary metabolites present in plants are known and structurally characterized by LC–MS, because commercially available standards are generally lacking, and this limits structural identification of individual components in plant LC–MS chromatograms (De Vos et al., 2007; Lehmann et al., 2009). Compounds like indole-3-acetic acid (IAA) and indole-3-acetaldoxime cannot be detected due to their low abundances in the ng/gfw range. To further the understanding of the biological significance of the altered metabolite profiles in mutant and wild-type, efforts were made to determine the chemical structure of those 23 metabolites that were significantly up- or down-regulated in sur2 compared to wild-type. This was accomplished for seven metabolites (Figure 4 and Table 4, metabolites marked with *) by combining accurate mass detection to identify the elemental composition, by UV–VIS spectral analyses to assess the presence of aromatic residues or other diagnostic features, and by mass fragmentation to obtain diagnostic fragment ions (Supplemental Figure 2).
The most significant differences identified in the metabolite content of sur2 and wild-type seedlings were found within the indole derivatives and phenylpropanoid metabolites (Table 4 and Figure 5). The indole derivatives indole-3-acetaldoxime glucoside (M5), indole-3-carboxy glucose (M2), and indole-3-acetic acid glucoside (M20) as well as the monolignol glucosides syringin (M8) and coniferin (M1) were up-regulated in the mutant, while the major phenylpropanoids sinapoyl malate (M3) and sinapoyl glucose (M10) were down-regulated. These results correlate with the demonstrated altered expression of the R2R3 MYB transcription factors in sur2 (Table 1) that serve as positive and negative regulators of these two pathways.
Figure 5.
Chemical Structures of the Metabolites Identified as Accumulating at Different Levels in the sur2 Mutant and Wild-Type Plants.
UDP Glucosyl Transferases Involved in Metabolite Homeostasis of sur2
MS2 spectra of the metabolites M1, M5, M6, M10, and M11 indicated that these were glucosides (Supplemental Figure 2). This assignment was based on the observation that soft fragmentation of these metabolites in each case provided a characteristic fragment ion with a mass of 185, indicative of a glucoside (Kristensen et al., 2005). Glucosylation provides a direct route to regulate homeostasis of secondary metabolites. The genome of A. thaliana encodes/contains 112 UDP-glucosyltransferases (UGTs) (Paquette et al., 2009, 2003) (www.p450.kvl.dk/UGT.shtml). The function of the majority of these UGTs remains unknown (e.g. Gachon et al., 2005) but the multigene family encoding the UGTs provides an array of enzymes to glycosylate a large number of low-molecular-mass compounds. Glycosylation by UGTs serves to modify and control the solubility and the chemical reactivity of a large number of plant secondary metabolites, to detoxify xenobiotics and to secure hormone homeostasis (Jones and Vogt, 2001; Vogt and Jones, 2000).
To identify the UGTs involved in the formation of the glucosides M1, M2, M5, M8, M11, and M20, a second microarray experiment was designed. A small focused microarray encompassing all A. thaliana UGTs was selected to perform the screen, because it has been shown that such microarray experiments are more sensitive to measure variation in gene expression of relatively lowly expressed genes compared to a global microarray (Kristensen et al., 2005). The UGT enzymes can be sorted into 14 subgroups according to the phylogenic study by Ross et al. (2001). Among the UGTs most affected in their expression in the sur2 mutant were UGTs within the D, L, E, and F groups (Table 3).
Group D contains 13 members and is known to be involved in stress responses (Langlois-Meurinne et al., 2005), suggesting that a stress signal in the sur2 mutant is responsible for their altered expression in sur2. Within group D, UGT73B3, UGT73B4, and UGT73B5 were up-regulated while UGT73C2 and UGT73C5 were down-regulated. The expression of the UGT73B subfamily has been extensively characterized. UGT73Bs are induced by abiotic stress such as oxidative stress, wounding, and UV light, and biotic stress such as upon infection by Pseudomonas syringae. Likewise, menadione induced oxidative stress of roots of A. thaliana resulted in early transcriptional up-regulation of UGT73B3, UGT73B4, and UGT73B5 (Lehmann et al., 2009). UGT73B3 and UGT73B5 knockout mutants are characterized as exhibiting a decreased resistance to P. syringae (Langlois-Meurinne et al., 2005). This suggests an involvement of the UGT73B subfamily in the formation of glucosidic metabolite(s) with an important function in plant defense. Within the subfamily of UGT73Cs, UGT73C5 has been shown to mediate inactivation of brassinosteroids by catalyzing 23-O-glucosylation of these hormones (Poppenberger et al., 2005).
Group L was the second most affected group, with six UGTs showing altered expression in the sur2 knockout mutant (Table 2). Several enzymes belonging to the L group have been characterized: UGT75C1 has been described as an anthocyanin 5-O-glucosyltransferase (Tohge et al., 2005) and UGT84A2 as a sinapic acid UDP glucosyltransferase (Sinlapadech et al., 2007) involved in the biosynthesis of the UV filter sinapoyl malate.
The group E was also affected in its expression with up-regulation of UGT72E2, UGT71C3 and UGT71C3, UGT72B1 and down-regulation of UGT71C1 and UGT71C2. The physiological function of most of the UGTs in Group E is unknown except for UGT72E2, which is involved in monolignol-4-glycosylation in the phenylpropanoid pathway (Lanot et al., 2006; Lim et al., 2005). The function of the remaining UGTs that were identified as being differentially expressed (Table 2) is currently unknown. In conclusion, UGTs involved in the phenylpropanoid pathway seem strongly affected in their expression. Because the physiological function of the majority of the up-regulated UGTs was unknown, an in vitro screen of the heterologously expressed up-regulated UGTs was carried out to determine which of these might catalyze the formation of the glucosidic metabolites observed to accumulate in sur2.
Screening for Activity towards Indole and Phenylpropanoid Derivates
UGTs are known for their relatively broad in vitro regiospecificity (e.g. Hansen et al., 2003; Lim et al., 2003; Modolo et al., 2007). As a consequence, it is often difficult to identify the physiological function of a UGT solely on the basis of its in vitro activity. Nevertheless, integrated approaches using transcriptomic and metabolomic experiments, supported by in vitro studies, have successfully identified the substrates of a number of UGTs (e.g. Lim et al., 2003; Osmani et al., 2008; Tohge et al., 2005). This is the approach also undertaken here.
To identify which UGTs are involved in the formation of the glucosides identified using MetAlign, the 13 most significantly up-regulated UGTs in the sur2 knockout mutant were assayed in vitro using radiolabeled UDP-glucose as a sugar donor in the presence of different commercially available indole and phenylpropanoid substrates (Table 2). Active enzyme preparations could not be achieved for all 13 UGTs and all possible substrates were not available. However, this approach provided an insight of the potential substrates and substrate specificity of those UGTs. Some UGTs are known to be able to glycosylate more than one substrate in in vitro assays as opposed to in planta substrate specificity, and, accordingly, a number of substrates have to be evaluated to identify strong leads for in planta substrates. As expected, UGT84A2 and UGT84A3 preferentially glucosylated sinapic acid, UGT72B1 preferentially glucosylated sinapoyl alcohol, while UGT73B3 was found to preferentially glucosylate coniferyl alcohol. Indole-3-carboxyaldehyde was not a good substrate for any of the UGTs tested. As glucosides of indole-3-carboxyaldehyde were not detected by the LC–MS analysis of sur2/SYP83B1 seedlings, this was to be expected. UGT75B1 was found to preferentially glucosylate indole-3-acetic acid and indole-3-carboxylic acid. While indole-3-acetic acid was only glucosylated by UGT75B1, indole-3- carboxylic acid was glucosylated by UGT73B2, UGT71C3, UGT74F1, UGT75B1, and UGT84A2. Based on previous published data, UGT84B1 has been identified as an IAA glucosyltransferase (Jackson et al., 2002), but was not included in our screening, as UGT84B1 was not identified as being up-regulated (Table 3). An additional in vitro screening including UGT84B1 and UGT73B5 identified indole-3-acetaldoxime as a good substrate for UGT84B1, while indole-3-acetaldoxime was not glucosylated by any of the other UGTs tested. UGT84B1 was included in this screen as it has previously been published as an IAA glucosyltransferase (Jackson et al., 2002), and it is phylogenetically closely related to UGT84A2 and UGT84A3, which was identified as up-regulated in sur2 (Table 3). That multiple UGTs were found to glucosylate several substrates in vitro emphasizes the possible pitfalls encountered in assigning in planta substrates for some UGTs based solely on an in vitro screen. As UGTs constitute a multigene family with many close paralogs, often with partially overlapping substrate specificities, analysis of single knockout lines cannot be used to pinpoint in planta substrates, especially with respect to detoxification.
DISCUSSION
The majority of the previous studies of the sur2 mutant have mainly focused on forward and reverse genetic approaches. Each study has emphasized a specific aspect of the complex phenotypic changes observed in this mutant. To gain a more coherent understanding of the molecular background for the phenotypic changes observed in sur2, we have characterized the mutant using a combination of transcriptomics and metabolomics approaches. As outlined in the Results, this approach to analyze sur2 revealed the induction of different hormonal-dependent processes, perturbation in indole and phenylpropanoid composition, alteration in cell wall metabolism, as well as induction of stress-related responses.
The Phenotype
The morphology of the sur2 mutant seedlings is not reminiscent of a clear-cut high auxin status. Rather, the slightly hyponastic petioles of the first true leaves and the thickened hypocotyls (Figure 1) are indicative of a high ethylene status. Indeed, we found that ACC-oxidase catalyzing the last step in ethylene production is up-regulated in the mutant. Exposure to auxin concentrations beyond the normal physiological range has long been known to promote ethylene biosynthesis, as IAA is known to activate ACC oxidase. As early as 1966, Burg and Burg proposed that effects observed due to excessive auxin levels might be brought about by enhanced levels of ethylene. The auxin influence on ethylene biosynthesis has since been substantiated by molecular data (e.g. Abel et al., 1996; Yip et al., 1992). Auxin generally promotes root branching, whereas high auxin concentrations are inhibitory to root elongation. The long primary roots with many side roots of the mutant seedlings may be indicative of a drought response. Ethylene acts agonistically with abscisic acid on root growth (Benschop et al., 2007). The role of abscisic acid in adaptation to drought stress is well established. However, abscisic acid responses have not previously been associated with the sur2 phenotype and none of the genes encoding enzymes known or assumed to be specifically involved in abscisic acid synthesis (Table 1 in Xiong and Zhu, 2003) is found among the 1000 most significantly differentially expressed genes (p-value < 0.008). This indicates that genes related to abscisic acid are exclusively found down-stream of abscisic acid sensing, or that sur2 responses to abscisic acid are not mediated through transcriptional regulation.
Auxin and Cell Wall Changes
High and variable levels of free auxin have been observed in the sur2 seedlings (Barlier et al., 2000; Delarue et al., 1998; Sorin et al., 2006). The activation of the GH3 and AUX/IAA auxin-early responsive proteins (Abel and Theologis, 1996) is in agreement with the high auxin content in sur2 but, as outlined above, the phenotype of the sur2 seedling cannot merely be ascribed to high auxin. Auxin-promoted growth of the mutant seedling hypocotyls might be expected to lead to enhanced transcription of genes involved in cell wall biogenesis. Two different observations enable us to rationalize why the transcription of cell wall genes remained relatively unaffected. First, while auxin is able to restore cell wall synthesis to normal levels following down-regulation, such as by stress, auxin treatment does not stimulate cell wall biosynthesis above normal levels in expanding cells (Ray, 1973). Second, the phenotype of the seedlings is not characterized by hypocotyls that elongate excessively at the expense of hypocotyl diameter as would have resulted from auxin-promoted growth. We conclude that these hypocotyls are not in a state of auxin-stimulated elongation. Rather, the hypocotyls are radially expanded as typically follows ethylene treatment.
Auxin Conjugation and Inactivation
In Arabidopsis, only ∼1% of the total pool of IAA is present as free IAA. The major portion is found as hydrolysable conjugates that may serve as storage forms of IAA or as irreversible conjugates (Tam et al., 2000). The main function of these conjugates is to regulate IAA activity as well as transport (Woodward and Bartel, 2005). The observed up-regulation of five members of the GH3 family indicates that sur2 is clearly trying to reduce the pool of free IAA. In agreement with our transcriptome analysis, GH3s protein levels were identified as up-regulated in a proteomic study of sur2 (Sorin et al., 2006).
Free IAA can also be conjugated to glucose by family 1 UDP-glucosyl transferases (Jackson et al., 2002). Based on in vitro studies, four UGTs belonging to group L have previously been characterized by their ability to form such conjugates in vitro (Jackson et al., 2002). Interestingly, among the UGTs that were up-regulated in sur2, four UGTs belonging to the group L were able to glucosylate IAA in vitro (Table 5). In agreement with the transcriptome data, indole-3-acetic acid glucoside (M20) was increased four-fold in sur2 (Table 3).
Modulation of auxin action can also be mediated by regulation of IAA transport. Flavonoids are generally known as light scavengers, but have also been reported as inhibitors of IAA transport (Besseau et al., 2007; Buer and Muday, 2004). Our transcriptome data suggest that sur2 does indeed activate the biosynthesis of flavonoids, since the transcription factor AtMYB12 and its target genes FH3 (AT3g51240, 1.5-fold p-value 0.001), F3′H (AT5G07990, 1.3-fold p-value 0.006), and FLS (AT5G08640, 1.5-fold p-value 0.0006) were up-regulated (Supplemental Data 1) (Mehrtens et al., 2005). In agreement with our transcriptomic data, AtMYB12 has been reported to be mainly expressed in roots and to strongly activate expression of F3H and FLS1 (Stracke et al., 2007).
Indole-3-Acetaldoxime Regulation in sur2
Despite the pivotal role of IAA, its biosynthesis has not been completely elucidated. In A. thaliana, a specific pathway that includes indole-3-acetaldoxime as an intermediate has been reported (Figure 2) (Woodward and Bartel, 2005). Our transcriptome data show that in the sur2 mutant, the production of indole-3-acetaldoxime is up-regulated as a result of increased transcription of MYB34 and its target genes CYP79B2 and CYP79B3, which encode the two cytochromes P450 that catalyze the conversion of tryptophan into indole-3-acetaldoxime. The reason why the biosynthetic genes converting tryptophan to indole-3-acetaldoxime are up-regulated at first appears counterproductive, as this increases the flux of indole-3-acetaldoxime into IAA, and thus would be expected to contribute to a more pronounced phenotype. A plausible hypothesis is that sur2/CY83B1 apparently tries to increase the level of indole glucosinolates to reduce the flux of indole-3-acetaldoxime into IAA by increasing the expression of CYP79B2 and CYPB3. Indole-3-acetaldoxime could not be directly monitored at the metabolic level due to its inherent instability and because of the minute amount expected to accumulate due to the high degree of channeling of these pathways (Jorgensen et al., 2005; Nielsen et al., 2008). However, a glucoside of indole-3-acetaldoxime (M5) was identified as significantly up-regulated (10 times) in sur2 (Figure 5 and Table 4), demonstrating the presence of UGT(s) able to glucosylate indole-3-acetaldoxime. Identification of a minute amount of the indole-3-acetaldoxime glucoside in the wild-type plant suggests a new level of regulation in this pathway, comprising the ability of plants to synthesize a pool of indole-3-acetaldoxime-glucoside that may be hydrolyzed upon demand. Based on our UGT in vitro screening, we identified UGT84B1 as the UGT that most efficiently catalyzes the conversion of indole-3-acetaldoxime to the corresponding glucoside (data not shown). UGT84B1 has previously been identified as an IAA glucosyl transferase (Jackson et al., 2002). The ability of CYP84B1 to glucosylate IAA as well as indole-3-acetaldoxime in vitro underpins the regiospecificity of UGTs and the difficulty in identifying physiological substrates based on in vitro screens (Osmani et al., 2008). It is clear that glucosylation of IAA is catalyzed by several UGTs in vitro (Jackson et al., 2002) (Table 2). However, our data suggest that UGT75B1 is preferentially responsible for auxin glucosylation under high-auxin conditions in seedlings. Whether the expression of different UGTs with the ability to glucosylate IAA is organ-specific or overlapping needs further investigation. Furthermore, it remains to be elucidated whether UGT84B1 preferentially glucosylates indole-3-acetaldoxime or IAA in planta.
Cross-Talk between Ethylene and Auxin in sur2
It is well know that auxin has a regulatory effect on ethylene production, and that it can be difficult to distinguish which morphological effects are related to auxin and which are related to ethylene. In our transcriptome analysis, the auxin response factors ARF7 and ARF19, known to control lateral root formation (Okushima et al., 2007), were identified as up-regulated in sur2-seedlings. In roots, ARF19 expression is known to be induced by both auxin and ethylene. Accordingly, arf19 mutants have ethylene-insensitive roots (Li et al., 2006). Similarly, PIN2/EIR1 (At3g23240) identified in our array data and first described as Ethylene Insensitive Root1 (Roman et al., 1995) was later described as being involved in auxin transport (Luschnig et al., 1998). Furthermore, crosses of sur2 with either wei2 or wei7 suppress the phenotype (Stepanova et al., 2005). WEI2 and WEI7 are induced by ethylene, and encode α and β subunits of anthranilate synthase, respectively, and hence are important for tryptophan and IAA biosynthesis.
sur2 Displays a Drought Response Phenotype and Transcriptome
The root phenotype of the sur2 mutant is indicative of a drought response. However, as the plantlets were grown on agar in closed containers, they were not exposed to water deficits in agreement with no up-regulation of ABA-synthesis. Nevertheless, several gene products that are all characterized as being induced by drought are up-regulated in the mutant. This suggests that ABA-dependent drought responses were induced neither by abscisic acid nor by water stress, but by intrinsic factors probably operating via cross-talk through ethylene signaling.
Cross-talk between drought signaling and ethylene has recently been investigated by Manavella et al. (2006), who overexpressed the sunflower Hahb-4 transcription factor in Arabidopsis. Hahb-4-expression was strongly stimulated by ethylene in the transgenic plants harboring the Hahb-4 gene under the control of its native promoter. The transformants were markedly drought-tolerant and the transcriptome significantly affected, with one of the observed effects being a down-regulation of genes involved in ethylene biosynthesis (Manavella et al., 2006). This is indicative of a negative feedback loop. Interestingly, the two A. thaliana transcription factors HB7 and HB12 were identified as putative Hahb-4 orthologs, and their presence among the up-regulated genes in our data supports the operation of such a negative feedback loop in the sur2 mutant.
Abscisic acid homeostasis is very important to properly control different physiological responses in planta. Abscisic acid inactivation occurs by two different pathways, either by 8′-hydroxylation or by glucosylation. The hydroxylation step was recently attributed to CYP707A1 and CYP707A3 (Kushiro et al., 2004; Saito et al., 2004). In agreement with this, our micro array data identified CYP707A1 as up-regulated. In adzuki bean seedlings, glucosylation of abscisic acid has been shown to be mediated by a specific UGT (Xu et al., 2002). Our UGT in vitro screening platform identified several UGTs that were able to glucosylate abscisic acid, including UGT73B1 (data not shown). Whether UGT73B1 is a UGT responsible for in planta glucosylation of abscicic acid is currently an open question, which requires further studies to be resolved. Based on these observations, it is likely that the endogenous concentration of free abscisic acid in sur2 is unaltered. Contrary wise, the sur2 mutant must experience a high ethylene status, because radial expansion of hypocotyls (Figure 1) is an IAA-induced ethylene-specific phenotype and no other hormone has been shown to interact agonistically or otherwise modify this process. The expression profile of genes involved in ethylene biosynthesis partly corroborates this suggestion. ACC-oxidase was significantly up-regulated in sur2 whereas the isoforms of ACC-synthase were not. It is usually assumed that ACC oxidase catalyzes the rate-limiting step in ethylene biosynthesis, but in silique maturation, for example, ethylene production is obviously controlled by the rate of conversion of ACC to ethylene (Child et al., 1998). Whether the ACC-oxidase is rate-limiting in sur2/CPY83B1 seedlings is not known.
Co-Regulation of the Indole Glucosinolate and Phenylpropanoid Pathways
In a previous study of the ref2/CYP83A1 mutant, an unexpected link was established between the pathways for aliphatic glucosinolate and phenylpropanoid synthesis (Hemm et al., 2003). The low levels of the phenylpropanoids sinapoyl malate and sinapoyl glucose in ref2/CYP83A1 was the result of a mutation in the glucosinolate pathway preventing the expression of the gene CYP83A1, the closest paralog of CYP83B1 (Figure 2). To explain this observation, it was proposed and verified in vitro that oximes inhibit caffeic acid-O-methyltransferase (COMT) enzymes involved in the phenylpropanoid pathway (Hemm et al., 2003). In parallel to the observations made with the ref2/CYP83A1 mutant, sur2 exhibits significant changes in phenylpropanoid content in comparison to wild-type seedlings. Identification of a 10-fold increase in the concentration of indole-3-acetaldoxime glucoside in sur2 strongly suggests that the free indole-3-acetaldoxime pool is also increased in the mutant. As shown for aliphatic oximes (Hemm et al., 2003), indole-3-acetaldoxime may also inhibit the flux into phenylpropanoids. Since the phenylpropanoid and indole glucosinolates pathways are both dependent on precursors obtained via the shikimate pathway (Figure 2), the flux of carbon through these two major pathways in A. thaliana must somehow be subjected to co-regulation to facilitate synthesis of balanced amounts of indole glucosinolates and phenylpropanoids. This contention is supported by the analysis of MYB transcription factors in sur2.
R2R3 MYB transcription factors have been grouped into 12 subgroups based on conserved motifs (Kranz et al., 1998). The MYB transcription factor AtMYB34 belongs to subgroup 12, and has been shown to activate the indole glucosinolates pathway (Celenza et al., 2005). MYB transcription factors of subgroup 4, including MYB 4, MYB7, which are up-regulated in sur2, contain a negative regulator motif. MYB4 is known to repress cinnamate 4 hydroxylase (CYP73) and thereby represses the phenylpropanoid pathway. MYB34 and MYB4 thereby provide a mechanism to co-regulate the two pathways. Similarly, five of the six MYB factors in subgroup 12: MYB 28, MYB29, MYB34, MYB51, and MYB76, have been characterized as regulators of glucosinolate biosynthesis (Celenza et al., 2005; Gigolashvili et al., 2007; Hirai et al., 2007). In a parallel manner, the subfamily 4 may serve as negative regulators of phenylpropanoid synthesis. Currently, only AtMYB4 has been clearly characterized as a negative regulator of cinnamic acid 4 hydroxylase (CYP73) (Jin et al., 2000), but the complexity of the phenylpropanoid pathway certainly needs several levels of regulation. The identification of the target genes of AtMYB7 and AtMYB32 will help to clarify the regulation of this pathway. In agreement with this hypothesis, Preston et al. (2004) suggests a negative regulation of dihydroflavonol-4-reductase and cyanidin synthase and up-regulation of caffeic acid-O-methyltransferase mediated by MYB32, belonging to subgroup 4. While three negative regulators, possibly controlling the flux of phenylpropanoids, were up-regulated, one involved in the activation of flavonoid synthesis, AtMYB12, was activated with its target genes flavone 3-hydroxylase–flavone synthase, revealing a reprogramming rather than a total blockage of the phenylpropanoid pathway. By activation of the first steps of the flavonoid pathway and down-regulation of the subsequent conversion to pelargonidin and cyanidin, the sur2 mutant apparently tries to increase the flavonoid content. However, the reduction of sinapoyl malate and sinapoyl glucose suggests a major reduction of the flux in the phenylpropanoid pathway in order to re-distribute shikimate flux into the indole glucosinolates pathway. Identification of the signals controlling expression of these transcription factors will help understand the complete relationship between the indole-glucosinolate and phenylpropanoid pathways.
Accumulation of Monolignol Glucosides in the Root of sur2
In sur2, the concentration of two monolignol glucosides identified as coniferin (M1) and syringin (M8) were up-regulated 13.9 and 2.7 times, respectively (Table 4 and Figure 5). Further analysis revealed that those two metabolites almost exclusively accumulated in the root (Table 4). UGT72E2 was the second most up-regulated UGT in the focused array (Table 3), and has previously been identified as the main monolignol glucosyltransferase expressed in roots of A. thaliana (Lanot et al., 2006). This demonstrates that the metabolism in roots is re-programmed independently of the metabolism in leaves, where a reduction in the phenylpropanoid pathway flux towards sinapate esters was observed. Interestingly, Hemm et al. (2003) have previously reported an accumulation of coniferin, syringin, and flavonoids in A. thaliana roots exposed to light and proposed a role of these secondary metabolites as defense compounds. The results obtained suggest that roots of sur2 are more light-sensitive than wild-type roots or that biosynthesis of monolignol glucosides is stimulated by light and ethylene in sur2.
Altered Biosynthesis of UV Filtering Compounds in sur2
Sinapoyl malate is the main UV filter present in leaves of A. thaliana and is derived from sinapoyl glucose. Both sinapoyl malate (M3) and sinapoyl glucose (M10) were identified among the main metabolites with a decreased content (2.5 and 1.7-fold, respectively) in sur2 (Table 4). A similar reduction of sinapoyl malate has previously been reported in 4-week-old sur2 mutant leaves (Hemm et al., 2003). In this context, it was unexpected to observe that UGT84A2 and UGT84A3, the two main UGTs involved in sinapoyl metabolism (Table 2) (Sinlapadech et al., 2007), were identified as up-regulated in sur2 (Table 2). This may reflect an attempt of the sur2 mutant to compensate for the lack of sinapoyl malate and sinapoyl glucose imposed by the reduced flux of carbon into phenylpropanoids mediated by MYB4. This demonstrates that UGT84A2 and UGT84A3 are under control of transcription factors independent of those controlling indole glucosinolates, sinapoyl malate and sinapoyl glucose synthesis.
Like sinapoyl malate, flavonoids play a role in protection against UV light. The biosynthesis of flavonoids is induced by light, and MYB transcription factors such as AtMYB12 have been identified as positive regulators of phenylpropanoid and flavonol synthesis (Mehrtens et al., 2005; Stracke et al., 2007). In agreement with this, myb11 myb12 myb111 triple knockout seedlings do not form flavonols, while accumulation of anthocyanins was not affected. Likewise, several genes of the phenylpropanoid and flavonoid pathways were down-regulated in the myb11 myb12 myb111 triple knockout mutant (Stracke et al., 2007), including several of the UGTs involved in flavonoid biosynthesis such as UGT75C1 involved in 5-O-glycosylation, and UGT78D1 and UGT78D3, which are closely related to UGT78D2 known to be involved in 3-O-glycosylation (Jones et al., 2003; Tohge et al., 2005). In sur2, the data obtained from the focused array demonstrate that MYB12 as well as its target genes F3H and FLS1 are induced (Supplementary Files). In conjunction with the overexpression of the above-mentioned UGTs (UGT75C1, UGT78D1, and UGT78D3) (Table 3), this demonstrates that the sur2 mutant re-directs part of the carbon flux within the phenylpropanoid pathway towards flavonoid biosynthesis. This serves to provide an alternative set of UV filters and thereby counteracts the low levels of sinapate esters present in sur2.
Concluding Remarks
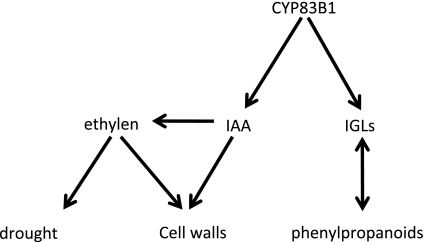
Our transcriptomic and metabolomic analyses of the sur2 mutant reveal several new aspects of this mutant, and documents that the sur2 phenotype cannot be merely ascribed to high IAA levels. The study illustrates the plasticity of plant metabolism (Figure 6). In the sur2 mutant, a very high number of genes are significantly altered in their expression. Nevertheless, the mutant is able to survive and to set seeds. The pathways related to the formation of indole and phenolic compounds are the main pathways altered as documented by major changes in the metabolome of sur2 (Figure 5). A number of MYB transcription factors orchestrate a coordinated regulation of these two pathways, rendering internal rearrangements of fluxes within the phenolic and glucosinolate pathways possible. The high auxin concentration in sur2 caused by the mutation is clearly beyond the physiological range and neither the morphology nor the transcriptome of the mutant seedlings is typically high-auxin. The phenotype shows that sur2 is pushed into a realm in which signaling via ethylene plays a predominant role. This observation corroborates the emerging evidence for ethylene signaling in drought responses and cross-talk to abscisic acid signal transduction. The drought response is set off neither by external stress nor by enhanced abscisic acid biosynthesis, but by a set of intrinsic factors of which the ethylene signaling has been documented in the present study.
Figure 6.
Diagram Displaying the Major Intrinsic Metabolomic Network in sur2 Mutant.
METHODS
Plant Culture Conditions and Collection
Plants were grown in controlled conditions in order to avoid stresses that could trigger changes in gene expression or metabolic biosynthetic or catabolic pathways. After surface sterilization and careful washing, seeds were sown on square Petri dishes (120 × 120 × 12 mm) containing 50 mL of agar (0.9%), Murashige Skoog medium with vitamins (1), and sucrose (2%). The Petri dishes were stored vertically at 4°C for 48 h in the dark to vernalize the seeds. The Petri dishes were sealed to avoid water loss and were kept in a vertical position during seedling growth (16 h light, 22°C, 110 μE light intensity). After 10 d, seedlings were collected at the end of the light period and directly frozen in liquid nitrogen.
Transcriptome Studies
Microarray experiments were carried out using two types of arrays. The first array was the global Agilent Arabidopsis 2 Oligo Microarray, containing 21 500 different 60-mer probes, and was purchased from Agilent Technologies. The second microarray was based on custom-designed manually annotated 50-mers and encompassed 453 probes representing the P450 family and family 1 UGTs as previously described (Ekstrom et al., 2004, 2005; Glawischnig et al., 2004; Kristensen et al., 2005). Total RNA from three biological replicates were extracted in a two-step procedure. Seedlings (200 mg) were ground in the presence of liquid nitrogen and 2 mL of Trizol (Sigma) were added and the mixture blended. Chloroform (0.4 mL) was added and the tubes were vortexed before separation of the two phases at 15 000 g. The water phase was carefully removed and mixed with an equal volume of isopropanol for precipitation. After centrifugation, the pellet was washed with 75% ethanol and dissolved in DEPC water. A second purification step was carried out using the Qiagen total RNA purification kit, following the manufacturer's protocol. Total RNA quality was checked by identification of distinct bands corresponding to ribosomal RNAs. Reverse transcription and labeling were performed using the Pronto ChipShot™ Labeling System (Promega) by following the manufacturer's protocol. Hybridization and washing were performed according to the Agilent protocol. Scanning of the slide was performed with an Affymetrix (Santa Clara, USA) GMS 418 array scanner varying laser-power and photomultiplier gain. Statistical analyses were performed on tiff files that exhibited few saturated spots.
Tiff files were analyzed with Imagene version 5.6 (BioDiscovery, Marina del Rey, CA).
Statistical analysis was based on three two-color dye swaps from three biological replicates (Supplemental File 3). Possible extra variation between replicates was accounted for by using the LIMMA model, which allows correlation of expression levels within each biological replicate and where information is drawn across genes by fixing the within-biological clone correlation (Smyth, 2005). No background was subtracted and log-expression data were LOESS-normalized on an array-by-array basis. Differentially expressed genes were identified by the modified t-test implemented in the LIMMA package. The LIMMA package uses a Benjamini-Hochberg correction to adjust for multiple testing. However, in this study, the exact p-values are of less importance than the ranking of the genes. A cut-off p-value < 0.001, corresponding to a p-value of 0.07 after adjustment for multiple testing, was arbitrarily chosen and selected 301 genes, which roughly corresponds to 1% of the genes. A more classical adjusted p-value of 0.005 would select 207 genes. Of the 301 up-regulated genes with a p-value cut-off of 0.001, a total number of 121 genes could be assigned to GO ontologies and used for BiNGO analysis (Supplemental File 4) using a hypergeometric test with a traditional Benjamini-Hochberg FDR correction and a post-correction selection significance level of 0.01. The dataset was also analyzed using SAM (Significance analysis of microarrays) (Tusher et al., 2001), and the ranking of genes based on LIMMA and SAM were comparable (data not shown) and, for simplicity, only the ranking based on LIMMA is shown.
Comparing the Transcriptome of the sur2 Mutant to Transcriptomes of Hormone-Treated WT Arabidopsis
Auxin: up-regulated genes among the 1000 most significantly differentially expressed genes were compared to Table 4 of Supplementary Data in Overvoorde et al. (2005; Supplemental File 1).
Ethylene: the top-1000 list was compared with the array and cDNA-AFLP data of De Paepe et al. (2004), and the up- and down-regulated genes separately with the 1164 up-regulated and 1306 down-regulated genes retrieved (Dr A.J.M. Peeters, personal communication) from the array experiment in Millenaar et al. (2006; Supplemental File 1).
Metabolomics
Extraction of semi-polar metabolites from individual seedlings was performed with boiling 85% methanol (vol./vol.) during 2 min. Ten sur2 seedlings and 10 10-day-old wild-type seedlings, shoots, and roots, respectively, were extracted, and after filtration (0.22 μm), the clarified methanol extracts were analyzed by LC–MS as described previously (Hansen et al., 2003; Kristensen et al., 2005). The recorded chromatograms were analyzed with MetAlign (www.pri.wur.nl/UK/products/MetAlign/) in order to align and normalize the two datasets. Only metabolites with intensity higher than two times the noise level and present in nine out of 10 chromatograms were retained and subjected to a t-test, 95% cut-off, to identify metabolites that accumulated significantly in the datasets (Supplemental File 2).
Metabolites were identified based on a combination of accurate mass, UV spectra, fragmentation patterns, as well as by comparison to the in vitro generated glucosides obtained by incubation of indole-3-carboxylic acid, indole-3-acetic acid, indole-3-carboxaldehyde, indole-3-acetaldoxime, abscisic acid, coniferyl alcohol, sinapoyl alcohol, and sinapic acid with a range of UGTs as described below. Accurate mass measurements of peaks of interest were determined on a Bruker micrOTOF-Q instrument (Bruker Daltonics, Bremen, Germany), coupled to an Agilent 1200 Series LC (Agilent Technologies, Germany). An XTerra MS C18 column (Waters, Milford, MA; 3.5 μm, 2.1 × 100 mm) was used at a flow rate of 0.2 mL min−1. The mobile phases were: A water; B acetonitrile. The gradient program was: 0–20 min, linear gradient 5–100% B; 20–23 min, isocratic 100% B. The mass spectrometer was run in electrospray mode, observing positive ions. Sodium formiate clusters created at the beginning and end of the chromatogram were used for calibration. Mass spectral data were handled with the native DataAnalysis software.
Expression of Glycosyltransferases
The glycosyltransferase enzyme preparations used for the in vitro glycosylation assays were produced either in the yeast Pichia pastoris or in Escherichia coli expressed with a HIS-tag. Expression in Pichia pastoris was exerted essentially as described in the EasySelect™ Pichia Expression Kit manual from Invitrogen™ (Carlsbad, CA), using strain KM71H and vector pPICZαA (without HIS-tag) (Invitrogen, Carlsbad, CA), except that expression was induced at 20°C for 5 d. The medium was treated with the weakly basic anion-exchanger Amberlite IRA-95 (Supelco), prior to a 50X concentration step and buffer-exchange (10 mM Tris, pH 8, 5 mM DTT) by Amicon Ultra with 30-kD cut-off (Millipore). For bacterial expression, E. coli strain XJb (DE3) Autolysis™ (Zymo Research) and expression vector pET30a+ (Novagen) were used. Enzymes from 250 mL bacterial cultures were purified using HIS-Select™ Nickel Affinity Gel (Sigma), according to the manufacturer's protocol, resulting in 1 mL final volume of purified enzyme. The enzymes were stored in 50% glycerol.
Activity Measurements Using the Glycosyltransferase Platform
The general assay mixture (total volume: 15 μL) for monitoring glucosylation contained 100 mM Tris, pH 8, 8.2 μM 14C-UDP-glucose (Amersham Biosciences), 100 μM substrate (15 mM stock in DMSO), 3 μg BSA, and 0.5–2 μg enzyme preparation. After incubation (30°C, 30 min), aliquots (5 μL) of the reaction mixture was applied onto TLC aluminum sheets (Silica gel 60 F254, Merck) and developed in a solvent system consisting of ethylacetate: acetone: dichloromethane: methanol: water (20:15:6:5:4 (vol./vol./vol./vol./vol.)). Radiolabeled glucosides formed, were monitored following exposure of the TLC plates on phosphor imager screens (Amersham Biosciences) and phosphor imager analysis (STORM 840 phosphor imager (Molecular Dynamics, Sunnyvale, CA). Structural elucidation of glucosides formed was carried out using LC–MS/MS analysis and aliquots withdrawn from incubation mixtures (50 μL) composed of 100 mM Tris, pH 8, 200 μM UDP-glucose, 100 μM substrate (15 mM stock in DMSO), 10 μg BSA, and 0.5–2 μg enzyme produced in E. coli. Negative controls without UDP-glucose were included. After incubation (30°C, 30 min), 50 μL MeOH was added, precipitate removed by centrifugation, and supernatants analyzed by LC–MS/MS.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by the Danish Agency for Science, Technology, and Innovation, The Danish Research Foundation to “PlaCe”, and by the Villum Kann Rasmussen Foundation to “Pro-Active Plants”. No conflict of interest declared.
Supplementary Material
Acknowledgments
Dr Majse Nafisi is thanked for kindly providing indole-3-acetaldoxime. Susanne Bidstrup is thanked for performing the real time PCR analysis. Emma O'Callahan is thanked for assisting in preparation of the manuscript.
References
- Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Feyereisen R. The involvement of two p450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 2001;127:108–118. doi: 10.1104/pp.127.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R. CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell. 2001;13:101–111. doi: 10.1105/tpc.13.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros ML, et al. LAF1, a MYB transcription activator for phytochrome A signaling. Genes & Development. 2001;15:2613–2625. doi: 10.1101/gad.915001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier I, et al. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl Acad. Sci. U S A. 2000;97:14819–14824. doi: 10.1073/pnas.260502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Fink GR. Ilr1, an amidohydrolase that releases active indole-3-acetic-acid from conjugates. Science. 1995;268:1745–1748. doi: 10.1126/science.7792599. [DOI] [PubMed] [Google Scholar]
- Bednarek P, Schneider B, Svatos A, Oldham NJ, Hahlbrock K. Structural complexity, differential response to infection, and tissue specificity of indolic and phenylpropanoid secondary metabolism in Arabidopsis roots. Plant Physiol. 2005;138:1058–1070. doi: 10.1104/pp.104.057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop JJ, Millenaar FF, Smeets ME, van Zanten M, Voesenek LACJ, Peeters AJM. Abscisic acid antagonizes ethylene-induced hyponastic growth in Arabidopsis. Plant Physiol. 2007;143:1013–1023. doi: 10.1104/pp.106.092700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M. Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell. 2007;19:148–162. doi: 10.1105/tpc.106.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, et al. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher C, Westphal L, Schmotz C, Prade E, Scheel D, Glaswischnig E. The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cystein-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell. 2009;21:1830–1845. doi: 10.1105/tpc.109.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK. The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell. 2004;16:1191–1205. doi: 10.1105/tpc.020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza JL, et al. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005;137:253–262. doi: 10.1104/pp.104.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child RD, Chauvaux N, John K, Ulvskov P, Van Onckelen HA. Ethylene biosynthesis in oilseed rape pods in relation to pod shatter. J. Exper. Bot. 1998;49:829–838. [Google Scholar]
- De Paepe A, Vuylsteke M, Van Hummelen P, Zabeau M, Van der Straeten D. Transcriptional profiling by cDNA–AFLP and microarray analysis reveals novel insights into the early response to ethylene in Arabidopsis. Plant J. 2004;39:537–559. doi: 10.1111/j.1365-313X.2004.02156.x. [DOI] [PubMed] [Google Scholar]
- De Vos RCH, Moco S, Lommen A, Keurentjes JJB, Bino RJ, Hall RD. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nature Protocols. 2007;2:778–791. doi: 10.1038/nprot.2007.95. [DOI] [PubMed] [Google Scholar]
- Delarue M, Prinsen E, Onckelen HV, Caboche M, Bellini C. Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 1998;14:603–611. doi: 10.1046/j.1365-313x.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- Ekstrom CT, Bak S, Rudemo M. Pixel-level signal modelling with spatial correlation for two-colour microarrays. Stat. Appl. Genet. Mol. Biol. 2005;4 doi: 10.2202/1544-6115.1112. Article 6. [DOI] [PubMed] [Google Scholar]
- Ekstrom CT, Bak S, Kristensen C, Rudemo M. Spot shape modelling and data transformations for microarrays. Bioinformatics. 2004;20:2270–2278. doi: 10.1093/bioinformatics/bth237. [DOI] [PubMed] [Google Scholar]
- Endt DV, Kijne JW, Memelink J. Transcription factors controlling plant secondary metabolism: what regulates the regulators? Phytochemistry. 2002;61:107–114. doi: 10.1016/s0031-9422(02)00185-1. [DOI] [PubMed] [Google Scholar]
- Gachon CMM, Langlois-Meurinne M, Saindrenan P. Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 2005;10:542–549. doi: 10.1016/j.tplants.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Berger B, Mock HP, Muller C, Weisshaar B, Fluegge UI. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA. Camalexin is synthesized from indole-3-acetaidoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2004;101:8245–8250. doi: 10.1073/pnas.0305876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R, Thornburg RW. DNA sequence of UDP glucose: indole-3-acetate β-D-glucosyl transferase from Arabidopsis thaliana (U81293) Plant Physiol. 1997;113:1004. [Google Scholar]
- Grubb CD, Zipp BJ, Ludwig-Muller J, Masuno MN, Molinski TF, Abel S. Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 2004;40:893–908. doi: 10.1111/j.1365-313X.2004.02261.x. [DOI] [PubMed] [Google Scholar]
- Hagemeier J, Schneider B, Oldham NJ, Hahlbrock K. Accumulation of soluble and wall-bound indolic metabolites in Arabidopsis thaliana leaves infected with Virulent or avirulent Pseudomonas syringae pathovar tomato strains. Proc. Natl Acad. Sci. U S A. 2001;98:753–758. doi: 10.1073/pnas.021551098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- Hansen CH, et al. CYP83b1 is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. J. Biol. Chem. 2001;276:24790–24796. doi: 10.1074/jbc.M102637200. [DOI] [PubMed] [Google Scholar]
- Hansen KS, et al. The in vitro substrate regiospecificity of recombinant UGT85B1, the cyanohydrin glucosyltransferase from Sorghum bicolor. Phytochemistry. 2003;64:143–151. doi: 10.1016/s0031-9422(03)00261-9. [DOI] [PubMed] [Google Scholar]
- Hemm MR, Ruegger MO, Chapple C. The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell. 2003;15:179–194. doi: 10.1105/tpc.006544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson E, et al. Homeodomain leucine zipper class I genes in Arabidopsis: expression patterns and phylogenetic relationships. Plant Physiol. 2005;139:509–518. doi: 10.1104/pp.105.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann KM. The shikimate pathway: early steps in the biosynthesis of aromatic-compounds. Plant Cell. 1995;7:907–919. doi: 10.1105/tpc.7.7.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl Acad. Sci. U S A. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Toledo-Ortiz G, Bender J, Quail PH. The photomorphogenesis-related mutant red1 is defective in CYP83B1, a red light-induced gene encoding a cytochrome P450 required for normal auxin homeostasis. Planta. 2004;219:195–200. doi: 10.1007/s00425-004-1211-z. [DOI] [PubMed] [Google Scholar]
- Jackson RG, et al. Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: phenotypic characterisation of transgenic lines. Plant J. 2002;32:573–583. doi: 10.1046/j.1365-313x.2002.01445.x. [DOI] [PubMed] [Google Scholar]
- Jin HL, et al. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Vogt T. Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta. 2001;213:164–174. doi: 10.1007/s004250000492. [DOI] [PubMed] [Google Scholar]
- Jones P, Messner B, Nakajima JI, Schaffner AR, Saito K. UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 2003;278:43910–43918. doi: 10.1074/jbc.M303523200. [DOI] [PubMed] [Google Scholar]
- Jorgensen K, et al. Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr. Opin. Plant Biol. 2005;8:280–291. doi: 10.1016/j.pbi.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Kalde M, Barth M, Somssich IE, Lippok B. Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Molecular Plant–Microbe Interactions. 2003;16:295–305. doi: 10.1094/MPMI.2003.16.4.295. [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell. 2002;14:343–357. doi: 10.1105/tpc.010362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 2006;11:109–112. doi: 10.1016/j.tplants.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Kranz HD, et al. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Kristensen C, et al. Metabolic engineering of dhurrin in transgenic Arabidopsis plants with marginal inadvertent effects on the metabolome and transcriptome. Proc. Natl Acad. Sci. U S A. 2005;102:1779–1784. doi: 10.1073/pnas.0409233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro T, et al. The Arabidopsis cytochrome P450CYP707A encodes ABA 8‘-hydroxylases: key enzymes in ABA catabolism. EMBO J. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzina V, Ekstrøm CT, Andersen SB, Nielsen JK, Olsen CE, Bak S. Identification of defense compounds in Barbarea vulgaris against the Herbivore Phyllotreta nemorum by an ecometabolomic approach. Plant Physiol. 2009 doi: 10.1104/pp.109.136952. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labavitch JM, Ray PM. Turnover of cell wall polysaccharides in elongating pea stem segments. Plant Physiol. 1974;53:669–673. doi: 10.1104/pp.53.5.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois-Meurinne M, Gachon CMM, Saindrenan P. Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol. 2005;139:1890–1901. doi: 10.1104/pp.105.067223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanot A, et al. The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 2006;48:286–295. doi: 10.1111/j.1365-313X.2006.02872.x. [DOI] [PubMed] [Google Scholar]
- LeClere S, Tellez R, Rampey RA, Matsuda SPT, Bartel B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 2002;277:20446–20452. doi: 10.1074/jbc.M111955200. [DOI] [PubMed] [Google Scholar]
- Lehmann M, et al. The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol. Plant. 2009;2:390–406. doi: 10.1093/mp/ssn080. [DOI] [PubMed] [Google Scholar]
- Li JS, Dai XH, Zhao YD. A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiol. 2006;140:899–908. doi: 10.1104/pp.105.070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim EK, Doucet CJ, Hou B, Jackson RG, Abrams SR, Bowles DJ. Resolution of (+)-abscisic acid using an Arabidopsis glycosyltransferase. Tetrahedron-Asymmetry. 2005;16:143–147. [Google Scholar]
- Lim EK, Higgins GS, Li Y, Bowles DJ. Regioselectivity of glucosylation of caffeic acid by a UDP-glucose: glucosyltransferase is maintained in planta. Biochem. J. 2003;373:987–992. doi: 10.1042/BJ20021453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommen A. MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal. Chem. 2009;81:3079–3086. doi: 10.1021/ac900036d. [DOI] [PubMed] [Google Scholar]
- Lu GH, Paul AL, McCarty DR, Ferl RJ. Transcription factor veracity: is GBF3 responsible for ABA-regulated expression of Arabidopsis Adh? Plant Cell. 1996;8:847–857. doi: 10.1105/tpc.8.5.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes & Development. 1998;12:2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclachlan G, Davies E, Fan DF. Induction of cellulase by 3-indoleacetic acid. In: Wightman F, Setterfield G, editors. Biochemistry and Physiology of Plant Growth Substances. Ottawa, Canada: Runge Press; 1968. pp. 443–453. [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Manavella PA, et al. Cross-talk between ethylene and drought signalling pathways is mediated by the sunflower Hahb-4 transcription factor. Plant J. 2006;48:125–137. doi: 10.1111/j.1365-313X.2006.02865.x. [DOI] [PubMed] [Google Scholar]
- Mehrtens F, Kranz H, Bednarek P, Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005;138:1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Naur P, Halkier BA. Arabidopsis mutants in the C–S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 2004;37:770–777. doi: 10.1111/j.1365-313x.2004.02002.x. [DOI] [PubMed] [Google Scholar]
- Millenaar FF, Okyere J, May ST, van Zanten M, Voesenek LACJ, Peeters AJM. How to decide? Different methods of calculating gene expression from short oligonucleotide array data will give different results. BMC Bioinformatics. 2006;7:137. doi: 10.1186/1471-2105-7-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolo LV, Blount JW, Achnine L, Naoumkina MA, Wang XQ, Dixon RA. A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol. Biol. 2007;64:499–518. doi: 10.1007/s11103-007-9167-6. [DOI] [PubMed] [Google Scholar]
- Muller D, Schmitz G, Theres K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell. 2006;18:586–597. doi: 10.1105/tpc.105.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi M, et al. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell. 2007;19:2039–2052. doi: 10.1105/tpc.107.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naur P, et al. CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 2003;133:63–72. doi: 10.1104/pp.102.019240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KA, Tattersall DB, Jones PR, Moller BL. Metabolon formation in dhurrin biosynthesis. Phytochemistry. 2008;69:88–98. doi: 10.1016/j.phytochem.2007.06.033. [DOI] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell. 2007;19:118–130. doi: 10.1105/tpc.106.047761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson ASB, Engstrom P, Soderman E. The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 2004;55:663–677. doi: 10.1007/s11103-004-1581-4. [DOI] [PubMed] [Google Scholar]
- Osmani SA, Bak S, Imberty A, Olsen CE, Moller BL. Catalytic key amino acids and UDP-sugar donor specificity of a plant glucuronosyltransferase, UGT94B1: molecular modeling substantiated by site-specific mutagenesis and biochemical analyses. Plant Physiol. 2008;148:1295–1308. doi: 10.1104/pp.108.128256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostin A, Kowalyczk M, Bhalerao RP, Sandberg G. Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol. 1998;118:285–296. doi: 10.1104/pp.118.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette S, Moller BL, Bak S. On the origin of family 1 plant glycosyltransferases. Phytochemistry. 2003;62:399–413. doi: 10.1016/s0031-9422(02)00558-7. [DOI] [PubMed] [Google Scholar]
- Paquette SM, Jensen K, Bak S. 2009. A web-based resource for the Arabidopsis P450, cytochromes b(5), NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases ( http://www.P450.kvl.dk). Phytochemistry, in press. [Google Scholar]
- Piotrowski M, et al. Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 2004;279:50717–50725. doi: 10.1074/jbc.M407681200. [DOI] [PubMed] [Google Scholar]
- Poppenberger B, et al. The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl Acad. Sci. U S A. 2005;102:15253–15258. doi: 10.1073/pnas.0504279102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 2004;40:979–995. doi: 10.1111/j.1365-313X.2004.02280.x. [DOI] [PubMed] [Google Scholar]
- Ray PM. Regulation of beta-glucan synthetase-activity by auxin in pea stem tissue. 2. Metabolic requirements. Plant Physiol. 1973;51:609–614. doi: 10.1104/pp.51.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 2000;3:423–434. doi: 10.1016/s1369-5266(00)00107-2. [DOI] [PubMed] [Google Scholar]
- Rodman JE, Karol KG, Price RA, Sytsma KJ. Molecules, morphology, and Dahlgren's expanded order Capparales. Systematic Bot. 1996;21:289–307. [Google Scholar]
- Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR. Genetic-analysis of ethylene signal-transduction in Arabidopsis- thaliana: 5 novel mutant loci integrated into a stress-response pathway. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002;43:1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim E-K, Bowles D. Higher plant glycosyltransferases. Genome Biol. 2001;2:reviews3004.3001–reviews3004.3006. doi: 10.1186/gb-2001-2-2-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, et al. Arabidopsis CYP707As encode (+)-abscisic acid 8‘-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004;134:1439–1449. doi: 10.1104/pp.103.037614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinlapadech T, Stout J, Ruegger MO, Deak M, Chapple C. The hyper-fluorescent trichome phenotype of the brt1 mutant of Arabidopsis is the result of a defect in a sinapic acid: UDPG glucosyltransferase. Plant J. 2007;49:655–668. doi: 10.1111/j.1365-313X.2006.02984.x. [DOI] [PubMed] [Google Scholar]
- Skirycz A, et al. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006;47:10–24. doi: 10.1111/j.1365-313X.2006.02767.x. [DOI] [PubMed] [Google Scholar]
- Smolen G, Bender J. Arabidopsis cytochrome P450 cyp83B1 mutations activate the tryptophan biosynthetic pathway. Genetics. 2002;160:323–332. doi: 10.1093/genetics/160.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. Limma: linear models for microarray data. Bioinformatics and Computational Biology Solutions using R and Bioconductor. 2005 [Google Scholar]
- Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 2006;61:897–915. doi: 10.1007/s11103-006-0057-0. [DOI] [PubMed] [Google Scholar]
- Sorin C, et al. Proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol. 2006;140:349–364. doi: 10.1104/pp.105.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Barsch GHA, Mehrtens F, Niehaus K, Weisshaar B. Differential regulation of closely related R2R3–MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3–MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Tam YY, Epstein E, Normanly J. Characterization of auxin conjugates in Arabidopsis: low steady-state levels of indole-9-acetyl-aspartate indole-3-acetyl-glutamate, and indole-3-acetyl-glucose. Plant Physiol. 2000;123:589–595. doi: 10.1104/pp.123.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikunov Y, et al. A novel approach for nontargeted data analysis for metabolomics: large-scale profiling of tomato fruit volatiles. Plant Physiol. 2005;139:1125–1137. doi: 10.1104/pp.105.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JPC, et al. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. PNAS. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero P, Labrador E. Inhibition of cell-wall autolysis and auxin-induced elongation of cicer-arietinum epicotyls by beta-galactosidase antibodies. Physiologia Plantarum. 1993;89:199–203. [Google Scholar]
- Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 2000;5:380–386. doi: 10.1016/s1360-1385(00)01720-9. [DOI] [PubMed] [Google Scholar]
- Winkler RG, Frank MR, Galbraith DW, Feyereisen R, Feldmann KA. Systematic reverse genetics of transfer-DNA-tagged lines of Arabidopsis: isolation of mutations in the cytochrome p450 gene superfamily. Plant Physiol. 1998;118:743–750. doi: 10.1104/pp.118.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA. Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 2002;7:263–270. doi: 10.1016/s1360-1385(02)02273-2. [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Annals Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu J-K. Regulation of abscisic acid biosynthesis. Plant Phys. 2003;133:29–36. doi: 10.1104/pp.103.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZJ, Nakajima M, Suzuki Y, Yamaguchi I. Cloning and characterization of the abscisic acid-specific glucosyltransferase gene from adzuki bean seedlings. Plant Physiol. 2002;129:1285–1295. doi: 10.1104/pp.001784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip WK, Moore T, Yang SF. Differential accumulation of transcripts for four tomato 1-aminocyclopropane-1-carboxylate synthase homologs under various conditions. Proc Natl Acad Sci USA. 1992;89:2475–2479. doi: 10.1073/pnas.89.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.