Abstract
Heterosis, or hybrid vigor, is one of the most important tools in plant breeding and has previously been demonstrated for plant freezing tolerance. Freezing tolerance is an important trait because it can limit the geographical distribution of plants and their agricultural yield. Plants from temperate climates increase in freezing tolerance during exposure to low, non-freezing temperatures in a process termed ‘cold acclimation’. Metabolite profiling has indicated a major reprogramming of plant metabolism in the cold, but it has remained unclear in previous studies which of these changes are related to freezing tolerance. In the present study, we have used metabolic profiling to discover combinations of metabolites that predict freezing tolerance and its heterosis in Arabidopsis thaliana. We identified compatible solutes and, in particular, the pathway leading to raffinose as crucial statistical predictors for freezing tolerance and its heterosis, while some TCA cycle intermediates contribute only to predicting the heterotic phenotype. This indicates coordinate links between heterosis and metabolic pathways, suggesting that a limited number of regulatory genes may determine the extent of heterosis in this complex trait. In addition, several unidentified metabolites strongly contributed to the prediction of both freezing tolerance and its heterosis and we present an exemplary analysis of one of these, identifying it as a hexose conjugate.
Keywords: Abiotic/environmental stress, cold acclimation, metabolomics, bioinformatics, biostatistics, Arabidopsis
INTRODUCTION
The term ‘heterosis’ (Shull, 1914) originally described the phenomenon of increased physiological performance of F1 hybrids in comparison to their parents in both animals and plants. From a genetical standpoint, heterosis can be either positive or negative, whereas for breeding purposes, only positive heterosis (i.e. higher performance) is of interest. Heterosis can be defined either as a positive or negative deviation of the F1 from the parental mean (mid-parent heterosis; MPH = F1 – (P1 + P2)/2). Although heterosis has been used extensively by breeders to increase the performance of crop plants (Lippman and Zamir, 2006), its molecular basis is not understood and no biomarkers have been identified that would allow reliable prediction of heterosis (see Birchler et al., 2003, 2006; Hochholdinger and Hoecker, 2007, for reviews). In an almost exclusively selfing species like Arabidopsis thaliana (Abbott and Gomes, 1989), accessions are largely homozygous and may be expected to exhibit inbreeding depression. Crossing such accessions leads to increased heterozygosity, which may result in heterosis. Indeed, heterosis in crosses between Arabidopsis accessions has been demonstrated for traits such as biomass accumulation (Barth et al., 2003; Meyer et al., 2004), phosphate uptake (Narang and Altmann, 2001) and freezing tolerance (Korn et al., 2008; Rohde et al., 2004).
Freezing tolerance is a primary factor that defines the geographic distribution of plants. In addition, it has a strong influence on the yield of crop plants in large parts of the world, where frost can lead to periodic catastrophic yield losses. Plants from temperate and cold climates, including many important crop species, increase in freezing tolerance during exposure to low, but non-freezing, temperatures in a process termed ‘cold acclimation’ (Smallwood and Bowles, 2002; Thomashow, 1999; Xin and Browse, 2000). Plant freezing tolerance is a multigenic, quantitative trait. Gene expression profiling with whole genome microarrays indicates that cold acclimation in the model plant Arabidopsis thaliana involves changes in the expression levels of several hundred genes (Hannah et al., 2005, 2006; Kaplan et al., 2007; Vogel et al., 2005), while metabolite profiling revealed changes in the content of a large part of cellular metabolites, including those involved in central metabolism (see Guy et al., 2008, for a recent review). It should be noted here that exposure to 4°C is not a lethal stress for Arabidopsis. Plants continue to grow, although at a much lower rate, and, eventually, flower, set seeds, and successfully complete their lifecycle under these conditions.
Attempts to understand the genetic and molecular basis of complex quantitative traits in plants have, in recent years, focused on the analysis of natural genetic variation. Such genetic variation between crop plant cultivars or between crop species and their wild relatives has also been identified as a promising tool to improve the yield and other agronomically important traits of crop plants (Gur and Zamir, 2004). Arabidopsis represents an ideal model for such investigations, as it is a geographically widely spread species containing diverse accessions with sufficient genetic variability to allow investigations of genotype × environment interactions (see Koornneef et al., 2004; Mitchell-Olds and Schmitt, 2006, for reviews). This is also true for freezing tolerance, where large phenotypic variability and clear correlations with both latitude of origin and habitat growth temperature have been shown (Hannah et al., 2006; McKhann et al., 2008; Zhen and Ungerer, 2008). Quantitative trait locus (QTL) mapping was successfully employed to gain insight into the molecular basis of the differences in acclimated freezing tolerance between the Arabidopsis accessions Cape Verde Islands (Cvi) and Landsberg erecta (Ler) (Alonso-Blanco et al., 2005). In addition, high-throughput profiling methods have been employed to unravel the molecular basis of natural genetic variation in other traits (see de Meaux and Koornneef, 2008, for a review). In particular, metabolic profiling has been applied to both tomato (Schauer et al., 2006) and Arabidopsis (Keurentjes et al., 2006; Meyer et al., 2007; Rowe et al., 2008) to elucidate the genetic basis of plant metabolism and its relationship to physiological performance. Predictive metabolites for biomass accumulation (Meyer et al., 2007) and for heterosis in biomass accumulation in Arabidopsis have been identified in recent studies (Gärtner et al., 2009). However, no metabolite profiling studies have been reported in relation to heterosis in plant freezing tolerance and no predictive metabolites have been reported for either freezing tolerance or heterosis in freezing tolerance.
Here, we have screened a selection of Arabidopsis accessions and their F1 progeny by metabolic profiling that have previously been shown (Korn et al., 2008) to differ widely in freezing tolerance and heterosis in freezing tolerance both before and after cold acclimation. Thus, we followed the same analytical strategy of monitoring relative changes in metabolite pools that led to the discovery of metabolic QTL in Arabidopsis (Keurentjes et al., 2006; Rowe et al., 2008) and tomato (Fridman et al., 2004; Schauer et al., 2006). By statistical methods, we identify combinations of metabolites that are predictive of leaf freezing tolerance and of heterosis in freezing tolerance, thus unraveling metabolites and metabolic pathways that may be functionally associated with these traits.
RESULTS
Changes in Metabolite Content during Cold Acclimation
We profiled a metabolome fraction enriched in primary metabolites (Supplemental Table 1) by routine gas chromatography–mass spectrometry (GC–MS) in five parental accessions (C24, Col-0, Co-2, Ler, Te) and eight F1 populations generated by manually crossing both C24 and Col-0 with the respective other four accessions (Korn et al., 2008). Plants were harvested either before or after 14 d of cold acclimation at 4°C.
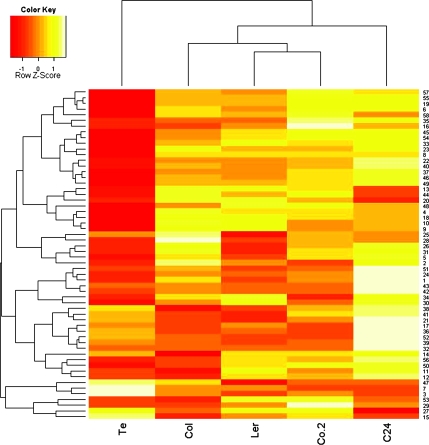
Figure 1 gives an overview of the changes in metabolite pool sizes in the parental accessions during cold acclimation. As observed in previous studies (Cook et al., 2004; Gray and Heath, 2005; Hannah et al., 2006; Kaplan et al., 2004, 2007), metabolism was largely reprogrammed upon exposure of plants to low temperature. The largest changes occurred in C24 and Co-2, the smallest in Te, with Ler and Col-0 intermediate. The same ranking was observed before (Hannah et al., 2006), attesting to the high degree of reproducibility of low temperature effects on metabolism noted previously (Guy et al., 2008).
Figure 1.
Hierarchical Clustering of Changes in Metabolite Pool Sizes in the Five Parental Accessions during 14 d of Cold Acclimation at 4°C.
Relative signal intensities for each metabolite were normalized to the mean intensity over all samples and Euclidean distance was used for clustering. Red indicates the smallest and yellow the biggest differences in metabolite content between samples from non-acclimated and cold-acclimated plants. The color key indicates the Z-scores of the distributions of these differences, namely the magnitude and direction of variation from the mean for every metabolite. Metabolites are identified by the ID numbers listed in Supplemental Table 1.
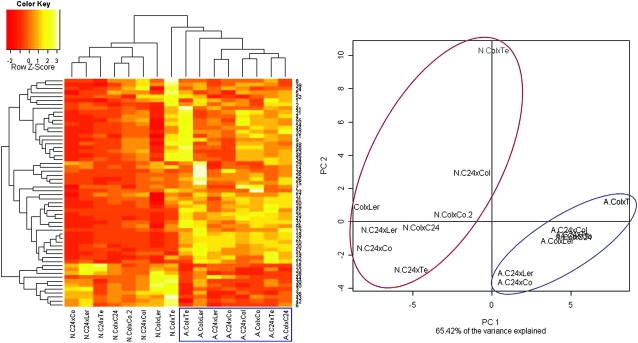
A similar extent of changes in metabolite pool sizes as observed in the parental accessions (Figure 1) was also evident in the crosses. Both hierarchical clustering and unsupervised principal component analysis (PCA) gave evidence for clearly distinct metabolic phenotypes comparing non-acclimated and cold-acclimated plants (Figure 2), again emphasizing the major reprogramming of plant metabolism in response to low temperature. In addition, both the parental accessions and the F1 crosses acquire increased leaf freezing tolerance during low temperature exposure. Acclimated and non-acclimated freezing tolerances vary strongly with genotype in parental accessions and F1 crosses, as determined in a previous study using an electrolyte leakage assay (Korn et al., 2008). Supplemental Table 2 lists the mean LT50 (freezing tolerance determined by electrolyte leakage measurements) values for all accessions and crosses used for metabolite profiling. However, the plants used for metabolite profiling were grown in separate experiments, independently of the plants used for the freezing tolerance measurements. Supplemental Table 2 also shows data from two earlier studies (Hannah et al., 2006; Rohde et al., 2004) on some of these genotypes that provide evidence for the excellent reproducibility of LT50 values over several years.
Figure 2.
Hierarchical Clustering and Principal Component Analysis (PCA) of the Metabolite Contents in all Crosses before (N) and After (A) Cold Acclimation.
Relative signal intensities for each metabolite were normalized to the mean intensity over all samples and Euclidean distance was used for clustering. Red indicates the lowest and yellow the highest metabolite content. The color key indicates the Z-scores of the distributions of the content of each metabolite, namely the magnitude and direction of variation from the mean. White indicates missing values. The cold-acclimated samples are highlighted by blue boxing. Metabolites are identified by the ID numbers listed in Supplemental Table 1. The PCA results are shown for the Principle Components (PC) 1 and 2, which together explain 65.4% of the total variance in the dataset. Non-acclimated samples are enclosed by a red line, acclimated samples by a blue line.
The availability of quantitative physiological (freezing tolerance expressed as LT50) and metabolic (metabolite pool sizes of 59 central metabolites consistently observable in all replicated experiments) phenotype data allowed statistical analyses to identify combinations of metabolites that predict freezing tolerance. For ranking metabolites according to their contribution to the prediction of freezing tolerance, we used the ‘variable importance in the projection’ (VIP; Eriksson et al., 2001). This calculates the contribution of each metabolite to freezing tolerance in a partial least squares model (Wold, 1975). To reduce the number of metabolites in the models, we retained only those with a high contribution. The threshold number of metabolites was determined by optimizing the predictive power in leave-one-out validation. Subsequently, the model with the highest predictive power was selected as described in detail recently (Gärtner et al., 2009). The statistical procedures that were used for the prediction model and the validation are outlined in a flowchart (Supplemental Figure 1) to illustrate this approach. All input and output variables used and generated in these analyses are listed in Table 1.
Table 1.
Response Variable, Samples, and Number of Input Variables Used to Train the Different Prediction Models and the Resulting Predictive Power in Cross-Validation.
| Response Y | Freezing tolerance C24-crosses | Freezing tolerance Col-crosses | MPH in freezing tolerance |
| Samples | C24-crosses: | Col-crosses | C24-crosses: |
| 4 NA + 4 ACC | (without Col×C24): 3 NA + 3 ACC | 4 NA + 4 ACC | |
| Col×C24: | Parental lines | Col-crosses: | |
| 1 NA + 1 ACC | (without C24): | 4 NA + 4 ACC | |
| Parental lines: | 4 NA + 4 ACC | ||
| 5 NA + 5 ACC | |||
| Sample size N | 20 | 14 | 16 |
| Variables with missing values | Maltitol (MVI) | Maltitol (MVI) | Maltitol (deleted) |
| Tryptophan (MVI) | Tryptophan (deleted) | Tryptophan (deleted) | |
| Unknown 44 (MVI) | |||
| Number p of variables X | 59 | 58 | 57 |
| PP of all p variables | 0.87 | 0.82 | 0.32 |
| Optimal number j.opt of variables | 20 | 14 | 17 |
| PP of j.opt selected variables | 0.91 | 0.93 | 0.85 |
MVI, missing value imputation (column-wise mean substitution for metabolites with not more than 10% missing values). Metabolites with more than 10% missing values were deleted from the analysis. PP, predictive power (Pearson-correlation between observed response and predicted response in leave-one-out validation).
These analyses revealed that 20 metabolites were sufficient to predict freezing tolerance in C24-crosses and the parental accessions, while 14 metabolites were sufficient to predict freezing tolerance in the Col-crosses and their corresponding parental accessions (Tables 1 and 2). In addition, the total of 21 highly predictive metabolites identified in the two independent crossing experiments (Table 2) showed a large overlap of 13 metabolites that appeared in the analysis of both experiments. The predictive power of the respectively optimal selection of metabolites was 0.91 for C24-crosses and 0.93 for Col-crosses (Table 1), which was clearly not inferior to the predictive power of combining all measured metabolites for the analysis. To test for the significance of the correlation between observed freezing tolerance and the freezing tolerance predicted from the optimal set of predictive metabolites in cross-validation (compare Supplemental Figure 1), we performed 5000 different random permutations of the observed response Y, freezing tolerance. Panels A and B in Supplemental Figure 2 correspond to the prediction models for freezing tolerance in the C24-crosses and Col-crosses, respectively. The correlation R between the actually observed response Y and the response vector YCV predicted in cross-validation is indicated in the figures (compare Table 1). The distance of R from the mean of the random correlations with YCV aggregates more than three standard deviations away from R and the estimated P-value is smaller than 0.001 in both cases.
Table 2.
Metabolites that Contribute to the Optimal Prediction Model for Freezing Tolerance.
| Metabolite number | Metabolite name | C24-crosses | Col-crosses |
| VIP | VIP | ||
| 3 | Fumaric acid | 2.2143 | 1.4084 |
| 7 | Succinic acid | 2.0124 | not selected |
| 8 | Fructose | 1.6657 | 1.7774 |
| 9 | Galactose | 1.6557 | 1.9322 |
| 10 | Glucose | 1.6072 | 1.2923 |
| 13 | Raffinose | 2.1335 | 2.7135 |
| 14 | Sucrose | 1.0619 | 1.2245 |
| 18 | Galactinol | 1.5055 | 2.1000 |
| 20 | Maltitol | Not selected | 1.1885 |
| 21 | Glycine | 1.0737 | Not selected |
| 22 | Proline [+CO2] | 1.1567 | Not selected |
| 23 | Proline | 1.5450 | 1.3678 |
| 30 | Dehydroascorbic acid dimer | 1.1416 | 1.3948 |
| 36 | Hexadecanoic acid | 0.9456 | Not selected |
| 38 | Itaconic acid | 0.9644 | Not selected |
| 39 | Ethanolamine | 1.0949 | Not selected |
| 48 | NA | 1.6438 | 1.6560 |
| 50 | NA | 0.9593 | 1.2818 |
| 51 | NA | 0.9857 | 1.2442 |
| 52 | NA | 1.0154 | Not selected |
| 59 | NA | 1.0534 | 1.5046 |
Data from both non-acclimated and acclimated plants were combined. Freezing tolerance data from electrolyte leakage measurements were taken from Korn et al. (2008) (compare Supplemental Table 2). The calculations were performed separately for Col-0- and C24-crosses and included the respective parental accessions, since the LT50 values for these groups were measured separately, leading to a small systematic shift in all LT50 values (C24-crosses: N = 20 (including C24×Col and Col×C24); Col-crosses: N = 14). Metabolite numbers refer to the numbers in Supplemental Table 1. VIP, variable importance in the projection. Higher VIP scores indicate greater importance of the selected metabolite in the prediction model. All 59 metabolites were included in the analysis for the C24-crosses, while for the Col-crosses, tryptophan was excluded because of more than 10% missing values.
On a physiological level, this analysis points to a crucial role of compatible solutes, such as sugars and proline, in determining plant freezing tolerance. In particular, metabolites of the raffinose biosynthetic pathway (galactinol, sucrose, raffinose, but not myo-inositol) make a substantial contribution to the prediction of the freezing tolerance phenotype. In addition, it is interesting to note that of the 13 metabolites highly ranked in both crossing experiments, four are structurally unknown, pointing to the presence of as yet unidentified metabolites in Arabidopsis that are, in combination with other metabolites, highly predictive for a complex trait such as freezing tolerance.
Heterosis in Metabolite Pool Sizes
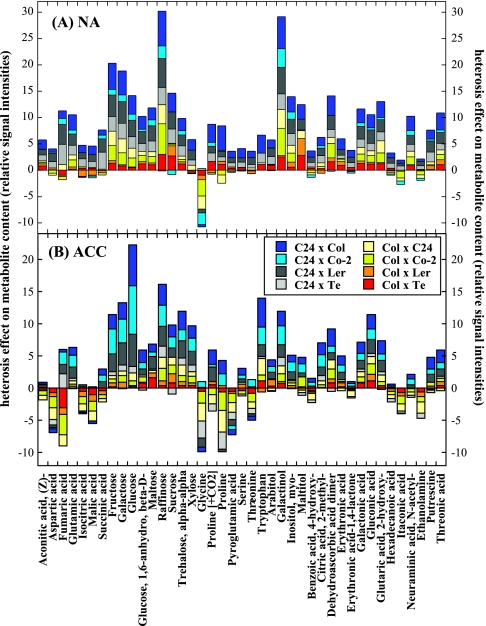
Heterotic effects on metabolite pool sizes were analyzed by comparing the mean metabolite level of the parental accessions to the metabolite content of the respective F1 plants (MPH). Figure 3 gives an overview of MPH in the pool sizes of all reliably identified metabolites (compare Supplemental Table 1) in all crosses calculated from the mean pool sizes over all three experiments. Supplemental Table 3 indicates the statistical significance of MPH for each metabolite and F1, tested separately for each experiment. At most, three experiments were available for each combination, resulting in 2377 statistical tests. In 742 cases, MPH was highly significant, with a threshold of 0.01 for the estimated FDR in the multiple testing set-up. With a threshold of 0.05 for the FDR, MPH was significant in 1061 cases. There were strongly and weakly affected metabolites and while most metabolites showed positive MPH in all crosses, some showed positive or negative MPH in different crosses and some mostly negative MPH, depending on the hybrid.
Figure 3.
Mid-Parent Heterosis (MPH) in the Metabolite Content of Leaf Tissue from Eight F1 Populations Before (NA; a) or After (ACC; b) 14 d of Cold Acclimation at 4°C.
MPH was calculated as F1 – (P1 + P2)/2 from the mean pool sizes over the three parallel experiments. For computing MPH of cold-acclimated plants, the mean pool sizes of the corresponding cold-acclimated parental plants were used.
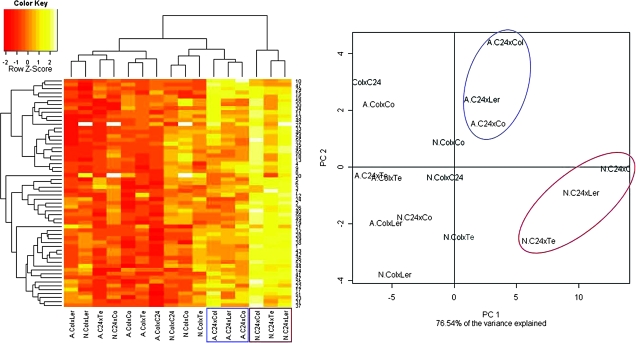
Figure 4 shows an analysis of the MPH levels in all metabolites by hierarchical clustering and PCA. Both analyses show that there were large differences in the extent of metabolic heterosis in different crosses. In contrast to the division between non-acclimated and acclimated plants that we obtained from their metabolic profiles (Figure 2), MPH of the metabolite pools was not obviously separating acclimated from non-acclimated plants (Figure 4). This implies that although cold acclimation is accompanied by a general shift in metabolism, there is no evidence for a principal shift in the extent of metabolic heterosis, which seems rather to be associated with genetic factors.
Figure 4.
Hierarchical Clustering and Principal Component Analysis (PCA) of the Mid-Parent Heterosis (MPH) in the Metabolite Content of Leaf Tissue from Eight F1 Populations Before (N) or After (A) 14 d of Cold Acclimation at 4°C.
Euclidian distance was used for clustering. Red indicates the smallest and yellow the largest MPH values. The color key indicates the Z-scores of the distributions of the MPH values in the metabolite pool sizes, namely the magnitude and direction of variation from the mean MPH for each metabolite. White indicates missing values. Metabolites are identified by the ID numbers listed in Supplemental Table 1. The PCA results are shown for the Principle Components (PC) 1 and 2, which together explain 76.5% of the total variance in the dataset. The two most extreme groups of non-acclimated and acclimated samples are highlighted in red and blue, respectively.
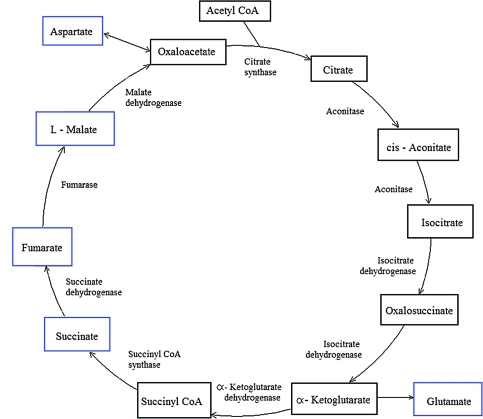
To identify the relationship between different metabolite pool sizes and metabolic pathways and the extent of heterosis in freezing tolerance, we performed the same type of statistical analysis as for freezing tolerance (Supplemental Figure 1). However, in this case, we used the heterosis in metabolite content as the input values to predict heterosis in freezing tolerance (Table 1). There was clear overlap between the two lists of highly predictive metabolites (compare Tables 2 and 3) with 13 of the 17 metabolites selected for prediction of heterosis in freezing tolerance in common with those selected for the prediction of freezing tolerance. Most of these were either compatible solutes (six) or unknown metabolites (four). Of the raffinose pathway, galactinol and raffinose were identified as highly predictive, while sucrose and myo-inositol were not in this list. Interestingly, the importance of a second metabolic pathway for heterosis in freezing tolerance was indicated by the presence of aspartic acid, fumaric acid, malic acid, succinic acid, and pyroglutamic acid (this GC peak contains a mixture of glutamine, glutamic acid, and pyroglutamic acid). These metabolites all belong to a central metabolic pathway, the tricarboxylic acid (TCA) cycle (Figure 5).
Table 3.
Metabolites that Contribute to the Optimal Prediction Model for MPH in the Freezing Tolerance of Arabidopsis.
| Number | Metabolite name | VIP |
| 2 | Aspartic acid | 1.3736 |
| 3 | Fumaric acid | 2.9162 |
| 6 | Malic acid | 0.9644 |
| 7 | Succinic acid | 0.9973 |
| 8 | Fructose | 1.4157 |
| 9 | Galactose | 1.4190 |
| 10 | Glucose | 2.6296 |
| 13 | Raffinose | 1.3597 |
| 16 | Xylose | 1.1764 |
| 18 | Galactinol | 1.2550 |
| 22 | Proline [+CO2] | 1.1315 |
| 23 | Proline | 1.3730 |
| 24 | Pyroglutamic acid | 1.0668 |
| 48 | NA | 2.2898 |
| 50 | NA | 0.9858 |
| 51 | NA | 0.9020 |
| 59 | NA | 1.0576 |
Data from both non-acclimated and acclimated plants were combined (N = 16; Table 1) and included significant and non-significant MPH values for both freezing tolerance and metabolites. Data for heterosis in freezing tolerance from electrolyte leakage measurements were taken from Korn et al. (2008). Metabolite numbers refer to the numbers in Supplemental Table 1. VIP, variable importance in the projection. Higher VIP scores indicate greater importance of the selected metabolite MPH in the prediction model. The metabolites maltitol and tryptophan were excluded from the analysis because of more than 10% missing values.
Figure 5.
Schematic Representation of the TCA Cycle.
Metabolite names are shown in the boxes and the names of the relevant enzymes inside the cycle. Metabolites boxed in blue were identified as contributing to the prediction of MPH in freezing tolerance (Table 2). Metabolites boxed in black were either not measured or were not selected in the prediction model for heterosis in freezing tolerance (cis-aconitate and isocitrate).
In cross-validation, the predictive power of the heterosis levels in the pool sizes of the selected 17 metabolites for MPH in freezing tolerance was 0.85, which is clearly superior to the predictive power of the heterosis levels of all quantified metabolites (Table 1). As in the case of freezing tolerance per se, a permutation test using 5000 different random permutations of the observed response Y indicated high significance of the correlation between observed MPH in freezing tolerance and the predictions for heterosis in freezing tolerance from metabolic heterosis data from cross-validation (Supplemental Figure 2C).
Characterization of the Mass Spectral Tag A196004 (Metabolite 48)
GC–MS-based metabolite profiling, like other metabolomic technologies, yields high numbers of not yet identified metabolic components (Bino et al., 2004). The structural elucidation of such metabolites represents one of the grand metabolomic challenges, as the chemical identification of each single substance is a complex and time-demanding task (Kopka, 2006). In GC–MS-based profiling experiments, these non-identified components are called mass spectral tags (MSTs). MSTs are archived by the publicly available Golm Metabolome Database (GMD, Kopka et al., 2005) for an evidence-based exchange of information in the international metabolomic field. Using preliminary MST identifiers, such as A196004 (Supplemental Table 1), GMD provides physicochemical information about MSTs, such as mass spectral fragmentation and chromatographic retention index (RI). The GMD reference data support both the recognition of such compounds in independent profiling experiments and the spectral interpretation prior to the tedious chemical elucidation of the underlying structure. In the following, we will shortly summarize the available information on MST A196004, which has been discovered as a potential biomarker in this study (metabolite 48 in Tables 2 and 3).
By comparison to previously established non-supervised libraries (Wagner et al., 2003) comprising mass spectra (MS) and retention indices (RI), MST A196004 was found to be present in Arabidopsis and tobacco leaf tissue and in tomato fruit. Therefore, A196004 does not represent a secondary product specific to Arabidopsis. Using a representative mass spectrum (Figure 6A), a search for the best MS match was performed, yielding 1-thioisopropyl-β-D-galactopyranoside, a salicylic acid glucopyranoside, disaccharides and with lower mass spectral agreement an epimeric set of hexonic acid-1,4-lactones, among others, gluconic acid-1,4-lactone. However, none of these compounds fulfilled the second identification criterion, namely a match of the RI property. The RI was calculated using n-alkane reference compounds (Strehmel et al., 2008). As direct matching failed, a classification based on mass spectral fragmentation will remain the final option. A196004 exhibited a mass shift upon 13C-labeling (Figure 6) of not more than six atomic mass units (amu) and showed all fragments typical of a glycoside. Furthermore, the RI indicates a higher chromatographic retention compared to possible glycosidic monomers, such as glucose, galactose, or mannose, but a much smaller RI than disaccharides. Taken together, this evidence indicates that A196004 may represent a small hexose conjugate. Unfortunately, the mass spectral fragmentation gives no clear evidence concerning the chemical nature of this moiety. The next steps of structural elucidation may be directed towards enrichment of the glycoside fraction and analysis of chemical cleavage products.
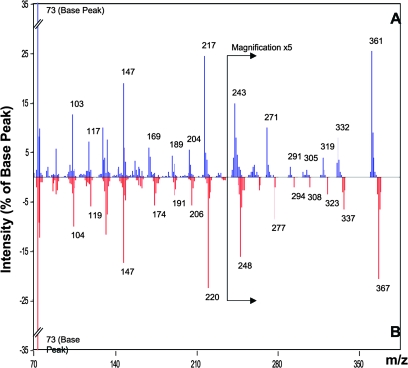
Figure 6.
Mass Spectral Tag (MST) Information of Analyte A196004 as Archived in the Golm Metabolome Database (GMD).
A196004 is represented in GMD by ambient mass spectra (A). In addition, a 13C-enriched variant (B) was obtained after 13CO2 in vivo labeling of Arabidopsis thaliana plants as described recently (Huege et al., 2007). The 13C-enriched variant proves metabolic origin of this MST and supports spectral interpretation by indicating the number of carbon atoms in each major mass fragment. Fragments that contain carbon atoms originating from the metabolite structure exhibit a respective mass shift; for example, the fragment at m/z 361 was shifted to 367 and thus contains six carbon atoms. Mass spectra were normalized to the base peak and are visualized by percent of base peak intensity.
DISCUSSION
Metabolite profiling of plant tissues by GC–MS quantifies a major part of the metabolites of central metabolism. Obviously, not all cellular metabolites can be detected with this method and recent results also show that secondary metabolites such as flavonols, that can be analyzed by LC–MS, may play a role in plant freezing tolerance (Korn et al., 2008). Previous metabolite profiling studies (Guy et al., 2008) indicated a major restructuring of plant metabolism during cold acclimation. The total magnitude of these metabolic changes, however, was not related to the increase in freezing tolerance of different Arabidopsis accessions during acclimation (Hannah et al., 2006). This is not surprising, considering the fact that plants need to adapt their metabolism not only to increase freezing tolerance, but also to assure growth and development after a drastic temperature shift. Due to these confounding effects and the multitude of metabolic changes, it has not been possible to identify metabolites that are of particular relevance to freezing tolerance. We therefore used a statistical method to identify groups of metabolites that together can accurately predict either freezing tolerance or MPH in freezing tolerance. In both cases, several substances were identified as important that are generally considered as compatible solutes.
Compatible solutes are synthesized by many organisms ranging from bacteria to animals and plants in response to various environmental stress conditions. This chemically heterogeneous group of compounds comprises, among others, amino acids such as proline and many sugars and sugar alcohols such as glucose, fructose, sucrose, raffinose, and galactinol (see Somero, 1992; Yancey et al., 1982, for reviews). Compatible solutes should have no adverse metabolic effects, even at very high concentrations, and stabilize sensitive cellular components under stress conditions. During freezing, they may act colligatively by decreasing the freezing point of the cytoplasm, thereby increasing the unfrozen cell volume in equilibrium with extracellular ice. In addition, they stabilize proteins by preferential exclusion from the hydration shell (Timasheff, 1993), assist refolding of unfolded polypeptides by chaperone proteins (Diamant et al., 2001), and stabilize membranes during freezing and drying (Crowe et al., 1990; Hincha et al., 2006).
Several of the metabolites forming a complete pathway involved in compatible solute biosynthesis, namely the raffinose pathway (Keller and Pharr, 1996; Peterbauer and Richter, 2001), were identified as contributing substantially to the prediction of both freezing tolerance and heterosis in freezing tolerance. Compatible solutes act rather non-specifically to increase stress tolerance and it has been suggested that they constitute a redundant cellular protection system (Hincha et al., 2005). It has been shown that neither a moderate increase in raffinose content through overexpression of a gene encoding the enzyme galactinol synthase nor the knockout of the gene encoding raffinose synthase had any measurable influence on Arabidopsis freezing tolerance (Zuther et al., 2004). However, from a recent metabolomic study comparing wild-type Arabidopsis plants under control, drought, and cold conditions with plants overexpressing the transcription factors DREB1A (CBF3) and DREB2A, it was also concluded that raffinose metabolism plays a crucial role in plant freezing tolerance (Murayama et al., 2009). Interestingly, in the knockout mutant plants that contained no raffinose, galactinol content was strongly increased (Zuther et al., 2004), suggesting that galactinol might be able to substitute for raffinose in protecting cells from freezing damage. Our approach of identifying combinations of metabolites with predictive power for a complex trait seems well suited to such redundant functional systems with several nonspecific components.
The second metabolic pathway that was identified through our analyses was the TCA cycle, which seemed to be specifically related to heterosis in freezing tolerance. Increased amounts of some TCA cycle intermediates during cold acclimation have been reported previously (Guy et al., 2008), but the functional significance of these increases is unclear, as are the molecular mechanisms underlying these changes in metabolite pool sizes. One crucial enzyme of the TCA cycle (α-ketoglutarate dehydrogenase) was found to be highly sensitive to oxidative stress, leading to a block in this metabolic pathway (Baxter et al., 2007). Whether this effect has contributed to the observed involvement of TCA cycle intermediates in the heterosis in freezing tolerance remains to be investigated. Also, the changes in TCA cycle intermediates could indicate either changes of flux into or from the TCA cycle for respiratory energy production or for biosynthetic processes coupled to TCA cycle activity. This has to be resolved by flux analysis and feeding of isotopically labeled precursors.
The coordinate involvement of metabolic pathways suggested by our analyses may indicate that heterosis could be related to effects on regulatory genetical elements, which might be identified as distinct loci through heterotic QTL mapping. The analysis of the underlying genes may lead to a new level of understanding of the phenomenon of heterosis. In addition, both combinations of metabolites as identified here and DNA polymorphisms to be identified through QTL mapping could be used in marker-assisted breeding approaches to improve the yield and stress tolerance of crop plants.
METHODS
Plant Material
We used Arabidopsis thaliana plants from the accessions C24, Coimbra-2 (Co-2), Columbia-0 (Col-0), Landsberg erecta (Ler), and Tenela (Te). The sources of the different seed stocks have been described in a recent publication (Schmid et al., 2006). Seeds for our experiments have been generated through single seed descent to assure genetic homogeneity of the plants (Törjek et al., 2003). F1 crosses were generated by manual pollination. Plants were grown in soil in a greenhouse at 16-h day length with light supplementation to reach at least 200 μE m−2 s−1 and a temperature of 20°C during the day, 18°C during the night until bolting. For cold acclimation, plants were transferred to a 4°C growth cabinet at 16-h day length with 90 μE m−2 s−1 for an additional 14 d (Hannah et al., 2006). Freezing damage was determined as electrolyte leakage after freezing of detached leaves to different temperatures as described previously (Hannah et al., 2006; Rohde et al., 2004). Between 12 and 24 plants were analyzed in each experiment from each genotype and treatment. All experiments were performed at least twice (Korn et al., 2008).
Metabolite Profiling by Gas Chromatography–Mass Spectrometry (GC–MS)
We have profiled primary metabolites (Supplemental Table 1) by gas chromatography–mass spectrometry (GC–MS) in five parental accessions (C24, Col-0, Co-2, Ler, Te) and eight F1 populations generated by crossing both C24 and Col-0 with the other four accessions (Korn et al., 2008). In three independent experiments, single mature leaves were harvested from 25 plants of each genotype, either before or after 14 d of cold acclimation at 4°C. Leaves were randomly pooled to generate five (experiments 1 and 2) to 10 (experiment 3) replicate samples for GC–MS analysis. Methods for the extraction of polar metabolites, GC–MS measurements, and metabolite identification and quantification were performed as previously published (Kopka et al., 2005; Lüdemann et al., 2008). All mass spectra and metabolite data will be made available upon request to either Alexander Erban (erban@mpimp-golm.mpg.de) or Joachim Kopka (kopka@mpimp-golm.mpg.de).
Statistical Methods
For all subsequent statistical analyses, the relative signal intensities for the detected metabolites were normalized to the mean intensity of all samples. Hierarchical clustering was performed using the hclust function in the software R (publicly available at www.r-project.org) that uses Euclidean distance as a measure of similarity between data points. The heatmap function in R was used to visualize the clustering results. Principal component analysis (PCA) was conducted employing the pcaMethods software package in R. This Probabilistic PCA (ppca) allows evaluation of incomplete datasets by estimating 10–15% missing values (Stacklies et al., 2007).
To train the prediction models, a partial least squares (PLS) regression (Wold, 1975) was performed by applying the function plsr within the R package pls. The variable importance in projection (VIP; Eriksson et al., 2001) was used to rank the predictor variables, namely the metabolites, according to their contribution to the response in the respective PLS model. Feature selection was adopted by optimizing the predictive power of the PLS model with respect to the number of predictor variables in the model (for details, see Gärtner et al., 2009). Here, the Pearson correlation between observed and in leave-one-out validation (n-fold cross-validation)-predicted response was consulted for determining the predictive power. To test the significance of this correlation, we compared it to the correlations between the predicted response and 5000 different random permutations of the observed response (see Supplemental Figures 1 and 2 and Table 1 for additional details of the statistical analyses).
To analyze the significance of MPH in the content of each metabolite in each cross, the R functions sam and sam2excel from the R package siggenes were used. The significance analysis of microarrays (SAM), originally conceived for gene expression data (Tusher et al., 2001), is a multiple testing method that estimates the false discovery rate (FDR). We considered the two class case for unpaired data assuming unequal variances and tested the respective metabolite levels in the F1 plants against the mean metabolite levels of the respective parents. The analysis was performed for a threshold of both 0.05 and 0.01 for the estimated FDR.
In case of the F1 plants, the data from all replicates could be used directly for the test. However, the parental means could not be calculated directly from the data from both parents, as these were not derived from paired samples. In some cases, even the number of replicate measurements was not the same for both parents, due to loss of samples during processing. Therefore, assuming independent normal distribution for each metabolite in every parental accession, 10 random numbers were generated following a normal distribution with mean and variation estimated from both parental metabolite levels (μ = (μ1 + μ2)/2 and var = (var1 + var2)/4). These 10 random numbers were then tested against the F1 metabolite levels using the SAM procedure. To compute the values of the test statistics that would be expected under the null hypothesis (Tusher et al., 2001), 1000 random permutations were generated. The number of tests was 2494, namely 59 metabolites were tested for eight crosses in three experiments for both the acclimated and non-acclimated case (not all crosses were available in each experiment); 117 tests were removed because there was not more than one non-missing value. All other missing values were replaced by a row-wise mean.
For all statistical analyses other than the SAM analysis, metabolite contents were averaged across the three experiments after normalization. Freezing tolerance data were also averaged over all measurements.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This project was supported in part by funds from the Max-Planck-Society.
Supplementary Material
Acknowledgments
M.K. gratefully acknowledges a Ph.D. scholarship from the Hans-Böckler-Stiftung. We are grateful to Thomas Altmann and Rhonda Meyer (IPK Gatersleben, Germany) for seed material. No conflict of interest declared.
References
- Abbott RJ, Gomes MF. Population genetic structure and outcrossing rate of Arabidopsis thaliana (L.) Heynh. Heredity. 1989;62:411–418. [Google Scholar]
- Alonso-Blanco C, Gomez-Mena C, Llorente F, Koornneef M, Salinas J, Martinez-Zapater JM. Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis. Plant Physiol. 2005;139:1304–1312. doi: 10.1104/pp.105.068510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S, Busimi AK, Utz HF, Melchinger AE. Heterosis for biomass yield and related traits in five hybrids of Arabidopsis thaliana L. Heynh. Heredity. 2003;91:36–42. doi: 10.1038/sj.hdy.6800276. [DOI] [PubMed] [Google Scholar]
- Baxter CJ, Redestig H, Schauer N, Repsilber D, Patil KR, Nielsen J, Selbig J, Liu J, Fernie AR, Sweetlove LJ. The metabolic response of heterotrophic Arabidopsis cells to oxidative stress. Plant Physiol. 2007;143:312–325. doi: 10.1104/pp.106.090431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bino RJ, et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 2004;9:418–425. doi: 10.1016/j.tplants.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Auger DL, Riddle NC. In search of the molecular basis of heterosis. Plant Cell. 2003;15:2236–2239. doi: 10.1105/tpc.151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S. Unraveling the genetic basis of hybrid vigor. Proc. Natl Acad. Sci. U S A. 2006;103:12957–12958. doi: 10.1073/pnas.0605627103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF. A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc. Natl Acad. Sci. U S A. 2004;101:15243–15248. doi: 10.1073/pnas.0406069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JH, Carpenter JF, Crowe LM, Anchordoguy TJ. Are freezing and dehydration similar stress vectors? A comparison of modes of interaction of stabilizing solutes with biomolecules. Cryobiology. 1990;27:219–231. [Google Scholar]
- de Meaux J, Koornneef M. The cause and consequences of natural variation: the genomic era takes off. Curr. Opin. Plant Biol. 2008;11:99–102. doi: 10.1016/j.pbi.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Diamant S, Eliahu N, Rosenthal D, Goloubinoff P. Chemical chaperones regulate molecular chaperones in vitro and cells under combined salt and heat stresses. J. Biol. Chem. 2001;276:39586–39591. doi: 10.1074/jbc.M103081200. [DOI] [PubMed] [Google Scholar]
- Eriksson L, Johansson E, Kettaneh-Wold N, Wold S. 2001. Multi- and Megavariate Data Analysis: Principles and Applications. (Umea, Sweden: Umetrics Academy) [Google Scholar]
- Fridman E, Carrari F, Liu Y-S, Fernie AR, Zamir D. Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science. 2004;305:1786–1789. doi: 10.1126/science.1101666. [DOI] [PubMed] [Google Scholar]
- Gärtner T, Steinfath M, Andorf S, Lisec J, Meyer RC, Altmann T, Willmitzer L, Selbig J. Improved heterosis prediction by combining information on DNA- and metabolic markers. PLoS One. 2009;4:e5220. doi: 10.1371/journal.pone.0005220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray GR, Heath D. A global reorganization of the metabolome in Arabidopsis during cold acclimation is revealed by metabolic fingerprinting. Physiol. Plant. 2005;124:236–248. [Google Scholar]
- Gur A, Zamir D. Unused natural variation can lift yield barriers in plant breeding. PLoS Biol. 2004;2:e245. doi: 10.1371/journal.pbio.0020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CL, Kaplan F, Kopka J, Selbig J, Hincha DK. Metabolomics of temperature stress. Physiol. Plant. 2008;132:220–235. doi: 10.1111/j.1399-3054.2007.00999.x. [DOI] [PubMed] [Google Scholar]
- Hannah MA, Heyer AG, Hincha DK. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 2005;1:e26. doi: 10.1371/journal.pgen.0010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah MA, Wiese D, Freund S, Fiehn O, Heyer AG, Hincha DK. Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol. 2006;142:98–112. doi: 10.1104/pp.106.081141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hincha DK, Popova AV, Cacela C. Effects of sugars on the stability of lipid membranes during drying. In: Leitmannova Liu A, editor. Advances in Planar Lipid Bilayers and Liposomes. Amsterdam: Elsevier; 2006. pp. 189–217. [Google Scholar]
- Hincha DK, Zuther E, Hundertmark M, Heyer AG. The role of compatible solutes in plant freezing tolerance: a case study on raffinose. In: Chen THH, Uemura M, Fujikawa S, editors. Cold Hardiness in Plants: Molecular Genetics, Cell Biology and Physiology. Wallingford, UK: CABI Publishing; 2005. pp. 203–218. [Google Scholar]
- Hochholdinger F, Hoecker N. Towards the molecular basis of heterosis. Trends Plant Sci. 2007;12:427–432. doi: 10.1016/j.tplants.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Huege J, Sulpice R, Gibon Y, Lisec J, Koehl K, Kopka J. GC–EI–TOF–MS analysis of in vivo-carbon-partitioning into soluble metabolite pools of higher plants by monitoring isotope dilution after (13CO2)-labelling. Phytochemistry. 2007;68:2258–2272. doi: 10.1016/j.phytochem.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 2004;136:4159–4168. doi: 10.1104/pp.104.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Sung DY, Zhao W, Popp M, Porat R, Guy CL. Transcript and metabolite profiling during cold acclimation of Arabidopsis reveals an intricate relationship of cold-regulated gene expression with modifications in metabolite content. Plant J. 2007;50:967–981. doi: 10.1111/j.1365-313X.2007.03100.x. [DOI] [PubMed] [Google Scholar]
- Keller F, Pharr DM. Metabolism of carbohydrates in sinks and sources: galactosyl-sucrose oligosaccharides. In: Zamski E, Schaffer AA, editors. Photoassimilate Distribution in Plants and Crops. New York: Marcel Dekker; 1996. pp. 115–184. [Google Scholar]
- Keurentjes JJB, Fu J, de Vos CHR, Lommen A, Hall RD, Bino RJ, van der Plas LHW, Jansen RC, Vreugdenhil D, Koornneef M. The genetics of plant metabolism. Nat. Genet. 2006;38:842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- Kopka J. Current challenges and developments in GC–MS based metabolite profiling technology. J. Biotechnol. 2006;124:312–323. doi: 10.1016/j.jbiotec.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Kopka J, et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- Korn M, Peterek S, Mock H-P, Heyer AG, Hincha DK. Heterosis in the freezing tolerance, and sugar and flavonoid contents of crosses between Arabidopsis thaliana accessions of widely varying freezing tolerance. Plant Cell Environ. 2008;31:813–827. doi: 10.1111/j.1365-3040.2008.01800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman ZB, Zamir D. Heterosis: revisiting the magic. Trends Genet. 2006;23:60–66. doi: 10.1016/j.tig.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Lüdemann A, Strassburg K, Erban A, Kopka J. TagFinder for the quantitative analysis of gas-chromatography–mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics. 2008;24:732–737. doi: 10.1093/bioinformatics/btn023. [DOI] [PubMed] [Google Scholar]
- McKhann HI, Gery C, Berard A, Leveque S, Zuther E, Hincha DK, de Mita S, Brunel D, Teoule E. Natural variation in CBF gene sequence, gene expression and freezing tolerance in the Versailles core collection of Arabidopsis thaliana. BMC Plant Biol. 2008;8:105. doi: 10.1186/1471-2229-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, et al. The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc. Natl Acad. Sci. U S A. 2007;104:4759–4764. doi: 10.1073/pnas.0609709104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, Törjek O, Becher M, Altmann T. Heterosis of biomass production in Arabidopsis: establishment during early development. Plant Physiol. 2004;134:1813–1823. doi: 10.1104/pp.103.033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 2006;441:947–952. doi: 10.1038/nature04878. [DOI] [PubMed] [Google Scholar]
- Murayama K, et al. Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 2009;150:1972–1980. doi: 10.1104/pp.109.135327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang RA, Altmann T. Phosphate acquisition heterosis in Arabidopsis thaliana: a morphological and physiological analysis. Plant Soil. 2001;134:91–97. [Google Scholar]
- Peterbauer T, Richter A. Biochemistry and physiology of raffinose family oligosaccharides and galactosyl cyclitols in seeds. Seed Sci. Res. 2001;11:185–197. [Google Scholar]
- Rohde P, Hincha DK, Heyer AG. Heterosis in the freezing tolerance of crosses between two Arabidopsis thaliana accessions (Columbia-0 and C24) that show differences in non-acclimated and acclimated freezing tolerance. Plant J. 2004;38:790–799. doi: 10.1111/j.1365-313X.2004.02080.x. [DOI] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell. 2008;20:1199–1216. doi: 10.1105/tpc.108.058131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, et al. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 2006;24:447–454. doi: 10.1038/nbt1192. [DOI] [PubMed] [Google Scholar]
- Schmid KJ, Törjek O, Meyer R, Schmuths H, Hoffmann MH, Altmann T. Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor. Appl. Genet. 2006;112:1104–1114. doi: 10.1007/s00122-006-0212-7. [DOI] [PubMed] [Google Scholar]
- Shull GH. Duplicate genes for capsule-form in Bursa bursa-pastoris. Zeitschr. indukt. Abstammungs- und Vererbungslehre. 1914;12:97–149. [Google Scholar]
- Smallwood M, Bowles DJ. Plants in a cold climate. Phil. Trans. R. Soc. Lond. B. 2002;357:831–847. doi: 10.1098/rstb.2002.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somero GN. Adapting to water stress: convergence on common solutions. In: Somero GN, Osmond CB, Bolis CL, editors. Water and Life. Berlin: Springer; 1992. pp. 3–18. [Google Scholar]
- Stacklies W, Redestig H, Scholz M, Walther D, Selbig J. pcaMethods: a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 2007;23:1164–1167. doi: 10.1093/bioinformatics/btm069. [DOI] [PubMed] [Google Scholar]
- Strehmel N, Hummel J, Erban A, Strassburg K, Kopka J. Estimation of retention time index thresholds for compound matching using routine gas chromatography–mass spectrometry based metabolite profiling experiments. J. Chromatog. B. 2008;871:182–190. doi: 10.1016/j.jchromb.2008.04.042. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Timasheff SN. The control of protein stability and association by weak interactions with water: how do solvents affect these processes? Annu. Rev. Biophys. Biomol. Struct. 1993;22:67–97. doi: 10.1146/annurev.bb.22.060193.000435. [DOI] [PubMed] [Google Scholar]
- Törjek O, Berger D, Meyer RC, Müssig C, Schmid KJ, Rosleff Sörensen T, Weisshaar B, Mitchell-Olds T, Altmann T. Establishment of a high-efficiency SNP-based framework marker set for Arabidopsis. Plant J. 2003;36:122–140. doi: 10.1046/j.1365-313x.2003.01861.x. [DOI] [PubMed] [Google Scholar]
- Tusher VR, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- Wagner C, Sefkow M, Kopka J. Construction and application of a mass spectral and retention time index database generated from GC/EI–TOF–MS metabolite profiles. Phytochemistry. 2003;62:887–900. doi: 10.1016/s0031-9422(02)00703-3. [DOI] [PubMed] [Google Scholar]
- Wold H. Soft modelling by latent variables: the nonlinear iterative partial least squares approach. In: Gani J, editor. Perspectives in Probability and Statistics. London: Academic Press; 1975. pp. 520–540. [Google Scholar]
- Xin Z, Browse J. Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ. 2000;23:893–902. [Google Scholar]
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with water stress: evolution of osmolyte systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- Zhen Y, Ungerer MC. Clinal variation in freezing tolerance among natural accessions of Arabidopsis thaliana. New Phytol. 2008;177:419–427. doi: 10.1111/j.1469-8137.2007.02262.x. [DOI] [PubMed] [Google Scholar]
- Zuther E, Büchel K, Hundertmark M, Stitt M, Hincha DK, Heyer AG. The role of raffinose in the cold acclimation response of Arabidopsis thaliana. FEBS Lett. 2004;576:169–173. doi: 10.1016/j.febslet.2004.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.