Abstract
Acyl carrier proteins involved in fatty acid biosynthesis have been shown to exhibit a high degree of conformational flexibility, in that they are able to sequester fatty acid intermediates between 4 and 18 carbons in length. This flexibility has been observed in X-ray and NMR structures of acyl carrier proteins attached to different fatty acids. NMR studies comparing decanoyl-ACP and stearoyl-ACP indicated that ACP exhibits more dynamic motions when bound to longer fatty acids. We have used complementary chemical and NMR methods as an approach to improving our understanding of the effect of fatty acid length on the dynamics of acyl carrier protein. A chemical assay of the accessibility of the acyl thioester to solvent revealed a positive correlation between chain length and rate of hydrolysis. Surprisingly, this linear correlation was biphasic, with accelerated hydrolysis observed for fatty acids longer than 15 carbons. To further understand the motions associated with this acceleration, we collected 15N relaxation dispersion data for 14:0-, 15:0-, and 16:0-ACP. The greatest dispersions were exhibited by residues that form the entrance to the fatty acid binding pocket. In addition, these dispersions were observed to increase with the length of the fatty acid. Because the exchange rates derived from fitting the data to a two-state model varied from residue to residue, a more complex motional model appears to be required to adequately explain the dynamics. Thus, acyl-ACP offers an interesting system for future investigations of complex protein motions on the micro- and millisecond time scales.
Acyl carrier proteins (ACPs)1 make up a class of small molecule carriers involved in the biosynthesis of a variety of molecules, including fatty acids (1), membrane oligosaccharides (2,3), polyketides (4,5), toxins (6), depsipeptides (7), and biofuels (8). Re-engineering of these systems offers prospects for developing a more efficient route for producing compounds of industrial and pharmaceutical interest. However, a better understanding of the binding and dynamic properties of acyl carrier proteins is required to realize this potential.
In these systems, ACP acts as an anchor that immobilizes small carboxylate-containing molecules for entrance into enzyme active sites. These molecules are held in place by a covalent thioester linkage to a phosphopantetheine moiety, which, in turn, is linked covalently to the protein at a conserved serine residue (S38 in spinach ACP). In some cases (type I ACPs), the ACP moiety is a domain of a large protein, and in other cases (type II ACPs), ACP is a discrete protein. In plants and bacteria, type II soluble ACP molecules participate in a variety of enzymatic steps, including the synthesis of saturated fatty acids, the desaturation of fatty acids, and the donation of fatty acids from ACP to fatty acid-containing molecules, such as phospholipids.
In both type I and type II systems, the phosphopantetheine moiety of the ACP contains a thiol group that can form a thioester linkage with a variety of carboxyl-containing compounds. This study focuses on the acyl carrier protein from spinach attached to fatty acids. A standard fatty acid naming convention is X:Y, where X is the number of carbons in the fatty acyl chain and Y is the number of desaturated carbon bonds. We confine our discussion to saturated fatty acids, for example, 14:0, which refers to the saturated 14-carbon myristic acid. Thus, 14:0-ACP here refers to spinach ACP with myristic acid covalently attached through the phosphopantetheine thiol group.
Motions involving ACP are essential to catalysis in both type I and type II systems. The type I ACP domain was not resolved in initial crystal structures of fatty acid synthase (FAS) (9). The authors proposed that the ACP domain was a swinging arm that shuttles the growing acyl chain among reaction centers. The ACP moiety has been resolved in subsequent X-ray crystallographic studies, but the conclusion has been that this structure represents a single conformation adopted by this multifunctional enzyme (10). Similarly, in type II (dissociated) fatty acid synthases and in many other enzymes that react with ACP-bound groups, the ACP must be able to associate and dissociate with a variety of protein partners and must be capable of exposing the fatty acyl chain to thioesterases and other enzymes.
Recent structural investigations of ACP proteins with covalently attached fatty acids revealed that fatty acids bind down the center of the four-helix bundle (11−14). Furthermore, this binding pocket can be remodeled to accommodate a variety of different fatty acid cargoes. NMR solution studies of spinach acyl ACP conducted in our laboratory revealed structural differences dependent on the length of the fatty acyl chain. Although ACP with an attached decanoate (10:0) was well-structured, ACP with an attached stearate (18:0) was partially unstructured with dynamic disorder affecting residues between F31 and D40. At the time, it was hypothesized that the large 18-carbon fatty acid could not be completely accommodated by the hydrophobic core of ACP, resulting in structural instability.
The study reported here is aimed at improving our understanding of the relationship between the fatty acid length and conformational stability of spinach acyl carrier protein. We have determined that slow time scale motions on the micro- and millisecond time scales are significantly affected by the length of the attached fatty acid, but faster time scale motions do not show a significant dependence on the length of the fatty acid. We determined the rates of hydrolysis of 12:0, 14:0, 15:0, 16:0, 17:0, and 18:0 fatty acids from acyl-ACPs. In addition, we examined the conformational dynamics of the protein backbone using 15N relaxation. We observed that longer fatty acids progressively destabilize the structure of ACP and increase micro- and millisecond motions in residues surrounding the fatty acid binding cavity.
Methods
Protein Production
Spinach acyl carrier protein used for NMR studies was produced from Escherichia coli grown in M9 medium containing [15N]ammonium chloride (Cambridge Isotope Laboratories, Andover, MA) as its sole nitrogen source. The 15N-labeled protein was isolated, purified, and acylated with fatty acids as previously described (15−17). Purified fatty acids were purchased from Sigma-Aldrich Corp. (St. Louis, MO).
NMR Spectroscopy
Samples of 15N-labeled 10:0-, 14:0-, 15:0-, 16:0-, and 18:0-ACP at a concentration of 1 mM were prepared as described previously (14). 15N T1, 15N T2, and 15N−1H heteronuclear NOE experiments (18) were performed on a Bruker (Billerica, MA) DMX 500 MHz NMR spectrometer with a room-temperature TXI probe and triple-axis gradients. The 15N T1 data were collected with relaxation delays of 10, 110, 190, 320, 400, 520, 630, and 750 ms. The 15N T2 data were collected with relaxation delays of 8, 32, 64, 96, 128, 192, and 248 ms. The 15N heteronuclear NOE data sets were acquired with the unenhanced and enhanced spectra interleaved and a 3 s proton excitation period for the NOE-enhanced spectrum. 15N relaxation dispersion experiments (19,20) were performed on a Bruker DMX 500 MHz NMR spectrometer with a room-temperature TXI Probe and triple-axis gradients. Relaxation curves were recorded with spin lock field strengths corresponding to τCP values of 0.5, 0.65, 1, 1.3, 2, 4, 10, and 15 ms. Different sets of relaxation delays were used with different τCP values: τCP = 0.5 ms (12*, 44, 86, 172*, and 222 ms); τCP = 0.65 ms (13, 65*, 130*, 195, and 254.8 ms); τCP = 1 ms (8*, 36, 76, 148*, and 220 ms); τCP = 1.3 ms (10.4, 67.6*, 130*, 192.4, and 254.8 ms); τCP = 2 ms (32, 72, 104*, 136*, and 208 ms); τCP = 4 ms (32, 64, 96*, 128*, and 192 ms); τCP = 10 ms (40, 80*, 120, 160*, and 200 ms); τCP = 15 ms (60*, 120*, 180, 240, and 300 ms). Values marked with an asterisk indicate that duplicate experiments were conducted to enable error estimation of relaxation rates.
The intensities of individual peaks were extracted using SPARKY 3 (Goddard and Kneller) and were fitted with Curvefit (available from A. Palmer at http://www.cumc.columbia.edu/dept/gsas/biochem/labs/palmer/software.html). 15N T1, 15N T2, and heteronuclear NOE were fit according to Lipari−Szabo model free formalism using the software Modelfree (available from A. Palmer). The choice of particular models to fit individual data points was automated using FastModelfree (21), available from P. Loria (http://xbeams.chem.yale.edu/∼loria/software.php). Relaxation dispersion data were fitted with CPMGfit (also available from A. Palmer) using the fast limit CPMG function.
Measurement of Hydrolysis Rates
Rates of hydrolysis, catalyzed by hydroxylamine, were determined for the acyl chains of 12:0-, 14:0-, 15:0-, 16:0-, 17:0-, and 18:0-ACP (22). Samples of 1 mM acyl-ACP were mixed with 200 mM hydroxylamine (pH 7.0) to yield final concentrations of 0.1 mM protein and 90 mM hydroxylamine. The solution was mixed by pipetting. Aliquots were removed immediately and at 10 min intervals thereafter. Each aliquot was diluted 10:1 with 5 mM DTNB, and the color was allowed to develop for 1 min (23). The concentration of free sulfhydryls was measured via the absorbance at 420 nm. Assays were conducted in triplicate. Pseudo-first-order hydrolysis rates were calculated using SigmaPlot (Cranes Software International, Bangalore, India).
Results
Slow and Not Fast Time Scale Motions Are Dependent on the Length of the Fatty Acid
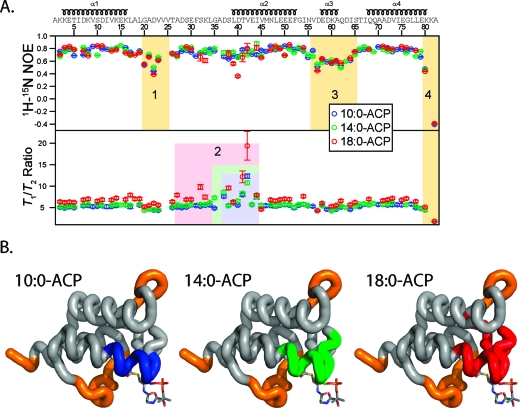
15N T1/T2 ratios and heteronuclear NOE enhancement ratios for 10:0-, 14:0-, and 18:0-ACP are shown in Figure 1. The heteronuclear NOE reveals fast time scale motions (a lower ratio indicates greater motion), and the T1/T2 ratio is responsive to both fast motions (indicated by a small ratio) and slow time scale motions. In contrast, stable regions of the protein exhibited large heteronuclear NOE ratios and median T1/T2 ratios. These included helices 1 (K3−K16) and 4 (Q68−E79), half of helix 2 (M46−F52), and a loop region between helices 1 and 2 (T26−K33 in 10:0- and 14:0-ACP). There are three segments of the protein that exhibit significant fast motions (as judged by heteronuclear NOE ratios of <0.6), and they are colored beige in Figure 1. These include a loop region between helices 1 and 2 (G20−V23), helix 3 and surrounding residues (V56−S65), and the C-terminus of the protein (K80−A82). Of particular interest is region 3, which contains helix 3. Although secondary structures are generally more stable than loop regions, this region is a particular exception. However, it is notable that heteronuclear NOE ratios are not generally dependent upon the length of the fatty acid.
Figure 1.
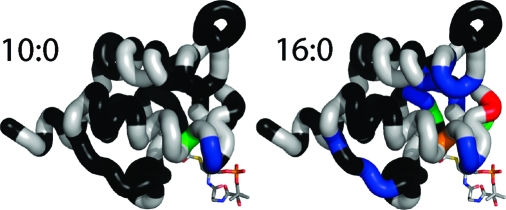
(A) 15N T1/T2 ratios and heteronuclear NOE enhancements plotted for all peptide backbone amide residues. These represent minimally processed relaxation data that provide insight into the motions of the peptide backbone. The heteronuclear NOE is sensitive to fast time scale motions of the peptide backbone; lower values indicate increased flexibility. The T1/T2 ratio is sensitive to both fast (pico- and nanosecond time scale motions) and slow (micro- and millisecond) time scale motions of the backbone. (B) Regions of motion shown in panel A plotted on the structure of 10:0-ACP. Residues exhibiting fast motions are colored orange. Regions exhibiting changes in T1/T2 ratio indicative of slow time scale motions are colored blue (10:0-ACP), green (14:0-ACP), and red (18:0-ACP).
In contrast, T1/T2 ratios reveal motions between helices 1 and 2 as well as along the first half of helix 2. Notably, the number of residues exhibiting elevated T1/T2 ratios increased with the length of the fatty acid. In 10:0-ACP, residues between D37 and I44 exhibited elevated T1/T2 ratios (blue box). For 14:0- and 18:0-ACP, the regions consisted of residues G35−I44 and A27−I44, respectively. These results suggest that more residues exhibit micro- and millisecond time scale motions in ACP as the length of the fatty acid increases from 10:0 to 14:0 to 18:0.
Figure 1 of the Supporting Information shows the results of Modelfree (24,25) analysis of the T1, T2, and heteronuclear NOE data. In accordance with the T1/T2 and heteronuclear NOE data, low generalized order parameters were observed between helices 1 and 2, at the C-terminus of the protein, and along helix 3, but there was no clear dependence of the generalized order parameter on the length of the fatty acid. Likewise, the Modelfree analysis reveals the presence of micro- and millisecond time scale motions along the first half of helix 2. However, the dependence of these motions upon the length of the fatty acid was not clear from the results of the Modelfree analysis. Notably, these residues are significantly broadened in 18:0-ACP (14). We chose to perform relaxation dispersion experiments to improve our understanding of the relationship between fatty acid length and these micro- and millisecond motions. To guide the choice of acyl-ACP variants used in these experiments, we used a chemical assay to reveal the flexibility of the fatty acid binding pocket.
Effect of Fatty Acid Length on Hydrolysis
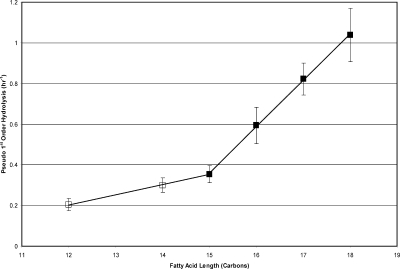
Figure 2 shows the pseudo-first-order rates for hydrolysis of the acyl chain from acyl-ACP for 12:0−18:0. In general, longer attached fatty acids resulted in faster hydrolysis. The differences in hydrolysis rate for the smaller fatty acids were generally small, and the differences in hydrolysis rates for 14:0- and 15:0-ACP were not statistically significant. However, the hydrolysis rate dramatically increased for fatty acids longer than 15:0. The curves of hydrolysis rate versus acyl chain length could be approximated by two separate linear relationships: the first for fatty acids 12:0−15:0 and the second for fatty acids 15:0−18.0. The inflection at 15:0 suggests that this is the largest fatty acid that can be easily accommodated within the spinach ACP binding pocket and that larger fatty acids lead to a larger exposure of the thioester bond and destabilization of the protein fold.
Figure 2.
Rates of hydrolysis of the thioester bond plotted as a function of fatty acyl chain length. The data were fitted to two straight lines [12:0−15:0 (◻) and 15:0−18:0 (◼)] to show the biphasic response.
Differential 15N Rates of Relaxation Dispersion
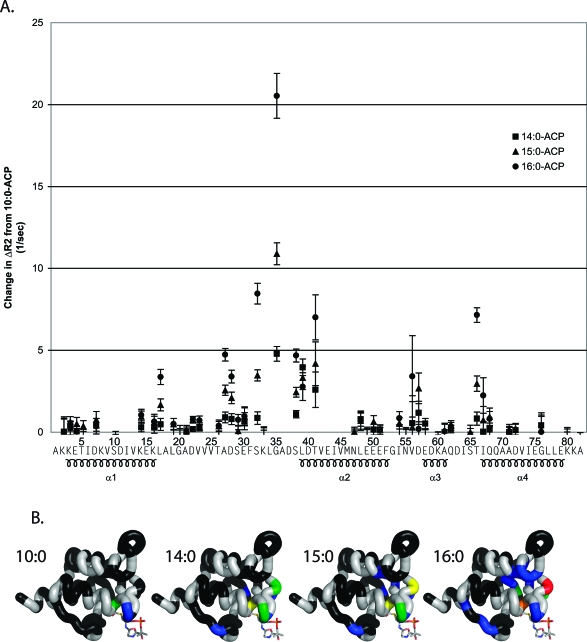
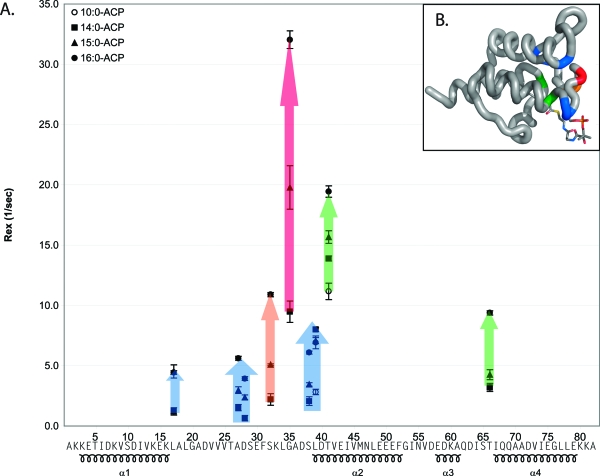
To improve our understanding of this destabilization, we used 15N relaxation dispersion to further examine the dynamics of 10:0-, 14:0-, 15:0-, and 16:0-ACP (Figure 3). The readout for 15N relaxation dispersion curves is the 15N HSQC spectrum. As a result, only residues corresponding to well-separated NMR peaks provide accurate relaxation curves. Residues 6, 9, 11, 12, 15, 18, 24, 25, 31, 34, 36, 37, 40, 44, 45, 52, 53, 59, 69, 70, 74, 77, 79, and 81 were excluded from the analysis on the basis of this criterion. The change in relaxation rate (ΔR2) was calculated as the difference between R2(τCP = 10) and R2(τCP = 0.5). ΔΔR2 was calculated as the difference in ΔR2 for either 14:0-, 15:0-, or 16:0-ACP and 10:0-ACP. Residues that experienced the largest changes in relaxation behavior surround the phosphopantetheine attachment site and form the entrance to the fatty acid binding pocket, including S32, G35, S38, L39, and T41. In addition, ΔR2 increased as a function of fatty acid length for all of these residues except for L39. Considering that these residues are significantly broadened in 18:0-ACP (14), it is likely that the relaxation dispersion is further increased for longer fatty acids in this region. Other regions exhibited small dispersions. In particular, V13 exhibits a small dispersion in 15:0-ACP, and residues V13−L17, which pack in the proximity of G35, exhibit dispersion in 16:0-ACP (Figure 3B). In addition, dispersions are observed for residues V56 and D57, but they are associated with large margins of error and do not progressively increase with the length of the fatty acid. Finally, residues T66 and I67 also form part of the entrance of the fatty acid binding cavity and exhibit dispersions that depend on the length of the fatty acid.
Figure 3.
(A) Change in relaxation dispersion plotted as a function of residue number. The change in relaxation dispersion was calculated by comparing R2(τCP = 10 ms) and R2(τCP = 0.5 ms). The change in dispersion was then subtracted from the dispersion for the corresponding residue in 10:0-ACP: 14:0-ACP (◼), 15:0-ACP (▲), and 16:0-ACP (●). (B) Relaxation dispersion shown on the structure of 10:0-ACP. Sections of backbone are colored according to the relaxation dispersion. Black indicates no significant dispersion, blue dispersion between 1 and 5 Hz, green dispersion between 5 and 10 Hz, yellow dispersion between 10 and 15 Hz, orange dispersion between 15 and 20 Hz, and red dispersion over 20 Hz.
Two-State Model Fitting of the Relaxation Data
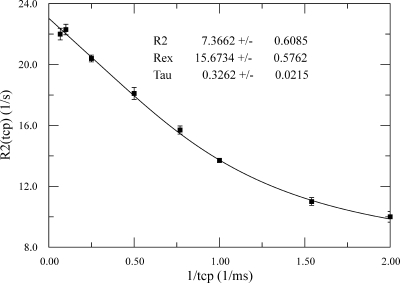
To improve our understanding of the dynamic motions of the peptide backbone, we analyzed the relaxation dispersion data using CPMGfit. Residues L17, A27, D28, S32, G35, S38, L39, T41, and T66 generated satisfactory relaxation dispersion curve fits. Although residue V56 exhibited relaxation dispersion, measurement errors prevented fitting of the relaxation dispersion curve. Figure 4 contains a representative relaxation curve fit for residue T41. Notably, the corrected R2 was 7.3 ± 0.6 s−1. This closely matches the R2 values observed for most residues that did not exhibit relaxation dispersion [between 7 and 8 s−1 (data not shown)]. This suggests that the motions in these residues are predominantly on the slower micro- to millisecond time scale rather than a faster nano- to picosecond time scale. The main exception to this is residue A82; it is typical for residues at the C-terminus of a protein to exhibit a small R2, indicating motions on a fast time scale.
Figure 4.
Relaxation data for residue T41 of 15:0-ACP analyzed by a two-site motion model using CPMGfit to fit the equation R2(τCP) = R2° + Rex{1 − (2 × τ/τCP) × tanh[1/(2 × τ/τCP)]}.
The exchange rates (Rex) obtained from the relaxation dispersion curve fits are shown in Figure 5. In general, the exchange rates (Rex) increased with the length of the acyl chain except for residue L39. Residue L17 exhibited an increase in Rex when the length of acyl chain increased from 14:0 to 15:0, but not from 15:0 to 16:0. The exchange rates for residues A27 and D28 increased approximately linearly with the length of the acyl chain. However, residues G35, S38, T41, and T66 exhibited a smaller change in Rex from 14:0 to 15:0 than from 15:0 to 16:0. For these residues, it appears that the Rex was approximately correlated with the fatty acid hydrolysis rate described above. Since there was a lack of correspondence between the Rex values of the different residues, the exchange rate analysis suggests that motions of 14:0-, 15:0-, and 16:0-ACP cannot be modeled as a simple conversion between two discrete structures.
Figure 5.
(A) Exchange rates (Rex) determined by CPMGfit plotted as a function of amino acid sequence: 10:0-ACP (○), 14:0-ACP (◼), 15:0-ACP (▲), and 16:0-ACP (●). Colored arrows indicate trends in the relaxation dispersion data. (B) Rex data plotted on the structure of 10:0-ACP, with the backbone colored according to the change in Rex from 10:0-ACP to 18:0-ACP shown in panel A.
Discussion
Dynamics of ACP
The dynamic nature of acyl carrier proteins was first recognized in NMR studies of E. coli ACP by the Prestegard group. Initial analysis revealed the presence of a select highly mobile residue based on COSY data (26), and initial NMR-derived structures of E. coli ACP were improved using refinement against two structures (27). Further studies revealed that spinach holo-ACP undergoes slow two-site exchange (28,29), and deuterium exchange of the amide backbone was used to estimate protein dynamics in E. coli ACP (30). These and other studies (11,31,32) established ACP as a model system for NMR investigations of structure and dynamics. One shortcoming of the early dynamics studies is that they utilized ACP in a poorly defined acylation state. In contrast, this work examines the effect of specifically designated long chain fatty acids on the structure of ACP. The results presented here reveal that ACP is a particularly dynamic molecule and that its motions are dependent on its interactions with the bound fatty acid. This provides new insight into the role of ACP as a carrier of growing fatty acyl chains in type I and II fatty acid synthases.
Effect of Bound Fatty Acids on ACP Dynamics
We have explored in detail the dynamic motions exhibited by spinach acyl carrier protein. Our initial experiments examined the backbone dynamics of 10:0-, 14:0-, and 18:0-ACP. As expected, residues forming helices 1, 2, and 4 are generally well-ordered, with few motions. However, helix 3 exhibited less stability. We previously observed that helix 3 reoriented to expand the fatty acid binding pocket of ACP, and this reorientation was accompanied by changes in the chemical shifts of the residues in this region (14). The results presented here provide additional insight into the motions that enable this behavior. Fast motions in this region do not significantly change as a consequence of a change in the length of the attached fatty acid, and we therefore propose that these motions are responsible for the flexibility of this helix and the ability of ACP to accommodate a variety of fatty acids in its hydrophobic core. Although Modelfree analysis did not reveal micro- and millisecond time scale motions in this region, analysis of the relaxation dispersion data revealed evidence of slower, acyl chain-dependent, motions.
The T1/T2 data and the Modelfree analysis suggested that residues between helices 1 and 2 as well as residues comprising the first half of helix 2 exhibited slow time scale motions. We have previously shown (14) that 15N HSQC peaks associated with these residues are significantly broadened in 18:0-ACP but not 10:0-ACP. To better assess these motions, we further analyzed these slow time scale motions using 15N relaxation dispersion as discussed below.
We also examined the stability of the thioester bond. The analysis of the dependence of the rate of hydrolysis of the phosphopantetheine−fatty acid thioester bond on the length of the fatty acyl chain was suggested by our previous observation that samples of 18:0-ACP degraded noticeably during the course of NMR experiments over the course of a week. In contrast, 10:0-ACP did not exhibit changes over a 2 month period (14). Because the hydrolysis releases a phosphopantetheine thiol, the Ellman assay was used as the detection method. However, the long time scale for thioester hydrolysis under standard solution conditions might lead to oxidation of the free thiol, which would complicate detection. Consequently, we used 90 mM hydroxylamine, a relatively strong nucleophile at pH 7 (33), to accelerate the hydrolysis of acyl-ACP to thiol-containing holo-ACP and produce a more conveniently measured rate (data not shown). At this concentration, hydroxylamine was in significant excess of ACP (1 mM) and the reaction could be successfully modeled with a pseudo-first-order rate constant.
At the inception of this study, we knew that 10:0-ACP had a relatively stable structure and a slow rate of hydrolysis (14). In contrast, 18:0-ACP was known to have a disrupted structure and a high rate of hydrolysis. In our study of the structure of acyl-ACPs, we postulated that the structure of ACP was flexible and capable of expanding to accommodate longer fatty acids but expected that there would be a limit to this flexibility. We hypothesized that fatty acids that exceed this limit would significantly disrupt the structure of ACP, because of evidence of the presence of an expanding pocket in the center of the ACP protein structure (14). In 10:0-ACP, this pocket was sufficient to fully sequester the fatty acid. However, the pocket was not sufficient to accommodate 18:0, corresponding to an increased hydrolysis rate. These results support this hypothesis. In particular, we determined that 15:0 represents the limit for complete chain accommodation in spinach ACP. Fatty acids larger than this lead to greater exposure of the labile thioester bond outside of the protection of the pocket.
Our choice of fatty acids to be used for NMR investigations of protein dynamics was based on the transition point identified from the hydrolysis experiments. We produced 15N-labeled 14:0-, 15:0-, and 16:0-ACPs for 15N NMR relaxation dispersion measurements. These experiments revealed motions in acyl-ACP on the micro- to millisecond time scale. Notably, the relaxation dispersion experiments revealed slow time scale residue motions that were not identified in the Modelfree analysis. Figure 3B depicts these regions in the context of the solution structure of 10:0-ACP (14). Residues that exhibit the greatest relaxation dispersion (S32, G35, S38, L39, T41, and T66) line the fatty acid binding cavity. These residues were previously predicted to undergo exchange broadening in 18:0-ACP (17), and these results confirm that prediction. Significantly, the dispersion for each of these residues increases with fatty acid length. This suggests that longer fatty acids progressively destabilize the protein structure surrounding the fatty acid binding pocket. Residues that form a second shell surrounding the fatty acid binding pocket exhibit smaller dispersions, including residues V13−L17, A27, D28, and I67. Also of interest is residue V56, which forms the closed end of the fatty acid binding pocket. Although V56 exhibits no dispersion in 10:0-ACP, it exhibits strong dispersion in 14:0-, 15:0-, and 16:0-ACP. We previously hypothesized that V56 and helix 3 serve as a hinge, allowing the expansion of the fatty acid binding pocket, and these results support the hypothesis.
Many of the residues exhibiting slow time scale motions in spinach ACP are conserved across plant and bacterial ACP sequences (Figure 2 of the Supporting Information). In particular, residues surrounding the phosphopantetheine attachment site, G35−T41, are highly conserved and exhibit the largest dispersions in 16:0-ACP. These residues also exhibit large backbone B factors in crystal structures of E. coli acyl-ACP (12). Other residues exhibiting dispersion lie in the proximity of the phosphopantetheine binding site. These include S32, while neighboring residue F31 is highly conserved but could not be analyzed due to overlapping peaks in the 15N HSQC spectrum. Motions in S32 may suggest destabilization of F31 and the hydrophobic core of ACP. Sequences surrounding unstable helix 3 are conserved. Furthermore, hydrophobic residues are found in the primary sequence positions corresponding to V56 and I67, and T66 is absolutely conserved. These results support the hypothesis that these residues form a conserved dynamic hinge, enabling modulation of the size of the fatty acid binding pocket. In agreement with our backbone dynamics data for helix 3, this region also exhibits elevated backbone B factors in the crystal structure of E. coli acyl-ACP (12). In 16:0-ACP, the hydrophobic core of ACP is partially destabilized, leading to motions in the conserved residue L17.
Applicability of a Two-State Exchange Model
In almost all cases, the relaxation dispersion profiles exhibited increased dispersion for longer acyl chains. However, fitting this motion to a two-state system proved to be unsatisfactory. Systems that follow a two-state exchange model, for example, DHFR and SH3 (34,35), were found to yield similar exchange rates for most residues analyzed. In contrast, our analysis led to a diversity of exchange rates. Additional 15N relaxation dispersion data collected at a second field strength (900 MHz) also did not fit a simple two-state model (data not shown). These results suggest that ACP undergoes more complex motions that are not properly modeled by two-site exchange. Such multiple conformational transitions may be required for ACP to accommodate fatty acids of various lengths (12,14).
Impact for Type II Fatty Acid Synthases
Differential stabilization of the fatty acid binding pocket may lead to different kinetics of enzymes involved in fatty acid metabolism and has the potential to change the fatty acid distribution within an organism. One potential example of this is the ACP-dependent enzyme Δ-9-desaturase (Δ9D), an enzyme that introduces a cis 9−10 double bond into 18:0-ACP. Analysis of the substrate specificity of Δ9D revealed a progressive decrease in the catalytic efficiency when using smaller substrates, including 16:0, or 14:0 (36). This was previously described in terms of partitioning of the fatty acid between an aqueous environment and a hydrophobic cleft in Δ9D. However, on the basis of the structure of ACP and the dynamics results presented here, the actual binding thermodynamics must be more complex as interactions of the fatty acyl chain with ACP must also be included. These results suggest that a mutated ACP containing a destabilized fatty acid binding pocket might increase the availability of shorter fatty acids, including 14:0 and 16:0. This may facilitate desaturation of shorter acyl chains bound to the mutated ACP if other aspects of the desaturation reaction do not depend on acyl chain length. The accessibility of acyl chains from ACP may similarly influence other enzymes whose function involves a transfer of the fatty acid from the ACP hydrophobic core to another active site cavity. For example, ACP variants with more stable fatty acid binding pockets may result in the production of longer fatty acids. The specialized AcpM from Mycobacterium tuberculosis involved in the synthesis of mycolic acid may be one example (37). The accessibility of ACP-bound fatty acids would likely also be affected by protein−protein interactions between enzymes and ACP. Thus, the effect of mutations of ACP on interactions with individual enzymes cannot be directly predicted. A fuller understanding of these interactions requires study of the effect of different ACPs as well as different fatty acids on ACP-dependent catalysis.
Impact for Type I ACPs
It is clear that conformational dynamics play an important role in the macromolecular machine FAS. This is underscored by the absence of electron density for ACP in the initial crystal structures of type I FAS (9,38). Further work on the yeast FAS revealed electron density for the ACP domain, although the authors noted that this region was less well-ordered than other parts of the structure (39). However, the ACP domain is generally similar in structure to spinach and E. coli ACP, and the authors identified surrounding disordered sequences that enable ACP to access multiple active sites within the complex.
The NMR experiments presented here cannot be applied directly to the entire type I fatty acid synthase. Instead, studies on the isolated ACP domain from FAS have shown it to fold into a helical bundle structure loosely resembling other ACPs (40). However, further studies of this isolated ACP have suggested that it does not bind the acyl chain within its hydrophobic core (41). These results may reflect the fact that the ACP has been removed from its traditional flanking sequences within the fatty acid synthase, which may be responsible for guiding the placement of the fatty acid. Nonetheless, it is also possible that ACP is utilized differently in type I and type II fatty acid synthases.
Acknowledgments
We gratefully acknowledge Marco Tonelli and Claudia Cornilescu for their assistance with the acquisition and interpretation of the relaxation dispersion data sets.
Supporting Information Available
Results of the Modelfree analysis and alignments of ACP sequences. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by National Institutes of Health (NIH) Grants R01 GM58667 (J.L.M.) and R01 GM50853 (B.G.F.). NMR data were collected at the National Magnetic Resonance Facility at Madison (NMRFAM) with support from NIH Grants P41 RR02301 (J.L.M.) and P41 GM66326 (J.L.M.).
Funding Statement
National Institutes of Health, United States
Footnotes
Abbreviations: ACP, acyl carrier protein; DTNB, dithionitrobenzene.
Supplementary Material
References
- Magnuson K.; Jackowski S.; Rock C. O.; Cronan J. E. (1993) Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57, 522–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therisod H.; Weissborn A.; Kennedy E. (1986) An essential function for acyl carrier protein in the biosynthesis of membrane-derived oligosaccharides of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 83, 7236–7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumley M.; Therisod H.; Weissborn A.; Kennedy E. (1992) Mechanisms of regulation of the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. J. Biol. Chem. 267, 11806–11810. [PubMed] [Google Scholar]
- Shen B.; Summers R. G.; Gramajo H.; Bibb M. J.; Hutchinson C. R. (1992) Purification and characterization of the acyl carrier protein of the Streptomyces glaucescens tetracenomycin C polyketide synthase. J. Bacteriol. 174, 3818–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers R. G.; Ali A.; Shen B.; Wessel W. A.; Hutchinson C. R. (1995) Malonyl-coenzyme A:acyl carrier protein acyltransferase of Streptomyces glaucescens: A possible link between fatty acid and polyketide biosynthesis. Biochemistry 34, 9389–9402. [DOI] [PubMed] [Google Scholar]
- Issartel J.; Koronakis V.; Hughes C. (1991) Activation of Escherichia coli prohaemolysin to the mature toxin by acyl carrier protein-dependent fatty acylation. Nature 351, 759–761. [DOI] [PubMed] [Google Scholar]
- Rusnak F.; Sakaitani M.; Drueckhammer D.; Reichert J.; Walsh C. T. (1991) Biosynthesis of the Escherichia coli siderophore enterobactin: Sequence of the entF gene, expression and purification of EntF, and analysis of covalent phosphopantetheine. Biochemistry 30, 2916–2927. [DOI] [PubMed] [Google Scholar]
- Scarth R.; Tang J. (2006) Modification of Brassica oil using conventional and transgenic approaches. Crop Sci. 46, 1225–1236. [Google Scholar]
- Jenni S.; Leibundgut M.; Maier T.; Ban N. (2006) Architecture of a fungal fatty acid synthase at 5 Å resolution. Science 311, 1263–1267. [DOI] [PubMed] [Google Scholar]
- Leibundgut M.; Jenni S.; Frick C.; Ban N. (2007) Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science 316, 288–290. [DOI] [PubMed] [Google Scholar]
- Jones P. J.; Cioffi E. A.; Prestegard J. H. (1987) [19F]-1H heteronuclear nuclear Overhauser effect studies of the acyl chain-binding site of acyl carrier protein. J. Biol. Chem. 262, 8963–8965. [PubMed] [Google Scholar]
- Roujeinikova A.; Simon W. J.; Gilroy J.; Rice D. W.; Rafferty J. B.; Slabas A. R. (2007) Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. J. Mol. Biol. 365, 135–145. [DOI] [PubMed] [Google Scholar]
- Roujeinikova A.; Baldock C.; Simon W. J.; Gilroy J.; Baker P. J.; Stuitje A. R.; Rice D. W.; Slabas A. R.; Rafferty J. B. (2002) X-ray crystallographic studies on butyryl-acp reveal flexibility of the structure around a putative acyl chain binding site. Structure 10, 825–835. [DOI] [PubMed] [Google Scholar]
- Zornetzer G. A.; Fox B. G.; Markley J. L. (2006) Solution structures of spinach acyl carrier protein with decanoate and stearate. Biochemistry 45, 5217–5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambalot R. H.; Walsh C. T. (1995) Cloning, overproduction, and characterization of the Escherichia coli holo-acyl carrier protein synthase. J. Biol. Chem. 270, 24658–24661. [DOI] [PubMed] [Google Scholar]
- Rock C. O.; Cronan J. E. J. (1979) Solubilization, purification, and salt activation of acyl-acyl carrier protein synthetase from Escherichia coli. J. Biol. Chem. 254, 7116–7122. [PubMed] [Google Scholar]
- Zornetzer G. A.; White R. D.; Markley J. L.; Fox B. G. (2005) Preparation of isotopically labeled spinach acyl-acyl carrier protein for NMR structural studies. Protein Expression Purif. 46, 446–455. [DOI] [PubMed] [Google Scholar]
- Farrow N. A.; Muhandiram R.; Singer A. U.; Pascal S. M.; Kay C. M.; Gish G.; Shoelson S. E.; Pawson T.; Forman-Kay J. D.; Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003. [DOI] [PubMed] [Google Scholar]
- Loria J. P.; Rance M.; Palmer A. G. III (1999) A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 121, 2331–2332. [Google Scholar]
- Loria J. P.; Rance M.; Palmer A. G. III (1999) A TROSY CPMG sequence for characterizing chemical exchange in large proteins. J. Biomol. NMR 15, 151–155. [DOI] [PubMed] [Google Scholar]
- Cole R.; Loria J. P. (2003) FAST-ModelFree: A program for rapid automated analysis of solution NMR spin-relaxation data. J. Biomol. NMR 26, 203–213. [DOI] [PubMed] [Google Scholar]
- Fenton S. S.; Fahey R. C. (1986) Analysis of biological thiols: Determination of thiol components of disulfides and thioesters. Anal. Biochem. 154, 34–42. [DOI] [PubMed] [Google Scholar]
- Ellman G. L. (1959) Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77. [DOI] [PubMed] [Google Scholar]
- Palmer A. G. III; Rance M.; Wright P. E. (1991) Intramolecular motions of a zinc finger DNA-binding domain from Xfin characterized by proton-detected natural abundance carbon-13 heteronuclear NMR spectroscopy. J. Am. Chem. Soc. 113, 4371–4380. [Google Scholar]
- Mandel A. M.; Akke M.; Palmer A. G. III (1995) Backbone dynamics of Escherichia coli ribonuclease HI correlations with structure and function in an active enzyme. J. Mol. Biol. 246, 144–163. [DOI] [PubMed] [Google Scholar]
- Kay L. E.; Holak T. A.; Prestegard J. H. (1988) AX3 spin system dynamics from forbidden cross peaks in double-quantum two-dimensional NMR experiments writh application to acyl carrier protein. J. Magn. Reson. 76, 30–40. [Google Scholar]
- Kim Y.; Prestegard J. H. (1989) A dynamic model for the structure of acyl carrier protein in solution. Biochemistry 28, 8792–8797. [DOI] [PubMed] [Google Scholar]
- Kim Y.; Ohlrogge J. B.; Prestegard J. H. (1990) Motional effects on NMR structural data. comparison of spinach and Escherichia coli acyl carrier proteins. Biochem. Pharmacol. 40, 7–13. [DOI] [PubMed] [Google Scholar]
- Kim Y.; Prestegard J. H. (1990) Demonstration of a conformational equilibrium in acyl carrier protein from spinach using rotating frame nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc. 112, 3707–3709. [Google Scholar]
- Andrec M.; Hill R. B.; Prestegard J. H. (1995) Amide exchange rates in Escherichia coli acyl carrier protein: Correlation with protein structure and dynamics. Protein Sci. 4, 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally H. U.; Spencer A. K.; Armitage I. M.; Prestegard J. H.; Cronan J. E. J. (1978) Acyl carrier protein from Escherichia coli: Characterization by proton and fluorine-19 nuclear magnetic resonance and evidence for restricted mobility of the fatty acid chain in tetradecanoyl-acyl-carrier protein. Biochemistry 17, 5377–5382. [DOI] [PubMed] [Google Scholar]
- Holak T. A.; Prestegard J. H.; Forman J. D. (1987) NMR-pseudoenergy approach to the solution structure of acyl carrier protein. Biochemistry 26, 4652–4660. [DOI] [PubMed] [Google Scholar]
- Simanenko Y.; Popov A.; Prokop'eva T.; Karpichev E.; Savelova V.; Suprun I.; Bunton C. (2002) Inorganic anionic oxygen-containing α-nucleophiles: Effective acyl group acceptors: Hydroxylamine ranks first among the α-nucleophile series. Russ. J. Org. Chem. 38, 1286–1298. [Google Scholar]
- McElheny D.; Schnell J. R.; Lansing J. C.; Dyson H. J.; Wright P. E. (2005) Defining the role of active-site loop fluctuations in dihydrofolate reductase catalysis. Proc. Natl. Acad. Sci. U.S.A. 102, 5032–5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzhnev D. M.; Salvatella X.; Vendruscolo M.; Di Nardo A. A.; Davidson A. R.; Dobson C. M.; Kay L. E. (2004) Low-populated folding intermediates of Fyn Sh3 characterized by relaxation dispersion NMR. Nature 430, 586–590. [DOI] [PubMed] [Google Scholar]
- Haas J. A.; Fox B. G. (1999) Role of hydrophobic partitioning in substrate selectivity and turnover of the Ricinus communis stearoyl acyl carrier protein Δ9 desaturase. Biochemistry 38, 12833–12840. [DOI] [PubMed] [Google Scholar]
- Wong H. C.; Liu G.; Zhang Y.; Rock C. O.; Zheng J. (2002) The solution structure of acyl carrier protein from Mycobacterium tuberculosis. J. Biol. Chem. 277, 15874–15880. [DOI] [PubMed] [Google Scholar]
- Maier T.; Jenni S.; Ban N. (2006) Architecture of mammalian fatty acid synthase at 4.5 Å resolution. Science 311, 1258–1262. [DOI] [PubMed] [Google Scholar]
- Lomakin I. B.; Xiong Y.; Steitz T. A. (2007) The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell 129, 319–332. [DOI] [PubMed] [Google Scholar]
- Reed M. A.; Schweizer M.; Szafranska A. E.; Arthur C.; Nicholson T. P.; Cox R. J.; Crosby J.; Crump M. P.; Simpson T. J. (2003) The type I rat fatty acid synthase ACP shows structural homology and analogous biochemical properties to type II ACPs. Org. Biomol. Chem. 1, 463–471. [DOI] [PubMed] [Google Scholar]
- Płoskoń E.; Arthur C. J.; Evans S. E.; Williams C.; Crosby J.; Simpson T. J.; Crump M. P. (2008) A mammalian type I fatty acid synthase acyl carrier protein domain does not sequester acyl chains. J. Biol. Chem. 283, 518–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.