Abstract
The divergence accumulated during the evolution of protein families translates into their internal organization as subfamilies, and it is directly reflected in the characteristic patterns of differentially conserved residues. These specifically conserved positions in protein subfamilies are known as “specificity determining positions” (SDPs). Previous studies have limited their analysis to the study of the relationship between these positions and ligand-binding specificity, demonstrating significant yet limited predictive capacity. We have systematically extended this observation to include the role of differential protein interactions in the segregation of protein subfamilies and explored in detail the structural distribution of SDPs at protein interfaces. Our results show the extensive influence of protein interactions in the evolution of protein families and the widespread association of SDPs with protein interfaces. The combined analysis of SDPs in interfaces and ligand-binding sites provides a more complete picture of the organization of protein families, constituting the necessary framework for a large scale analysis of the evolution of protein function.
Keywords: functional residues, protein family evolution, protein function, protein–protein interfaces, specificity determining positions
The structure of protein families is shaped by the sequence divergence accumulated as a consequence of speciation, gene duplication, and deletion events, as well as by the evolutionary selective pressure exerted on each protein in accordance with the corresponding 3D structure and the specific function performed (1, 2). The balance between genomic rearrangements and selective pressure to increase the functional repertoire available to organisms leads to the appearance of new subfamilies in evolutionary time (3).
There are many aspects of protein function that contribute to the evolution of the family organization. These may include the global conservation of catalytic mechanisms (in the case of enzymes), specific binding to substrates and cofactors, as well as the interaction with other proteins in processes such as cell signaling, the regulation of reactions and the formation of macromolecular complexes. Interestingly, even though specific protein interactions certainly are an important part of protein function, the organization of protein families in relation to the specific interactions of different subfamilies remains a poorly studied aspect of functional specificity.
Multiple sequences alignments (MSAs) provide essential information on the evolution of protein families. The positions in MSAs can be interpreted in terms of the amino acid changes allowed or disallowed during evolution, and therefore useful information at the residue level can be inferred from them (4). The most obvious example is the study of fully conserved positions that pinpoint important residues for the structure and function of the family members (5).
A subtler pattern of conservation is represented by the positions differentially conserved within subfamilies. A commonly accepted working hypothesis is that whereas fully conserved positions are related to functional features common to all the members of the family, these other residues are related to functional specificity (e.g., binding of different cofactors). For this reason, they have been termed “specificity determining positions” (SDPs). A variety of computational methods have been used to detect conserved positions and SDPs in MSAs (6–12); for a review see ref. 13. Moreover, the implication of SDPs in determining the differential binding to substrates and interaction partners has been experimentally followed up in a number of cases (14–16).
Despite these efforts, fundamental questions regarding the association between subfamilies, SDPs, and function remain largely unexplored at the systematic level. Notwithstanding, the information currently available on protein sequences, structures, functions, and interactions opens the door to performing more comprehensive studies of the relationships between family organization and functional divergence (17). Indeed, such studies can involve biochemical function and protein interaction specificity. Similarly, they can take into account the associated conservation at the molecular signatures level (SDPs) in fundamental regions corresponding to ligand-binding sites and protein interaction sites.
To carry out a unified analysis of subfamilies and associated SDPs, we have developed a protocol based on multiple correspondence analysis (MCA) (18) that can detect both entities simultaneously. Here we apply this methodology to the largest possible dataset of eukaryotic protein families for which it was possible to compile reliable information on catalytic activity, ligand binding, and protein interactions. The results are interpreted in terms of the relationship between the internal structure of protein families, their functional properties, and specific molecular signatures, with particular attention to the analysis of protein interaction sites.
Results
Functions in Protein Families: Biochemical and Protein Interaction Specificity.
This work evaluates the influence of functional constraints on protein family evolution by studying the functional features associated with their subfamily organization and their corresponding SDPs. For this purpose, we developed a multivariate-based protocol capable of detecting protein subfamilies and SDPs in a concomitant way and applied it to a collection of eukaryotic Pfam families (see Methods). In this section, the internal organization of protein families in subfamilies was analyzed on a collection of cases for which functional information is available regarding (i) their catalytic mechanism as defined in the Enzyme Commission (EC) classes, and (ii) the specificity of protein interactors for Saccharomyces cerevisiae and Homo sapiens, as inferred from “small scale” experiments, which provides a sound basis for the definition of interaction specificities within protein families (see Methods).
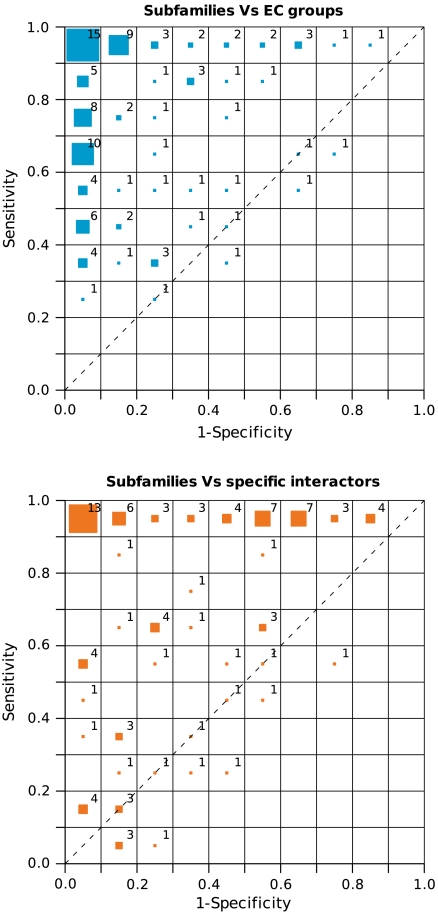
When 149 families with a representative number of EC labels and 72 families with a representative number of identified interactions were analyzed (Fig. 1 and Table S2), there was a general agreement between the subfamilies and the two functional labels considered: EC classes and specific interactors. Similar results were obtained for a larger set of families when compared to classes based on SwissProt IDs equivalences (Fig. S1). Indeed, this correspondence between functional classes and subfamilies can be observed in the receiver operating characteristic (ROC) space (Fig. 1; see Methods). In these plots a sensitivity of 1.0 implies that all the proteins with the same functional label belong to the same subfamily, and a specificity of 1.0 implies that all the proteins in a subfamily have the same label. Therefore, a perfect agreement would be represented in the Upper Left (0.0, 1.0) corner. Fig. 1 shows that most of the families displayed very good specificity and sensitivity, reflecting a good agreement between their organization and the functional labels. This agreement held true for the two datasets of EC and interaction labels, indicating that differential protein interaction patterns are integrated in a coherent manner within the subfamily composition, at a level similar to that of the better characterized biochemical functions represented by the EC classification. Similar results were obtained for other methodologies explicitly reporting subfamilies (see SI Text and Fig. S2).
Fig. 1.
Correspondence between the different subfamilies, and the EC groups (Top) and specific interactors (Bottom) for each protein family represented in the ROC space, where the distribution of the families is shown as a bidimensional histogram. The size of the colored boxes in each bin of the ROC space represents the percentage of protein families they contain, whereas the number shows the actual percentage. For the sake of simplicity, percentage values are rounded to the nearest integer (so that they may not add up to 100).
Examples of the Sequence Spaces Defined by a Protein Family and Their Relationship with Functional Specificities and the Associated Positions
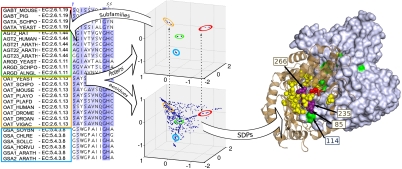
The procedure used to automatically define the sequence subfamilies (see Methods) is illustrated for the class III aminotransferases family (Pfam PF00202) in Fig. 2. The agreement between the EC numbers (beside the protein names) and the subfamilies defined as clusters in the sequence space can be observed.
Fig. 2.
Workflow implemented to simultaneously detect the protein subfamilies and those residues responsible for such segregation (SDPs). This process is depicted for the class III aminotransferase family (Pfam PF00202). The homodimeric structure of the human ornithine aminotransferase (PDB 1oat) bound to Pyridoxal-5’-phosphate (in red spheres) is represented. The two subunits of the complex are shown as brown cartoon representation and with a gray surface. SDPs are highlighted in a yellow/violet spacefill and with a green surface. The figure was generated with Pymol (pymol.sourceforge.com).
As an example of the relationship between the subfamilies and their interaction specificity, we report the results for the E2F/TDP family of transcription factors (Pfam PF02319), whose members show a number of different interaction specificities. The protocol we used clearly distinguished between the E2F and TDP types. The biological meaning of such a division can indeed be inferred from the full list of positive and negative interactions extracted for this family (see SI Text and Fig. S3). Thus, it was evident how these groupings reflected the different ability of E2F proteins to form homodimers or heterodimers interacting with TDPs, leading to different DNA binding properties (19).
The positions in the alignment responsible for the segregation are classified simultaneously with the detection of the subfamily composition. For instance, the “residue space” of the class III aminotransferase family above mentioned reflects the natural equivalence between the protein and the residue spaces (Fig. 2) with the SDPs corresponding to the various subfamilies at equivalent positions. In addition, most of the SDPs map to the interaction surface in the 3D structure of the homodimer, and are close to the ligand-binding site (Fig. 2 and Movie S1). The SDPs include 3 positions that have been experimentally mutated, demonstrating their implication in determining substrate specificity (20). Despite not directly contacting the substrate, one of these positions has been experimentally shown to be one of the main determinants of substrate specificity (corresponding to residue 85 in PDB 1oat). Interestingly, this position is in contact with another SDP (114 of the other chain) located at the homodimerization interface. These results point to the possibility that modifications to homodimerization could regulate the interaction with the ligand and hence, determine the substrate specificity.
Relationship Between SDPs and Functional Regions.
The relationship between SDPs and functional regions was investigated in terms of their structural proximity to (i) ligand-binding sites of small molecules and (ii) protein interaction sites. Ligand-binding sites are conceptually associated to biochemical functions, typically corresponding to the EC numbers analyzed in the previous section. Similarly, protein interaction sites are also related to the protein interaction specificity analyzed above. As explained in Methods, we gathered reliable structural information for 208 Pfam (21) protein domain families with a known ligand-binding site and for 276 families with detectable interaction regions defined from complexes of known structure (Table S2).
We analyzed the distribution of the Cβ-Cβ atom distances (Fig. S4) between SDPs, ligand-binding sites, and interaction surfaces, averaged per family and per structurally redundant group (see Methods). SDPs were significantly closer to the “functional regions” (median 9.4 ± 5.3 Å for sites and 7.6 ± 6.0 Å for interfaces) than the average of the positions (background, 11.8 ± 4.0 Å and 9.1 ± 4.8 Å, respectively). For comparison, the conserved positions (defined as > 90% identity) were also close to the functional regions (7.9 ± 4.4 Å and 7.2 ± 5.5 Å) and on average, even closer than the SDPs. These differences were associated with a p-value lower than 1e-13 for SDPs and < 1e-15 for the conserved positions when considering ligand binding, whereas the corresponding figures for the interaction surfaces were < 1e-9 for SDPs and < 1e-15 for the conserved positions (Wilcoxon test for paired data; see Methods).
The shift of SDPs and the conserved residues towards binding sites was significant, even taking into account that the current knowledge of the functional regions is not complete. That is, the sites structurally solved are a subset of all the biological catalytic/active sites, and the interfaces reported in the 3D structures of protein complexes represent a fraction of the actual interactions of a given protein. Additionally, positions close to a ligand-binding site but far from an interface would be considered as “noninterface residues” (Fig. S5A), and conversely for binding sites. This means that we are overestimating the number of “negatives” by excluding a number of actual binding sites.
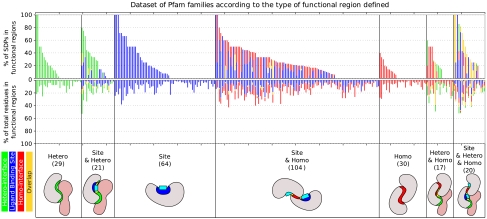
A more demanding experiment is to not just evaluate whether a given SDP is close to a functional region but whether it is part of the region itself (annotated as a site or being part of the interface, Fig. 3). For this purpose, we calculated whether a given set of positions (SDPs or conserved) is “enriched” at annotated sites or interface residues, and we applied a Wilcoxon rank sum test to the list of the enrichment values corresponding to the structurally nonredundant set of Pfam groups (see Methods). Both SDPs and conserved positions were clearly enriched in (i) annotated sites, (ii) interface residues, and (iii) in the combination of sites and interfaces as a whole (Table 1). It can also be observed that those enrichments were generally higher for binding sites than for protein interaction surfaces. These results were consistent when SDPs reported by different methodologies were considered (see SI Text and Table S3). All these tests were done assuming the existence of sequence information alone, because the SDPs and conserved positions were only extracted from the MSAs. When these tests were restricted to the positions in the surface of the proteins [similarly to (7)], the aforementioned enrichments of SDPs and conserved positions increased their significance (Table 1).
Fig. 3.
Percentage of SDPs in the functional regions (Top) compared to the corresponding percentage of protein residues in these functional regions (medium) in each Pfam family. The data are grouped according to the type of functional region detected in each family (Bottom), whereas the number of families in each category is shown in parentheses. Ligand-binding sites shown in blue, heterodimeric interfaces in green, homodimeric interface in red, and their combinations in yellow. The intrachain interfaces have been omitted for the sake of simplicity.
Table 1.
Results of the Wilcoxon rank sum tests evaluating the enrichment of SDPs and the conserved positions at the annotated sites.
|
p-value |
Site and total interface |
Site |
Total interface |
Hetero |
Homo |
Intra |
|
| Out of total | SDPs | 1.67E-05 | 4.25E-04 | 1.89E-02 | 1.75E-02 | 5.13E-01 | 4.99E-01 |
| Cons | 1.92E-27 | 4.73E-20 | 1.92E-09 | 5.32E-02 | 9.09E-03 | 4.29E-07 | |
| Given surface | SDPs | 1.00E-07 | 2.79E-04 | 4.02E-04 | 1.56E-02 | 8.75E-01 | 1.13E-01 |
| Cons | 2.65E-30 | 1.21E-18 | 2.54E-15 | 1.19E-03 | 1.27E-04 | 1.60E-10 | |
| Median difference |
Site + total interface |
Site |
Total interface |
Hetero |
Homo |
Intra |
|
| Out of total | SDPs | 5.32% | 3.97% | 2.00% | 4.77% | -0.01% | 0.00% |
| Cons | 15.25% | 14.60% | 6.34% | 1.84% | 1.99% | 6.76% | |
| Given surface | SDPs | 8.69% | 5.53% | 4.28% | 7.40% | 1.32% | 1.30% |
| Cons | 22.73% | 18.66% | 11.71% | 5.60% | 4.30% | 11.27% | |
Functional Association of SDPs in Proteins with Both Ligand-Binding Sites and Protein Interaction Regions.
Once the relationship between SDPs and ligand or protein interaction sites was statistically established in the previous section, we assessed whether there is a preference for the involvement of SDPs between them. Here we focused on the 168 families for which both types of regions are known, and we analyzed the joint distribution of the distances from the SDPs to the sites and interfaces (Fig. S5A). To disentangle this association, we assigned importance to the amount of SDPs that were close to one type of region but far from the other, or close to both regions at the same time. This approach provided a viewpoint complementary to that of the previous tests that quantified these tendencies corrected by the respective region sizes.
If we take a typical contact distance of 8 Å between Cβ atoms, we could define four regimes: (i) positions that were contacting to both sites and interfaces (approximately 24% SDPs/approximately 27% conservation), (ii) positions that contacted sites but not interfaces (approximately 16%/approximately 22%), (iii) those that contacted interfaces but not sites (approximately 29%/approximately 26%), and (iv) finally positions that were not in direct contact with either (approximately 25%/approximately 22%). This distribution shows that the distance of SDPs is similar for ligand and protein interaction sites. An analogous joint distribution for the averaged distances per family can also be found in Fig S5B.
To complement these figures, we assessed the number of families that contain SDPs in one and/or the other type of functional region (Fig. S6). This analysis should provide a qualitative indication of the type of region responsible for the functional diversity within a family. These results seem to indicate that the protein families do not have a preference for their SDPs being part of one or the other type of region, but they have a tendency to have SDPs at least in one of them.
Involvement of SDPs in Intra, Homo-, and Heterocomplexes.
To further characterize the involvement of SDPs in protein interactions, we analyzed the enrichment of SDPs at intra (between domains of the same protein), homo-, and heterointerfaces independently (Fig. 3). Thus, different subsets of families were considered according to these types of interfaces (170, 171, and 87 cases, respectively; Table S2). The equivalent enrichment tests corresponding to these interaction types were assessed (Table 1, the same test as described above). We observed that the enrichment for SDPs was significant at interfaces for heterocomplexes (p < 0.05). However, such enrichment was not evident in a significant number of cases for the other types of interfaces when considered in isolation. In contrast, there was a statistical association between the completely conserved positions and the three types of interfaces. Again, these results were consistent when SDPs reported by different methodologies were considered (see SI Text and Table S3). When the test was restricted to the positions in the surface, the enrichments remain significant and they have better p-values.
In terms of the E2F/TDP family of transcription factors previously mentioned, there was a clear relationship between the SDPs and the determination of interaction specificity. The hetero-complex E2F/TDP bound to DNA (Fig. S3) had a number of SDPs in each chain that were located at the interacting surface, complementing the other SDPs that were in contact with DNA, and highlighting their potential role in determining specific heteromeric recognition.
Discussion
The current expansion in the information available on the sequences, structures, functions, and interactions of proteins makes it possible to assess concepts and relationships that have been discussed for years in the absence of hard data. In this work we present a large scale assessment of how protein families are organized in functional terms, and of how the residues associated with this organization (SDPs) relate to the fundamental functional regions that correspond to the ligand binding and protein interaction sites. Of special interest is the study of the relationship between differential protein interactions and both subfamilies and SDPs, an analysis that is presented here in a unique and exhaustive way.
To analyze both the entities (subfamilies and SDPs) involved in this study in a uniform and self-consistent manner, we used a protocol based on multiple correspondence analysis (MCA). MCA is a multivariate descriptive technique that is conceptually related to but essentially different from the principal components analysis (PCA) we have used in our previous works (6). The power of the multivariate approach is that it enables the significant sources of information within a MSA to be disentangled, in such a way that the subfamily structure and the corresponding SDPs are determined simultaneously. The rationale is that the positions determine the separation of the sequences and, at the same time, the sequence separation weights the contribution of the positions to such segregation. Therefore, it makes it possible to analyze both entities simultaneously. The results obtained with this approach were qualitatively similar to those produced by four other methods dedicated to the detection of SDPs. The quality of the results together with the capacity of MCA to produce a simultaneous classification of residues and subfamilies, made it particularly adequate for the analysis proposed here (see SI Text).
By applying this methodology to the large set of eukaryotic protein families available in the Pfam database at the domain level, we obtained a robust and large dataset of subfamilies and SDPs. This dataset gave us the opportunity to systematically study the distribution of sequences and key residues in relation to two major aspects of their biological function: (i) the biochemical function associated to their catalytic binding activity, and (ii) their specific binding to other proteins.
We show how the specificity of protein interactions is correlated with the internal organization of the protein families, at a level similar to that which might be expected for the better characterized biochemical functions (22, 23). Indeed, the clear relationship between subfamilies and both the protein interaction and biochemical classes quantitatively supports the generally assumed functionally driven divergence between subfamilies (22–24). These observations could be interpreted in the context of the acquisition of new functions after gene duplication [subfunctionalization versus neofunctionalization, see (25)].
Additionally, we characterized the set of protein residues robustly connected to the subfamily structure (SDPs) using a representative set of reliably identified bound small ligands (26) and a set of protein interaction sites inferred from the corresponding protein structures. Our results show that SDPs fit very well with the organization of binding sites, measured both in distance distributions and in rigorous enrichments. At the same time, we show how SDPs accumulate in protein interaction regions (Fig. 3), indicating their possible role in the selection of interacting partners. Whereas functional specificity in the context of differential ligand binding has been examined extensively, an equivalent study of differential interactions was missing in the literature. This might be due to the prevalence of detailed biochemical studies for individual proteins (enzymes) in contrast to the more recent interest in protein interactions and networks. Our results suggest that functional modulation via differential interactions could be a more widespread phenomenon than previously suspected.
To better study the relationship between ligand binding and interaction sites, we concentrated the analysis on the SDPs of families in which both types of regions have been experimentally detected. The relevant number of families in which SDPs map to both types of regions could either point to a concerted action of these residues in both functions, or to their role as compensating mutations that allow the evolution of one region at the expense of the other. This possibility has indeed been previously illustrated for the class III aminotransferase family, where we see how SDPs linking the ligand-binding site with the homodimerization interface determine the specificity of the protein. At this moment, it is interesting to point out that the definition of SDPs is to some degree related with the concept of “correlated mutations” (27), and that the physical proximity of SDPs to protein-binding regions resembles the distributions of correlated positions (28–31).
We further characterized the distribution of SDPs in protein interaction regions by analyzing their distribution in intra- (between domains of the same protein), homo-, and heteromeric interfaces. In the first two types, we did not detect a significant enrichment of SDPs, even if in specific cases SDPs were clearly distributed at the homodimeric interface and they are potentially implicated in modulating the specificity of protein interactions, as in the class III aminotransferase family. In contrast, the analysis showed how fully conserved residues in protein families are statistically involved in both homomeric and intraprotein interfaces, consistently with previous analysis (32, 33). The specific involvement of SDPs in the heteromeric interfaces detected is similar to that observed for conserved residues, which are also significant in heteromeric interfaces, although less than in the case of homomeric and intraprotein interfaces. Unfortunately, the number of protein families with different subfamily members known to complex with different interactors is still not sufficiently large to carry out a more detailed study. The observation of the individual cases allows to propose that binding specificity evolves by selecting key residues differentially conserved in the subfamilies as pivotal points indicative of binding with their effectors (see, for example, the case of the E2F/TDP family in Results).
The persistence of SDPs in protein interaction interfaces in combination with their relationship to ligand-binding sites suggests that the previous success using SDPs to guide protein docking, to predict functional sites and to design mutants (14–16, 30) is far from anecdotal. This work demonstrates the crucial role of protein interactions in protein evolution driven by functional specificity and it extends the conceptual framework of SDPs to a more comprehensive definition of protein function.
Methods
Dataset of Protein Families.
We started with the whole Pfam-A database of multiple sequence alignments for protein families (21) (release 22.0) and filtered it, without realigning them. The complete details of the filtering process are given in SI Text but it mainly involved using only eukaryotic sequences with evidence at protein or transcript level [following Uniprot/SwissProt (34)], removing “gappy” sequences (> 30% gaps), removing redundancy (> 95% ID), removing outliers (< 40% ID), removing gappy columns (> 10% gaps), and ignoring alignments with < 12 sequences or < 25 positions. Our final dataset contains 1262 domains (Table S2).
Detection of Subfamilies and SDPs Within a MSA.
To carry out the joint functional analysis of subfamilies and associated SDPs we have developed a protocol able to detect both entities together in a concomitant way (Fig. 2). MCA provides the framework for this protocol (18). MCA is a multivariate descriptive technique that can be viewed as an equivalent to PCA when dealing with qualitative/binary data (35). MCA provides the orthogonal decomposition of the sources of variation within the initial MSA. These sources are disentangled by each of the principal axes, which can be prioritized through their associated eigenvalues. It allows to evaluate the statistical confidence of each dimension for being informative by means of a nonparametric Wilcoxon test (36). Sequences and residues are then represented in equivalent spaces where their natural association is revealed.
An unsupervised k-means clustering algorithm as implemented in ref. 37 was performed on the space of sequences. Clustering solutions are gathered for a prespecified number of groups ranging from 2 to 1/4 of the number of proteins (with a maximum of 50) and the solution maximizing the CHindex (38) is selected. This procedure automatically identifies the putative groups of proteins that are regarded as different subfamilies within the MSA (Fig. 2 and SI Text). Protein subfamilies are then linked with the corresponding regions in the space of residues to automatically assign the set of residues that uniquely characterizes each group. Positions within the MSA whose residues follow the subfamily segregation are defined as the SDPs of the family. Full details of the mathematical procedure are given in SI Text.
Analysis of the Functional Organization of Protein Subfamilies.
This analysis is done in terms of EC code and interaction specificity. The EC classification is taken from the UniprotKB database (34). EC groups are defined as proteins sharing the 4 digits of the EC code.
Highly reliable protein interactions for the two most complete eukaryotic interactomes (S. cerevisiae and H. sapiens) are taken from the small scale experiment of the Database of Interacting Proteins (39) core datasets. Negative (noninteracting) sets are constructed for these two organisms with pairs of proteins for which (i) both members are manually annotated in the Kyoto Encyclopedia of Genes and Genomes (40) as belonging to two different pathways or (ii) both members do not share subcellular location experimentally determined, as annotated in MIPS (41) and eSLDB (42) (see SI Text). As a further requirement to define a noninteracting pair, we check that it has not been reported in BIOGRID (release 2.0.49) (43), a general repository for interaction datasets that considers high-throughput experiments.
The agreement between subfamilies and EC groups or differential interactions is assessed in terms of specificity/sensitivity with a ROC analysis. For each Pfam, we calculate the sensitivity as TP/(TP + FN), and specificity as TN/(TN + FP), where TP (True Positives) is the number of protein pairs that agree both in “functional label” and subfamily; TN (true negatives) the number of protein pairs that disagree both in functional label and subfamily; FN (false negatives) the number of protein pairs that agree in functional label but disagree in subfamily and FP (false positives) the number of protein pairs that share subfamily but not functional label. Only cases for which TP + FN > 0, FP + TN > 0, TP + FP > 0, and FN + TN > 0 were considered.
For EC code, defining pairs of proteins with the same or different functional labels is trivial. However, for the interactions the situation is slightly more complicated. For each pair of proteins within the MSA, we calculate a shared interactors ratio as P++/(P++ + P+-), where P++ is the number of interacting partners common to both proteins and P+- is the number of partners interacting with one protein and “noninteracting” with the other. Whereas in the case of EC for each pair of proteins we have a binary (0/1) value representing whether the two proteins belong to the same EC group or not, for the interactions this value is continuous from 0.0 (no interactors shared) to 1.0 (all the interactors shared). Consequently, we calculate specificity and sensitivity for each family with the formulas above adapted to this new continuous value: TP = Σ(SIR) over the protein pairs in the same subfamily, TN = Σ(1 - SIR) over the protein pairs where both proteins are in different subfamily, FN = Σ(SIR) over the protein pairs in different subfamily, and FP = Σ(1 - SIR) over the protein pairs in the same subfamily.
The groups of proteins known to interact with the same protein are smaller than those labeled with the same EC, which tend to be rather large. As a consequence of this difference in granularity, the analysis of protein families based on their interactions tends to show higher sensitivity, whereas the one based on their EC code higher specificity.
Structural Information About Ligand-Binding Sites and Interfaces.
All the sequences within a Pfam family with structural information available are aligned to sequences of the PDB crystal structures. Only structures with a structural domain assigned by the Structural Classification of Proteins (SCOP) (44) were considered. Structural domains with < 80% alignment overlap with the corresponding Pfam domain are removed. Additionally, Pfam families mapping to > 1 SCOP superfamily are disregarded. Finally, Pfam families mapping to the same SCOP superfamily are considered structurally redundant groups. These groups are used to further perform the structurally nonredundant analyses (see below) to avoid bias due to uneven SCOP superfamily representation within the Pfam family database.
Ligand binding and catalytic residues are retrieved from FireDB database (26). FireDB integrates data from the close atomic contacts in PDB structures and reliably annotated catalytic residues from the Catalytic Site Atlas (45). The dataset of PDB complexes are retrieved from the 3D complex database (May 25, 2008) (46). From these data, cases annotated as errors by the PiQSi manual curation effort (May 25, 2008) (47) are disregarded. Protein chains with < 60 residues were removed to avoid interactions with protein fragments and protein–peptide interactions. Protein–protein interaction sites are defined according to the standard criteria based on change of accessibility upon interaction (48). Surface residues are those with relative accessible surface area (RSA) of 5% or more. RSA is calculated with the Naccess program (see SI Text). Residues in interaction surfaces are defined as those that fulfill the accessibility criteria only when the chain is considered in isolation. Interaction surfaces are classified in homo-, hetero-, and intrainteractions depending on whether they involve two chains representing the same protein (according to the Swissprot AC), two different proteins, or two structural domains of the same protein [according to SCOP (44)].
Finally, for each MSA only one of their structures is selected as a family’s representative and the structural information of the others (ligand-binding residues and protein binding sites) is projected on it.
Enrichment Tests.
The sets of SDPs and conserved residues (identity > 90%) are compared to the sets of binding residues using an enrichment test. This test is intended to quantify the improvement of the predictions with respect to the corresponding background. For each family, we calculate the proportion of SDPs annotated as part of a binding region and the proportion of the whole protein annotated. The enrichment is defined as the difference between these two percentages. Enrichments are averaged within each structurally redundant group of Pfam families. Finally, a Wilcoxon rank sum test is performed on the whole list of enrichments (one per SCOP superfamily). This test assesses a p-value for the null hypothesis that there is no difference between the target subset of positions and a random one. The procedure is repeated for the different types of positions (SDPs and conserved) and the different types of binding sites (ligand-binding, hetero-, homo-, and intrainteractions), as well as for combinations of them.
Supplementary Material
Acknowledgments.
We thank the members of the Structural Biology and Biocomputing Programme (CNIO), especially G. Lopez for his help on the FireDB data retrieval, and the Computational Systems Biology Group (CNB-CSIC) for interesting discussions and support, as well as A. Wilkins and O. Lichtarge from the Baylor College of Medicine for providing the ET predictions, and B. Reva, Y. Antipin, and C. Sander from the Memorial Sloan–Kettering Center for providing the CEO software. Work at CNIO was in part funded by the CSD2007-00050 (CONSOLIDER) and BIO2007-66855 projects from the Spanish Ministry for Education and Science, the EU project LSHG-CT-2004-503567 (ENFIN) and the Spanish National Institute for Bioinformatics (INB, www.inab.org). Work at CNB-CSIC was in part funded by the BIO2006-15318 and PIE 200620I240 projects from the Spanish Ministry for Education and Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908044107/DCSupplemental.
References
- 1.Koehl P, Levitt M. Improved recognition of native-like structures using a family of designed sequences. Proc Natl Acad Sci USA. 2002;99(2):691–696. doi: 10.1073/pnas.022408799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orengo CA, Thornton JM. Protein families and their evolution-a structural perspective. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 3.Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 4.Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–366. doi: 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
- 5.Valdar WS. Scoring residue conservation. Proteins. 2002;48(2):227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 6.Casari G, Sander C, Valencia A. A method to predict functional residues in proteins. Nat Struct Biol. 1995;2:171–178. doi: 10.1038/nsb0295-171. [DOI] [PubMed] [Google Scholar]
- 7.Lichtarge O, Bourne HR, Cohen FE. An Evolutionary Trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 8.Hannenhalli SS, Russell RB. Analysis and prediction of functional sub-types from protein sequence alignments. J Mol Biol. 2000;303:61–76. doi: 10.1006/jmbi.2000.4036. [DOI] [PubMed] [Google Scholar]
- 9.del Sol Mesa A, Pazos F, Valencia A. Automatic Methods for Predicting Functionally Important Residues. J Mol Biol. 2003;326(4):1289–1302. doi: 10.1016/s0022-2836(02)01451-1. [DOI] [PubMed] [Google Scholar]
- 10.Pazos F, Sternberg MJE. Automated prediction of protein function and detection of functional sites from structure. Proc Natl Acad Sci USA. 2004;101(41):14754–14759. doi: 10.1073/pnas.0404569101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pazos F, Rausell A, Valencia A. Phylogeny-independent detection of functional residues. Bioinformatics. 2006;22(12):1440–1448. doi: 10.1093/bioinformatics/btl104. [DOI] [PubMed] [Google Scholar]
- 12.Reva B, Antipin Y, Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 2007;8(11):R232. doi: 10.1186/gb-2007-8-11-r232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pazos F, Bang J-W. Computational Prediction of Functionally Important Regions in Proteins. Curr Bioinform. 2006;1(1):15–23. [Google Scholar]
- 14.Bauer B, et al. Effector recognition by the small GTP-binding proteins Ras and Ral. J Biol Chem. 1999;274(25):17763–17770. doi: 10.1074/jbc.274.25.17763. [DOI] [PubMed] [Google Scholar]
- 15.Morillas M, et al. Identification of conserved amino acid residues in rat liver carnitine palmitoyltransferase I critical for malonyl-CoA inhibition. Mutation of methionine 593 abolishes malonyl-CoA inhibition. J Biol Chem. 2003;278(11):9058–9063. doi: 10.1074/jbc.M209999200. [DOI] [PubMed] [Google Scholar]
- 16.Cordente AG, et al. Redesign of carnitine acetyltransferase specificity by protein engineering. Modification of methionine564 broadens the specificity to longer acyl-CoAs as substrates. J Biol Chem. 2004;279:33899–33908. doi: 10.1074/jbc.M402685200. [DOI] [PubMed] [Google Scholar]
- 17.Petrey D, Honig B. Is protein classification necessary? Toward alternative approaches to function annotation. Curr Opin Struct Biol. 2009;19(3):363–368. doi: 10.1016/j.sbi.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenacre M, Blasius J. Multiple correspondence analysis and related methods. Berlin/Heidelberg: Springer; [Google Scholar]
- 19.Wu CL, Zukerberg LR, Ngwu C, Harlow E, Lees JA. In vivo association of E2F and DP family proteins. Mol Cell Biol. 1995;15(5):2536–2546. doi: 10.1128/mcb.15.5.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markova M, Peneff C, Hewlins MJ, Schirmer T, John RA. Determinants of substrate specificity in omega-aminotransferases. J Biol Chem. 2005;280(43):36409–36416. doi: 10.1074/jbc.M506977200. [DOI] [PubMed] [Google Scholar]
- 21.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32(Database issue):D138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devos D, Valencia A. Practical limits of function prediction. Proteins. 2000;41:98–107. [PubMed] [Google Scholar]
- 23.Rost B. Enzyme function less conserved than anticipated. J Mol Biol. 2002;318(2):595–608. doi: 10.1016/S0022-2836(02)00016-5. [DOI] [PubMed] [Google Scholar]
- 24.Sjölander K. Phylogenomic inferrence of protein molecular function: Advances and challenges. Bioinformatics. 2004;20(2):170–179. doi: 10.1093/bioinformatics/bth021. [DOI] [PubMed] [Google Scholar]
- 25.He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez G, Valencia A, Tress M. FireDB—a database of functionally important residues from proteins of known structure. Nucleic Acids Res. 2007;35:D219–223. doi: 10.1093/nar/gkl897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Göbel U, Sander C, Schneider R, Valencia A. Correlated mutations and residue contacts in proteins. Proteins. 1994;18:309–317. doi: 10.1002/prot.340180402. [DOI] [PubMed] [Google Scholar]
- 28.Madaoui H, Guerois R. Coevolution at protein complex interfaces can be detected by the complementarity trace with important impact for predictive docking. Proc Natl Acad Sci USA. 2008;105(22):7708–7713. doi: 10.1073/pnas.0707032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yeang CH, Haussler D. Detecting coevolution in and among protein domains. PLoS Comput Biol. 2007;3(11):e211. doi: 10.1371/journal.pcbi.0030211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tress M, et al. Scoring docking models with evolutionary information. Proteins. 2005;60(2):275–280. doi: 10.1002/prot.20570. [DOI] [PubMed] [Google Scholar]
- 31.Pazos F, Helmer-Citterich M, Ausiello G, Valencia A. Correlated mutations contain information about protein-protein interaction. J Mol Biol. 1997;271(4):511–523. doi: 10.1006/jmbi.1997.1198. [DOI] [PubMed] [Google Scholar]
- 32.Jones S, Thornton JM. Principles of protein–protein interactions. Proc Natl Acad Sci USA. 1996;1(93):13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones S, Thornton JM. Prediction of protein-protein interaction sites using surface patches. J Mol Biol. 1997;272:121–132. doi: 10.1006/jmbi.1997.1234. [DOI] [PubMed] [Google Scholar]
- 34.Consortium U. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37(Database issue):D169–174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lebart L, Morineau A, Warwick KM. Multivariate Descriptive Statistical Analysis. New York: John Wiley & Sons; p. 175. [Google Scholar]
- 36.Miller I, Miller M. Mathematical Statistics. London: Prentice Hall International; 1998. [Google Scholar]
- 37.de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 38.Calinski T, Harabasz J. A Dendrite Method for Cluster Analysis. Comm Stat. 1974;3(1):1–27. [Google Scholar]
- 39.Xenarios I, et al. DIP, the Database of Interacting Proteins: A research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mewes H, Albermann K, Heumann K, Liebl S, Pfeiffer F. MIPS: A database for protein sequences, homology data and yeast genome information. Nucleic Acids Res. 1997;25(1):28–30. doi: 10.1093/nar/25.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierleoni A, Martelli PL, Fariselli P, Casadio R. eSLDB: Eukaryotic subcellular localization database. Nucleic Acids Res. 2007;35:D208–212. doi: 10.1093/nar/gkl775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stark C, et al. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andreeva A, et al. SCOP database in 2004: Refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32(Database issue):D226–229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: A resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32(Database issue):D129–133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy ED, Pereira-Leal JB, Chothia C, Teichmann SA. 3D complex: A structural classification of protein complexes. PLoS Comput Biol. 2006;2(11):e155. doi: 10.1371/journal.pcbi.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levy ED. PiQSi: Protein quaternary structure investigation. Structure. 2007;15(11):1364–1367. doi: 10.1016/j.str.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 48.Valdar WS, Thornton JM. Protein–protein interfaces: Analysis of amino acid conservation in homodimers. Proteins. 2001;42(1):108–124. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.