Chibby (Cby) acts with 14-3-3 to regulate β-catenin localization in the canonical Wnt pathway. We show that Cby harbors functional NLS and NES motifs, and shuttles between the nucleus and cytoplasm. Cby distribution at steady state is controlled by an intricate cooperation between 14-3-3, CRM1 and importin-α, which impacts on β-catenin signaling.
Abstract
In the canonical Wnt pathway, β-catenin acts as a key coactivator that stimulates target gene expression through interaction with Tcf/Lef transcription factors. Its nuclear accumulation is the hallmark of active Wnt signaling and is frequently associated with cancers. Chibby (Cby) is an evolutionarily conserved molecule that represses β-catenin–dependent gene activation. Although Cby, in conjunction with 14-3-3 chaperones, controls β-catenin distribution, its molecular nature remains largely unclear. Here, we provide compelling evidence that Cby harbors bona fide nuclear localization signal (NLS) and nuclear export signal (NES) motifs, and constitutively shuttles between the nucleus and cytoplasm. Efficient nuclear export of Cby requires a cooperative action of the intrinsic NES, 14-3-3, and the CRM1 nuclear export receptor. Notably, 14-3-3 docking provokes Cby binding to CRM1 while inhibiting its interaction with the nuclear import receptor importin-α, thereby promoting cytoplasmic compartmentalization of Cby at steady state. Importantly, the NLS- and NES-dependent shuttling of Cby modulates the dynamic intracellular localization of β-catenin. In support of our model, short hairpin RNA–mediated knockdown of endogenous Cby results in nuclear accumulation of β-catenin. Taken together, these findings unravel the molecular basis through which a combinatorial action of Cby and 14-3-3 proteins controls the dynamic nuclear-cytoplasmic trafficking of β-catenin.
INTRODUCTION
The canonical Wnt/β-catenin signaling pathway plays diverse roles in embryonic development and in maintenance of organs and tissues in adults (Wodarz and Nusse, 1998; Pinto and Clevers, 2005; Klaus and Birchmeier, 2008). In recent years, dysregulation of this signaling cascade has been linked to the pathogenesis of a range of human diseases (Moon et al., 2004; Clevers, 2006). In particular, aberrant activation of Wnt/β-catenin signaling is found in a variety of human malignancies including melanoma and colon and hepatocellular carcinomas (Polakis, 2000; Klaus and Birchmeier, 2008). Therefore, the Wnt/β-catenin pathway has gained recognition as an attractive molecular target for anticancer therapeutics (Barker and Clevers, 2006; Takemaru et al., 2008).
In the canonical Wnt pathway, β-catenin plays a central role, acting as a transcriptional coactivator (Takemaru, 2006). In the absence of Wnt ligands, β-catenin is sequentially phosphorylated by casein kinase 1 (CK1) and glycogen synthase kinase 3 (GSK3) in the so-called “destruction complex,” containing the tumor suppressors Axin and Adenomatous polyposis coli (APC). This constantly targets β-catenin for ubiquitin-mediated proteasomal degradation, keeping the cytosolic pool of β-catenin low (Kimelman and Xu, 2006; Macdonald et al., 2007). On engagement with the seven transmembrane Frizzled (Fz) receptors and the low-density lipoprotein receptor–related protein (LRP) coreceptors, LRP5 and LRP6, Wnts trigger recruitment of Axin to the plasma membrane, resulting in inhibition of β-catenin phosphorylation and degradation (Cadigan and Liu, 2006; Huang and He, 2008). Consequently, β-catenin accumulates in the cytoplasm and then translocates into the nucleus where it forms a complex with the T-cell factor/lymphoid enhancer factor (Tcf/Lef) family of transcription factors, leading to activation of target genes (Stadeli et al., 2006; Willert and Jones, 2006). β-Catenin also functions in cell adhesion by binding to type I cadherins to mediate actin filament assembly via α-catenin at the plasma membrane (Takemaru, 2006). Although β-catenin resides in multiple subcellular compartments, the cellular and molecular bases controlling its dynamic intracellular trafficking remain poorly defined.
Chibby (Cby) is a small protein of 14.5 kDa that is highly conserved throughout evolution from fly to human (Takemaru et al., 2003). Cby directly interacts with the C-terminal activation domain of β-catenin and represses β-catenin–mediated transcriptional activation by competing with Tcf/Lef factors for β-catenin binding (Takemaru et al., 2003, 2009). More recently, we proposed a second mode of action for Cby in inhibiting β-catenin signaling (Li et al., 2008). We showed that 14-3-3 chaperone proteins specifically recognize serine 20 within the N-terminal 14-3-3–binding motif of Cby when phosphorylated by Akt kinase, leading to sequestration of Cby into the cytoplasm. Importantly, Cby and 14-3-3 form a stable tripartite complex with β-catenin to facilitate its partition into the cytoplasm. However, the nuclear-cytoplasmic shuttling nature of Cby and precise mechanisms underlying the nuclear export of β-catenin by Cby and 14-3-3 have not been fully understood at the molecular level.
14-3-3 proteins constitute a family of highly conserved dimeric proteins, comprised of seven isoforms in mammals (β, γ, ε, σ, ζ, τ, and η; Aitken, 2006; Morrison, 2009). In most cases, 14-3-3 proteins bind to one of the two high-affinity consensus sequences within the target protein, RSXpSXP (mode I) and RXXXpSXP (mode II), where pS represents phosphoserine (Muslin et al., 1996; Yaffe et al., 1997). More than 200 different 14-3-3–binding partners have been identified (Jin et al., 2004; Meek et al., 2004) but the vast majority of these interactions are largely uncharacterized to date. The 14-3-3 family members serve as chaperon-like adaptor molecules that play important roles in a wide variety of cellular processes, including intracellular protein targeting by modulating nuclear import or export of their ligands (Muslin and Xing, 2000; Eckardt, 2001). Exactly how 14-3-3 proteins trigger changes in subcellular localization of their targets remains to be elucidated. Several lines of evidence indicate that 14-3-3 causes cytoplasmic sequestration of the phosphatase Cdc25 and histone deacetylase (HDAC) 4, at least in part, by inhibiting their interaction with the nuclear import receptor importin-α (Kumagai and Dunphy, 1999; Yang et al., 1999; Grozinger and Schreiber, 2000). In contrast, 14-3-3 binding induces nuclear translocation of myopodin in myoblasts by facilitating its association with importin-α (Faul et al., 2005). Thus, it seems likely that the final destinations of 14-3-3-ligand complexes are determined by the ligand per se rather than 14-3-3, through either masking or exposing the nuclear localization signal (NLS) or nuclear export signal (NES) sequence of ligands.
In the present study, we have investigated the nuclear-cytoplasmic shuttling properties of Cby. Cby contains functional NLS and NES motifs that physically interact with importin-α and the nuclear export receptor CRM1 (chromosomal region maintenance protein 1), respectively. Cby NLS mutants predominantly localize to the cytoplasm, whereas Cby NES mutants reside in the nucleus, exhibiting colocalization with β-catenin. Intriguingly, 14-3-3–bound Cby favors a complex formation with CRM1 but not that with importin-α, providing a molecular basis accounting for the cytoplasmic redistribution of Cby by 14-3-3 proteins. In good agreement with our model, knockdown of endogenous Cby expression by short hairpin RNA (shRNA) leads to marked nuclear accumulation of β-catenin. Our findings therefore suggest that Cby constitutively cycles between the nucleus and cytoplasm and regulates the dynamic nuclear-cytoplasmic trafficking and signaling activity of β-catenin.
MATERIALS AND METHODS
Expression Constructs and Bacterial Protein Expression
Expression plasmids for Flag-tagged CbyWT and CbyS20A, maltose-binding protein (MBP)-CbyWT, β-catenin-Myc, β-cateninS33Y-Flag. and HA-14-3-3ζ have been described previously (Takemaru et al., 2003; Li et al., 2008). The HA-CRM1 construct was kindly provided by Dr. J. A. Diehl (Abramson Family Cancer Research Institute; Benzeno and Diehl, 2004). The His-RanQ69L-GTP plasmid was a gift from Dr. M. Dasso (NIH). The bacterial expression vectors for glutathione S-transferase (GST)-importin-α and GST-importin-β were kind gifts from Dr. N. Reich (Stony Brook University; Liu et al., 2005) and Dr. Y. Yoneda (Osaka University, Japan; Nagoshi and Yoneda, 2001), respectively. N-terminally Flag-tagged Cby point mutants were created with the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using the Flag-CbyWT vector as a template. The HA-importin-α3 expression plasmid was constructed by digesting the GST-importin-α3 vector with BamHI and SalI, and ligating the cDNA insert into pcDNA5-HA (Invitrogen, Carlsbad, CA). To generate the CbyWT-C-Flag and CbyΔNLS2-C-Flag constructs, C-terminally Flag-tagged Cby cDNA fragments were PCR-amplified using a reverse primer containing a Flag sequence at the 3′ end, digested with EcoRI and XhoI, and subcloned into pCS2+. For Flag-CbyΔNLS2, GFP-CbyWT and GFP-CbyΔNLS2 or MBP-CbyΔNLS2 and MBP-CbyR76A/R78A/R79A, a corresponding cDNA insert, was amplified by PCR and subcloned into pCS2+, pCS2+GFP-BB, or pMAL-c2E (New England Biolabs, Beverly, MA). We verified all constructs by DNA sequencing. GST and MBP fusion proteins were expressed in Escherichia coli BL21 cells and purified using GST·Bind (Novagen, Madison, WI) and amylose (New England Biolabs) beads, respectively, according to the manufacturer's instructions.
Cell Culture and Transfection
Human embryonic kidney (HEK) 293, HEK293T. and Cos7 cell lines were purchased from ATCC (Manassas, VA) and propagated in DMEM with 10% FBS and 100 U/ml penicillin-streptomycin. Mouse embryonic fibroblasts (MEFs) were prepared from mouse embryos at embryonic day 12.5 as described previously (Li et al., 2007). For transient transfection, cells were seeded onto 6- or 12-well tissue culture dishes, cultured overnight, and then transfected using Lipofectamine 2000 (Invitrogen) or Expressfect (Denville Scientific, Denville, NJ) according to the manufacturer's instructions. Empty vector was added to adjust the total amount of DNA to be the same in each transfection. For generation of Cby-knockdown cell lines, HEK293 cells were transfected with a human Cby-specific SureSilencing shRNA or negative control scrambled shRNA plasmid (SABiosciences, Frederick, MD), and selected with 2.5 μg/ml puromycin (Invitrogen). Puromycin-resistant single colonies were isolated, grown, and tested for knockdown of Cby protein levels by Western blotting. Three independent stable cell lines expressing control shRNA or Cby shRNA with maximum knockdown were used to examine β-catenin localization, and similar trends were observed using all the cell lines.
Coimmunoprecipitation and Western Blot Analyses
Transfected HEK293T cells were washed twice with PBS and collected into ice-cold lysis buffer containing 20 mM Tris-HCl, pH 8.0, 135 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 10% glycerol, and complete protease inhibitor cocktail (Roche, Indianapolis, IN). Cell lysates were then cleared by centrifugation at 12,000 rpm for 10 min at 4°C. The supernatants were collected and processed for coimmunoprecipitation and immunoblotting experiments as previously described (Li et al., 2007, 2008). The primary antibodies used were as follows: rabbit anti-Cby (Takemaru et al., 2003); mouse anti-Flag M2 (Sigma-Aldrich, St. Louis, MO); rat anti-HA (Roche); mouse anti-GST (Novagen); mouse anti-MBP (New England Biolabs); rabbit anti-CRM1 and rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA); mouse anti-GAPDH (Biodesign International, Saco, ME). All HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
In Vitro Pulldown Experiments
Equal amounts of bacterially expressed and purified MBP or MBP-Cby derivatives were mixed with GST, GST-importin-α, or GST-importin-β in protein-binding buffer (20 mM HEPES, pH 7.9, 100 mM NaCl, 6 mM MgCl2, 0.5 mM EDTA, 0.1% NP-40. and 20% glycerol) and incubated for 1 h on ice. For the binding assays in Figure 6B, equal amounts of GST or GST-importin-α3 was incubated with HEK293T cell lysates expressing Flag-Cby variants or HA-14-3-3ζ for 1 h on ice as indicated. On addition of 10 μl (packed volume) of amylose beads (New England Biolabs) or GST·Bind resin (Novagen), the mixture was rotated for additional 1 h at 4°C and washed extensively with protein-binding buffer. The bound materials were then eluted with SDS sample buffer, separated on SDS-PAGE, and examined by Western blotting.
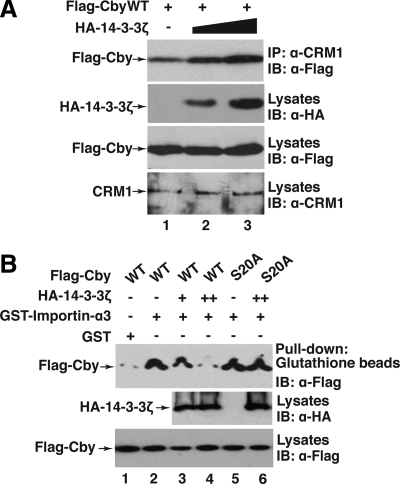
Figure 6.
14-3-3 facilitates the Cby-CRM1 interaction but blocks the Cby-Importin-α interaction. (A) 14-3-3 promotes Cby binding to CRM1. Flag-CbyWT and increasing amounts of HA-14-3-3ζ were coexpressed in HEK293T cells, and cell lysates immunoprecipitated with anti-CRM1 antibody, followed by Western blot analysis with anti-Flag antibody. Immunoblotting of cell lysates with the indicated antibodies was performed to monitor protein levels. (B) 14-3-3 inhibits Cby binding to importin-α. Bacterially produced GST or GST-importin-α3 was incubated with cell lysates from HEK293T cells expressing Flag-tagged CbyWT or CbyS20A incapable of 14-3-3 binding and from those expressing HA-14-3-3ζ. The samples were then pulled down with glutathione beads, separated by SDS-PAGE, and immunoblotted with anti-Flag antibody.
Immunofluorescence Microscopy
Immunofluorescence microscopy was performed essentially as described previously (Li et al., 2008). The primary antibodies used were as follows: mouse anti-Flag M2 (Sigma-Aldrich); rat anti-HA (Roche); mouse anti-Myc (Invitrogen); rabbit anti-Cby (Takemaru et al., 2003); mouse anti-β-catenin (BD Transduction Laboratories, Lexington, KY). Fluorescent FITC- and TRITC-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories. Immunostained cells were analyzed by a Leica DM5000 fluorescent microscope (Deerfield, IL) with an HCX PL Fluotar 100×/1.30 NA oil objective lens. All images were acquired at room temperature with a Leica DFC300 FX camera using Leica Application Suite software version 2.8.1. To quantify subcellular localization, independent transfections were performed at least three times, and a minimum of 100 cells was counted and scored. In cotransfections, only cells expressing both proteins were scored.
TopFlash Assays
HEK293T cells were seeded onto 12-well plates the day before transfection. Cells were transfected with appropriate combinations of plasmids in triplicate. Luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and a Berthold luminometer (Berthold Technologies, Bad Wildbad, Germany) as previously described (Li et al., 2007, 2008). An expression plasmid for Renilla luciferase (pRL-TK) was cotransfected to normalize transfection frequency.
RESULTS
The C-Terminal NLS Is Necessary for Nuclear Import of Cby
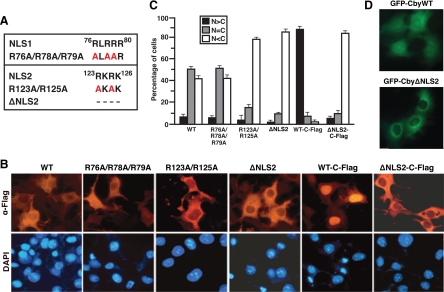
Human Cby is a 14.5-kDa protein comprised of 126 amino acids, and contains two highly conserved putative NLS motifs spanning residues 76–80 (NLS1) and 123–126 (NLS2; Takemaru et al., 2003; Figure 1A). To investigate the functionality of these NLSs, we introduced a series of alanine substitutions for conserved basic residues within the NLSs by site-directed mutagenesis (Figure 1A). Western blotting revealed that all the Cby NLS mutants were stably expressed at levels comparable to the wild-type (WT) protein upon overexpression in mammalian cultured cells (data not shown). Consistent with our previous observation (Li et al., 2008), when expressed in Cos7 cells, N-terminally Flag-tagged CbyWT primarily localized to the cytoplasm or both the nucleus and cytoplasm, especially in cells with high levels of Cby expression (Figure 1, B and C). A Cby NLS1 mutant, carrying alanine substitutions for three arginine residues at positions 76, 78, and 79 (R76A/R78A/R79A), displayed no major changes in subcellular distribution in comparison with CbyWT. In sharp contrast, point mutations within NLS2 (R123A/R125A) or deletion of the entire NLS2 sequence (ΔNLS2) dramatically shifted Cby localization to the cytoplasm. To confirm the importance of NLS2 in nuclear translocation of Cby, we expressed N-terminally green fluorescent protein (GFP)-tagged CbyWT or CbyΔNLS2 in Cos7 cells and examined their intracellular localization. As shown in Figure 1D, GFP-CbyWT showed a distribution pattern similar to that of Flag-CbyWT, whereas GFP-CbyΔNLS2 was predominantly cytoplasmic. These results clearly establish that the NLS2 motif, but not NLS1, is crucial for mediating nuclear entry of Cby.
Figure 1.
Cby harbors a functional NLS at the C-terminal end. (A) Cby contains two putative NLSs (NLS1 and NLS2). Cby NLS mutants generated by alanine substitutions are shown. CbyΔNLS2 lacks the last four C-terminal amino acids of the protein. (B) The C-terminal NLS2 is critical for nuclear import of Cby. Cos7 cells were transiently transfected with an expression vector encoding Flag-tagged at the N- or C-terminus (C-Flag) of WT or mutant Cby as indicated. Cells were fixed 24 h after transfection, followed by immunostaining with anti-Flag antibody. Nuclei were stained with DAPI. (C) Quantitative analysis of the results in B. The subcellular localization of Flag-tagged Cby was scored as follows: N>C, predominantly nuclear; N = C, evenly distributed between the nucleus and cytoplasm; N<C, predominantly cytoplasmic. Error bars, means ± SD of three independent experiments. (D) Fluorescence imaging of GFP-Cby. Cos7 cells were transfected with a plasmid expressing GFP-CbyWT or GFP-CbyΔNLS2, and observed under fluorescence microscopy.
During the course of our study, we found that C-terminally Flag-tagged CbyWT (CbyWT-C-Flag) displays almost complete nuclear accumulation (Figure 1, B and C), most likely due to a conformational change that exposes the normally buried C-terminal NLS2. Essentially, similar results were obtained for C-terminally Myc- or GFP-tagged CbyWT (data not shown). Deletion of the NLS2 sequence again resulted in a predominant cytoplasmic distribution of the protein. Quantification of Cby localization revealed that <10% of cells expressing CbyΔNLS2-C-Flag exhibited clear nuclear immunofluorescence, whereas ∼90% of cells expressing CbyWT-C-Flag showed discrete nuclear staining (Figure 1C). These findings further support the functional importance of NLS2 in translocation of Cby into the nucleus.
Importin-α Physically Interacts with Cby NLS2 to Mediate Its Nuclear Entry
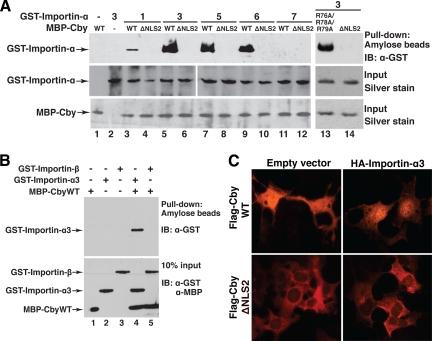
In the classical nuclear import pathway, importin-α plays a pivotal role by directly binding to NLS-containing proteins, linking them to importin-β (Macara, 2001; Goldfarb et al., 2004; Lange et al., 2007). Importin-β in turn docks the ternary import complex at the nuclear pore complex (NPC) and facilitates its translocation into the nucleus. In mammals, there are at least six importin-α family members with distinct substrate specificity (Kohler et al., 1999; Liu et al., 2005). To examine if any of these importin-α proteins directly recognize Cby, we performed in vitro pulldown assays using bacterially expressed and purified MBP-Cby and GST fusion proteins with different importin-α members (1, 3, and 5–7). The N-terminal importin-β-binding (IBB) domain of importin-α contains an internal autoinhibitory NLS that associates with its own central Armadillo repeat region when it is not bound to an NLS cargo and/or importin-β (Moroianu et al., 1996; Kobe, 1999). Thus, the IBB domain was deleted from the GST-importin-α proteins to eliminate this autoinhibition. MBP-CbyWT, MBP-CbyΔNLS2, or MBP-CbyR76A/R78A/R79A was incubated with individual GST-importin-α family members and pulled down using amylose resin. Subsequently, the bound proteins were separated by SDS-PAGE, followed by Western blotting with anti-GST antibody. As shown in Figure 2A, CbyWT specifically bound to importin-α3, importin-α5 as well as importin-α6 with high affinity. On the other hand, CbyWT showed only a weak interaction with importin-α1 and no interaction with importin-α7. Importantly, deletion of Cby NLS2 completely abrogated its binding to importin-α proteins, whereas the triple NLS1 mutant (R76A/R78A/R79A) interacted with importin-α3 as efficiently as CbyWT (lane 13). In some scenarios, importin-β has been shown to directly bind to NLS-containing proteins and mediate their nuclear import independently of importin-α (Palmeri and Malim, 1999; Truant and Cullen, 1999). However, importin-β was unable to interact with CbyWT in our in vitro pulldown assays (Figure 2B). Taken together, our data demonstrate that Cby specifically interacts with multiple importin-α family members through the C-terminal NLS2 motif.
Figure 2.
Importin-α binds to Cby and facilitates its nuclear entry. (A) Cby interacts with multiple importin-α isoforms through the NLS2 motif. Bacterially expressed and purified MBP-CbyWT, MBP-CbyΔNLS2, or MBP-CbyR76A/R78A/R79A was incubated with GST fusion proteins with different importin-α members (1, 3, and 5–7). The complexes were then pulled down with amylose beads and subjected to Western blotting using anti-GST antibody. Input corresponds to the same amounts of fusion proteins used in each reaction mixture, visualized by silver staining to confirm that equal amounts of recombinant proteins were used. (B) Importin-β does not interact with Cby. In vitro pulldown assays were performed using GST-importin-β as described in A. Ten percent of each reaction mixture was resolved on a separate SDS-PAGE, followed by immunoblotting with anti-GST and anti-MBP antibodies to verify that equal amounts of recombinant proteins were used. (C) Overexpression of importin-α promotes nuclear localization of CbyWT but not that of CbyΔNLS2. Cos7 cells were cotransfected with an expression construct for Flag-tagged CbyWT or CbyΔNLS2 and a negative control empty vector or an HA-importin-α3 expression plasmid and immunostained with anti-Flag antibody.
To evaluate the functional relevance of the Cby-importin-α interaction, we tested whether ectopic expression of importin-α influences Cby localization within cells. To this end, Flag-tagged CbyWT or CbyΔNLS2 was transiently coexpressed with HA-importin-α3 in Cos7 cells, and immunostained for Cby using anti-Flag antibody. On coexpression with HA-importin-α3, significant levels of Flag-CbyWT were observed in the nucleus (Figure 2C, top right panel), whereas the predominant cytoplasmic localization of Flag-CbyΔNLS2 remained unaffected (bottom right panel). Of note, when ectopically expressed in Cos7 cells, HA-importin-α3 was distributed throughout the cytoplasm and nucleus in most cells (Supplemental Figure S1). Therefore, we conclude that Cby NLS2 at the C-terminus functions as a bona fide NLS required for its nuclear translocation through interaction with importin-α.
Cytoplasmic Localization of Cby Requires a Functional NES in Its N-Terminal Region
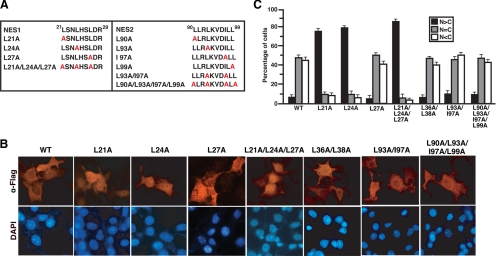
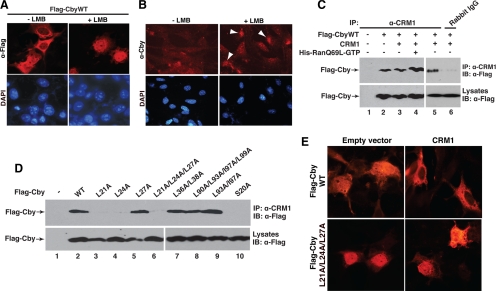
Given the fact that Cby is present in both the nucleus and cytoplasm, Cby might be constantly shuttling in and out of the nucleus. Consistent with this notion, we found that Cby harbors two predicted NESs (NES1, 21L-SN-L-HS-L-d-R29; NES2, 90L-LR-L-KVD-I-L-L99; critical hydrophobic residues are shown in bold), which show close matches to the classical NES consensus sequence, Φ-(X2–3)-Φ-(X2–3)- Φ-X-Φ (Φ = L, I, V, F, M; X is any amino acid; Figure 3A; la Cour et al., 2004; Kutay and Guttinger, 2005). To assess if the putative NESs in Cby are responsible for its nuclear export, we generated a series of N-terminally Flag-tagged Cby mutants carrying leucine- or isoleucine-to-alanine substitutions within these motifs by site-directed mutagenesis (Figure 3A). When expressed in mammalian cultured cells, expression levels of Cby NES1 mutants were slightly reduced, but the other Cby mutants were found to be present at levels similar to CbyWT as revealed by Western blotting (data not shown). Next, the Cby NES mutants were transiently expressed in Cos7 cells, and their subcellular distribution evaluated by immunofluorescence microscopy (Figure 3, B and C). Strikingly, three Cby NES1 mutants (L21A, L24A, and L21A/L24A/L27A) showed a dramatic shift in their intracellular localization toward the nuclear compartment. The triple point mutant CbyL21A/L24A/L27A frequently had even more discrete nuclear staining. Conversely, no appreciable changes were observed with CbyL27A. The other Cby mutants carrying alanine substitutions within the NES2 motif (L90A, L93A, I97A, L99A, L93A/I97A, and L90A/L93A/I97A/L99A) or outside of the NES sequences (L36A/L38A) manifested localization patterns similar to that of CbyWT (Figure 3, B and C; data not shown). Collectively, these results demonstrate that NES1 but not NES2 is a genuine NES necessary for nuclear export of Cby.
Figure 3.
Identification of a functional NES of Cby. (A) Cby carries two potential NESs in its N-terminal region (NES1 and NES2). Shown are the Cby NES point mutants used in this study. (B) The NES1 motif mediates nuclear export of Cby. Cos7 cells were transiently transfected with an expression plasmid for Flag-tagged CbyWT or various NES mutant derivatives. Cells were fixed 24 h after transfection and immunostained with anti-Flag antibody. Nuclei were stained with DAPI. (C) Quantitative analysis of the results in B. The subcellular localization of Flag-Cby was scored as in Figure 1C. Error bars, means ± SD of three independent experiments.
Cby Exits the Nucleus via the CRM1-Dependent Nuclear Export Pathway
CRM1/Exportin1 is the major nuclear export receptor for classical NES-containing proteins (Macara, 2001; Pemberton and Paschal, 2005; Hutten and Kehlenbach, 2007). This, together with our identification of a functional NES in Cby, raises the possibility that Cby is exported from the nucleus via the CRM1-mediated pathway. To test this hypothesis, we used leptomycin B (LMB), a potent inhibitor of CRM1-dependent nuclear export (Kumagai and Dunphy, 1999; Brunet et al., 2002), to examine whether it can block nuclear export of Cby. As we predicted, LMB treatment of Cos7 cells expressing Flag-CbyWT led to a significant redistribution of Cby to an almost exclusive nuclear localization (Figure 4A). Similar results were obtained for endogenous Cby in MEFs (Figure 4B), and for exogenous Cby expressed in HEK293T cells (Supplemental Figure S2). Cby, therefore, continuously shuttles between the nucleus and cytoplasm, and transits out of the nucleus in a CRM1-dependent manner.
Figure 4.
CRM1 mediates nuclear export of Cby. (A) LMB treatment causes nuclear accumulation of Cby. Cos7 cells were transfected with a Flag-Cby expression vector, treated with methanol (−LMB) or 40 nM LMB (+LMB) for 5 h, and immunostained with anti-Flag antibody for Cby. Nuclei were counterstained with DAPI. (B) Enrichment of endogenous Cby protein in the nucleus upon LMB treatment. MEFs were treated with methanol (−LMB) or 100 nM LMB (+LMB) for 5 h, and immunostained with anti-Cby antibody. White arrowheads point to nuclear Cby. Nuclei were visualized by DAPI. (C) Cby physically interacts with CRM1. Cell lysates from HEK293T transfected with the indicated combinations of expression plasmids for Flag-Cby and CRM1 were incubated in the presence or absence of recombinant His-RanQ69L-GTP and immunoprecipitated using rabbit anti-CRM1 antibody or control rabbit IgG, followed by Western blot analysis with anti-Flag antibody. (D) CRM1 binds to the NES1 motif in the N-terminal region of Cby. HEK293T cells were transfected with Flag-tagged CbyWT or individual NES variants, and cell lysates were immunoprecipitated with anti-CRM1 antibody and immunoblotted with anti-Flag antibody. Note that, to compensate for protein levels, the amounts of DNA for CbyL21A, CbyL24A, CbyL27A, and CbyL21A/L24A/L27A were appropriately increased for transfection, and hence roughly similar expression levels were observed for all the Cby mutants (bottom panel). (E) Forced expression of CRM1 triggers cytoplasmic relocation of CbyWT but not that of the triple NES1 mutant CbyL21A/L24A/L27A. Cos7 cells were cotransfected with an expression plasmid for Flag-tagged CbyWT or CbyL21A/L24A/L27A and a control empty or CRM1 vector, followed by immunostaining with anti-Flag antibody.
CRM1 mediates nuclear export of numerous cellular proteins by directly binding to their NES (Macara, 2001; Pemberton and Paschal, 2005). Hence, we asked if Cby forms a complex with CRM1 using coimmunoprecipitation (CoIP) experiments. Cell lysates from HEK293T cells expressing Flag-CbyWT alone or in combination with CRM1 were immunoprecipitated with control rabbit IgG or rabbit anti-CRM1 antibody, and immunoblotted with anti-Flag antibody. As shown in Figure 4C, Cby specifically interacted with endogenous CRM1 (cf. lane 2 with lanes 1 and 6). Endogenous CRM1 appears to be abundant as exogenous expression of CRM1 did not significantly increase the amount of Cby coprecipitating with CRM1 (lane 3). Ran-GTP binds to CRM1 and stabilizes a CRM1-NES complex (Fornerod et al., 1997; Bogerd et al., 1998). In accordance with this, the RanQ69L-GTP mutant defective in GTP hydrolysis (Hetzer et al., 2000) slightly, but reproducibly, augmented the Cby-CRM1 interaction (lane 4).
To determine whether the CRM1-Cby interaction occurs through the NESs of Cby, we tested binding of CRM1 to various Cby NES mutants using CoIPs (Figure 4D). The three Cby NES1 mutants predominantly localized to the nucleus (L21A, L24A, and L21A/L24A/L27A; Figure 3) failed to associate with CRM1 (lanes 3, 4, and 6). On the contrary, CbyL27A, both Cby NES2 mutants (L93A/I97A and L90A/L93A/I97A/L99A) and the control non-NES variant (L36A/L38A), showing normal subcellular distribution, effectively interacted with CRM1 (lanes 5 and 7–9). Curiously, CbyS20A incapable of 14-3-3 binding (Li et al., 2008) was unable to bind to CRM1 (lane 10). Consistent with the CoIP results, forced expression of CRM1 sequestered CbyWT into the cytoplasm, while having no or little effect on the predominant nuclear localization of the triple NES1 mutant CbyL21A/L24A/L27A (Figure 4E). Taken together, these findings argue that the NES1 motif mediates Cby nuclear export through its direct association with CRM1.
The Efficient Nuclear Export of Cby Requires a Cooperative Action of CRM1 and 14-3-3
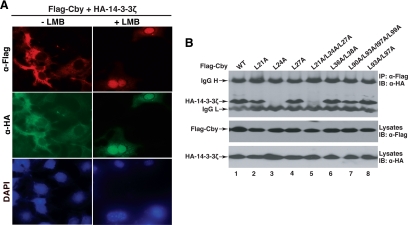
Consistent with our previous observations (Li et al., 2008), when coexpressed with 14-3-3, Cby is primarily sequestered in the cytoplasm where it colocalizes with 14-3-3 (Figure 5A). Notably, LMB treatment resulted in nuclear accumulation of both Cby and 14-3-3 proteins. This suggests that Cby constantly shuttles between the nucleus and cytoplasm even in the presence of excess 14-3-3 and that the primary role of 14-3-3 is to shift the equilibrium of Cby localization toward the cytoplasm rather than causing cytoplasmic retention of Cby.
Figure 5.
14-3-3 binding alone is not sufficient for nuclear export of Cby. (A) LMB treatment induces nuclear accumulation of Cby and 14-3-3. Cos7 cells were cotransfected with Flag-Cby and HA-14-3-3ζ, treated with LMB as described in the legend to Figure 4A, and doubly immunostained with anti-Flag and anti-HA antibodies. Nuclei were stained with DAPI. (B) Interactions between Cby NES mutants and 14-3-3. Cell lysates from HEK293T cells expressing Flag-tagged CbyWT or individual NES mutants and HA-14-3-3ζ were immunoprecipitated with anti-Flag antibody and detected with anti-HA antibody. Note that CbyL21A fails to bind to CRM1 (Figure 4D, lane 3) but retains the ability to interact with 14-3-3, and yet predominantly localizes to the nucleus (Figure 3, B and C). IgG H and IgG L denote IgG heavy and light chains, respectively.
To begin to understand the molecular basis underlying the steady-state cytoplasmic compartmentalization of Cby by 14-3-3 proteins, we first assessed binding of various Cby NES mutants to 14-3-3 using CoIP assays. For this purpose, HEK293T cells were transiently cotransfected with expression vectors for HA-14-3-3ζ and individual Cby NES mutants, and cell lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-HA antibody. As shown in Figure 5B, Cby mutants carrying a leucine-to-alanine substitution at position 24 (L24A and L21A/L24A/L27A) completely lost their ability to interact with 14-3-3ζ (lanes 3 and 5), demonstrating that leucine 24, which lies just C-terminal to the 14-3-3–binding motif, strongly influences 14-3-3 binding. This is consistent with a prior report that mutations in amino acids C-terminal to the 14-3-3–binding motif in Cdc25 severely impair 14-3-3 binding (Uchida et al., 2006). The other Cby NES mutants efficiently bound to 14-3-3ζ. Among them, CbyL21A is of particular interest as it fails to bind to CRM1 (Figure 4D), thereby accumulating in the nucleus (Figure 3), despite its ability to form a stable complex with 14-3-3. These data indicate that 14-3-3 binding alone is not sufficient to promote Cby nuclear exclusion and that binding of both CRM1 and 14-3-3 ensures efficient nuclear exit of Cby.
14-3-3 Establishes the Steady-State Cytoplasmic Localization of Cby by Enhancing the Interaction of Cby with CRM1 and Inhibiting Cby Binding to Importin-α
Once bound to 14-3-3 proteins, Cby is predominantly localized to the cytoplasm at steady state (Figure 5A; Li et al., 2008). How is this dynamic equilibrium of Cby distribution accomplished? Interestingly, we noted that the functional NES1 (21LSNLHSLDR29) partially overlaps with the adjacent 14-3-3–binding motif (16RKSASLS22) in the N-terminal region of Cby protein. Additionally, the 14-3-3-binding–deficient mutant CbyS20A is unable to interact with CRM1 (Figure 4D, lane 10). These lines of circumstantial evidence prompted us to hypothesize that 14-3-3 docking might provoke a conformational change of Cby, allowing high-affinity recognition of the NES1 motif by CRM1. To test this possibility, we coexpressed a constant amount of Flag-CbyWT and increasing amounts of HA-14-3-3ζ in HEK293T cells, and cell lysates were immunoprecipitated with anti-CRM1 antibody, followed by Western blotting with anti-Flag antibody. As shown in Figure 6A, expression of 14-3-3ζ enhanced the interaction between Cby and CRM1 in a dose-dependent manner.
Previous studies have shown that 14-3-3 proteins eliminate binding of Cdc25 and HDAC4 to importin-α, thereby blocking their nuclear import (Kumagai and Dunphy, 1999; Yang et al., 1999; Grozinger and Schreiber, 2000). Hence, we examined whether 14-3-3 binding affects the Cby-importin-α interaction using in vitro binding assays (Figure 6B). Bacterially produced GST or GST-importin-α3 was incubated with cell lysates from HEK293T cells expressing Flag-tagged CbyWT or 14-3-3-binding–defective CbyS20A and from those expressing HA-14-3-3ζ, and pulled down using glutathione beads. The bound protein complexes were then resolved by SDS-PAGE and subjected to Western blotting with anti-Flag antibody. Increasing amounts of 14-3-3ζ effectively interfered with CbyWT binding to importin-α, while having essentially no effect on the CbyS20A-importin-α interaction. These observations suggest that 14-3-3 proteins facilitate nuclear export of Cby while inhibiting its nuclear import, resulting in cytoplasmic accumulation of Cby in a steady-state equilibrium.
Nuclear-Cytoplasmic Shuttling of Cby Influences Subcellular Distribution and Signaling Activity of β-Catenin
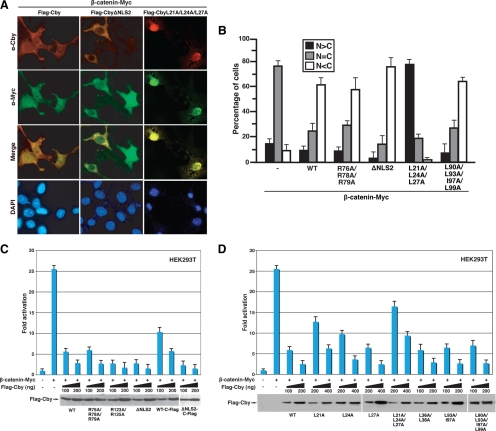
Cby and 14-3-3 form a stable tripartite complex with β-catenin, causing β-catenin to partition into the cytoplasm, thereby antagonizing β-catenin signaling (Li et al., 2008). To elucidate if the nuclear-cytoplasmic shuttling of Cby modulates β-catenin subcellular distribution, we examined β-catenin localization after coexpression with the cytoplasmic CbyΔNLS2 or nuclear CbyL21A/L24A/L27A mutant. On ectopic expression in mammalian cultured cells, a stabilized form of β-catenin (β-catenin-Myc) is typically distributed throughout the nucleus and cytoplasm (Figure 7B; Li et al., 2008). Forced expression of CbyWT causes a dramatic shift in β-catenin localization toward the cytoplasm (Figure 7, A and B) by cooperating with endogenous 14-3-3 (Li et al., 2008). Strikingly, expression of cytoplasmic CbyΔNLS2 resulted in sequestration of β-catenin mainly in the cytoplasm, whereas that of nuclear CbyL21A/L24A/L27A trapped β-catenin in the nucleus (Figure 7, A and B). As anticipated, the triple NLS1 mutant (R76A/R78A/R79A) and the quadruple NES2 mutant (L90A/L93A/I97A/L99A) produced an effect similar to that of CbyWT. These results support the notion that Cby shuttling controls the intracellular distribution of β-catenin.
Figure 7.
Cby shuttling controls β-catenin localization and function. (A) CbyΔNLS2 sequesters β-catenin in the cytoplasm, whereas CbyL21A/L24A/L27A traps β-catenin in the nucleus. Cos7 cells were cotransfected with stabilized β-catenin-Myc and Flag-tagged CbyWT, CbyΔNLS2, or CbyL21A/L24A/L27A, and double-stained with anti-Cby (red) and anti-Myc (green) antibodies. Nuclei were visualized with DAPI. (B) Quantitative analysis of the results in A. The subcellular distribution of β-catenin-Myc was scored as described in the legend to Figure 1C. Error bars, means ± SD of three independent experiments. (C and D) Effects of Cby NLS mutants (C) or Cby NES mutants (D) on β-catenin–mediated transcriptional activation were evaluated by TopFlash assays. HEK293T cells were transfected with 10 ng of TopFlash luciferase reporter with or without 10 ng of an expression vector for stabilized β-catenin (β-catenin-Myc) and the indicated amounts of a Flag-tagged Cby expression plasmid. Luciferase activity was measured 24 h after transfection and normalized to Renilla luciferase activity used as an internal control. Transfections were carried out in triplicate, and the means ± SD are shown. The basal TopFlash value was set as 1. Western blot analysis with anti-Flag antibody showed that Cby proteins were expressed at similar levels (bottom panels). Note that, to compensate protein levels, higher amounts of DNA for CbyL21A, CbyL24A, CbyL27A, and CbyL21A/L24A/L27A were used for transfection.
Next, we sought to address whether the nuclear-cytoplasmic shuttling of Cby influences β-catenin signaling activity using Tcf/Lef luciferase reporter (TopFlash) assays (Korinek et al., 1997; Takemaru et al., 2003). First, we evaluated the effects of Cby NLS mutations on β-catenin signaling (Figure 7C). Transfection of stabilized β-catenin-Myc stimulated TopFlash activity 26-fold, and CbyWT potently repressed it in a dose-dependent manner. As expected, the triple NLS1 mutant (R76A/R78A/R79A) behaved similarly to CbyWT. Intriguingly, Cby NLS2 mutants defective in nuclear entry (R123A/R125A and ΔNLS2) inhibited β-catenin–mediated TopFlash activation slightly more potently than CbyWT by trapping β-catenin in the cytoplasmic compartment (Figure 7, A and B). In contrast, exclusively nuclear C-terminally Flag-tagged CbyWT (CbyWT-C-Flag) displayed a reduced ability to inhibit TopFlash activity. CbyΔNLS2-C-Flag, however, repressed β-catenin signaling as efficiently as CbyΔNLS2.
We also assessed the ability of Cby NES mutants to inhibit β-catenin signaling using TopFlash assays. We found that Cby NES1 mutants incapable of nuclear export (L21A, L24A. and L21A/L24A/L27A) reproducibly manifested a reduced ability to repress β-catenin signaling (Figure 7D), presumably due to their failure to export nuclear β-catenin into the cytoplasm (Figure 7, A and B). Conversely, the other Cby mutants showing no overt localization defects (L27A, L36A/L38A, L93A/I97A, and L90A/L93A/I97A/L99A) suppressed TopFlash activation by β-catenin to an extent similar to CbyWT. These data concur with our dual mechanism model in which Cby antagonizes β-catenin signaling in both the nucleus and cytoplasm (Li et al., 2008), and also imply that cytoplasmic Cby confers a more potent repressive activity than nuclear Cby in regulating β-catenin signaling.
Knockdown of Endogenous Cby Results in Nuclear Accumulation of β-Catenin
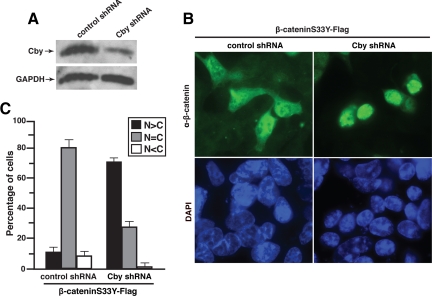
To investigate whether endogenous Cby modulates β-catenin localization, we generated HEK293 cell lines stably expressing shRNA against Cby or negative control scrambled shRNA. As shown in Figure 8A, the levels of endogenous Cby protein were effectively reduced by Cby shRNA expression. To determine the effect of Cby knockdown on β-catenin localization, we transiently expressed stabilized β-catenin (β-cateninS33Y-Flag) in Cby and control shRNA-expressing cells, and evaluated its subcellular distribution by fluorescence microscopy. Intriguingly, β-catenin predominantly localized to the nucleus in Cby-knockdown cells as opposed to its diffuse nuclear and cytoplasmic distribution in control cells (Figure 8B). Quantification of β-catenin localization revealed that ∼10% of control cells displayed nuclear immunofluorescence, whereas roughly 70% of Cby-knockdown cells exhibited clear nuclear staining (Figure 8C). These results provide further evidence to support our model in which Cby controls nuclear export of β-catenin.
Figure 8.
Cby knockdown leads to nuclear accumulation of β-catenin. (A) Reduction of endogenous Cby protein levels by shRNA. Total cell lysates from HEK293 stable cell lines expressing control scrambled shRNA or Cby shRNA were immunoblotted with antibodies against Cby or GAPDH (loading control) as indicated. (B) β-Catenin accumulates in the nucleus in Cby-knockdown cells. Control or Cby shRNA-expressing HEK293 cells were transiently transfected with stabilized β-cateninS33Y-Flag and stained with anti-β-catenin antibody 17 h after transfection. Nuclei were labeled with DAPI. Similar results were obtained using anti-Flag antibody. (C) Quantitative analysis of the results in B. The subcellular distribution of β-catenin was scored as described in the legend to Figure 1C. Error bars, means ± SD of three independent experiments.
DISCUSSION
The Wnt/β-catenin signaling pathway has been implicated in numerous aspects of development and disease. Wnt components are often mutated or otherwise dysregulated in a variety of human cancers. In particular, greater than 70% of colon cancer patients are known to harbor activating mutations in the Wnt/β-catenin pathway (Polakis, 2000; Lustig and Behrens, 2003). These mutations ultimately lead to stabilization and nuclear migration of the key coactivator β-catenin, resulting in changes in gene expression that initiates tumor formation. Accordingly, the nuclear and cytoplasmic trafficking of β-catenin is of considerable clinical importance and can be exploited as a therapeutic target for cancers and other diseases associated with altered Wnt signaling, but remains poorly defined.
In this study, we have shown that Cby possesses the intrinsic NLS (123RKRK126) and NES (21LSNLHSLDR29) motifs that physically interact with importin-α and CRM1, respectively. Cby NLS2 mutants fail to bind to importin-α and thus localize to the cytoplasm, whereas Cby NES1 mutants are incapable of binding to CRM1 and accumulate in the nucleus. In support of these findings, Cby constitutively travels between the nucleus and cytoplasm.
14-3-3 family members control the subcellular localization of their target proteins through several distinct molecular mechanisms (Muslin and Xing, 2000; Morrison, 2009). 14-3-3 itself, however, contains neither NLS nor NES (Kumagai and Dunphy, 1999; Brunet et al., 2002). Thus, final destinations of 14-3-3-ligand complexes entirely depend on the NLS and/or NES sequences that are intrinsic to their ligands. A series of elegant studies have demonstrated that 14-3-3 binding causes cytoplasmic enrichment of the mitotic activator Cdc25 phosphatase by masking a nearby NLS and blocking the interaction of Cdc25 with importin-α (Kumagai and Dunphy, 1999; Yang et al., 1999). However, not all 14-3-3–bound cargoes are destined for cytoplasmic sequestration. For instance, 14-3-3 has been shown to promote nuclear accumulation of myopodin by facilitating importin-α binding to myopodin NLSs that are, in this case, positioned distantly from its 14-3-3–binding sites (Faul et al., 2005). Furthermore, interactions between CRM1 and NES-containing proteins can also be influenced by 14-3-3 (Seimiya et al., 2000). In the case of Cby, we found that 14-3-3 enhances the Cby-CRM1 interaction while interfering with the Cby-importin-α interaction, thereby achieving the cytoplasmic sequestration of Cby at a steady-state equilibrium. It is noteworthy that Cby enters the nucleus even in the presence of excess 14-3-3 (Figure 5A). This clearly suggests that 14-3-3 binding induces a shift in the distribution equilibrium of Cby toward the cytoplasm rather than retaining Cby in the cytoplasmic compartment. Our model for the regulatory action of 14-3-3 on Cby closely parallels the one proposed for Cdc25 as it is also capable of migrating into the nucleus even when 14-3-3 is overexpressed (Kumagai and Dunphy, 1999).
Curiously, Cby NES1 (21LSNLHSLDR29) is juxtaposed to its 14-3-3–binding motif (16RKSASLS22) in a partially overlapping manner. Thus, it is tempting to speculate that 14-3-3 docking triggers a conformational change in Cby that exposes the NES1 motif to permit CRM1 to recognize and mediate nuclear export. Indeed, overexpression of 14-3-3 facilitates Cby binding to CRM1 (Figure 6A). In further support of this idea, CRM1 fails to interact with 14-3-3-binding–defective CbyS20A (Figure 4D, lane 10). Moreover, 14-3-3 binding alone is not sufficient to transport Cby out of the nucleus (Figures 3 and 5B). These results therefore indicate that Cby's ability to interact with both 14-3-3 and CRM1 is necessary for efficient Cby nuclear exit. This is reminiscent of the nuclear export model proposed for the Forkhead transcription factor FoxO1 in which both intrinsic NES sequences and 14-3-3 binding ensure its rapid nuclear export (Brunet et al., 2002).
Once Cby has been exported out of the nucleus, Cby may remain associated with 14-3-3 proteins in the cytoplasm as 14-3-3 inhibits binding of Cby to importin-α (Figure 6B). The fact that Cby can reenter the nucleus even in the presence of excess 14-3-3 (Figure 5A) suggests the presence of a cellular phosphatase that dephosphorylates Cby at serine 20 before its binding to importin-α and reimport into the nucleus. Previous work has revealed an intricate mechanism regulating 14-3-3 release from Cdc25 that involves the PP2A/B56δ phosphatase (Margolis et al., 2006).
Cby antagonizes β-catenin signaling through two distinct molecular mechanisms (Li et al., 2008). In the nucleus, Cby competes with Tcf/Lef transcription factors for binding to β-catenin. In addition, Cby associates with 14-3-3 to form a stable trimolecular complex with β-catenin to promote nuclear exclusion of β-catenin. Consistent with this model, we found that the cytoplasmic CbyΔNLS2 mutant sequesters β-catenin in the cytoplasm (Figure 7A). In this context, we postulate that endogenous 14-3-3 is part of the complex because CbyΔNLS2 retains the ability to interact with 14-3-3 proteins (Supplemental Figure S3). On the contrary, the nuclear triple NES1 mutant (L21A/L24A/L27A) traps β-catenin almost exclusively in the nucleus (Figure 7A) as previously observed with 14-3-3-binding-deficient CbyS20A (Li et al., 2008). Both cytoplasmic Cby NLS2 mutants and nuclear Cby NES1 mutants are capable of blocking β-catenin–dependent transcriptional activation (Figure 7, C and D). Cytoplasmic Cby appears to be more potent than nuclear Cby in repressing β-catenin signaling. One potential explanation is that in the nucleus, Tcf/Lef factors might gain access to β-catenin freed from nuclear Cby as they compete with Cby for β-catenin binding. On the other hand, cytoplasmic Cby sequesters β-catenin in the cytoplasmic compartment away from Tcf/Lef transcription factors, eliciting the more potent repressive effect. Likewise, nuclear and cytoplasmic APC differentially regulate β-catenin signaling activity (Neufeld et al., 2000). These observations underscore the crucial role of the localization signals within Cby in controlling the subcellular localization and signaling activity of β-catenin. In further support of our model, knockdown of endogenous Cby by shRNA causes nuclear accumulation of β-catenin (Figure 8).
Besides the Cby-14-3-3 pathway, the negative regulators of β-catenin, APC and Axin, have been shown to shuttle between the nucleus and cytoplasm, and facilitate nuclear export of β-catenin in a CRM1-dependent manner (Henderson, 2000; Rosin-Arbesfeld et al., 2000; Cong and Varmus, 2004; Wiechens et al., 2004). The APC- and Axin-directed pathways may be coupled with proteasomal degradation of β-catenin. At present, the exact fate of Cby-14-3-3–bound β-catenin is unknown. Interestingly, ectopic expression of 14-3-3ζ was found to increase the steady-state level of β-catenin in the cytoplasm without overtly affecting nuclear β-catenin levels (Tian et al., 2004). Thus, it is possible that the trimeric Cby-14-3-3–β-catenin complex remains stable in the cytoplasm. This may serve as a reservoir of signaling competent β-catenin that is readily available for release in response to upstream signals. Clearly, further experiments will be required to delineate the final outcome of the Cby-14-3-3 nuclear export pathway.
In summary, we have unambiguously identified functionally active NLS and NES motifs in Cby. A balance between these subcellular trafficking signals, in cooperation with 14-3-3, regulates β-catenin intracellular distribution and signaling activity. Altogether, these data provide novel insights into the molecular mechanisms by which Cby and 14-3-3 proteins control the dynamic nuclear-cytoplasmic trafficking of β-catenin.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. N. Reich, J. A. Diehl and M. Dasso for reagents. This work was supported by an American Diabetes Association Junior Faculty Award (107JF42) and National Institutes of Health, NICHD-ARRA Grant HD06020401 to F.-Q. L. and NIH/NIDDK Grant R01 DK073191 to K.-I.T.
Abbreviations used:
- Cby
Chibby
- CoIP
coimmunoprecipitation
- CRM1
chromosomal region maintenance 1
- GST
glutathione S-transferase
- HEK
human embryonic kidney
- Lef
lymphoid enhancer factor
- LMB
leptomycin B
- MBP
maltose-binding protein
- NES
nuclear export signal
- NLS
nuclear localization signal
- shRNA
short hairpin RNA
- Tcf
T-cell factor
- WT
wild type.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-05-0437) on November 25, 2009.
REFERENCES
- Aitken A. 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Barker N., Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat. Rev. Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- Benzeno S., Diehl J. A. C-terminal sequences direct cyclin D1-CRM1 binding. J. Biol. Chem. 2004;279:56061–56066. doi: 10.1074/jbc.M411910200. [DOI] [PubMed] [Google Scholar]
- Bogerd H. P., Echarri A., Ross T. M., Cullen B. R. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J. Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A., Kanai F., Stehn J., Xu J., Sarbassova D., Frangioni J. V., Dalal S. N., DeCaprio J. A., Greenberg M. E., Yaffe M. B. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 2002;156:817–828. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan K. M., Liu Y. I. Wnt signaling: complexity at the surface. J. Cell Sci. 2006;119:395–402. doi: 10.1242/jcs.02826. [DOI] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Cong F., Varmus H. Nuclear-cytoplasmic shuttling of Axin regulates subcellular localization of beta-catenin. Proc. Natl. Acad. Sci. USA. 2004;101:2882–2887. doi: 10.1073/pnas.0307344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt N. A. Transcription factors dial 14-3-3 for nuclear shuttle. Plant Cell. 2001;13:2385–2389. doi: 10.1105/tpc.13.11.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faul C., Huttelmaier S., Oh J., Hachet V., Singer R. H., Mundel P. Promotion of importin alpha-mediated nuclear import by the phosphorylation-dependent binding of cargo protein to 14-3-3. J. Cell Biol. 2005;169:415–424. doi: 10.1083/jcb.200411169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I. W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Grozinger C. M., Schreiber S. L. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B. R. Nuclear-cytoplasmic shuttling of APC regulates beta-catenin subcellular localization and turnover. Nat. Cell Biol. 2000;2:653–660. doi: 10.1038/35023605. [DOI] [PubMed] [Google Scholar]
- Hetzer M., Bilbao-Cortes D., Walther T. C., Gruss O. J., Mattaj I. W. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell. 2000;5:1013–1024. doi: 10.1016/s1097-2765(00)80266-x. [DOI] [PubMed] [Google Scholar]
- Huang H., He X. Wnt/beta-catenin signaling: new (and old) players and new insights. Curr. Opin. Cell Biol. 2008;20:119–125. doi: 10.1016/j.ceb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S., Kehlenbach R. H. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 2007;17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Jin J., et al. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr. Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Kimelman D., Xu W. beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene. 2006;25:7482–7491. doi: 10.1038/sj.onc.1210055. [DOI] [PubMed] [Google Scholar]
- Klaus A., Birchmeier W. Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 1999;6:388–397. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- Kohler M., Speck C., Christiansen M., Bischoff F. R., Prehn S., Haller H., Gorlich D., Hartmann E. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W. G. Binding of 14-3-3 proteins and nuclear export control the intracellular localization of the mitotic inducer Cdc25. Genes Dev. 1999;13:1067–1072. doi: 10.1101/gad.13.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Guttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- la Cour T., Kiemer L., Molgaard A., Gupta R., Skriver K., Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Design Select. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Q., Mofunanya A., Harris K., Takemaru K. Chibby cooperates with 14-3-3 to regulate beta-catenin subcellular distribution and signaling activity. J. Cell Biol. 2008;181:1141–1154. doi: 10.1083/jcb.200709091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Q., Singh A. M., Mofunanya A., Love D., Terada N., Moon R. T., Takemaru K. Chibby promotes adipocyte differentiation through inhibition of beta-catenin signaling. Mol. Cell. Biol. 2007;27:4347–4354. doi: 10.1128/MCB.01640-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., McBride K. M., Reich N. C. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc. Natl. Acad. Sci. USA. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig B., Behrens J. The Wnt signaling pathway and its role in tumor development. J. Cancer Res. Clin. Oncol. 2003;129:199–221. doi: 10.1007/s00432-003-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara I. G. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald B. T., Semenov M. V., He X. SnapShot: Wnt/beta-catenin signaling. Cell. 2007;131:1204. doi: 10.1016/j.cell.2007.11.036. [DOI] [PubMed] [Google Scholar]
- Margolis S. S., et al. Role for the PP2A/B56delta phosphatase in regulating 14-3-3 release from Cdc25 to control mitosis. Cell. 2006;127:759–773. doi: 10.1016/j.cell.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek S. E., Lane W. S., Piwnica-Worms H. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J. Biol. Chem. 2004;279:32046–32054. doi: 10.1074/jbc.M403044200. [DOI] [PubMed] [Google Scholar]
- Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat. Rev. Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- Moroianu J., Blobel G., Radu A. The binding site of karyopherin alpha for karyopherin beta overlaps with a nuclear localization sequence. Proc. Natl. Acad. Sci. USA. 1996;93:6572–6576. doi: 10.1073/pnas.93.13.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. K. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19:16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- Muslin A. J., Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- Nagoshi E., Yoneda Y. Dimerization of sterol regulatory element-binding protein 2 via the helix-loop-helix-leucine zipper domain is a prerequisite for its nuclear localization mediated by importin beta. Mol. Cell. Biol. 2001;21:2779–2789. doi: 10.1128/MCB.21.8.2779-2789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld K. L., Zhang F., Cullen B. R., White R. L. APC-mediated downregulation of beta-catenin activity involves nuclear sequestration and nuclear export. EMBO Rep. 2000;1:519–523. doi: 10.1093/embo-reports/kvd117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeri D., Malim M. H. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol. Cell. Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L. F., Paschal B. M. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Pinto D., Clevers H. Wnt control of stem cells and differentiation in the intestinal epithelium. Exp. Cell Res. 2005;306:357–363. doi: 10.1016/j.yexcr.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Rosin-Arbesfeld R., Townsley F., Bienz M. The APC tumour suppressor has a nuclear export function. Nature. 2000;406:1009–1012. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- Seimiya H., Sawada H., Muramatsu Y., Shimizu M., Ohko K., Yamane K., Tsuruo T. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 2000;19:2652–2661. doi: 10.1093/emboj/19.11.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadeli R., Hoffmans R., Basler K. Transcription under the control of nuclear Arm/beta-catenin. Curr. Biol. 2006;16:R378–R385. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Takemaru K., Fischer V., Li F. Q. Fine-tuning of nuclear-catenin by Chibby and 14-3-3. Cell Cycle. 2009;8:210–213. doi: 10.4161/cc.8.2.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru K., Ohmitsu M., Li F. Q. An oncogenic hub: beta-catenin as a molecular target for cancer therapeutics. Handb. Exp. Pharmacol. 2008;186:261–284. doi: 10.1007/978-3-540-72843-6_11. [DOI] [PubMed] [Google Scholar]
- Takemaru K., Yamaguchi S., Lee Y. S., Zhang Y., Carthew R. W., Moon R. T. Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway. Nature. 2003;422:905–909. doi: 10.1038/nature01570. [DOI] [PubMed] [Google Scholar]
- 53.Takemaru K.-I. Catenin, beta. UCSD-Nature Molecule Pages. 2006 doi: 10.1038/mp.a000506.01. [Google Scholar]
- Tian Q., Feetham M. C., Tao W. A., He X. C., Li L., Aebersold R., Hood L. Proteomic analysis identifies that 14-3-3zeta interacts with beta-catenin and facilitates its activation by Akt. Proc. Natl. Acad. Sci. USA. 2004;101:15370–15375. doi: 10.1073/pnas.0406499101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant R., Cullen B. R. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol. Cell. Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S., Kubo A., Kizu R., Nakagama H., Matsunaga T., Ishizaka Y., Yamashita K. Amino acids C-terminal to the 14-3-3 binding motif in CDC25B affect the efficiency of 14-3-3 binding. J. Biochem. 2006;139:761–769. doi: 10.1093/jb/mvj079. [DOI] [PubMed] [Google Scholar]
- Wiechens N., Heinle K., Englmeier L., Schohl A., Fagotto F. Nucleo-cytoplasmic shuttling of Axin, a negative regulator of the Wnt-beta-catenin Pathway. J. Biol. Chem. 2004;279:5263–5267. doi: 10.1074/jbc.M307253200. [DOI] [PubMed] [Google Scholar]
- Willert K., Jones K. A. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- Wodarz A., Nusse R. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 1998;14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., Cantley L. C. The structural basis for 14-3-3, phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Yang J., Winkler K., Yoshida M., Kornbluth S. Maintenance of G2 arrest in the Xenopus oocyte: a role for 14-3-3-mediated inhibition of Cdc25 nuclear import. EMBO J. 1999;18:2174–2183. doi: 10.1093/emboj/18.8.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.