Abstract
Changes in telomere chromatin have been linked to cellular senescence, but the underlying mechanisms and impact on lifespan are unclear. We found that inactivation of the Sas2 histone acetyltransferase delays senescence in Saccharomyces cerevisiae telomerase (tlc1) mutants through a homologous recombination-dependent mechanism. Sas2 acetylates histone H4 lysine 16 (H4K16), and telomere shortening in tlc1 mutants was accompanied by a selective and Sas2-dependent increase in subtelomeric H4K16 acetylation. Further, mutation of H4 lysine 16 to arginine, which mimics constitutively deacetylated H4K16, delayed senescence and was epistatic to sas2 deletion, indicating that deacetylated H4K16 mediates the delay caused by sas2 deletion. Sas2 normally prevents the Sir2/3/4 heterochromatin complex from leaving the telomere and spreading to internal euchromatic loci. Senescence was delayed by sir3 deletion, but not sir2 deletion, indicating that senescence delay is mediated by release of Sir3 specifically from the telomere repeats. In contrast, sir4 deletion sped senescence and blocked the delay conferred by sas2 or sir3 deletion. We thus show that manipulation of telomere chromatin modulates senescence caused by telomere shortening.
Keywords: chromatin, SAS2, senescence, Sir complex, telomere
Introduction
Ageing in humans is accompanied by the shortening of telomeres, the nucleoprotein complexes that cap the ends of chromosomes, and several lines of evidence indicate that such shortening contributes to age-related pathology (Aubert and Lansdorp, 2008). Telomeres are generally maintained in a heterochromatic state, which in mammals is mediated by the posttranslational modification of histones and by DNA methylation (Baur et al, 2001; Garcia-Cao et al, 2004; Gonzalo et al, 2006; Blasco, 2007). In Saccharomyces cerevisiae, telomere heterochromatin is mediated principally by the silent information regulator (Sir) complex, composed of Sir2, 3 and 4 (Grunstein, 1997). Yeast telomeres are composed of irregular DNA repeats having the consensus 5′-[(TG)0–6TGGGTGTG(G)]n-3′ and are non-nucleosomal. The repeats are instead bound by Rap1, to which Sir3 and Sir4 can bind independently or cooperatively (Wright et al, 1992; Moretti and Shore, 2001; Luo et al, 2002; Liaw and Lustig, 2006; Feeser and Wolberger, 2008). Sir4, which is also bound to Ku at the telomere terminus, recruits Sir2 to the complex, but Sir2 is not required for the binding of Sir3 or Sir4 to Rap1 (Hoppe et al, 2002; Luo et al, 2002; Roy et al, 2004). The histone deacetylase activity of Sir2 targets lysine 16 within the N-terminal histone H4 tails (H4K16) of nearby subtelomeric nucleosomes, and because the unacetylated H4 tail is a principal binding site for Sir3, Sir2-mediated deacetylation enables propagation of the Sir complex from the telomere repeats to subtelomeric sequences (Imai et al, 2000; Carmen et al, 2002; Hoppe et al, 2002; Luo et al, 2002). In contrast, acetylation of histone H4K16 by the Sas2 acetyltransferase limits this propagation by interfering with the binding of Sir3, thus setting a boundary separating telomeric and subtelomeric heterochromatin from internal euchromatin (Kimura et al, 2002; Suka et al, 2002; Shia et al, 2006; Xu et al, 2006; Altaf et al, 2007; Lafon et al, 2007). Although acetylation of the histone H4 tail opens chromatin and thus generally activates gene expression, Sas2 enforces the silencing of reporter genes inserted at subtelomeric loci or at the silent mating loci HML and HMR (Xu et al, 1999b, 2006; Shogren-Knaak et al, 2006; Zou and Bi, 2008). At least in the case of the telomere, this is because the Sas2-enforced boundary prevents loss of Sir proteins (which are at a limiting cellular concentration, particularly in the case of Sir3 (Wiley and Zakian, 1995; Lustig et al, 1996; Maillet et al, 1996)) from telomeric and subtelomeric sites (Kimura et al, 2002; Suka et al, 2002).
Telomere length is normally maintained by the constitutive expression of the telomerase enzyme in yeast, but tlc1 mutants, lacking the RNA template component of telomerase, lose telomere length with cell division and thus gradually lose replicative potential (Lundblad and Szostak, 1989; Singer and Gottschling, 1994). Extensive shortening causes telomeres to uncap and signal cell cycle checkpoint-mediated arrest, termed senescence. This provides a model to study senescence caused by telomere shortening, and is distinct from other models of ageing in yeast, including those based on the number of daughters budded from each mother cell (‘replicative ageing') or based on the time cells can survive in stationary phase (‘chronological ageing'). Although most cells lacking telomerase senesce, rare survivors escape and maintain their telomeres using homologous recombination (HR)-based break-induced replication mechanisms (Lundblad and Blackburn, 1993; Le et al, 1999; Enomoto et al, 2002; IJpma and Greider, 2003; Lydeard et al, 2007). HR not only enables the growth of survivors, but also helps slow the rate of senescence before survivor formation, apparently by aiding the repair of replication-associated telomere damage (Lundblad and Blackburn, 1993; Le et al, 1999; Azam et al, 2006; Lee et al, 2007; Abdallah et al, 2009). Notably, the HR mechanisms that slow senescence and those that support survivor formation are not identical, with mutations such as the deletion of POL32 (encoding a non-essential subunit of DNA polymerase δ) having no effect on the rate of senescence, but completely blocking the formation of survivors (Lydeard et al, 2007).
Recent evidence indicates an important interplay between telomere length and chromatin state. As telomeres shorten in mice lacking telomerase (mTerc−/− mutants), they lose DNA and histone modifications characteristic of heterochromatin (cytosine methylation and trimethylation of histone H3K9 and histone H4K20) and accumulate marks characteristic of open chromatin (acetylation of H3K9 and H4K20) (Benetti et al, 2007). Deletion of Sirt6, encoding a mammalian Sir2 homologue that targets H3K9, in mice causes both premature age-related pathology and telomere loss events (Michishita et al, 2008). Human senescent cells develop extensive heterochromatic regions on a genome-wide scale, visualized as senescence-associated heterochromatin foci (SAHF), but telomeres seem to be excluded from SAHF indicating possible loss of telomere heterochromatin at senescence (Ye et al, 2007). However, the mechanisms underlying these telomere length-related chromatin changes are unclear, as are the prospects for extending telomere-limited lifespan on the basis of their manipulation.
Here, we provide the first demonstration that senescence driven by telomere shortening can be delayed through modulation of telomere chromatin. Our findings show that disruption of histone acetylation through loss of Sas2 promotes loss of Sir3 from the telomere and thus enables HR-dependent telomere maintenance and delay of senescence.
Results
Loss of the SAS-I complex results in delayed senescence caused by telomere shortening
We used S. cerevisiae cells lacking telomerase to begin to identify chromatin factors modulating the rate of senescence caused by telomere shortening. Diploid cells heterozygous for wild type and deletion alleles of TLC1 and for wild type and deletion alleles of a candidate chromatin regulatory factor were sporulated, and the senescence rates were compared between haploid progeny lacking TLC1 alone or also lacking the chromatin factor. This allowed for controlled comparisons of senescence rates because all mutants had inherited telomeres of similar size and from the same epigenetic environment. Senescence was monitored by plotting the extent of growth after each day of serial reculturing versus population doublings (PDs) from spore germination, as carried out earlier (Johnson et al, 2001; Lee et al, 2007). PD rather than time was used as our metric because telomere shortening caused by the end-replication problem and other replication-related events is related to cell division and not time, and moreover, the use of PD prevents mutations that merely alter the rate of cell division from being mistakenly interpreted as modulating the rate of senescence of tlc1 mutants. As observed earlier, the absolute lifespan of a strain of any given genotype varied somewhat between experiments, perhaps reflecting slightly different growth conditions, the different combinations of heterozygous alleles in the parental diploids or the natural variation in steady-state telomere lengths among different TLC1+ cells. However, this variability does not affect our interpretations because all of our comparisons involved cells descended from the same diploid parent and compared within the same experiment. For these reasons, we emphasize that reliable comparisons of senescence rates cannot be made between experiments.
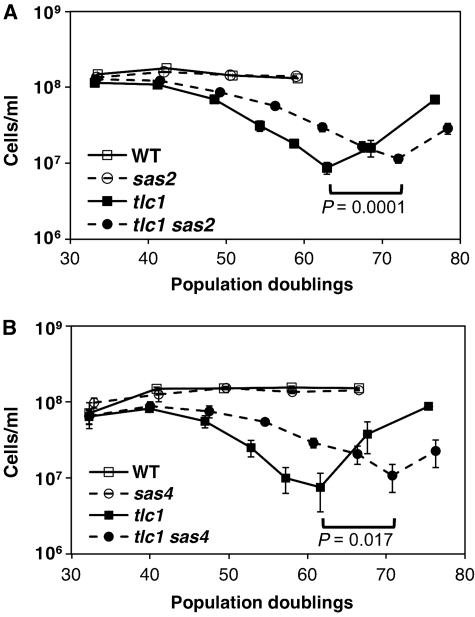
The results of our screen will be published separately (AC, MLK and FBJ, in preparation), but early in the course of screening, we found that although the deletions of 13 of 14 loci investigated either had no effect or were deleterious, deletion of SAS2 delayed senescence by an average of 10 PDs (Figure 1A). As mutations that slow age-related processes identify mechanisms that are rate limiting for longevity more readily than do mutations that speed such processes, we decided to investigate how sas2 deletion slows senescence. Sas2 is a member of the conserved MYST family of histone acetyltransferases and is the catalytic component of the SAS-I complex, comprising Sas2, 4 and 5, which functions principally to acetylate H4K16 (Xu et al, 1999a, 1999b; Shia et al, 2005). Senescence was also delayed in tlc1 sas4 mutants (Figure 1B), indicating that loss of the SAS-I complex explains the slowed senescence. Importantly, the delay did not result from a slowed growth rate because sas2 mutants grow similarly to wild type (Supplementary Figure 1A) and because senescence rates are plotted versus PD instead of time (as an aside, when plotted versus time, the extension is still apparent; Supplementary Figure 1B). Further, the extension did not seem to be caused by the early appearance of survivors because the growth rate of tlc1 sas2 mutants still reached a nadir similar to tlc1 mutants (but at a later PD) and because Southern analysis of telomere length and structure revealed no evidence of early survivors (see Figure 3A; Supplementary Figure 2). Thus, extension corresponded to a shift in both senescence and survivor formation to later PDs.
Figure 1.
Loss of the SAS-I complex delays senescence of tlc1 mutants. (A) Senescence rates were measured in liquid culture by serially passaging spore products of the indicated genotypes derived from a TLC1/tlc1 SAS2/sas2 diploid. The means and s.e.m.s for 14 independent spore products per genotype are shown. (B) Effect of sas4 deletion on replicative senescence was determined as in (A), except that four spore products per genotype were used.
Figure 3.
Deletion of SAS2 does not change the overall rate of telomere attrition or the checkpoint responses to uncapped telomeres. (A) Detailed in Materials and methods, mean telomere lengths and s.e.m.s were calculated from Phosphorimager scans. Fourteen pairs of tlc1 and tlc1 sas2 mutants (each set from one tetrad; samples from Figure 1A) were senesced in liquid culture and samples at the indicated PDs were analysed. Note that senescent tlc1 and tlc1 sas2 mutants were harvested approximately two PDs before lowest point of growth (i.e. maximal senescence) to avoid contribution of survivors to the mean telomere length. The y axis represents the average telomere repeat tract length, calculated by subtracting the approximate length of Y' sequences between the XhoI site and the base of the telomere repeats (900 bp), and the x axis represents the number of PDs at which cells were harvested. (B) Effect of DNA damage agents (MMS and HU) on growth at 30°C was compared by spot assay of the indicated numbers of wild type and sas2 mutant cells. A sensitive strain, rad52, was used as a control. (C) TLC1/tlc1 TEL1/tel1 SAS2/sas2 diploids were sporulated and the senescence of tlc1, tlc1 sas2, tlc1 tel1 and tlc1 sas2 tel1 mutants were compared. (D) The effect of sas2 deletion on telomere uncapping was determined in spot assays of cdc13-1 mutants at permissive (24°C), semi-permissive (28°C) and non-permissive (30°C) temperatures. (E) Rad53 phosphorylation levels were compared in cdc13-1 and cdc13-1 sas2 mutants shifted for the indicated times to the non-permissive temperature (30°C). An HA-tag was fused at the 3′ end of the native RAD53 ORF, and immunoblots were probed with anti-HA antibodies. Tubulin was used as a loading control. A full-colour version of this figure is available at The EMBO Journal Online.
Deletion of SAS2 delays senescence through an HR-dependent pathway
Earlier studies have shown that HR has a critical function in the maintenance of telomeres during senescence as well as in survivors, although the type of HR mechanisms are distinct in the two settings (Lundblad and Blackburn, 1993; Le et al, 1999; Teng and Zakian, 1999; Lee et al, 2007; Lydeard et al, 2007). We, therefore, tested whether extension by sas2 deletion depends on HR by examining the rate of senescence in tlc1 sas2 compared with tlc1 sas2 rad52 or tlc1 sas2 rad51 mutants. Rad52 is required generally for HR and functions in strand annealing and invasion into homologous targets, whereas Rad51 has more restricted functions in HR, principally involving strand invasion. As found earlier, tlc1 rad52 and tlc1 rad51 mutants senesced rapidly, and tlc1 rad52 mutants did not form survivors (Figure 2A and B). The delay of senescence by sas2 deletion was lost in the absence of Rad52 or Rad51 suggesting that loss of Sas2 slows senescence through an HR-based mechanism. Sas2 has a modest function in repressing the silent mating type locus HML (which carries the α mating type allele), and a minor function in repressing HMR (Xu et al, 2006). It was thus conceivable that sas2 mutant MAT a haploids might coexpress levels of a and α genes sufficient to indirectly stimulate an HR-dependent pathway because the co-expression of a and α genes in haploids upregulates HR (Lee et al, 1999). However, the same delay of senescence by sas2 deletion was observed in MAT a and MAT α haploids (not shown) and, moreover, was also observed in a different strain background (JKM111), in which both silent mating loci are deleted (Supplementary Figure 1C). Therefore, such indirect upregulation of HR does not explain delayed senescence by sas2 deletion.
Figure 2.
Delayed senescence through sas2 deletion depends on HR. (A) TLC1/tlc1 RAD52/rad52 SAS2/sas2 diploids were sporulated and the senescence of tlc1, tlc1 sas2, tlc1 rad52 and tlc1 sas2 rad52 mutants were compared. (B) Same as in (A) except that rad51 deletion was tested in place of rad52 deletion. The means and s.e.m.s for five (A) and six (B), respectively, independent spore products per genotype are shown. P-values for the differences between tlc1 and tlc1 sas2 mutants in the absence of Rad52 (A) and Rad51 (B) were 0.844 and 0.302, respectively. (C) Analysis of sequenced telomeres. Chromosome I-L telomeres from tlc1 and tlc1 sas2 mutants matched for PD or extent of senescence were PCR amplified, cloned and sequenced. Telomeres are sorted by length with the non-recombined tracks in black and the recombinant tracts in grey. (D) Summary of the data in (C), showing the percent of telomeres that were recombined and P-values compared with tlc1 cells, calculated using a one-tailed Fisher's exact test. A full-colour version of this figure is available at The EMBO Journal Online.
To test more directly whether sas2 deletion stimulates telomere HR, examples of telomere I-L from tlc1 and tlc1 sas2 mutants were cloned and sequenced. Yeast telomere repeats are imperfect and their sequences differ among telomeres. In a clone of senescing tlc1 cells, the sequence of a shortening telomere is typically fixed, but recombination events can occasionally append new sequences to its terminus (Teixeira et al, 2004). The tlc1 mutants had significantly lower levels of recombinant telomeres than tlc1 sas2 mutants matched either for PD or extent of senescence (Figure 2C and D; Supplementary Figure 3), indicating that sas2 deletion increases telomere recombination.
Telomere attrition rates and checkpoint responses are not affected by sas2 deletion
To evaluate the effect of sas2 deletion on the global rate of telomere attrition, we used Southern analysis to measure telomere lengths during senescence of 14 pairs of tlc1 or tlc1 sas2 mutants (each pair derived from the same tetrad). Bulk telomere lengths shortened at the same rate in tlc1 and PD matched tlc1 sas2 mutants (Figure 3A). However, overall telomere lengths continued to shorten in tlc1 sas2 mutants and reached shorter lengths at senescence than in tlc1 mutants (Figure 3A; Supplementary Figure 2; P=0.035). Therefore, sas2 deletion does not delay senescence by simply slowing the overall rate of telomere shortening, but rather allows cells to continue dividing to the point at which overall telomere lengths become shorter.
The greater extent of telomere shortening in tlc1 sas2 mutants could be explained by a diminished sensitivity of sas2 mutants to telomere uncapping, thus allowing cells to divide longer with uncapped telomeres and continue to shorten their telomeres. Although Sas2 has not been directly implicated in DNA damage response (DDR) pathways, several recent findings indicate that it might have such a function. These include (1) acetylation of H4K16 at short telomeres (see Figure 4B below) and at DNA double strand breaks (DSBs) during repair by HR (Tamburini and Tyler, 2005), (2) a function for hMOF, the human orthologue of Sas2, in ATM activation after ionizing radiation (Gupta et al, 2005) and (3) a function for the histone H3K79 methylase Dot1 (which cooperates with Sas2 to establish the boundary separating telomere heterochromatin from internal euchromatic loci) in activating checkpoint responses at uncapped telomeres (Altaf et al, 2007; Lazzaro et al, 2008). To investigate the contribution of Sas2 to DDR, sas2 mutants were tested for growth inhibition caused by genotoxic agents including hydroxyurea (HU), methane methylsulfonate (MMS), 4-nitroquinoline-1-oxide (4NQO) or UV or γ irradiation. Deletion of SAS2 did not confer increased sensitivity to these agents, indicating that DDRs are not affected globally (Figure 3B; Supplementary Figure 4), consistent with earlier findings (Osada et al, 2001).
Figure 4.
Telomere shortening results in increased SAS2-dependent H4K16 acetylation at subtelomeric regions. (A) Maps of telomeres VI-R and V-L showing probes used to interrogate ChIPed samples; the distances of the probes from the base of the telomere repeats are indicated. (B, C) ChIPs were performed on the following mutants: pre-senescent tlc1 and tlc1 sas2 senescent tlc1, PD-matched tlc1 sas2 or senescent tlc1 sas2; PDs for each sample are indicated in parentheses. Chromatin was precipitated using H4K16Ac, H3K14Ac or H3 antibodies. The y axis represents ratios of H4K16Ac (B) or H3K14Ac (C) levels over total H3 levels. The means and s.e.m.s for three independent quantitative PCR (Q-PCR) experiments are shown, and similar results were obtained in four other experiments using independent biological replicates.
To test the contribution of Sas2 to telomere-specific damage, we first compared its effects during senescence to those of Tel1, a homologue of the mammalian ATM checkpoint kinase. Deletion of TEL1 has been found to slow senescence, and Tel1 is thought to enhance recognition of a critically shortened telomere by the Mec1 kinase, a homologue of mammalian ATR, causing arrest (Ritchie et al, 1999; Abdallah et al, 2009). sas2 and tel1 deletions each slowed senescence of tlc1 cells by about 10 PDs, and their effects were additive, with senescence slowed by ∼20 PDs in tlc1 sas2 tel1 mutants (Figure 3C). Thus, Sas2 and Tel1 most likely affect different processes, consistent with a checkpoint-independent effect of sas2 deletion. As a further test, we used cdc13-1 temperature-sensitive mutants, which at non-permissive temperature (30°C) lose functional Cdc13; normally Cdc13 caps telomeres by protecting the C-rich strand from exonucleolytic attack (Garvik et al, 1995; Lin and Zakian, 1996; Nugent et al, 1996; Zubko et al, 2004). Loss of Sas2 did not affect growth rates of cdc13-1 mutants at permissive and non-permissive temperatures (Figure 3D, 24°C versus 30°C) indicating that Sas2 does not have a significant function in cell cycle arrest after telomere uncapping. Further, Sas2 did not affect the kinetics and extent of Rad53 phosphorylation, a key indicator of checkpoint responses to uncapped telomeres (Jia et al, 2004), after instantaneous telomere uncapping in cdc13-1 mutants shifted to 30°C (Figure 3E; Supplementary Figure 5). However, loss of Sas2 at semi-permissive temperature (28°C) significantly improved cdc13-1 growth (Figure 3D), similar to its contribution to slowed senescence in tlc1 mutants. Importantly, just as in tlc1 mutants, the improved growth that we observed in cdc13-1 mutants was RAD52 dependent. We, therefore, interpret these findings as indicating that Sas2 does not signal the telomere DDR, but under conditions in which cells are at risk for telomere dysfunction and are still replicating their DNA (i.e. tlc1 mutants undergoing senescence or cdc13-1 mutants growing at semi-permissive temperature), sas2 deletion can facilitate HR-dependent telomere maintenance to prevent or repair critical telomere loss events.
Loss of H4K16 acetylation mediates delayed senescence caused by loss of Sas2
The main acetylation target of Sas2 is H4K16 at the boundary between telomeric heterochromatin and internal euchromatin (Kimura et al, 2002; Suka et al, 2002; Shia et al, 2006), but it also weakly acetylates H3K14 and has some functions at other heterochromatic regions, including the silent mating loci and ribosomal DNA (rDNA) (Meijsing and Ehrenhofer-Murray, 2001; Sutton et al, 2003; Oki et al, 2004; Xu et al, 2006). To identify Sas2 targets affected at senescence, we performed chromatin immunoprecipitation (ChIP) on pre-senescent and senescent tlc1 mutants as well as pre-senescent, PD-matched and senescent tlc1 sas2 mutants. Subtelomeric H4K16 acetylation increased in senescent tlc1 mutants at the two different chromosomes examined compared with their pre-senescent counterparts, peaking around 3–10 kb inwards from the telomere repeats (Figure 4A and B). The increased acetylation depended on Sas2 and was not apparent immediately internal (0.1 kb) to the telomere repeats, perhaps because association of the Sir complex with telomere repeats stabilizes adjacent nucleosome-bound Sir complexes. Subtelomeric H3K14 acetylation levels, although also dependent on Sas2, did not change with senescence in tlc1 mutants (Figure 4C). There was no clear trend in H4K16 acetylation within the silent mating loci and rDNA at senescence (Supplementary Figure 6).
To test whether abrogation of the increased H4K16 acetylation explained the senescence delay conferred by sas2 deletion, we examined senescence in cells in which the native H4 locus was replaced by plasmid-based H4 or H4 in which K16 was substituted with arginine (16R), a constitutive mimic of unacetylated lysine (Figure 5A and B). In 16R cells, sas2 deletion no longer slowed senescence, indicating that extension depends on loss of H4K16 acetylation. In contrast, sas2 deletion conferred full delay on cells with an H3K14R allele (Supplementary Figure 7). To determine whether the H4K16R allele is sufficient to confer slowed senescence, a TLC1/tlc1 diploid carrying both 16K and 16R plasmids was sporulated and senescence of tlc1 haploids carrying 16K or 16R was compared. Remarkably, the 16R allele delayed senescence significantly and by about five PD (Figure 5C), albeit not to the same extent as sas2 deletion (cf. Figure 5A). However, two factors may explain the diminished effectiveness of 16R in the experiment. First, 16R is not chemically identical to unacetylated 16K, cannot be interconverted between unacetylated and acetylated states and would replace 16K genome wide instead of only at Sas2-targeted regions; together these might be suboptimal for slowed senescence. Second, the haploids inherit a mixture of 16K and 16R histones, which are gradually replaced by newly synthesized H4 encoded by the plasmid-based allele they carry, and this might diminish the apparent difference between the strains. Overall, these findings support H4K16 as the key target of Sas2 for setting the pace of senescence.
Figure 5.
Loss of acetylation at H4K16 is epistatic to sas2 mutation. (A) sas2 delays senescence in the presence of a plasmid-based wild-type copy of H4K16. TLC1/tlc1 SAS2/sas2 diploids carrying plasmid-based H4K16 (16K) as their only source of histone H4 were sporulated and growth monitored as in Figure 1. (B) sas2 deletion does not slow senescence in tlc1 mutants carrying H4K16R (16R) as their only source of histone H4. (C) Senescence rates of the indicated tlc1 mutants from a TLC1/tlc1 SAS2/sas2 diploid carrying both plasmid-based 16K and 16R H4 alleles were compared.
Deletion of SIR3 delays senescence and is epistatic to sas2 deletion and the H4K16R allele
Given the function of Sas2 in retaining the Sir complex at telomere ends (Kimura et al, 2002; Suka et al, 2002), we wondered whether modulation of Sir complex activities had a function in delayed senescence. Association of the Sir complex with subtelomeric nucleosomes through Sir3 binding to the H4 tail is normally strongly blocked by Sas2-dependent acetylation of H4K16 (Altaf et al, 2007; Onishi et al, 2007). Although Sas2 activity prevents Sir3 binding to nucleosomes, the TG repeats of the telomere are non-nucleosomal and Sir3 instead associates indirectly with the repeats through the telomere repeat-binding protein, Rap1, and through Sir4, which itself binds Rap1 as well as Ku at the telomere terminus (Moretti et al, 1994; Moretti and Shore, 2001; Roy et al, 2004). Further, cellular Sir3 levels are limiting (Wiley and Zakian, 1995; Hecht et al, 1996; Lustig et al, 1996; Maillet et al, 1996). Therefore, sas2 deletion has two relevant consequences. First, the absence of acetylation on H4K16 opens binding sites for Sir3 at internal loci, and second, by doing so it depletes Sir3 from the telomere repeats and subtelomere. Thus, sas2 deletion might delay senescence by enabling Sir3 to occupy beneficial internal chromosomal targets or by relieving a lifespan-inhibitory effect of Sir3 at the telomere or it could even be independent of Sir3. The first, second and third models predict that sir3 deletion should speed, slow or have no effect on the senescence of tlc1 mutants, respectively. We found that sir3 deletion delayed senescence (Figure 6A), supporting the second model that loss of Sir3 from the telomere might explain the delay by sas2 deletion. Indeed, the senescence profiles of tlc1 sir3, tlc1 sas2 and tlc1 sas2 sir3 mutants were nearly identical (Figure 6A), indicating that sas2 and sir3 deletions are equivalent for this phenotype.
Figure 6.
Loss of Sir3 delays senescence in the same manner as loss of Sas2. (A) TLC1/tlc1 SAS2/sas2 SIR3/sir3 diploids were sporulated and the growth of six independent spore products per genotype was measured. Note that individual or combined deletion of sas2 and sir3 had no effect on the growth rate of TLC1+ cells. The P-value for the difference between tlc1 and tlc1 sas2 (P=0.026), tlc1 sir3 (P=0.0024) or tlc1 sas2 sir3 (P=0.027) at maximal senescence is indicated by the least significant value. (B) Sir3 ChIP showing that deletion of SAS2 depletes Sir3 from telomere proximal sites in senescent tlc1 mutants. The y axis shows ChIP levels normalized to input. sir3 null cells were used to control for non-specific antibody binding, and probes were as in Figure 4A. (C) sir3 deletion does not delay senescence of tlc1 mutants carrying H4K16R. TLC1/tlc1 SIR3/sir3 SAS2/sas2 diploids, carrying plasmid-based H4K16R as their only source of histone H4, were sporulated and the growth of the indicated spore products (N=6 per genotype) were monitored as in Figure 1. (D) H4K16 acetylation remains high in sir3 mutants. ChIP experiment as in Figure 4A comparing H4K16 acetylation between tlc1 and tlc1 sir3 mutants at early PDs or senescence. The means and s.e.m.s for three independent Q-PCR experiments are shown. A full-colour version of this figure is available at The EMBO Journal Online.
To further explore whether sas2 deletion and loss of H4K16 acetylation delay senescence by depleting Sir3 from the telomere, we performed the following experiments. First, we examined Sir3 binding through ChIP and confirmed that it is depleted by sas2 deletion at telomeres in both TLC1+ and senescent tlc1 cells (Figure 6B; Supplementary Figure 8). Second, we determined the epistatic relationship of delayed senescence by sir3 deletion to the H4K16R allele. As histone H4K16R binds Sir3 avidly (Onishi et al, 2007), cells expressing only this form of H4 should enhance Sir3 binding at non-telomere sites and thus deplete Sir3 from the telomere repeats. Similar to sas2 deletion, sir3 deletion had no effect on senescence in cells with H4K16R and indeed combining sas2 sir3 deletions has no effect on senescence in tlc1 mutants carrying H4K16R (Figure 6C). Therefore, sas2 and sir3 deletions likely delay senescence because they relieve an inhibitory activity of Sir3 at the telomere and not at other nucleosome-containing loci. Third, we compared H4K16Ac levels by ChIP in tlc1 and tlc1 sir3 mutants early after loss of telomerase or just before maximal senescence. Deletion of SIR3 caused a general increase in H4K16Ac levels early after loss of telomerase, and moreover, these high levels were generally maintained at senescence (Figure 6D). Therefore, in cells lacking Sir3, slowed senescence does not require decreased H4K16Ac. Interestingly, H4K16Ac levels actually increased near the base of the telomere repeats in tlc1 sir3 mutants at senescence, consistent with our ChIP data indicating that Sir3 remains bound at the distal telomere in senescent tlc1 mutants. Lastly, we examined the effect of deletion of the Dot1 histone methyltransferase on senescence. Dot1 cooperates with Sas2 to establish the boundary separating telomeric heterochromatin from internal euchromatin by methylation of H3K79, which inhibits binding by Sir3 (Onishi et al, 2007; Buchberger et al, 2008). The tlc1 dot1 mutants senesced approximately five PDs later than tlc1 mutants (Supplementary Figure 9). This delay is smaller than that conferred by sas2 deletion, but this difference is explained by the well-established fact that Dot1 has a less prominent function than Sas2 in preventing the spread of Sir3 past the boundary (Katan-Khaykovich and Struhl, 2005; Onishi et al, 2007; Yang et al, 2008). Together, these observations indicate that the mechanism by which sas2 deletion and diminished subtelomeric H4K16Ac levels slow senescence is through release of Sir3 from the telomere and not through effects independent of Sir3.
Sir2, Sir3 and Sir4 have distinct functions during senescence
The Sir2 H4K16 deacetylase enables the propagation of the Sir2/3/4 complex from the Rap1-bound telomere repeats into subtelomeric nucleosomes by deacetylating H4K16, and thus providing a binding site for Sir3 (Onishi et al, 2007). If delayed senescence by sas2 or sir3 deletion is caused by the depletion of Sir3 from the telomere repeats, deletion of SIR2 should have no significant effect on senescence because Sir2 is not required for the binding of Sir3 at the telomere repeats, where Sir3 is bound to Rap1 (Hoppe et al, 2002; Luo et al, 2002; Rusche et al, 2002). Indeed, we found that tlc1 sir2 mutants have the same senescence profile as tlc1 mutants (Figure 7A, and data not shown). Furthermore, tlc1 sas2 sir2 mutants had slowed senescence similar to tlc1 sas2 mutants, indicating that delay of senescence by sas2 deletion does not depend on Sir2. The lack of effect of sir2 deletion on the senescence rates of tlc1 and tlc1 sas2 mutants points to a function for Sir3 at the telomere repeats themselves, rather than at the subtelomeric nucleosomes immediately adjacent to the telomere repeats.
Figure 7.
Contribution of Sir2 and Sir4 to delayed senescence by sas2 deletion. (A) TLC1/tlc1 SAS2/sas2 SIR2/sir2 diploids were sporulated and the growth of four independent spore products per genotype was measured. P-values for the difference between tlc1 and tlc1 sir2 mutants were not significant (P=0.15). (B, C) Delayed senescence depends on SIR4. Senescence rates were examined similarly to (A) except with sir4 in the absence of SAS2 (B) or SIR3 (C). The difference in PDs at senescence was significant for tlc1 versus tlc1 sir4 (P=0.0015) and for tlc1 versus tlc1 sir3 sir4 (P=0.0385), but not for tlc1 sir4 versus tlc1 sir3 sir4 (P=0.35). A full-colour version of this figure is available at The EMBO Journal Online.
We next examined Sir4 and found that sir4 deletion actually sped the senescence of tlc1 mutants (Figure 7B and C). Further, sir4 deletion effectively blocked the slowing of senescence conferred by sas2 or sir3 deletion. Thus, each protein of the Sir2/3/4 complex functions differently during senescence, and extension through loss of Sas2 or Sir3 depends on the presence of Sir4.
Discussion
Here, we show for the first time that modulation of chromatin can delay senescence driven by telomere dysfunction. Deletion of SAS2, encoding a histone H4K16 acetyltransferase, delayed both senescence and the onset of survivor formation. Telomeres in senescent tlc1 mutants seemed to become less heterochromatic, because the acetylation of subtelomeric H4K16 increased at senescence. This acetylation was Sas2 dependent, and the senescence delay conferred by sas2 deletion depended on the deacetylation of H4K16. Nonetheless, lifespan extension was not a direct effect of H4K16 deacetylation, but rather seemed to be a consequence of removal of Sir3 from the telomere repeats, thus facilitating an HR-dependent mechanism of telomere repair.
We suggest the following model for the interplay of Sas2, Sir3, Sir4 and H4K16 during the senescence of tlc1 mutants (Figure 8) and detail our reasoning in the sections below. Telomere shortening is accompanied by Sas2-dependent acetylation of H4K16 and loss of Sir proteins from subtelomeric locations, but retention of Sir proteins at the shortened telomere repeats (Figure 8A). In tlc1 sas2 mutants, subtelomeric H4K16 acetylation is lost, allowing Sir3 to leave the telomere repeats, which allows for efficient HR-based maintenance of telomeres and thus delays senescence (Figure 8B). In tlc1 sir3 mutants, there is no Sir3 at the telomere repeats to begin with, and senescence is similarly delayed (Figure 8C). Sir4 binds to Rap1 (and Ku) at the telomere repeats in a Sir2 and Sir3-independent manner, and facilitates the HR mechanism.
Figure 8.
Model for the interactions among telomere length, H4K16 acetylation, Sas2, Sir3 and Sir4 during senescence in (A) tlc1 mutants, (B) tlc1 sas2 mutants and (C) tlc1 sir3 mutants. Indicated are Rap1, Sas2, Sir3, Sir4, the acetylation state of H4K16, the receptivity of the telomere to HR and the rate of senescence (normal or delayed). Sir2 is omitted for simplicity. See text for details. A full-colour version of this figure is available at The EMBO Journal Online.
Loss of Sas2 leads to delayed senescence through the activation of HR-dependent mechanisms
The delay in senescence conferred by sas2 deletion did not correlate with an overall slowing in the rate of telomere shortening. Rather, the extent of overall telomere shortening was greater in senescent tlc1 sas2 than in tlc1 mutants. Delayed senescence was apparently not a consequence of a blunted DDR, because sas2 deletion did not affect growth inhibition caused by DNA damaging agents, was not epistatic to tel1 deletion during senescence and did not affect the kinetics of Rad53 phosphorylation or growth after uncapping of telomeres in cdc13-1 mutants when shifted to non-permissive temperature. Delayed senescence depended on Rad52 and Rad51, which have central functions in HR and have been earlier shown to slow senescence (Le et al, 1999), and sas2 deletion caused higher levels of telomere recombination during senescence. We found earlier that replication-associated telomere HR, apparently between sister telomeres, increases as telomeres shorten. Cells in which telomere HR had occurred gave rise to viable progeny, indicating that the telomere HR may contribute to the repair of telomere damage (possibly stalled replication forks, by allowing template switch events that bypass impediments to replication), and thus slow senescence by minimizing rare, but dramatic telomere loss events (Lee et al, 2007). Loss of a single telomere is sufficient to cause checkpoint arrest in yeast (Sandell and Zakian, 1993), and although such loss would not have a significant effect of the overall length profile given the contribution of the other 31 telomeres, it would prevent the continued shortening of other telomeres despite sufficient reserve length in a senescing tlc1 mutant cell. Avoidance of rare loss events would prevent defects at a single telomere from halting cell division, allow cells to continue dividing and thus lead to greater shortening among all telomeres at senescence. This model is in line with the greater extent of telomere shortening seen within senescent tlc1 sas2 cells.
As they are replicated unidirectionally, yeast telomeres are particularly reliant on HR-dependent mechanisms that rescue damaged or stalled replication forks (Marians, 2000; Branzei and Foiani, 2007), especially in cells lacking telomerase (Chavez et al, 2009). Although a damaged fork at an internal genomic site can be rescued by convergence with a neighbouring fork, no such neighbour exists for the fork that replicates the telomere terminus. In cells with telomerase, collapse and breakage of the damaged fork would lead to a dramatically shortened telomere that could be repaired by telomerase; in the absence of telomerase, the only recourse for avoiding dramatic telomere shortening would be to repair the fork through HR (Supplementary Figure 10). This explains how mutations that impair HR (e.g. rad52Δ or sgs1Δ) accelerate senescence, and how a mutation that improves HR-dependent repair (e.g. sas2Δ) delays senescence. Interestingly, sas2 deletion also improved the growth of cdc13-1 mutants at semi-permissive temperature in a Rad52-dependent manner. Addinall et al (2008) recently observed similar bypass of cdc13-1 mutant arrest by deletion of SAS-I complex factors, but did not probe Rad52 dependence. As these conditions of partial telomere uncapping in dividing cells might lead to telomere replication difficulties (Miller et al, 2006), we suggest that telomere HR might similarly explain the rescue by sas2 deletion in this setting. The association of the SAS-I complex with the CAF-I and Asf1 histone chaperones, which assemble nucleosomes onto replicating DNA, supports a function for Sas2 in the regulation of replication-associated processes (Meijsing and Ehrenhofer-Murray, 2001). Notably, rare but dramatic telomere loss events occur in fibroblasts obtained from people with Werner syndrome (Crabbe et al, 2004), which is characterized by premature features of ageing and is caused by lack of the WRN RecQ-family DNA helicase, which functions in HR pathways (Saintigny et al, 2002). An increase in these rare loss events also seems to explain the rapid senescence of tlc1 mutants when the Sgs1 RecQ helicase is inactivated (Lee et al, 2007). Similar telomere loss events occur in human cells lacking the Sirt6 histone H3K9 deacetylase, which is required to recruit WRN to telomeres (Michishita et al, 2008). Thus, chromatin and RecQ-family helicases can apparently cooperate to maintain telomeres, and it will be interesting to determine whether a similar relationship exists between H4K16 acetylation and telomere maintenance by these helicases and other HR factors.
We emphasize that the HR mechanisms that slow senescence in tlc1 mutants, and which are augmented in tlc1 sas2 mutants, may be different from those operating to allow survivor formation. Indeed, sas2 deletion did not simply cause early survivor formation, in contrast to the early survivor formation reported earlier for tlc1 mutants lacking the histone H1-like protein encoded by HHO1 (Downs et al, 2003). Rad52 was recently proposed to have a function in telomere protection during senescence that is inhibited by Tel1 and does not involve synthesis of new telomere DNA (Abdallah et al, 2009), for example through stabilization of stalled telomere replication forks. Our finding that deletions of TEL1 and SAS2 delay senescence in a non-epistatic manner suggests that each deletion impacts a different pathway, and thus Rad52 might function differently in each pathway. The increased telomere recombination in tlc1 sas2 cells suggests that the Sas2-inhibited pathway does involve the acquisition of new telomere repeat tracts at shortened telomeres. We favour the idea that the increased telomere recombination explains the slowed senescence, but it is possible that it is merely correlated with it. Further, the Rad51 and Rad52 dependence of delayed senescence might reflect senescence in tlc1 rad51 and tlc1 rad52 mutants that is so rapid as to preclude benefit from sas2 deletion, rather than reflecting a bona fide function for HR in sas2Δ cells. It is difficult to completely rule out this alternative interpretation, but we note that the defect in tlc1 rad51/52 mutants is mild enough to permit over 55 PD before senescence. In addition, in the cdc13-1 model, in which sas2 deletion also confers a Rad52-dependent growth benefit at semi-permissive temperature, the growth defect of cdc13-1 rad52 mutants is no worse than that of cdc13-1 mutants (Figure 3C), and thus the Rad52 dependence for improved telomere maintenance need not reflect a growth defect.
We also note that our results are compatible with either a telomere specific or with a genome-wide function for Sas2 in the regulation of HR. However, we suspect that Sas2 preferentially affect HR near telomeres because (1) acetylation of H4K16 by Sas2 occurs predominantly at subtelomeric sites (Shia et al, 2006), (2) sas2 mutants had normal sensitivity to several DNA damaging agents and (3) the mechanism by which sas2 deletion slows senescence seems to be through removal of Sir3 from the telomere repeats. As Sir3 has no apparent direct function in global HR (Lee et al, 1999), if Sas2 does affect global HR, then it likely does so using mechanisms that are different from those that delay senescence in tlc1 sas2 mutants.
Delayed senescence by sas2 deletion functions through the loss of acetylation at H4K16
Sas2 can acetylate K16 of histone H4 and, more weakly, K14 of histone H3 (Sutton et al, 2003). Analysis by ChIP revealed that H4K16 levels increase selectively at subtelomeres during senescence of tlc1 mutants and that the increases were Sas2 dependent, suggesting that sas2 deletion slowed senescence through the loss of acetylation at H4K16. The reasons for increased subtelomeric H4K16 acetylation at senescence are not yet clear. It is possible that the shortening of the telomere repeats and the consequent loss of Rap1 nucleation sites causes a decrease in the local concentration of Sir proteins at telomeres and subsequently leads to fewer Sir proteins spreading into the subtelomeric region, thus opening sites on subtelomeric nucleosomes to acetylation by Sas2. Consistent with this view, at senescence, Rap1, Sir3 and Sir2 were each found to lose their normal localization in foci at the nuclear periphery and to become more diffusely nuclear (Straatman and Louis, 2007). Alternatively, shortened telomeres, which are sensed as DSBs, might increase H4K16 acetylation in the same manner as generic DSBs during HR-mediated repair (Tamburini and Tyler, 2005), and thus loss of Sir protein binding would be a consequence rather than a cause of increased acetylation. A third possibility is that because senescence activates a stress response (Nautiyal et al, 2002), and because other stresses induce phosphorylation of Sir3 and promote its loss from subtelomeric regions (Martin et al, 1999; Mills et al, 1999; Ai et al, 2002), Sir3 might be phosphorylated during senescence and be similarly lost. Regardless, our findings that an H4K16R mutation delays senescence and is epistatic to sas2 deletion indicate that H4K16 is the key target of Sas2 for setting the pace of senescence.
Differential activities of Sir2, 3 and 4 during senescence
Deletion of Sas2 or the H4K16R mutation causes loss of the Sir complex from the telomere and relocalization to internal loci (Kimura et al, 2002; Suka et al, 2002). We, therefore, examined functions for Sir proteins during senescence in tlc1 mutants. Sir3 deletion slowed senescence in a manner epistatic with sas2 deletion, indicating that sas2 deletion delays senescence through the loss of Sir3 from the telomere and not through translocation of Sir3 to internal loci because such a mechanism would be dependent on Sir3 rather than being inhibited by it. Of note, the delayed senescence conferred by sir3 deletion, in the setting of functional Sas2 and fully acetylated H4K16, indicates that loss of H4K16 acetylation through sas2 deletion does not lead to delayed senescence per se, but is rather the means by which Sir3 is depleted from the telomere. The epistasis of sir3 deletion and H4K16R mutation for senescence rate supports this view. Thus, modulation of senescence through manipulation of subtelomeric nucleosomal chromatin occurs through effects on the non-classical (i.e. non-nucleosomal) heterochromatin of the telomere repeats. In addition, the fact that sir3 and sas2 deletions are equivalent is consistent with the model that sas2 deletion maximally disrupts the heterochromatin boundary and thus depletes Sir3 from the telomere.
Sir4 had a different function than Sir3 during senescence and was required to prevent rapid senescence and for deletion of SIR3 or SAS2 to slow senescence. Sir4 can bind Rap1 and Ku at the telomere repeats independent of Sir2 and Sir3, and among the Sir proteins is uniquely capable of anchoring telomeres to the nuclear envelope (NE) (Hediger et al, 2002; Taddei et al, 2004). This involves interactions with Esc1 and Mps3, each of which localizes outside of nuclear pore complexes (NPCs) (Taddei et al, 2004; Bupp et al, 2007; Schober et al, 2009). Telomeres were recently found to translocate to NPCs at senescence (Khadaroo et al, 2009). The relationship of subnuclear localization to choice of HR repair pathway seems to be complex and is incompletely understood (Nagai et al, 2008; Bystricky et al, 2009; Oza et al, 2009; Schober et al, 2009), but we speculate that the critical function of Sir4 during senescence may be to regulate telomere localization at the NE and thus the choice of an HR pathway that slows senescence but does not drive survivor formation. It may be that the beneficial effect of Sir3 loss from telomeres is to facilitate this function of Sir4.
Sir2 was neither required for a normal rate of senescence nor the delayed senescence conferred by sas2 deletion. These findings indicate that Sir3 inhibits lifespan at the telomere repeats, in which its interactions with Rap1 and Sir4 do not require Sir2. As sir2 deletion might be expected to increase free Sir3 levels (by increasing H4K16 acetylation and thus blocking Sir3 binding to nucleosomes), but has no effect on the rate of senescence, we infer that Sir3 is bound to terminal repeats at saturating levels and thus sir2 deletion does not increase Sir3 levels at the telomere repeats. The Sir2 family of proteins regulates the pace of ageing in many organisms (Guarente, 2007). This regulation ranges from lifespan inhibition to extension, for example limiting lifespan during the chronological ageing of yeast, but extending the replicative lifespan of yeast mother cells (Kaeberlein et al, 1999; Fabrizio et al, 2005). Our findings further emphasize the importance of molecular context in determining the effects of this important family of lifespan regulators.
An earlier study of the function of the Sir proteins in telomerase mutants found that individual deletion of the different SIR genes each caused early survivor formation (Lowell et al, 2003). We have never observed this for SIR2 or SIR3, but it is possible that the different strain background (W303) and experimental design might explain the different results. Strains used in the earlier study apparently generated a high rate of petite mutants and caused the authors to use rho0 strains (lacking mitochondrial DNA) for most of their experiments, but we did not observe petite mutants in our experiments. Furthermore, comparisons were made between haploid SIR+ and sirΔ strains from which telomerase was deleted, rather than making comparisons between spore products derived from the same parental diploid as we did in our study; because sir3Δ and sir4Δ mutations shorten steady-state telomere lengths in TLC1+ cells (Palladino et al, 1993), this would cause more rapid senescence and thus early survivor formation, and in addition differences in telomere chromatin preceding the loss of telomerase might also have affected the results.
In summary, we have identified two factors, Sas2 and Sir3, that impact telomere chromatin and when removed, delay the senescence of yeast lacking telomerase. Similar to murine cells lacking telomerase (Benetti et al, 2007), the nucleosomal regions of telomeres in yeast telomerase mutants seem to become less heterochromatic at senescence as indicated by the increased acetylation of subtelomeric H4K16. The status of telomere chromatin in senescent human cells is largely unexplored, but the fact that telomeres are excluded from SAHF suggests that they too might lose heterochromatin features (Ye et al, 2007). It will be informative to characterize telomere chromatin dynamics in senescent human cells as well as determine the function of the mammalian Sas2 orthologue, hMOF (Lafon et al, 2007), in this setting.
Materials and methods
Yeast strains and plasmids
Yeast were cultured at 30°C in YPAD or SC-complete medium (Amberg and Burke, 2005). All senescence experiments were performed in the BY4741/4742 background (Brachmann et al, 1998), except for Supplementary Figure 1C, which was in JKM111 (Moore and Haber, 1996). Unless otherwise specified, all alleles are deletions. See Supplementary Table 1 for strain details. Double and triple mutants were constructed by deleting one copy of TLC1 in diploids heterozygous for genes of interest or by crossing haploid mutants of interest with tlc1 mutants at early PDs after loss of telomerase, followed by serial streaking to equilibrate telomere lengths. The cdc13-1 allele was introduced through pop-in/pop-out recombination using pVL451 (kindly provided by V Lunblad, Salk Institute, through J Haber). All strains were genotyped using auxotrophy or drug resistance along with PCR-based verification. Plasmid pWD23 carrying the H4K16R mutation or pWD37 (H3K14R) was generated by QuickChange (Stratagene) site-directed mutagenesis. The haploid strains expressing the H4K16R or H3K14R mutant as the sole source of H4 or H3, respectively, were made by transforming the mutant plasmid into FY1716, followed by selection on synthetic-complete medium containing 5-fluoroorotic acid. An HA tag was inserted at the RAD53 locus as described (Longtine et al, 1998).
Senescence assays
Each senescence assay was performed starting with freshly dissected spore products at approximately PD 25. Spores (N=4 per genotype, unless otherwise noted) were serially passaged in liquid culture (either YPAD or SC complete; held constant within any experiment) for 22 h. For each passage, cells were counted using a Coulter Counter and 1.4 × 106 cells were diluted into 7 ml of fresh medium. Strains carrying the wild type or the mutant histone H4 plasmids were cultured in SC-Ura or SC-Trp media, respectively, for plasmids maintenance. The total cell counts for each culture were used to determine the number of PDs (PD=log2(final/starting concentration)). P-values for differences between PDs at maximal senescence (lowest point of growth) were calculated using an unpaired two-sample two-tailed t-test.
Chromatin immunoprecipitation
ChIP experiments were performed essentially as described earlier (Strahl-Bolsinger et al, 1997), and full details are provided in Supplementary Data. Briefly, 108 cells were harvested and disrupted mechanically. Chromatin was immunoprecipitated using rabbit anti-H4K16Ac or anti-H3K14Ac, rabbit anti-total H3 or rabbit anti-Sir3 (kind gift of D Moazed). Purified DNA was quantified using Q-PCR. Primers are listed in Supplementary Table 2. All signals were first normalized to input using standard curves.
Telomere PCR and sequencing
Telomere PCR was performed as described in Lee et al, 2007 with the following modifications; genomic DNA from tlc1 mutants at 50% senescence or tlc1 sas2 mutants matched by degree of senescence or number of PDs was denatured and C-tailed. The telomere ends of chromosome I-L were amplified in the presence of 0.5 M betaine and 50–600 bp fragments were cloned using the StrataClone PCR Cloning kit (Stratagene). Telomere sequences were aligned and compared using MegAlign software. Sequences are available on request.
Additional experimental details are described in Supplementary Data.
Supplementary Material
Supplementary Figures
Supplementary Materials and Methods
Supplementary Tables 1 and 2
Review Process File
Acknowledgments
We thank the members of the Johnson laboratory, Peter Adams, Ronen Marmorstein, Jim Haber, David Schultz and Jamie Hayden for advice and discussions, Leonid Kozak for designing Q-PCR analysis software and Danesh Moazed for kindly providing the rabbit anti-Sir3 antibody. This work was supported by NIH grants R01-AG021521 and P01-AG031862. Alejandro Chavez initiated screen and identified delayed senescence in tlc1 sas2 mutants, and was supported by NIH T32-AG000255.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abdallah P, Luciano P, Runge KW, Lisby M, Geli V, Gilson E, Teixeira MT (2009) A two-step model for senescence triggered by a single critically short telomere. Nat Cell Biol 11: 988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall SG, Downey M, Yu M, Zubko MK, Dewar J, Leake A, Hallinan J, Shaw O, James K, Wilkinson DJ, Wipat A, Durocher D, Lydall D (2008) A genomewide suppressor and enhancer analysis of cdc13-1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics 180: 2251–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai W, Bertram PG, Tsang CK, Chan TF, Zheng XF (2002) Regulation of subtelomeric silencing during stress response. Mol Cell 10: 1295–1305 [DOI] [PubMed] [Google Scholar]
- Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Cote J (2007) Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell 28: 1002–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ (2005) Methods in Yeast Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Aubert G, Lansdorp PM (2008) Telomeres and aging. Physiol Rev 88: 557–579 [DOI] [PubMed] [Google Scholar]
- Azam M, Lee JY, Abraham V, Chanoux R, Schoenly KA, Johnson FB (2006) Evidence that the S.cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence. Nucleic Acids Res 34: 506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur JA, Zou Y, Shay JW, Wright WE (2001) Telomere position effect in human cells. Science 292: 2075–2077 [DOI] [PubMed] [Google Scholar]
- Benetti R, Garcia-Cao M, Blasco MA (2007) Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet 39: 243–250 [DOI] [PubMed] [Google Scholar]
- Blasco MA (2007) The epigenetic regulation of mammalian telomeres. Nat Rev Genet 8: 299–309 [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2007) Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amst) 6: 994–1003 [DOI] [PubMed] [Google Scholar]
- Buchberger JR, Onishi M, Li G, Seebacher J, Rudner AD, Gygi SP, Moazed D (2008) Sir3-nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae. Mol Cell Biol 28: 6903–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp JM, Martin AE, Stensrud ES, Jaspersen SL (2007) Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol 179: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K, Van Attikum H, Montiel MD, Dion V, Gehlen L, Gasser SM (2009) Regulation of nuclear positioning and dynamics of the silent mating type loci by the yeast Ku70/Ku80 complex. Mol Cell Biol 29: 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmen AA, Milne L, Grunstein M (2002) Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J Biol Chem 277: 4778–4781 [DOI] [PubMed] [Google Scholar]
- Chavez A, Tsou AM, Johnson FB (2009) Telomeres do the (un)twist: helicase actions at chromosome termini. Biochim Biophys Acta 1792: 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J (2004) Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306: 1951–1953 [DOI] [PubMed] [Google Scholar]
- Downs JA, Kosmidou E, Morgan A, Jackson SP (2003) Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol Cell 11: 1685–1692 [DOI] [PubMed] [Google Scholar]
- Enomoto S, Glowczewski L, Berman J (2002) MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol Biol Cell 13: 2626–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD (2005) Sir2 blocks extreme life-span extension. Cell 123: 655–667 [DOI] [PubMed] [Google Scholar]
- Feeser EA, Wolberger C (2008) Structural and functional studies of the Rap1 C-terminus reveal novel separation-of-function mutants. J Mol Biol 380: 520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA (2004) Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 36: 94–99 [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA (2006) DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 8: 416–424 [DOI] [PubMed] [Google Scholar]
- Grunstein M (1997) Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol 9: 383–387 [DOI] [PubMed] [Google Scholar]
- Guarente L (2007) Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol 72: 483–488 [DOI] [PubMed] [Google Scholar]
- Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK (2005) Involvement of human MOF in ATM function. Mol Cell Biol 25: 5292–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M (1996) Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96 [DOI] [PubMed] [Google Scholar]
- Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM (2002) Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol 12: 2076–2089 [DOI] [PubMed] [Google Scholar]
- Hoppe GJ, Tanny JC, Rudner AD, Gerber SA, Danaie S, Gygi SP, Moazed D (2002) Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol Cell Biol 22: 4167–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IJpma AS, Greider CW (2003) Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol Biol Cell 14: 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Jia X, Weinert T, Lydall D (2004) Mec1 and Rad53 inhibit formation of single-stranded DNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics 166: 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FB, Marciniak RA, McVey M, Stewart SA, Hahn WC, Guarente L (2001) The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J 20: 905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13: 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y, Struhl K (2005) Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J 24: 2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadaroo B, Teixeira MT, Luciano P, Eckert-Boulet N, Germann SM, Simon MN, Gallina I, Abdallah P, Gilson E, Geli V, Lisby M (2009) The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat Cell Biol 11: 980–987 [DOI] [PubMed] [Google Scholar]
- Kimura A, Umehara T, Horikoshi M (2002) Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet 32: 370–377 [DOI] [PubMed] [Google Scholar]
- Lafon A, Chang CS, Scott EM, Jacobson SJ, Pillus L (2007) MYST opportunities for growth control: yeast genes illuminate human cancer gene functions. Oncogene 26: 5373–5384 [DOI] [PubMed] [Google Scholar]
- Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M (2008) Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J 27: 1502–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S, Moore JK, Haber JE, Greider CW (1999) RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152: 143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kozak M, Martin J, Pennock E, Johnson FB (2007) Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol 5: e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Paques F, Sylvan J, Haber JE (1999) Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol 9: 767–770 [DOI] [PubMed] [Google Scholar]
- Liaw H, Lustig AJ (2006) Sir3 C-terminal domain involvement in the initiation and spreading of heterochromatin. Mol Cell Biol 26: 7616–7631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA (1996) The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci USA 93: 13760–13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lowell JE, Roughton AI, Lundblad V, Pillus L (2003) Telomerase-independent proliferation is influenced by cell type in Saccharomyces cerevisiae. Genetics 164: 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH (1993) An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 73: 347–360 [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Luo K, Vega-Palas MA, Grunstein M (2002) Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev 16: 1528–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig AJ, Liu C, Zhang C, Hanish JP (1996) Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol Cell Biol 16: 2483–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE (2007) Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448: 820–823 [DOI] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM (1996) Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev 10: 1796–1811 [DOI] [PubMed] [Google Scholar]
- Marians KJ (2000) Replication and recombination intersect. Curr Opin Genet Dev 10: 151–156 [DOI] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM (1999) Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97: 621–633 [DOI] [PubMed] [Google Scholar]
- Meijsing SH, Ehrenhofer-Murray AE (2001) The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev 15: 3169–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF (2008) SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452: 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP (2006) Semi-conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828 [DOI] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L (1999) MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell 97: 609–620 [DOI] [PubMed] [Google Scholar]
- Moore JK, Haber JE (1996) Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 16: 2164–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D (1994) Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev 8: 2257–2269 [DOI] [PubMed] [Google Scholar]
- Moretti P, Shore D (2001) Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol Cell Biol 21: 8082–8094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ (2008) Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal S, DeRisi JL, Blackburn EH (2002) The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 99: 9316–9321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Oki M, Valenzuela L, Chiba T, Ito T, Kamakaka RT (2004) Barrier proteins remodel and modify chromatin to restrict silenced domains. Mol Cell Biol 24: 1956–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D (2007) Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell 28: 1015–1028 [DOI] [PubMed] [Google Scholar]
- Osada S, Sutton A, Muster N, Brown CE, Yates JR III, Sternglanz R, Workman JL (2001) The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev 15: 3155–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL (2009) Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev 23: 912–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser SM (1993) SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75: 543–555 [DOI] [PubMed] [Google Scholar]
- Ritchie KB, Mallory JC, Petes TD (1999) Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol 19: 6065–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Meier B, McAinsh AD, Feldmann HM, Jackson SP (2004) Separation-of-function mutants of yeast Ku80 reveal a Yku80p-Sir4p interaction involved in telomeric silencing. J Biol Chem 279: 86–94 [DOI] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J (2002) Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol Biol Cell 13: 2207–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ Jr (2002) Homologous recombination resolution defect in werner syndrome. Mol Cell Biol 22: 6971–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA (1993) Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75: 729–739 [DOI] [PubMed] [Google Scholar]
- Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM (2009) Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev 23: 928–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia WJ, Li B, Workman JL (2006) SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev 20: 2507–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia WJ, Osada S, Florens L, Swanson SK, Washburn MP, Workman JL (2005) Characterization of the yeast trimeric-SAS acetyltransferase complex. J Biol Chem 280: 11987–11994 [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847 [DOI] [PubMed] [Google Scholar]
- Singer MS, Gottschling DE (1994) TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Straatman KR, Louis EJ (2007) Localization of telomeres and telomere-associated proteins in telomerase-negative Saccharomyces cerevisiae. Chromosome Res 15: 1033–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev 11: 83–93 [DOI] [PubMed] [Google Scholar]
- Suka N, Luo K, Grunstein M (2002) Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet 32: 378–383 [DOI] [PubMed] [Google Scholar]
- Sutton A, Shia WJ, Band D, Kaufman PD, Osada S, Workman JL, Sternglanz R (2003) Sas4 and Sas5 are required for the histone acetyltransferase activity of Sas2 in the SAS complex. J Biol Chem 278: 16887–16892 [DOI] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM (2004) Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J 23: 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK (2005) Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol 25: 4903–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Teng SC, Zakian VA (1999) Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol 19: 8083–8093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley EA, Zakian VA (1995) Extra telomeres, but not internal tracts of telomeric DNA, reduce transcriptional repression at Saccharomyces telomeres. Genetics 139: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JH, Gottschling DE, Zakian VA (1992) Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev 6: 197–210 [DOI] [PubMed] [Google Scholar]
- Xu EY, Kim S, Replogle K, Rine J, Rivier DH (1999a) Identification of SAS4 and SAS5, two genes that regulate silencing in Saccharomyces cerevisiae. Genetics 153: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu EY, Kim S, Rivier DH (1999b) SAS4 and SAS5 are locus-specific regulators of silencing in Saccharomyces cerevisiae. Genetics 153: 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu EY, Zawadzki KA, Broach JR (2006) Single-cell observations reveal intermediate transcriptional silencing states. Mol Cell 23: 219–229 [DOI] [PubMed] [Google Scholar]
- Yang B, Britton J, Kirchmaier AL (2008) Insights into the impact of histone acetylation and methylation on Sir protein recruitment, spreading, and silencing in Saccharomyces cerevisiae. J Mol Biol 381: 826–844 [DOI] [PubMed] [Google Scholar]
- Ye X, Zerlanko B, Zhang R, Somaiah N, Lipinski M, Salomoni P, Adams PD (2007) Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol Cell Biol 27: 2452–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Bi X (2008) Positive roles of SAS2 in DNA replication and transcriptional silencing in yeast. Nucleic Acids Res 36: 5189–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko MK, Guillard S, Lydall D (2004) Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics 168: 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Materials and Methods
Supplementary Tables 1 and 2
Review Process File