Abstract
MicroRNAs (miRNA) are ∼21 nucleotide-long non-coding small RNAs, which function as post-transcriptional regulators in eukaryotes. miRNAs play essential roles in regulating plant growth and development. In recent years, research into the mechanism and consequences of miRNA action has made great progress. With whole genome sequence available in such plants as Arabidopsis thaliana, Oryza sativa, Populus trichocarpa, Glycine max, etc., it is desirable to develop a plant miRNA database through the integration of large amounts of information about publicly deposited miRNA data. The plant miRNA database (PMRD) integrates available plant miRNA data deposited in public databases, gleaned from the recent literature, and data generated in-house. This database contains sequence information, secondary structure, target genes, expression profiles and a genome browser. In total, there are 8433 miRNAs collected from 121 plant species in PMRD, including model plants and major crops such as Arabidopsis, rice, wheat, soybean, maize, sorghum, barley, etc. For Arabidopsis, rice, poplar, soybean, cotton, medicago and maize, we included the possible target genes for each miRNA with a predicted interaction site in the database. Furthermore, we provided miRNA expression profiles in the PMRD, including our local rice oxidative stress related microarray data (LC Sciences miRPlants_10.1) and the recently published microarray data for poplar, Arabidopsis, tomato, maize and rice. The PMRD database was constructed by open source technology utilizing a user-friendly web interface, and multiple search tools. The PMRD is freely available at http://bioinformatics.cau.edu.cn/PMRD. We expect PMRD to be a useful tool for scientists in the miRNA field in order to study the function of miRNAs and their target genes, especially in model plants and major crops.
INTRODUCTION
MicroRNAs (microRNA) are ∼21 nucleotide-long endogenetic non-coding small RNAs and function as post-transcriptional regulators in eukaryotes. The processing of miRNA is well studied and is comprised of a discrete series of steps (1). miRNA genes are transcribed and excised into miRNAs, then miRNA is recruited by RISC (RNA-induced silencing complex, including Argonaute and other proteins) which combines with mRNA to inhibit or degrade the target mRNA (1) and thus translation is interrupted.
MiRNAs play essential roles in regulating plant growth and development (2). Research into miRNA function on target genes has progressed at a very rapid rate in plants. As examples, Archak et al. (3) discovered that miRNAs may participate in diverse functions including transcription, catalysis, binding and transporter activity. Some miRNAs related to abiotic stress responses such as cold stress and nutrient deprivation were identified in Arabidopsis thaliana using transcriptome analysis (4,5); Lu et al. (6) analyzed miRNA regulatory roles in the response of Populus trichocarpa to the stressful environment incurred over their long-term growth; Morin et al. conducted comparative analyses on the conservation of miRNAs between Pinus contorta and Oryza sativa and discovered that important RNA silencing processes were highly developed in the earliest spermatophytes (7).
Recently, more and more miRNAs have been identified in plant genomes. Jones-Rhoades et al. (8) developed comparative genomic approaches to systematically identify both miRNAs and target genes, which enlarged the miRNAs family and the number of target genes in A. thaliana. Using high-throughput sequencing, Rajagopalan et al. (9) identified 38 new miRNAs in A. thaliana. Using a similar approach, a large number of miRNAs were recently identified in rice (10). Using support vector machine classification based on intra-genomic matching of potential miRNAs and their targets, Lindow et al. (11) predicted ∼1200, ∼2100 and ∼2500 miRNA candidate genes in A. thaliana, O. sativa and P. trichocarpa, respectively. Using EST analysis, Zhang et al. (12,13) identified numerous new miRNAs in many species. Finally, Ramanjulu Sunkar and Guru Jagadeeswaran (14) identified miRNAs in a large number of plant species.
With the discovery of a large number of miRNAs and their target genes, some public resources have been constructed: miRBase (http://microrna.sanger.ac.uk/), provides a set of precursor and mature miRNAs discovered in many plants (15); ASRP (http://asrp.cgrb.oregonstate.edu/) lists miRNAs and their target genes in Arabidopsis (16); CSRDB (http://sundarlab.ucdavis.edu/smrnas/) is a collection of miRNAs identified in maize and rice (17); Rfam (http://rfam.sanger.ac.uk/) provides secondary structures of miRNA precursors in many plant species (18,19). However, there are limitations to the current databases in this field. For example, miRBase only provides miRNA sequence data and annotation in 25 plant species; Rfam only contains miRNA precursor sequences from less than 50 plant species; the ASRP integrates miRNAs, precursor, target genes and genome browser only for Arabidopsis. It is necessary to develop a plant miRNA database through integration of the large amount of information about plant miRNAs available to the public. For this purpose, we developed a plant microRNA database (PMRD) to integrate the available data pertinent to plant miRNAs available from public resources. Uniquely, we provided miRNA expression profiles in the PMRD, including our local generated rice oxidative stress microarray data and the recent published microarray data generated from poplar, Arabidopsis, tomato, maize and rice.
DATABASE CONTENT
miRNA data collection
The majority of our data was gleaned from miRBase, Rfam and literature published in recent years. The number of mature miRNAs that we immediately retrieved from the miRBase was 1931, 73 miRNAs came from Rfam, and other miRNAs were from the literature. As shown in Table 1, we integrated 8433 non-redundant miRNA sequences and their original resources. Annotation of miRNAs was mainly dependent on information present in miRBase, but in some particular instances, the annotation included information about the corresponding author who discovered the miRNA. Detailed information concerning the variety of miRNAs in PMRD is presented in Table 1.
Table 1.
Species and sources for raw miRNA data
| Species | Number of miRNAs |
References | ||
|---|---|---|---|---|
| Total | Experimental | Computational | ||
| Aegilops speltoides | 1 | 1 | (12,13) | |
| Aegilops tauschii | 2 | 2 | (19) | |
| Alliaria petiolata | 1 | 1 | (19) | |
| Allium cepa | 7 | 7 | (12,13) | |
| Annona cherimola | 1 | 1 | (19) | |
| Antirrhinum majus | 1 | 1 | (19) | |
| Arabidopsis arenosa | 1 | 1 | (19) | |
| Arabidopsis cebennensis | 1 | 1 | (19) | |
| Arabidopsis halleri | 1 | 1 | (19) | |
| Arabidopsis thaliana | 1427 | 207 | 1220 | (2,4,8,9,11–13,16,41–58) |
| Arabis hirsuta | 1 | 1 | (19) | |
| Arachis hypogaea | 1 | 1 | (59) | |
| Australopyrum velutinum | 1 | 1 | (19) | |
| Avena sativa | 1 | 1 | (19) | |
| Barbarea vulgaris | 1 | 1 | (19) | |
| Boechera stricta | 1 | 1 | (14) | |
| Brachypodium distachyon | 12 | 12 | (25) | |
| Brachypodium sylvaticum | 1 | 1 | (19) | |
| Brassica napus | 45 | 45 | (50,60,61) | |
| Brassica oleracea | 9 | 1 | 8 | (14,50,62) |
| Brassica rapa | 20 | 20 | (14,50,62) | |
| Camelina microcarpa | 1 | 1 | (19) | |
| Capsella rubella | 1 | 1 | (19) | |
| Capsicum annuum | 3 | 3 | (12,13) | |
| Carica papaya | 2 | 1 | 1 | (14,63) |
| Chlamydomonas reinhardtii | 84 | 84 | (64,65) | |
| Citrus clementina | 1 | 1 | (14) | |
| Citrus sinensis | 5 | 5 | (12,13) | |
| Citrus × paradisi × Poncirus trifoliata | 4 | 4 | (14) | |
| Conringia orientalis | 1 | 1 | (19) | |
| Corylus avellana | 1 | 1 | (19) | |
| Cycas rumphii | 1 | 1 | (12,13) | |
| Descurainia sophia | 1 | 1 | (19) | |
| Dictyostelium discoideum | 2 | 2 | (66) | |
| Erysimum cheiri | 1 | 1 | (19) | |
| Eschscholzia californica | 1 | 1 | (12,13) | |
| Festuca arundinacea | 1 | 1 | (14) | |
| Festuca glaucescens × Lolium multiflorum | 1 | 1 | (19) | |
| Fourraea alpina | 1 | 1 | (19) | |
| Glycine clandestina | 3 | 3 | (12,13,22,23) | |
| Glycine max | 166 | 80 | 86 | (12,13,20–23,59) |
| Glycine soja | 3 | 3 | (22,23) | |
| Gossypium arboreum | 2 | 2 | (12,13) | |
| Gossypium herbaceum | 4 | 4 | (32) | |
| Gossypium hirsutum | 53 | 53 | (30–32) | |
| Gossypium raimondii | 11 | 11 | (12,13,30,32) | |
| Hedyotis centranthoides | 3 | 3 | (12,13) | |
| Hedyotis terminalis | 1 | 1 | (12,13) | |
| Helianthus annuus | 3 | 3 | (12,13) | |
| Henrardia persica | 1 | 1 | (19) | |
| Heteranthelium piliferum | 1 | 1 | (19) | |
| Hordeum vulgare | 1 | 1 | (14) | |
| Hordeum vulgare subsp. Spontaneum | 1 | 1 | (12,13) | |
| Hordeum vulgare subsp. Vulgare | 16 | 16 | (12,13) | |
| Ipomoea batatas | 1 | 1 | (12,13) | |
| Ipomoea nil | 5 | 5 | (12,13) | |
| Lactuca sativa | 3 | 3 | (12,13) | |
| Liriodendron tulipifera | 2 | 2 | (12,13) | |
| Lotus japonicus | 8 | 8 | (12,13,22,59) | |
| Lupinus luteus | 1 | 1 | (12,13) | |
| Malcolmia maritima | 1 | 1 | (19) | |
| Malus x domestica | 3 | 3 | (12,13) | |
| Manihot esculenta | 1 | 1 | (14) | |
| Medicago truncatula | 76 | 36 | 40 | (13,20,22,59,67–69) |
| Nasturtium officinale | 1 | 1 | (19) | |
| Nicotiana benthamiana | 4 | 4 | (12,13) | |
| Nicotiana sylvestris | 1 | 1 | (19) | |
| Nicotiana tabacum | 1 | 1 | (12,13) | |
| Nuphar advena | 3 | 3 | (12,13) | |
| Oenocarpus bataua | 1 | 1 | (19) | |
| Oryza barthii | 1 | 1 | (19) | |
| Oryza brachyantha | 1 | 1 | (19) | |
| Oryza nivara | 1 | 1 | (19) | |
| Oryza rufipogon | 6 | 6 | (19) | |
| Oryza sativa | 2540 | 269 | 2271 | (2,3,6–8,10–13,28,52–55,69–78) |
| Panicum virgatum | 1 | 1 | (14) | |
| Pennisetum ciliare | 1 | 1 | (14) | |
| Pennisetum glaucum | 1 | 1 | (12,13) | |
| Peridictyon sanctum | 1 | 1 | (19) | |
| Persea americana | 1 | 1 | (12,13) | |
| Petunia x hybrida | 1 | 1 | (19) | |
| Phaseolus coccineus | 2 | 2 | (12,13) | |
| Phaseolus vulgaris | 2 | 2 | (14,59) | |
| Phleum pratense | 1 | 1 | (19) | |
| Physcomitrella patens | 282 | 281 | 1 | (2,12,13,70,79–82) |
| Picea glauca | 5 | 5 | (12,13) | |
| Picea sitchensis | 3 | 3 | (12,13) | |
| Pinus taeda | 40 | 24 | 16 | (6,7,12,13,77) |
| Populus tremula | 4 | 4 | (12,13) | |
| Populus tremula × Populus tremuloides | 6 | 6 | (14) | |
| Populus tremuloides | 7 | 7 | (12,13) | |
| Populus trichocarpa | 2780 | 77 | 2703 | (2,6,11,13,83,84) |
| Populus trichocarpa × Populus deltoides | 5 | 5 | (12–14) | |
| Prunus armeniaca | 2 | 2 | (12,13) | |
| Prunus persica | 3 | 3 | (14) | |
| Prunus salicina | 1 | 1 | (19) | |
| Ricinus communis | 24 | 24 | (19) | |
| Rorippa indica | 1 | 1 | (19) | |
| Saccharum officinarum | 32 | 32 | (14,20) | |
| Saccharum sp | 2 | 2 | (12,13) | |
| Schedonorus arundinaceus | 1 | 1 | (12,13) | |
| Secale cereale | 1 | 1 | (12,13) | |
| Selaginella moellendorffii | 64 | 64 | (79) | |
| Sesamum indicum | 2 | 2 | (12,13) | |
| Sibara virginica | 1 | 1 | (19) | |
| Solanum demissum | 1 | 1 | (19) | |
| Solanum lycopersicum | 37 | 21 | 16 | (12–14,34,85) |
| Solanum tuberosum | 14 | 14 | (12–14) | |
| Sorghum bicolor | 76 | 76 | (12–14,20,27,86) | |
| Sorghum propinquum | 2 | 2 | (12,13) | |
| Theobroma cacao | 1 | 1 | (12,13) | |
| Thlaspi arvense | 1 | 1 | (19) | |
| Trifolium repens | 1 | 1 | (19) | |
| Triticum aestivum | 85 | 71 | 14 | (12,13,24,25) |
| Triticum monococcum | 2 | 2 | (19) | |
| Triticum turgidum | 2 | 2 | (12,13) | |
| Triticum urartu | 2 | 2 | (19) | |
| Vigna unguiculata | 2 | 2 | (22,59) | |
| Vitis vinifera | 142 | 142 | (13,22,87) | |
| Zea mays | 207 | 2 | 205 | (12,13,20,26–29) |
| Zinnia elegans | 3 | 3 | (12,13) | |
The curation of miRNAs that are not included in miRBase or that have not yet been named was performed using a special format that includes information about the corresponding author. An example is osa-miRf10000-akr, a rice miRNA discovered by Anders Krogh’s group (11).
Recently in PMRD, there are 1427, 2540 and 2780 mature miRNA entries for Arabidopsis, rice and poplar, respectively, while in miRBase the entries for these three species are 207, 415 and 237, respectively. The other miRNAs we curated from these three species mainly come from Lindow et al. (11). There are 166 soybean entries in PMRD, of which 79 sequences came from miRBase (15,20–22), and the others were from Zhang et al. (13,23). For wheat we deposited 85 miRNAs in PMRD, of which 32 came from miRBase (15,24) and the others were from Yao et al. (24), Zhang et al. (13) and Wei et al. (25). As for maize, 207 entries were created in PMRD, including 98 from miRBase (13,15,20,26–28) and 109 from Zhang et al. (13,29). For cotton we deposited 53 mature sequences in PRMD, in which 13 items were from miRBase (15,30), and the others were from Qiu et al. (31) and Zhang et al. (32). In total, there were 8433 miRNAs taken from 121 plant species integrated into PMRD.
miRNA expression profiling
In our PMRD database, we had already added data concerning miRNA expression profiling for several public miRNA expression data sets including maize miRNA-seq results (33), poplar cold stress-responsive miRNAs (6) (sample data shown in Supplementary Figure S1), tomato miRNA expression in leaf tissue (34), Arabidopsis stress (cold, salt and osmotic stresses) regulated miRNAs (35), maize miRNAs responding to salt stress in roots (36), miRNA expression profiles generated from Arabidopsis grown at different temperatures (GEO: GSE11535), and rice miRNAs responding to drought stress (GEO: GSE5986), etc.
For example, in the data set for poplar cold stress, there were 75 probes for 168 miRNAs, and a total of 21 samples including seven time points collected in triplicate. As shown in Supplementary Figure S1, the ratio represented the fold-change of poplar ptc-miR474b expression between cold treatment and control. We highlighted time points demonstrating a significant change in miRNA expression using P ≤ 0.01 as the cut-off.
In addition, we added our local rice oxidative stress related microarray data into PMRD. In order to investigate differences in expression levels of miRNAs in response to oxidative stress in rice, we conducted a series of experiments using the miRPlants_10.1 small RNA chip (LC Sciences, Houston, TX). One-week-old 9311 (rice Indica variety) seedling plants were incubated in a solution containing 10 μM methyl viologen (MV) (paraquat, an herbicide that induces oxidative stresses in plants) using water as a mock-treatment control, for 24 h. A total of four samples were examined in these experiments: 9311-CK01 and 9311-CK02 were controls and 9311-MV01 and 9311-MV02 were two samples treated with MV. Quantile normalization (37) was used to analyze the miRNA expression data. Within the total of 653 probes related to 1773 miRNAs representing 14 species in the chip, 18 probes showed significant differential expression (P ≤ 0.05, listed in Table 2) between MV treatment and mock, including 12 probes that were up-regulated and 6 probes that were down-regulated after MV treatment. Detailed relative expression levels of each probe are presented in the Expression page in the PMRD database. In this article, we present the example of osa-miR159a as Supplementary Figure S2.
Table 2.
Differentially expressed rice miRNAs after methyl viologen (MV) treatment
| Probe ID | Log2 ratio (MV/mock) | P-value | Target_Seq |
|---|---|---|---|
| ptc-miR474c | 2.17 | 3.67E−06 | CAAAAGCUGUUGGGUUUGGCUGGG |
| ptc-miR474b | 2.11 | 4.13E−05 | CAAAAGUUGUUGGGUUUGGCUGGG |
| ptc-miR474a | 2.08 | 1.49E−05 | CAAAAGUUGCUGGGUUUGGCUGGG |
| tae-miR1125 | 1.66 | 1.12E−02 | AACCAACGAGACCAACUGCGGCGG |
| ath-miR404 | 1.37 | 2.51E−05 | AUUAACGCUGGCGGUUGCGGCAGC |
| ath-miR854a | 0.80 | 7.00E−04 | GAUGAGGAUAGGGAGGAGGAG |
| osa-miR169b | 0.77 | 4.30E−02 | CAGCCAAGGAUGACUUGCCGG |
| ppt-miR903 | 0.76 | 4.31E−02 | GCUACUUCGGCGGGACAAGAGC |
| osa-miR529b | 0.68 | 2.29E−03 | GAGAAGAGAGAGAGUACAGC |
| osa-miR820a | 0.66 | 7.61E−03 | CGGCCUCGUGGAUGGACCAGG |
| ppt-miR900-5p | 0.65 | 4.60E−02 | UCCCAGGUACAAGAACACAGC |
| ppt-miR395 | 0.60 | 5.00E−03 | CUGAAGCGUUUGGGGGAAGG |
| osa-miR159f | −0.76 | 5.86E−04 | CUUGGAUUGAAGGGAGCUCUA |
| osa-miR396a | −0.81 | 2.72E−02 | UUCCACAGCUUUCUUGAACUG |
| osa-miR159a | −0.83 | 1.19E−04 | UUUGGAUUGAAGGGAGCUCUG |
| osa-miR397b | −0.99 | 6.99E−03 | UUAUUGAGUGCAGCGUUGAUG |
| tae-miR1120 | −1.08 | 2.36E−02 | ACAUUCUUAUAUUAUGAGACGGAG |
| osa-miR396c | −1.22 | 1.36E−03 | UUCCACAGCUUUCUUGAACUU |
miRNA target gene, genome browser and stem-loop information
In order to construct a collection of miRNA target genes, we mainly focused on Arabidopsis, rice, poplar, soybean, cotton, Medicago and maize. The target genes collection was curated from literature published in recent years. We also listed the location of interaction sites between miRNAs and target genes in Arabidopsis and rice [predicted by psRNATarget server (38,39)] in our database. Furthermore, we added information concerning miRNA action on target genes in Arabidopsis, rice and soybean into our local genome browser. We also provided the chromosomal location, secondary structure and dot matrix [generated by RNAfold (40)] of miRNA precursor sequences.
ACCESS TO THE DATABASE
The PMRD website was constructed using Hypertext Markup Language (HTML), perl CGI (http://www.perl.com), and the MySQL 4.0 (http://www.mysql.com) database engine. The detailed architecture of the database tables is presented in Supplementary Figure S3. These tables are divided into three sections: the first section concerns miRNAs, the second section is about target genes, and the last is a presentation of miRNA expression profiles. The whole database is run on a server managed by the LINUX operating system.
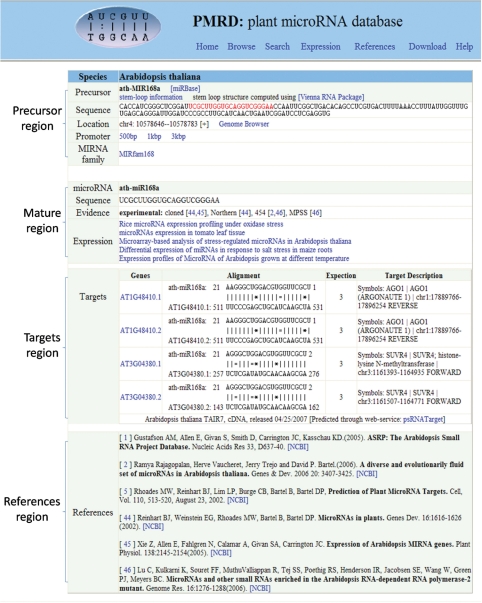
The PMRD database web interface enables users to view and analyze each miRNA through either the Browse page or Search page. Detail information page (shown as Figure 1) displays basic information for each unique miRNA. The evidence for each entry is either experimental or computational. We listed the methods used for experimentally demonstrated miRNAs and the programs applied for computational prediction. Within the total 8433 miRNAs in PMRD, 1297 of them were experimentally demonstrated and 7136 of them were from computational predictions (the detailed number shown in Table 1). Users can view a miRNA display in the local Genome Browser (shown as Supplementary Figure S4) in which users can extract information about Arabidopsis, rice and soybean. Through PMRD, users can also inspect miRNA expression profiles, target gene list and predicted interaction sites between miRNA and cDNA. In additional, the secondary structure of miRNA precursor could be predicted by using the prediction tool in our PMRD database. All miRNA sequences are available for downloading.
Figure 1.
Sample page for detailed information regarding each unique miRNA. Contents of this page are divided into four parts (miRNA ath-MIR168a was used as an example): the first section concerns miRNA precursor, including sequence, stem-loop information, miRNA family and location in the whole genome; the second section is about the mature miRNA, including sequence, evidence and expression patterns; the third section concerns target genes: for Arabidopsis and rice, the target genes and sites were predicted by psRNATarget server (38) in default parameters; for poplar, soybean, cotton, medicago and maize, the listed genes were collected from the references. The last section includes the related references. The miRNA family and the evidence were based on the definition from miRBase, Rfam and the references.
Currently, we have collected thousands of miRNA sequences from Arabidopsis, rice and poplar in our database, but for other species such as soybean, maize, wheat, cotton and sorghum, etc., the identified miRNAs are limited. Since this field is progressing rapidly due to new deep sequencing technologies, many more plant miRNAs will be discovered, as well as their target genes and expression profiles. The PMRD will be updated and maintained in order to integrate the newest plant miRNA data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China grants (2008AA02Z312 and 2006CB100105). Funding for open access charge: Ministry of Science and Technology of China (2008AA02Z312 and 2006CB100105).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Ms Wenying Xu for critical suggestions. They also thank Dongxia Yao and Qiang Wei for their help on taxonomic browse.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 3.Archak S, Nagaraju J. Computational prediction of rice (Oryza sativa) miRNA targets. Genomics Proteomics Bioinformatics. 2007;5:196–206. doi: 10.1016/S1672-0229(08)60007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sunkar R, Chinnusamy V, Zhu J, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X, Wang G, Sutoh K, Zhu JK, Zhang W. Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim. Biophys. Acta. 2008 doi: 10.1016/j.bbagrm.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Lu S, Sun YH, Chiang VL. Stress-responsive microRNAs in Populus. Plant J. 2008;55:131–151. doi: 10.1111/j.1365-313X.2008.03497.x. [DOI] [PubMed] [Google Scholar]
- 7.Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 2008;18:571–584. doi: 10.1101/gr.6897308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK. Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol. 2008;8:25. doi: 10.1186/1471-2229-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindow M, Jacobsen A, Nygaard S, Mang Y, Krogh A. Intragenomic matching reveals a huge potential for miRNA-mediated regulation in plants. PLoS Comput. Biol. 2007;3:e238. doi: 10.1371/journal.pcbi.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15:336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- 14.Sunkar R, Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 2008;8:37. doi: 10.1186/1471-2229-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gustafson AM, Allen E, Givan S, Smith D, Carrington JC, Kasschau KD. ASRP: the Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2005;33:D637–640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson C, Bowman L, Adai AT, Vance V, Sundaresan V. CSRDB: a small RNA integrated database and browser resource for cereals. Nucleic Acids Res. 2007;35:D829–833. doi: 10.1093/nar/gkl991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dezulian T, Palatnik J, Huson D, Weigel D. Conservation and divergence of microRNA families in plants. Genome Biol. 2005;6:P13. [Google Scholar]
- 21.Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O. Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics. 2008;9:160. doi: 10.1186/1471-2164-9-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Li P, Cao X, Wang X, Zhang A, Li X. Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochem. Biophys. Res. Commun. 2009;378:799–803. doi: 10.1016/j.bbrc.2008.11.140. [DOI] [PubMed] [Google Scholar]
- 23.Zhang B, Pan X, Stellwag EJ. Identification of soybean microRNAs and their targets. Planta. 2008;229:161–182. doi: 10.1007/s00425-008-0818-x. [DOI] [PubMed] [Google Scholar]
- 24.Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.) Genome Biol. 2007;8:R96. doi: 10.1186/gb-2007-8-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei B, Cai T, Zhang R, Li A, Huo N, Li S, Gu YQ, Vogel J, Jia J, Qi Y, et al. Novel microRNAs uncovered by deep sequencing of small RNA transcriptomes in bread wheat (Triticum aestivum L.) and Brachypodium distachyon (L.) Beauv. Funct. Integr. Genomics. 2009;9:532–541. doi: 10.1007/s10142-009-0128-9. [DOI] [PubMed] [Google Scholar]
- 26.Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- 27.Maher C, Timmermans M, Stein L, Ware D. Identifying microRNAs in plant genomes. Proc. IEEE CSB. 2004;6:718–723. [Google Scholar]
- 28.Xue LJ, Zhang JJ, Xue HW. Characterization and expression profiles of miRNAs in rice seeds. Nucleic Acids Res. 2009;37:916–930. doi: 10.1093/nar/gkn998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B, Pan X, Anderson TA. Identification of 188 conserved maize microRNAs and their targets. FEBS Lett. 2006;580:3753–3762. doi: 10.1016/j.febslet.2006.05.063. [DOI] [PubMed] [Google Scholar]
- 30.Khan Barozai MY, Irfan M, Yousaf R, Ali I, Qaisar U, Maqbool A, Zahoor M, Rashid B, Hussnain T, Riazuddin S. Identification of micro-RNAs in cotton. Plant Physiol. Biochem. 2008;46:739–751. doi: 10.1016/j.plaphy.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Qiu CX, Xie FL, Zhu YY, Guo K, Huang SQ, Nie L, Yang ZM. Computational identification of microRNAs and their targets in Gossypium hirsutum expressed sequence tags. Gene. 2007;395:49–61. doi: 10.1016/j.gene.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 32.Zhang B, Wang Q, Wang K, Pan X, Liu F, Guo T, Cobb GP, Anderson TA. Identification of cotton microRNAs and their targets. Gene. 2007;397:26–37. doi: 10.1016/j.gene.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Elling AA, Li X, Li N, Peng Z, He G, Sun H, Qi Y, Liu XS, Deng XW. Genome-Wide and Organ-Specific Landscapes of Epigenetic Modifications and Their Relationships to mRNA and Small RNA Transcriptomes in Maize. Plant Cell. 2009;21:1053–1069. doi: 10.1105/tpc.109.065714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Zeng R, Chen J, Liu X, Liao Q. Identification of conserved microRNAs and their targets from Solanum lycopersicum Mill. Gene. 2008;423:1–7. doi: 10.1016/j.gene.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 35.Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14:836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding D, Zhang L, Wang H, Liu Z, Zhang Z, Zheng Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann. Botany. 2008;103:29–38. doi: 10.1093/aob/mcn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu J, He X. Enhanced quantile normalization of microarray data to reduce loss of information in gene expression profiles. Biometrics. 2007;63:50–59. doi: 10.1111/j.1541-0420.2006.00670.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuster P, Fontana W, Stadler PF, Hofacker IL. From sequences to shapes and back: a case study in RNA secondary structures. Proceedings. 1994;255:279–284. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- 41.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adai A, Johnson C, Mlotshwa S, Archer-Evans S, Manocha V, Vance V, Sundaresan V. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 44.Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arteaga-Vazquez M, Caballero-Perez J, Vielle-Calzada JP. A family of microRNAs present in plants and animals. Plant Cell. 2006;18:3355–3369. doi: 10.1105/tpc.106.044420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franco-Zorrilla JM, Del Toro FJ, Godoy M, Perez-Perez J, Lopez-Vidriero I, Oliveros JC, Garcia-Casado G, Llave C, Solano R. Genome-wide identification of small RNA targets based on target enrichment and microarray hybridizations. Plant J. 2009;59:840–850. doi: 10.1111/j.1365-313X.2009.03904.x. [DOI] [PubMed] [Google Scholar]
- 47.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 48.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Kutter C, Schob H, Stadler M, Meins F, Jr, Si-Ammour A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell. 2007;19:2417–2429. doi: 10.1105/tpc.107.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu C, Kulkarni K, Souret FF, MuthuValliappan R, Tej SS, Poethig RS, Henderson IR, Jacobsen SE, Wang W, Green PJ, et al. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang XJ, Reyes JL, Chua NH, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 56.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 57.Mette MF, van der Winden J, Matzke M, Matzke AJ. Short RNAs can identify new candidate transposable element families in Arabidopsis. Plant Physiol. 2002;130:6–9. doi: 10.1104/pp.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou X, Wang G, Zhang W. UV-B responsive microRNA genes in Arabidopsis thaliana. Mol. Syst. Biol. 2007;3:103. doi: 10.1038/msb4100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jagadeeswaran G, Zheng Y, Li YF, Shukla LI, Matts J, Hoyt P, Macmil SL, Wiley GB, Roe BA, Zhang W, et al. Cloning and characterization of small RNAs from Medicago truncatula reveals four novel legume-specific microRNA families. New Phytol. 2009;184:85–98. doi: 10.1111/j.1469-8137.2009.02915.x. [DOI] [PubMed] [Google Scholar]
- 60.Wang L, Wang MB, Tu JX, Helliwell CA, Waterhouse PM, Dennis ES, Fu TD, Fan YL. Cloning and characterization of microRNAs from Brassica napus. FEBS Lett. 2007;581:3848–3856. doi: 10.1016/j.febslet.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 61.Xie FL, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM. Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett. 2007;581:1464–1474. doi: 10.1016/j.febslet.2007.02.074. [DOI] [PubMed] [Google Scholar]
- 62.He XF, Fang YY, Feng L, Guo HS. Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett. 2008;582:2445–2452. doi: 10.1016/j.febslet.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 63.Porter BW, Aizawa KS, Zhu YJ, Christopher DA. Differentially expressed and new non-protein-coding genes from a Carica papaya root transcriptome survey. Plant Sci. 2008;174:38–50. [Google Scholar]
- 64.Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- 66.Hinas A, Reimegard J, Wagner EG, Nellen W, Ambros VR, Soderbom F. The small RNA repertoire of Dictyostelium discoideum and its regulation by components of the RNAi pathway. Nucleic Acids Res. 2007;35:6714–6726. doi: 10.1093/nar/gkm707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou ZS, Huang SQ, Yang ZM. Bioinformatic identification and expression analysis of new microRNAs from Medicago truncatula. Biochem. Biophys. Res. Commun. 2008;374:538–542. doi: 10.1016/j.bbrc.2008.07.083. [DOI] [PubMed] [Google Scholar]
- 68.Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics. 2008;9:593. doi: 10.1186/1471-2164-9-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guddeti S, Zhang DC, Li AL, Leseberg CH, Kang H, Li XG, Zhai WX, Johns MA, Mao L. Molecular evolution of the rice miR395 gene family. Cell Res. 2005;15:631–638. doi: 10.1038/sj.cr.7290333. [DOI] [PubMed] [Google Scholar]
- 70.Arazi T, Talmor-Neiman M, Stav R, Riese M, Huijser P, Baulcombe DC. Cloning and characterization of micro-RNAs from moss. Plant J. 2005;43:837–848. doi: 10.1111/j.1365-313X.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- 71.Sunkar R, Girke T, Jain PK, Zhu JK. Cloning and characterization of microRNAs from rice. Plant Cell. 2005;17:1397–1411. doi: 10.1105/tpc.105.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y, Li W, Jin YX. Computational identification of novel family members of microRNA genes in Arabidopsis thaliana and Oryza sativa. Acta Biochim Biophys. Sin. 2005;37:75–87. [PubMed] [Google Scholar]
- 73.Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008;18:1456–1465. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, German R, Thatcher SR, Maher C, Zhang L, Ware D, et al. Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs) Proc. Natl Acad. Sci. USA. 2008;105:4951–4956. doi: 10.1073/pnas.0708743105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lacombe S, Nagasaki H, Santi C, Duval D, Piegu B, Bangratz M, Breitler JC, Guiderdoni E, Brugidou C, Hirsch J, et al. Identification of precursor transcripts for 6 novel miRNAs expands the diversity on the genomic organisation and expression of miRNA genes in rice. BMC Plant Biol. 2008;8:123. doi: 10.1186/1471-2229-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu B, Li P, Li X, Liu C, Cao S, Chu C, Cao X. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 2005;139:296–305. doi: 10.1104/pp.105.063420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu S, Sun YH, Amerson H, Chiang VL. MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. 2007;51:1077–1098. doi: 10.1111/j.1365-313X.2007.03208.x. [DOI] [PubMed] [Google Scholar]
- 78.Luo YC, Zhou H, Li Y, Chen JY, Yang JH, Chen YQ, Qu LH. Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Lett. 2006;580:5111–5116. doi: 10.1016/j.febslet.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 79.Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fattash I, Voss B, Reski R, Hess WR, Frank W. Evidence for the rapid expansion of microRNA-mediated regulation in early land plant evolution. BMC Plant Biol. 2007;7:13. doi: 10.1186/1471-2229-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Talmor-Neiman M, Stav R, Frank W, Voss B, Arazi T. Novel micro-RNAs and intermediates of micro-RNA biogenesis from moss. Plant J. 2006;47:25–37. doi: 10.1111/j.1365-313X.2006.02768.x. [DOI] [PubMed] [Google Scholar]
- 82.Cho SH, Addo-Quaye C, Coruh C, Arif MA, Ma Z, Frank W, Axtell MJ. Physcomitrella patens DCL3 is required for 22-24 nt siRNA accumulation, suppression of retrotransposon-derived transcripts, and normal development. PLoS Genet. 2008;4:e1000314. doi: 10.1371/journal.pgen.1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science (New York, NY) 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 84.Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL. Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008;18:1602–1609. doi: 10.1101/gr.080127.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bedell JA, Budiman MA, Nunberg A, Citek RW, Robbins D, Jones J, Flick E, Rholfing T, Fries J, Bradford K, et al. Sorghum genome sequencing by methylation filtration. PLoS Biol. 2005;3:e13. doi: 10.1371/journal.pbio.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]