Abstract
PrimerBank (http://pga.mgh.harvard.edu/primerbank/) is a public resource for the retrieval of human and mouse primer pairs for gene expression analysis by PCR and Quantitative PCR (QPCR). A total of 306 800 primers covering most known human and mouse genes can be accessed from the PrimerBank database, together with information on these primers such as Tm, location on the transcript and amplicon size. For each gene, at least one primer pair has been designed and in many cases alternative primer pairs exist. Primers have been designed to work under the same PCR conditions, thus facilitating high-throughput QPCR. There are several ways to search for primers for the gene(s) of interest, such as by: GenBank accession number, NCBI protein accession number, NCBI gene ID, PrimerBank ID, NCBI gene symbol or gene description (keyword). In all, 26 855 primer pairs covering most known mouse genes have been experimentally validated by QPCR, agarose gel analysis, sequencing and BLAST, and all validation data can be freely accessed from the PrimerBank web site.
INTRODUCTION
Quantitative Polymerase Chain Reaction (QPCR) has become a commonly used method for precise determination of gene expression and evaluating DNA microarray data (1,2). The main advantages of this technique are its unparalleled dynamic range, being able to detect >107-fold differences in expression, and the potential to amplify very small amounts of DNA template, down to a single copy (3–5). QPCR products can be detected by two general methods: one utilizing various types of fluorescence containing hybridization probes (6–20) and the other utilizing SYBR Green I dye fluorescence (21–23). Hybridization probes are designed to be target specific and can thus minimize nonspecific amplification, but can be difficult to design and costly (5). The SYBR Green I method is the most simple and inexpensive QPCR method and has become the most commonly used for gene expression analysis (21,22). SYBR Green I dye intercalation into double-stranded DNA, such as PCR products, results in detectable fluorescence, corresponding to the amount of PCR product generated in each cycle (23). QPCR amplification plots provide information for relative quantification between samples and on the amount of initial DNA template (24–26). Dissociation curves, generated after the QPCR step, can give information on the specificity of the reaction (27).
We have developed a database, named PrimerBank (http://pga.mgh.harvard.edu/primerbank/), for the retrieval of human and mouse primer pairs for gene expression analysis by PCR and QPCR. PrimerBank primers can work with SYBR Green I detection methods and the primer design was based on an algorithm that had been previously used for oligonucleotide probe design for DNA microarrays (28). Nonspecific amplification of nontarget sequences is a common problem encountered in PCR and QPCR experiments. So, for the PrimerBank primer design, various filters for cross-reactivity were used to reduce nonspecific amplification (29). Furthermore, all primers have been designed to work under a high annealing temperature of 60°C. At least one primer pair represents each gene, and in many cases alternative primer pairs have been designed. See Table 1 for information on primers contained in PrimerBank. In addition, we have previously experimentally validated 26 855 primer pairs, which cover most known mouse genes (30). We found that PrimerBank primers can amplify specifically the genes for which they have been designed (82.6% success rate based on visualization of one band of the expected size by agarose gel analysis). The reproducibility of the QPCR technique and the uniformity of amplification using PrimerBank primers were also analyzed (30). Furthermore, the amplification efficiency of the QPCR using PrimerBank primers was determined and it was found that for 13 primer pairs tested it ranged from 79% to 96%, using an analytical method. The same 13 PrimerBank primer pairs as above were used and one-way ANOVA (ANalysis Of VAriance) analysis was done using each primer pair in a series of titration QPCRs of template DNA, in order to determine if amplification efficiencies were similar between different PrimerBank primer pairs. The efficiencies were found to be similar between these primer pairs (P = 0.7338 i.e. P > 0.05) (30). Since PrimerBank primers have been designed to be used under the same annealing temperature, high-throughput QPCR in parallel is facilitated, as an alternative approach to DNA microarrays (31,32) for the study of gene expression.
Table 1.
Primers that can be retrieved from PrimerBank
| Number of primers | Number of genes covered | Organism | Number of experimentally validated primer pairs |
|---|---|---|---|
| 306 800 | 61 425 | All organisms | 26 855 |
| 138 918 | 27 684 | Mus musculus | 26 855 |
| 167 882 | 33 741 | Homo sapiens |
Primers stored in the PrimerBank database have been designed to represent most known human and mouse genes. A total of 26 855 primer pairs, representing 27 684 mouse genes, have been experimentally validated by QPCR, agarose gel electrophoresis, sequencing and BLAST and all experimental validation data can be viewed from the PrimerBank web site.
PRIMER DESIGN
Oligonucleotide probe sequence design for DNA microarrays has become the subject of many studies using a number of algorithms (33–46). Most of these algorithms use BLAST (47) to identify regions of the gene from which oligonucleotide sequences can be selected (48). PCR primer design can be based on these algorithms, since BLAST is used in design of both DNA microarray probes and PCR primers. The PrimerBank primer design was based on a successful approach that had been previously used for the prediction of oligonucleotide probes for DNA microarrays (28). However, the PrimerBank primer design differs by the addition of filters that are considered to be important for primer specificity (29).
All gene sequence information was obtained from the NCBI protein database (http://www.ncbi.nlm.nih.gov/entrez/) (49). DNA-coding sequences were retrieved and the redundant sequences were clustered using the DeRedund program (28). Low-complexity regions, which may contribute to primer cross-reactivity (50), were excluded using the program DUST (51). If primers contained six or more identical contiguous bases they were rejected, so that more complicated sequences could be chosen. Furthermore, no primers were selected from low-quality regions of sequence (29). Primers were designed to represent at least once each gene, and most known human and mouse genes were covered. See Table 2 for the statistics of primer pair design with respect to gene representation.
Table 2.
PrimerBank mouse primer pair design and validation
| Mouse primer pairs or genes | Number of mouse genes/primer pairs validated | Percentage (%) of total primer pairs |
|---|---|---|
| Total number of primer pairs | 26 855 | 100 |
| Total number of genes represented | 27 684 | – |
| Total number of genes not represented | 1165 | – |
| Primer pairs with no redundancy | 23 700 | 88.2 |
| Primer pairs with two target genes | 2534 | 9.4 |
| Primer pairs with >2 target genes | 621 | 2.3 |
| Total number of successful primer pairs based on all validation criteria | 17 483 | 65.1 |
| Total number of successful primer pairs based on agarose gel electrophoresis | 22 189 | 82.6 |
| Total number of successful primer pairs based on BLAST analysis | 19 453 | 72.4 |
| Total number of failed primer pairs by QPCR (due to no amplification) | 1745 | 6.5 |
A total of 26 855 primer pairs were synthesized, which correspond to a higher number of 27 684 mouse gene targets since some of these primer pairs amplify the same sequence from two genes or gene isoforms. Primers were not designed for another 1165 mouse genes, mainly because of low sequence quality. The average mouse gene has 1293 bp; however, the average length for these genes is 435 bp and most are ‘unknown’ or RIKEN sequences.
In many cases, coding regions were scanned from the 5′- to the 3′-end until three suitable primer pairs were found (in these cases the PrimerBank IDs of the primers contain ‘a1’, ‘a2’ or ‘a3’, the ‘a1’ primer pair being most 5′ and the ‘a3’ being most 3′). Two general methods can be used for cDNA library preparation: the oligo(dT) and random priming methods. Oligo(dT) priming during cDNA preparation can result in reduced coverage of the 5′-end of sequences, since some 3′ UTRs can be very long (3). Random priming can result in the highest coverage of the 5′-end and this method was used for our cDNA preparations (3,30). Because of this higher coverage at the 5′-end, the most 5′primers were experimentally validated (see ‘Database generation and content’ section below). Also, primers were designed irrespective of their location on exons. In order to prevent any nonspecific amplification of any contaminating genomic DNA, primers can be designed to be located on exon boundaries; however, in many cases it was not possible to design primers located on exon boundaries that fulfilled all the design criteria, since some transcripts consist of a single exon. See Table 3 for the statistics of primer location with respect to exons.
Table 3.
Analysis of PrimerBank primer pair genomic position
| F primer | R primer | Number of PrimerBank mouse primer pairs |
|---|---|---|
| Analyzed by BLAST | Analyzed by BLAST | 26 854 |
| Matched to genome sequences | Matched to genome sequences | 19 668 |
| Located on exon–exon boundary | Located on exon–exon boundary | 311 |
| Located on exon | Located on exon–exon boundary | 1576 |
| Located on exon–exon boundary | Located on exon | 1425 |
| Located on exon | Located on exon | 16 356 |
| Located on the same exon | Located on the same exon | 11 235 |
Mouse genome sequences from the UCSC genome browser were downloaded and the primer pair sequences were matched by BLASTn to the genome sequences, in order to identify the location of 26 854 mouse forward (F) and reverse (R) primer pairs with respect to exons. A total of 19 668 primer pairs matched to sequences that were downloaded from the genome browser. The remaining sequences did not match probably due to differences in genomic information.
PrimerBank primers have been designed to have uniform length and GC% properties (29). All PrimerBank primers are 19–23 nt, with a preferred length of 21 nt. This length is optimal for gene-specific sequences and minimizes cross-reactivity. Also, this length is optimized to reduce costs if primers are synthesized in large sets. Primers have similar GC% from 35% to 65% in order to ensure uniform priming. The algorithm used for primer design also evaluated the ΔG value for the last five residues at the 3′-end of the primers and a threshold value of −9 kcal/mol was adopted for primer rejection. This was done in order to minimize nonspecific amplification, since the 3′ part of the primer contributes most to nonspecific primer extension, especially if the binding of these residues is relatively stable (52).
The melting temperature (Tm) determines the optimal annealing temperature. Various methods exist to determine the Tm (53–55). We used the nearest neighbor method (55) based on which all primer Tms are between 60°C and 63°C. Thus, a high annealing temperature can be used for these primers, reducing nonspecific amplification, which is a frequent problem in PCR experiments. All primers were designed to amplify short amplicons of 150–350 bp and occasionally, if this requirement could not be satisfied due to other design constraints, 100–800 bp amplicons were accepted. Short amplicons can be amplified more easily and the PCR efficiency of these reactions is higher.
Our main filter for cross-reactivity was the rejection of primers containing contiguous residues also found in other sequences. We have found that a filter cutoff rejecting perfect 15-mer matches was the most stringent feasible filter (28). So, if a repetitive 15-mer was present in the primer, it was rejected (by comparing every possible 15-mer in the primer sequence to both strands of all known sequences in the design space). In order to determine if there is any cross-reactivity, BLAST searches for sequence similarity were carried out against all known sequences in the design space and primers accepted were required to have BLAST scores of <30 (28). Additional filters were applied to compensate for templates from noncoding RNAs, which are very abundant when using random priming (29).
In order to reduce self-complementarity, no contiguous 5-mer match was allowed anywhere between a primer and its complementary sequence (29). A BLAST similarity search for the primer sequence was carried out on the complementary strand and the score was required to be <18. To prevent primer dimers, primers were rejected if the four residues at the 3′-end of the primer were found in its complementary sequence. Complementarity in the forward and reverse primers in a pair was also evaluated in order to prevent heterodimer formation.
The filters used for primer selection were very stringent and this was reflected by the fact that 99.5% of primers were rejected. Of the rejected primers, 50.7% had too high or too low Tm values, 28.7% cross-hybridized to nontarget sequences, 19.8% were rejected because of sequence self-complementarity, 0.5% were from low-complexity regions and 0.3% were rejected because of other properties (GC content and end stability).
DATABASE GENERATION AND CONTENT
We have stored the primer sequences in the PrimerBank database (http://pga.mgh.harvard.edu/primerbank/) together with other information about the primers such as Tm, location on the transcript and expected amplicon size. Furthermore, we have experimentally validated 26 855 of these primer pairs, corresponding to 27 684 mouse transcripts, by QPCR, agarose gel electrophoresis, sequencing and BLAST analysis (30). Random priming was used to prepare the cDNA library from a commercial universal mouse composite total RNA preparation, which contains RNA from a panel of 11 different mouse cell types for a good representation of the majority of mouse genes. The cDNA library prepared was used as the template in all QPCRs for the high-throughput validation and the same PCR conditions were used for all reactions. QPCR amplification plots and dissociation curves were analyzed and the presence of a single band of the expected size was determined by agarose gel analysis. Sequences obtained from these PCR products were BLAST analyzed as batch sets against the NCBI database (47). For the identification of successful samples, the main parameters considered were the alignment length of the PCR product sequence to the expected, the expected sequence match position to the sequence returned by NCBI BLASTn and the percent identity of the two sequences. The success rate of the high-throughput PrimerBank primer validation experiments was high, observed both from agarose gel and BLAST analysis, and 17 483 primer pairs were found to be successful based on all validation criteria. See Table 2 for data obtained from the high-throughput experimental validation procedure.
Because of no amplification, 1745 samples (6.5%) failed. Several were found to belong to olfactory receptors, vomeronasal receptors, transcription factors and low abundance transcripts while others were of unknown function or RIKEN sequences. To determine whether the templates for the failed primer pairs were present in the cDNA sample used, we tested these primers using genomic DNA template (30). From these experiments, we found that these primer pairs had originally failed in most cases because their respective templates were not present or present in very low amounts in the source of DNA template used and not because of poor primer design. Specific tissues may be used as sources of cDNA templates where expression of the genes of interest is known, for increased amplification success. Furthermore, we determined the uniformity of amplification using fully validated PrimerBank primer pairs i.e. primer pairs that had been successful in all steps of the validation procedure and found that amplification using PrimerBank primers was relatively uniform (30).
WEB INTERFACE
Search tools
The PrimerBank database can be searched for primers for a gene of interest using any of the following search terms: GenBank accession number, NCBI protein accession number, NCBI gene ID, PrimerBank ID, NCBI gene symbol or keyword (gene description). Search results show the primer sequences and some information on the primers, such as the expected amplicon size and Tm. The cDNA and amplicon sequences as well as the experimental validation data can be viewed from the PrimerBank search result web pages, by clicking on the appropriate links. All validation data can be retrieved from the PrimerBank web site, since the criteria of the users for success or fail may be different from our validation criteria. In addition, users can use a BLAST tool that can be found on the PrimerBank homepage, to find any primers contained in the PrimerBank database that would amplify their sequence of interest. Users can also BLAST analyze the sequence obtained from the PCR product generated during the high-throughput experimental validation procedure by using a BLAST tool to query the NCBI database, which can be found on the validation data web page.
Sample search results
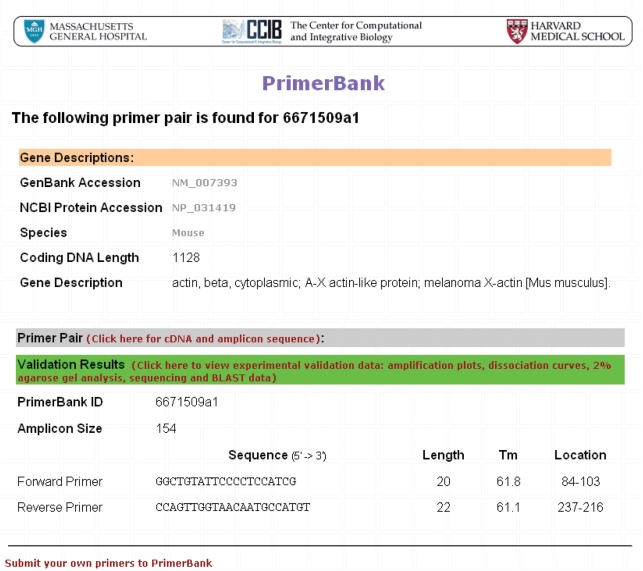
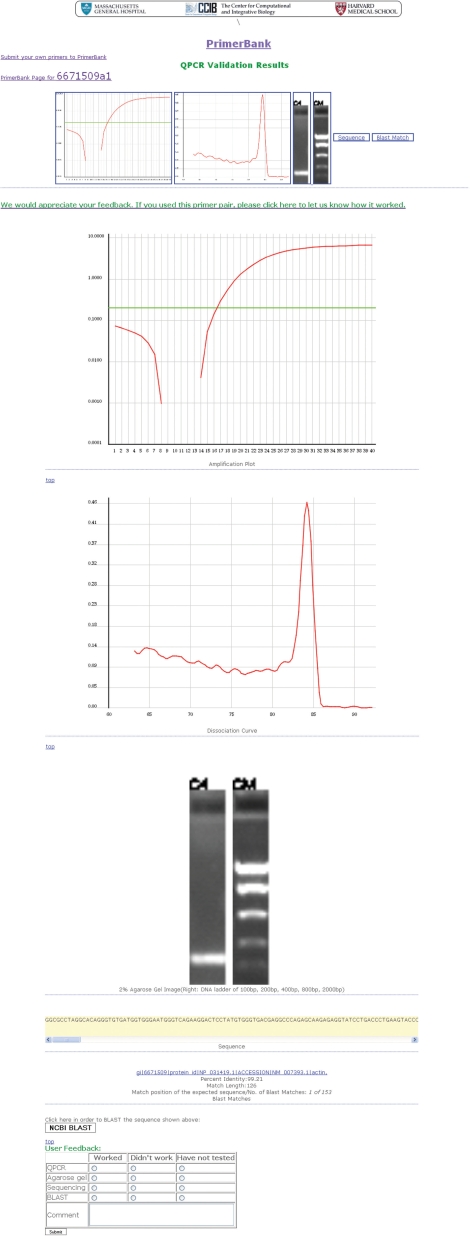
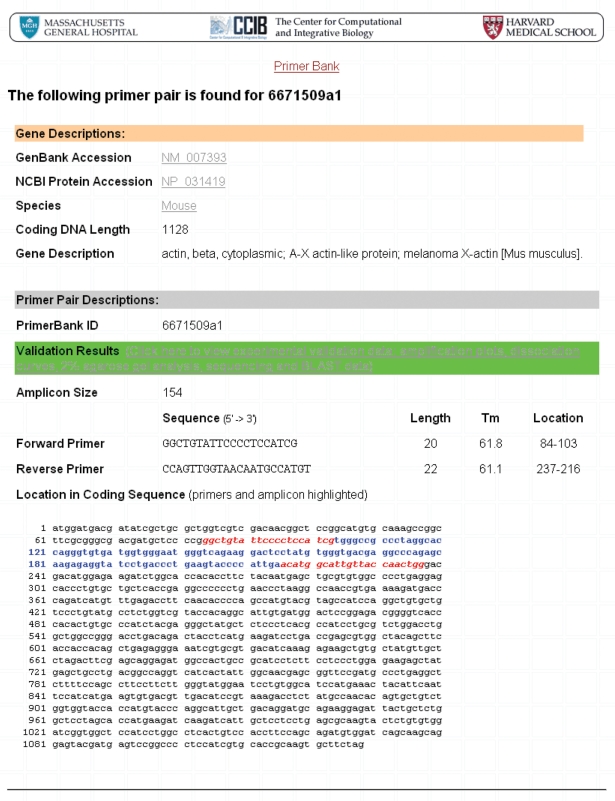
The results obtained from a primer pair search (mouse beta-actin primer pair; PrimerBank ID: 6671509a1) can be seen in Figures 1–3, as an example. Primer sequences for mouse beta-actin can be viewed together with information on the primer Tms and expected amplicon size (Figure 1). Users can click on the cDNA and amplicon sequence link shown on the primer information web page to view the full cDNA sequence and highlighted location of primers on this (Figure 2). In order to view the experimental validation data, users can click on the validation results link shown on the primer information web page and on the cDNA and amplicon sequence web page. On the experimental validation web page, the QPCR amplification plot, followed by the dissociation curve and agarose gel data can be seen (Figure 3). The sequence obtained follows and it is possible for users to scroll through this to view it in its entirety. A summary of the BLAST results obtained is shown below the sequencing result, including the percent (%) identity of the sequence obtained to the expected sequence, the match length of the two sequences and the match position of the expected sequence out of the total number of sequences to which the queried sequence matched to (Figure 3).
Figure 1.
PrimerBank search results for beta-actin primers. The primer search function can be found on the PrimerBank homepage. The database was searched for mouse beta-actin primer pairs by PrimerBank ID (6671509a1) and the search results obtained are shown here. The primer sequences, lengths, Tms, location of primers on the amplicon and expected amplicon size can be seen.
Figure 3.
Experimental validation data for beta-actin primers. All experimental validation data are seen here and can be viewed from PrimerBank by clicking on the validation results links found on the primer information web page (seen in Figure 1) and on the cDNA and amplicon sequence web page (seen in Figure 2). The QPCR amplification plot, the dissociation curve and agarose gel data can be seen followed by the sequence obtained, which users can scroll through. A summary of the BLAST results obtained can be seen below the sequencing result. A BLAST tool can be seen below the BLAST results, which can be used to directly BLAST the sequence obtained against the NCBI database. Also, a feedback table can be seen, where users can provide information about their experimental data using the primers.
Figure 2.
cDNA and amplicon sequence for beta-actin primers. The full cDNA and amplicon sequences as well as the highlighted location of primers on the amplicon are seen here and can be viewed from PrimerBank by clicking on the cDNA and amplicon sequence link found on the primer information web page (seen in Figure 1).
Primer statistics
Users can click on the ‘Primer Statistics’ tab found on the PrimerBank homepage to view some statistics of the primers currently contained in the database.
Protocols
The QPCR and reverse transcription protocols used for the high-throughput primer validation procedure can be found on the PrimerBank web site, as well as a troubleshooting guide, under the ‘PCR Protocol’ tab. These protocols may be used with all PrimerBank primer pairs, unless specific protocols are required.
Comments
Users can provide their comments on the PrimerBank web site or primer design by clicking on the ‘Comments’ tab seen on the homepage and filling out the ‘Comments’ box. Also, users can input information regarding their experimental use of the validated primer pairs and add any comments in a feedback table that can be seen on the validation data web page.
Primer submission
Users can recommend their own primer pairs for human and mouse genes by clicking on the ‘Primer submission’ tab seen on the homepage. In order to do this, users must provide their name, name of institution, email address and a password (optionally) the first time when they submit primers. If the submitted primers conform to the PrimerBank standards they will be added to the database.
DISCUSSION
A large number of tools and resources are available that can be used for designing PCR primers for various applications (56–67). In addition, databases exist that contain primers for PCR and QPCR, which have been submitted by researchers, but only a few thousand of these primer pairs have been experimentally validated (68,69). Also, primers contained in these databases have not been designed to work under the same PCR conditions. Therefore, it would be required to validate the primers for the gene(s) of interest and optimize the PCR conditions in order to use these primers. The PrimerBank database contains experimentally validated primers for most known mouse genes (17 483 primer pairs were successful based on all validation criteria) and all primers work under the same PCR conditions. PrimerBank thus provides a resource for researchers who are interested in validating DNA microarray data or high-throughput QPCR.
FUNDING
National Institutes of Health Program for Genomic Applications (grant U01 HL66678). Funding for open access charge: Center for Computational and Integrative Biology, Massachusetts General Hospital.
Conflict of interest statement. None declared.
REFERENCES
- 1.Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 2.Arikawa E, Sun Y, Wang J, Zhou Q, Ning B, Dial SL, Guo L, Yang J. Cross-platform comparison of SYBR Green real-time PCR with TaqMan PCR, microarrays and other gene expression measurement technologies evaluated in the MicroArray Quality Control (MAQC) study. BMC Genomics. 2008;9:328. doi: 10.1186/1471-2164-9-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR – a perspective. J. Mol. Endocrinol. 2005;34:597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
- 4.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 5.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 6.Cardullo RA, Agrawal S, Flores C, Zamecnik PC, Wolf DE. Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 1988;85:8790–8794. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holland P, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′ to 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl Acad. Sci. USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee LG, Connell CR, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 10.Emig M, Saussele S, Wittor H, Weisser A, Reiter A, Willer A, Berger U, Hehlmann R, Cross NC, Hochhaus A. Accurate and rapid analysis of residual disease in patients with CML using specific fluorescent hybridization probes for real time quantitative RT-PCR. Leukemia. 1999;13:1825–1832. doi: 10.1038/sj.leu.2401566. [DOI] [PubMed] [Google Scholar]
- 11.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 12.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 13.Marras SAE, Kramer FR, Tyagi S. Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal. 1999;14:151–156. doi: 10.1016/s1050-3862(98)00018-7. [DOI] [PubMed] [Google Scholar]
- 14.Whitcombe D, Theaker J, Guy SP, Brown T, Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat. Biotechnol. 1999;17:804–807. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- 15.Thelwell N, Millington S, Solinas A, Booth J, Brown T. Mode of action and application of Scorpion primers to mutation detection. Nucleic Acids Res. 2000;28:3752–3761. doi: 10.1093/nar/28.19.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svanvik N, Westman G, Wang D, Kubista M. Light-up probes: thiazole orange-conjugated peptide nucleic acid for detection of target nucleic acid in homogeneous solution. Anal. Biochem. 2000;281:26–35. doi: 10.1006/abio.2000.4534. [DOI] [PubMed] [Google Scholar]
- 17.Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, et al. 3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solinas A, Brown LJ, McKeen C, Mellor JM, Nicol J, Thelwell N, Brown T. Duplex Scorpion primers in SNP analysis and FRET applications. Nucleic Acids Res. 2001;29:e96. doi: 10.1093/nar/29.20.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, Luan G, Guo Q, Liang J. A new class of homogeneous nucleic acid probes based on specific displacement hybridization. Nucleic Acids Res. 2002;30:e5. doi: 10.1093/nar/30.2.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nazarenko I, Lowe B, Darfler M, Ikonomi P, Schuster D, Rashtchian A. Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore. Nucleic Acids Res. 2002;30:e37. doi: 10.1093/nar/30.9.e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittwer CT, Hermann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
- 22.Morrison TB, Weis JJ, Wittwer CT. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques. 1998;24:954–958, 960, 962. [PubMed] [Google Scholar]
- 23.Zipper H, Brunner H, Bernhagen J, Vitzthum F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 2004;32:e103. doi: 10.1093/nar/gnh101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higuchi R, Dollinger G, Walsh PS, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology. 1992;10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 25.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 26.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L, et al. The real-time polymerase chain reaction. Mol. Aspects Med. 2006;27:95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Seed B. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics. 2003;19:796–802. doi: 10.1093/bioinformatics/btg086. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A comprehensive collection of experimentally validated primers for polymerase chain reaction quantitation of murine transcript abundance. BMC Genomics. 2008;9:633. doi: 10.1186/1471-2164-9-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schinke-Braun M, Couget JA. Expression profiling using affymetrix genechip probe arrays. Methods Mol. Biol. 2007;366:13–40. doi: 10.1007/978-1-59745-030-0_2. [DOI] [PubMed] [Google Scholar]
- 32.Lee NH, Saeed AI. Microarrays: an overview. Methods Mol. Biol. 2007;353:265–300. doi: 10.1385/1-59745-229-7:265. [DOI] [PubMed] [Google Scholar]
- 33.Chang P-C, Peck K. Design and assessment of a fast algorithm for identifying specific probes for human and mouse genes. Bioinformatics. 2003;19:1311–1317. doi: 10.1093/bioinformatics/btg162. [DOI] [PubMed] [Google Scholar]
- 34.Rouillard J-M, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chou H-H, Hsia A-P, Mooney DL, Schnable PS. Picky: oligo microarray design for large genomes. Bioinformatics. 2004;20:2893–2902. doi: 10.1093/bioinformatics/bth347. [DOI] [PubMed] [Google Scholar]
- 36.Reymond N, Charles H, Duret L, Calevro F, Beslon G, Fayard J-M. ROSO: optimizing oligonucleotide probes for microarrays. Bioinformatics. 2004;20:271–273. doi: 10.1093/bioinformatics/btg401. [DOI] [PubMed] [Google Scholar]
- 37.Li X, He Z, Zhou J. Selection of optimal oligonucleotide probes for microarrays using multiple criteria, global alignment and parameter estimation. Nucleic Acids Res. 2005;33:6114–6123. doi: 10.1093/nar/gki914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordberg EK. YODA: selecting signature oligonucleotides. Bioinformatics. 2005;21:1365–1370. doi: 10.1093/bioinformatics/bti182. [DOI] [PubMed] [Google Scholar]
- 39.Rouchka EC, Khalyfa A, Cooper NGF. MPrime: efficient large-scale multiple primer and oligonucleotide design for customized gene microarrays. BMC Bioinformatics. 2005;6:175. doi: 10.1186/1471-2105-6-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rimour S, Hill D, Militon C, Peyret P. GoArrays: highly dynamic and efficient microarray probe design. Bioinformatics. 2005;21:1094–1103. doi: 10.1093/bioinformatics/bti112. [DOI] [PubMed] [Google Scholar]
- 41.Wernersson R, Nielsen HB. OligoWiz 2.0–integrating sequence feature annotation into the design of microarray probes. Nucleic Acids Res. 2005;33:W611–W615. doi: 10.1093/nar/gki399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, Ying X. Mprobe 2.0: computer-aided probe design for oligonucleotide microarray. Appl. Bioinformatics. 2006;5:181–186. doi: 10.2165/00822942-200605030-00006. [DOI] [PubMed] [Google Scholar]
- 43.Schretter C, Milinkovitch MC. OligoFaktory: a visual tool for interactive oligonucleotide design. Bioinformatics. 2006;22:115–116. doi: 10.1093/bioinformatics/bti728. [DOI] [PubMed] [Google Scholar]
- 44.Hu G, Llinas M, Li J, Preiser PR, Bozdech Z. Selection of long oligonucleotides for gene expression microarrays using weighted rank-sum strategy. BMC Bioinformatics. 2007;8:350. doi: 10.1186/1471-2105-8-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S-H, Lo C-Z, Tsai M-C, Hsiung CA, Lin C-Y. The unique probe selector: a comprehensive web service for probe design and oligonucleotide arrays. BMC Bioinformatics. 2008;9(Suppl. I):S8. doi: 10.1186/1471-2105-9-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leparc GG, Tuchler T, Striedner G, Bayer K, Sykacek P, Hofacker IL, Kreil DP. Model-based probe set optimization for high-performance microarrays. Nucleic Acids Res. 2009;37:e18. doi: 10.1093/nar/gkn1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Lemoine S, Combes F, Le Crom S. An evaluation of custom microarray applications: the oligonucleotide design challenge. Nucleic Acids Res. 2009;37:1726–1739. doi: 10.1093/nar/gkp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 51.Hancock JM, Armstrong JS. SIMPLE34: an improved and enhanced implementation for VAX and Sun computers of the SIMPLE algorithm for analysis of clustered repetitive motifs in nucleotide sequences. Comput. Appl. Biosci. 1994;10:67–70. doi: 10.1093/bioinformatics/10.1.67. [DOI] [PubMed] [Google Scholar]
- 52.Rychlik W. Priming efficiency in PCR. Biotechniques. 1995;18:84–86. 88–90. [PubMed] [Google Scholar]
- 53.Breslauer KJ, Frank R, Blocker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugimoto N, Nakano S, Yoneyama M, Honda K. Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res. 1996;24:4501–4505. doi: 10.1093/nar/24.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.SantaLucia JJ. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pesole G, Liuni S, Grillo G, Belichard P, Trenkle T, Welsh J, McClelland M. GeneUp: a program to select short PCR primer pairs that occur in multiple members of sequence lists. Biotechniques. 1998;25:112–117. doi: 10.2144/98251bc02. 120–123. [DOI] [PubMed] [Google Scholar]
- 57.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Misener S, Krawetz SA, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ, USA: Humana Press Inc.; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 58.Podowski RM, Sonnhammer ELL. MEDUSA: large scale automatic selection and visual assessment of PCR primer pairs. Bioinformatics. 2001;17:656–657. doi: 10.1093/bioinformatics/17.7.656. [DOI] [PubMed] [Google Scholar]
- 59.Gordon PMK, Sensen CW. Osprey: a comprehensive tool employing novel methods for the design of oligonucleotides for DNA sequencing and microarrays. Nucleic Acids Res. 2004;32:e133. doi: 10.1093/nar/gnh127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wrobel G, Kokocinski F, Lichter P. AutoPrime: selecting primers for expressed sequences. Genome Biol. 2004;5:PII. [Google Scholar]
- 61.Marshall OJ. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20:2471–2472. doi: 10.1093/bioinformatics/bth254. [DOI] [PubMed] [Google Scholar]
- 62.Andersson A, Bernander R, Nilsson P. Dual-genome primer design for construction of DNA microarrays. Bioinformatics. 2005;21:325–332. doi: 10.1093/bioinformatics/bti001. [DOI] [PubMed] [Google Scholar]
- 63.Wu X, Munroe DJ. EasyExonPrimer: automated primer design for exon sequences. Appl. Bioinformatics. 2006;5:119–120. doi: 10.2165/00822942-200605020-00007. [DOI] [PubMed] [Google Scholar]
- 64.Fernandes R, Skiena S. MultiPrimer: a system for microarray PCR primer design. Methods Mol. Biol. 2007;402:305–314. doi: 10.1007/978-1-59745-528-2_15. [DOI] [PubMed] [Google Scholar]
- 65.Kim N, Lee C. QPRIMER: a quick web-based application for designing conserved PCR primers from multigenome alignments. Bioinformatics. 2007;23:2331–2333. doi: 10.1093/bioinformatics/btm343. [DOI] [PubMed] [Google Scholar]
- 66.Li K, Brownley A, Stockwell TB, Beeson K, McIntosh TC, Busam D, Ferriera S, Murphy S, Levy S. Novel computational methods for increasing PCR primer design effectiveness in directed sequencing. BMC Bioinformatics. 2008;9:191. doi: 10.1186/1471-2105-9-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.You FM, Huo N, Gu YQ, Luo M-C, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD. BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinformatics. 2008;9:253. doi: 10.1186/1471-2105-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cui W, Taub DD, Gardner K. qPrimerDepot: a primer database for quantitative real time PCR. Nucleic Acids Res. 2007;35:D805–D809. doi: 10.1093/nar/gkl767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lefever S, Vandesompele J, Speleman F, Pattyn F. RTPrimerDB: the portal for real-time PCR primers and probes. Nucleic Acids Res. 2009;37:D942–D945. doi: 10.1093/nar/gkn777. [DOI] [PMC free article] [PubMed] [Google Scholar]