Abstract
Bioactive peptides, including DNA-binding, endosomal release and cell targeting peptides, have been integrated into synthetic gene carriers to improve delivery efficiencies by enabling the vectors to overcome barriers to gene delivery. Our overall goal is to develop multifunctional, peptide-based polymers that incorporate motifs to condense DNA and facilitate sequential trafficking steps. One approach is to polymerize vinyl-terminated peptides by radical polymerization. In this work, cationic oligolysine peptides were designed to contain vinyl termini with internal reducible linkers. These peptides were copolymerized with HPMA to form biodegradable, DNA-condensing copolymers for gene delivery. The polymerization conditions were optimized by varying the initiator to monomer ratios, macromonomer to comonomer ratios, and reactant concentrations. The synthesized copolymers were shown to possess several important properties required for in vivo gene delivery applications, including (i) efficient DNA binding and condensation, (ii) the ability to stabilize particles against salt-induced aggregation, (iii) the ability to resist extracellular polyplex unpackaging, (iv) biocompatibility and the potential to be degraded into nontoxic components after cellular uptake, and (v) efficient delivery of plasmid to cultured cells.
Introduction
Polymeric gene delivery vectors have been widely studied over the past two decades as alternatives to viral vectors. Though there have been significant advances in the field of polymer-based delivery during this time, the limited efficiency of these vectors remains a weakness. Bioactive peptides can be incorporated into the delivery vehicles to aid in overcoming different barriers to delivery, and the functionalities of the peptides that have been used include DNA binding, targeting, and endosomal escape (1–3). Though many investigators have generated peptide-functionalized polymers by conjugating peptides to polymers, several groups have recently demonstrated polymerization of peptide monomers with an acrylamide comonomer (4–8). In this work, we apply this polymerization technique to synthesize peptide-based copolymers for use as gene delivery agents. These unique polymers possess integrated properties of DNA packaging, stability, and biodegradability in a single material without the need for further modification.
Cationic polymers used to deliver DNA can be highly toxic, so it is important to consider the interplay between the potential toxicity and transfection efficiency of the polymer when designing a new polycation. It has been shown that both the efficacy and toxicity of nonviral gene delivery vehicles are directly correlated to their molecular weight, leading to a situation where it is difficult to develop a system with both high delivery efficiency and low toxicity (9). To engineer a solution to this problem, investigators have crosslinked low molecular weight components using reducible crosslinkers to increase effective molecular weight for DNA condensation while ensuring that the cationic components can degrade into less toxic low molecular weight peptides or polymers (9–15). Reducible linkers are also attractive for gene delivery applications because such linkers enable the design of copolymers that maintain stability extracellularly, when a high molecular weight is needed to retain the DNA in a packaged state, and degrade intracellularly, when a low molecular weight is desired to facilitate unpackaging of the DNA for transcription.
The peptide-based polymers presented here contain a linker between the polymerizable vinyl group and the peptide. A stable linker and a reducible linker were both used in order to evaluate the effect of polymer degradation on transfection efficiency and cell toxicity. A cationic oligolysine peptide was selected for polymerization due to its ability to package DNA through electrostatic interactions. Wadhwa et al. first studied the dependence of DNA condensation and transfection efficiency on the length of the oligolysine peptide used to condense the DNA (16). The authors found that peptides with 13 or more lysine residues bound tightly to DNA, condensed the DNA into small polyplexes around 100 nm in diameter, and mediated transfection. However, peptides possessing 8 lysine residues or fewer were drastically weakened in their ability to bind DNA and mediate effective transfection (16). The materials presented here contain oligolysines with 11 lysine residues. This length was selected so that the polymerized material would potentially mediate strong DNA binding, while the free peptides could easily be displaced after cleavage from the HPMA backbone, thus facilitating DNA unpackaging.
In summary, we report here the synthesis and evaluation of HPMA-peptide copolymers for gene delivery applications. First, a series of copolymers was synthesized in order to determine the effect of the polymerization conditions on the polymer properties. The copolymers were characterized for their molecular weight, composition, and ability to condense DNA. The most promising copolymers were studied further, and polyplexes of these copolymers and DNA were assessed for their size, ability to resist unpackaging, salt stability, transfection efficiency, and cellular toxicity in a series of in vitro experiments. Several copolymers were identified with transfection efficiencies similar to that of PLL but with much lower cellular toxicities. This copolymer system is modular, and this model system can be expanded to include other peptide monomers that further increase the gene delivery efficiency of the copolymers.
Experimental Section
Materials
gWiz-Luciferase plasmid (gWiz-Luc, Endotoxin-free) was purchased from Aldevron. All other materials were purchased from Sigma-Aldrich and used without further purification unless otherwise stated.
Synthesis of Fmoc-AEDP
N-(9-fluorenylmethoxycarbonyl)-protected 3-[(2-aminoethyl)dithio] propionic acid (Fmoc-AEDP) was synthesized by a modified protocol based on published methods of Fields and coworkers (17). Briefly, 3 mmol of 9-fluorenylmethyl N-succinimidyl carbonate (Fmoc-OSu) was dissolved in 15 mL of dimethoxyethane (DME). Two mmol of 3-[(2-Aminoethyl)dithio] propionic acid (AEDP, from Pierce) and 4.4 mmol of sodium bicarbonate were dissolved in 5 mL of dH2O and then added dropwise to the 15 mL of Fmoc-OSu in DME while stirring. The reaction was allowed to proceed overnight at RT with stirring in the dark. Reaction completion was checked by TLC using a running buffer of 97% chloroform/3% glacial acetic acid (v/v) with the following spots: AEDP (Rf = 0), Fmoc (Rf = 1), Fmoc-AEDP (Rf = 0.95). The reaction was filtered through a 0.2 μm pore size PVDF filter and the DME was removed by rotary evaporation. Forty mL of 0.1 N HCl was added to the remaining solution and stirred for 1 h. Forty mL of chloroform was added and the solution was stirred for 4 h. The reaction mixture was extracted with two washes of 0.1 N HCl and two washes of dH2O. The organic phase was isolated and filtered, the chloroform was removed by rotary evaporation, and a slightly yellow, oily substance was recovered.
Synthesis of DNP-labeled K11 and Unlabeled K11 Peptides
One gram of Fmoc-Lys(Boc)-NovaSyn® TGT resin (0.2 mmol Lysine, from Novabiochem) was swelled in dimethylformamide (DMF) and the N-terminal amine groups of the Lys preloaded onto the resin were deprotected by two incubations for 10 min in 20% (v/v) piperidine in DMF. Four equiv each of Lysine-dinitrophenyl (Lys-DNP, from Novabiochem) and O-(7-Azabenzotriazole-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU, from GenScript Corp) were dissolved in 5 mL of 0.4 M N-methylmorpholine (NMM) in DMF. The activator and amino acid solution was then added to the resin and the coupling reaction was allowed to proceed for 1 h at RT. The resin was washed with DMF and the remainder of the peptide was synthesized using standard Fmoc-based solid phase peptide synthesis (SPPS) conditions with 4 equiv of lysine amino acid (Fmoc-Lys(Boc), from Protein Technologies) and 4 equiv of HATU on a PS3 peptide synthesizer (Protein Technologies). Ten additional Lys residues were added for a total of eleven Lys residues with one Lys-DNP residue on the K11-DNP peptide. The purity of the peptide was assessed by HPLC and the mass was measured using an Esquire electrospray ion trap mass spectrometer (ESI-MS) from Bruker Daltonics (Supporting Information). The unlabeled K11 peptide was synthesized in exactly the same way, except without adding the Lys-DNP residue.
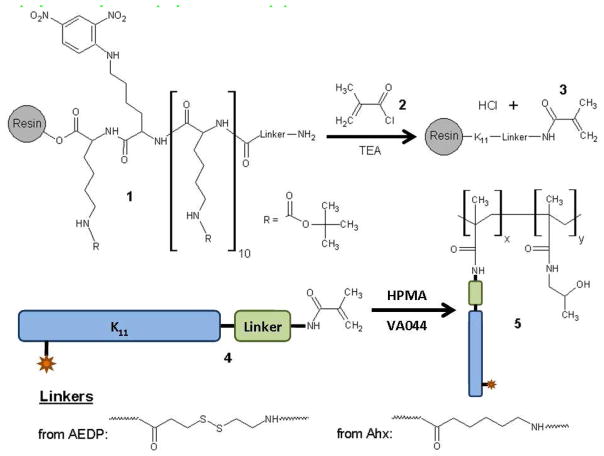
Synthesis of DNP-labeled K11-AEDP-MC and K11-Ahx-MC Modified Peptides (4)
Four equiv each of Fmoc-AEDP and HATU dissolved in 5 mL of 0.4 M NMM in DMF were added to N-terminally deprotected K11-DNP-resin containing 0.1 mmol peptide. The coupling reaction was allowed to proceed for 1 hr at RT. The resin was washed with DMF and then deprotected by twice incubating the resin with 20% piperidine in DMF for 5 min at RT, yielding K11-AEDP 1 (with the linker coming from AEDP). The peptide was then methacryloylated as shown in Scheme 1, following a protocol similar to that of O’Brien-Simpson et al. (4). The minimum amount of anhydrous DMF required to fluidize the resin was added and the resin was transferred to a reaction flask kept on ice under nitrogen. Triethylamine (TEA, 20 equiv) was injected into the flask, followed by dropwise addition of 10 equiv of methacryloyl chloride 2 (MC). The reaction was stirred for 1 h on ice and then 1 h at RT in O2-free conditions, yielding compound 3. The resin was washed and the peptide cleaved by incubation with a cleavage cocktail of 95% trifluoroacetic acid (TFA), 2.5% triisopropylsilane (TIPS), and 2.5% dH2O for 3 h at RT. The cleaved peptide K11-AEDP-MC 4 was precipitated in diethyl ether three times and the pellet was dried, yielding a chalky yellow solid, and stored at −20°C. The synthesis of K11-Ahx-MC 4 was performed in the same way, except that four equiv of Fmoc-6-aminohexanoic acid (Fmoc-Ahx, from Novabiochem) was used instead of Fmoc-AEDP to generate a peptide monomer that does not contain a cleavable disulfide linkage. The purity of the peptides was assessed by HPLC and found to be >90% (Supporting Information). Desired products were confirmed by mass spectrometry using an Esquire ESI-MS from Bruker Daltonics (Supporting Information).
Scheme 1.
Methacryloylation of linker-conjugated peptides 1 and synthesis of copolymers 5 by radical polymerization of peptide monomers 4 and HPMA
Copolymerization of HPMA with Peptide Monomer
For each copolymer, 10 mg peptide monomer 4 was used in the feed at the mol% shown in Table 1 with the required amount of HPMA monomer (Polysciences, Inc). The weight of the initiator 2,2′-azobis[2-(2-imidazolin-2-yl)propane]dihydrochloride (VA044, from Wako Specialty Chemicals) used for each polymerization was calculated as a weight percent of HPMA monomer, as indicated in Table 1. For a given copolymerization, the appropriate amounts of peptide monomer, HPMA monomer, and VA044 initiator were dissolved in the amount of degassed reaction buffer (6 M guanidine hydrochloride, 2 mM EDTA, 0.5 M Tris base, buffered to pH 8.3 with HCl) to give the final total monomer concentration indicated in Table 1. The reaction solutions were transferred to glass reaction vials, bubbled with N2 gas for 15 min, and then sealed and heated at 44°C for 48 h to generate copolymers 5. Four separate copolymer series were investigated, as shown in Table 1, where series A, B, and C used the non-cleavable K11-Ahx-MC 4 in the copolymers while series D used the disulfide-linked K11-AEDP-MC 4. Series A varied the wt% of initiator while series B and C varied the mol% of peptide monomer and total monomer concentration.
Table 1.
Reaction conditions and molecular properties of the copolymer library
| Copolymer | Initiator wt%a | Peptide Monomer mol% feed | Total Monomer Conc (mg/mL) | Mw (kDa) | PDI | Peptide mol% in copolymer | Peptide wt% in copolymer | % Yieldb |
|---|---|---|---|---|---|---|---|---|
| A1 | 1 | 5 | 85.0 | 64.3 | 3.6 | 2.86 | 28.1 | 38 |
| A2 | 3 | 5 | 85.0 | 55.7 | 4.2 | 2.95 | 28.7 | 76 |
| A3 | 5 | 5 | 85.0 | 53.6 | 4.4 | 3.02 | 29.3 | 81 |
| A4 | 7 | 5 | 85.0 | 47.9 | 4.8 | 3.12 | 30.0 | 79 |
| B1 | 5 | 1 | 85.0 | 213.3 | 8.3 | 0.63 | 7.7 | 76 |
| B2 | 5 | 1 | 56.7 | 94.0 | 7.0 | 0.40 | 5.1 | 91 |
| C1 | 5 | 10 | 85.0 | 21.2 | 3.1 | 4.37 | 37.8 | 65 |
| C2 | 5 | 10 | 42.5 | 13.2 | 2.2 | 3.94 | 35.3 | 54 |
| D1 | 5 | 5 | 85.0 | 28.4 | 3.9 | 3.87 | 35.5 | 80 |
The initiator wt% was calculated as a weight percent of HPMA monomer.
The percent yield is presented as the weight of polymer recovered divided by the total weight of the monomers added to the reaction.
Purification and Characterization of Copolymers
The copolymers were dialyzed against dH2O using a 7 kDa MWCO membrane and purified by HPLC to remove any unreacted monomers. After organic solvent removal in vacuo and lyophilization to recover a fluffy yellow solid, the copolymers were dissolved at 5 mg/mL in running buffer (0.3 M sodium acetate buffered to pH 4.4 with acetic acid) for analysis by gel permeation chromatography (GPC) as described by Hennink and coworkers (18). A Viscotek GPCmax, which was controlled by Viscotek OmniSEC software, was used with an OHpak SB-G guard column (Shodex) in line with an OHpak SB-804 HQ column (Shodex) and a refractive index (RI) detector (Viscotek). The molecular weights of the copolymers were determined by using the OmniSEC software to compare the elution time of the copolymer to a standard curve generated from the elution times of narrow-polydispersity poly(ethylene oxide) TSK standards (Tosoh) of different molecular weights. The percent incorporation of peptide into the copolymer was determined by comparing to a standard curve of DNP absorbance at 360 nm (A360).
Polymer Degradation Studies
Copolymers A4 and D1 were diluted to 5 mg/mL in HEPES Buffered Glucose (HBG; 20 mM HEPES and 5% glucose (w/v) buffered to pH 7.4 with NaOH) and analyzed by HPLC. HPLC was performed using a Jupiter 4 μm Proteo 90 Å (250 × 4.6 mm) reversed phase analytical column (Phenomenex) and a gradient from 98% Buffer A (H2O + 0.01% TFA) to 30% Buffer B (Acetonitrile + 0.01% TFA) developed over 25 minutes. The same copolymers were then diluted to 5 mg/mL in HBG containing the reducing agent dithiothreitol (DTT, final concentration of 10 mM) and incubated for 5, 30, or 60 min before being analyzed by HPLC with detection at 215 nm.
Polyplex Formulation and Characterization
The gWiz-Luc plasmid was diluted in HBG to a concentration of 0.1 mg/mL and mixed with an equal volume of polymer (also diluted in HBG) at the desired lysine to DNA phosphate (N/P) ratio. For particle sizing measurements, all polyplexes were formed at N/P = 5. After mixing, the polyplexes were allowed to form for 5 minutes at room temperature. 20 μL of each polyplex sample (containing 1 μg DNA) was mixed with either 80 μL HBG or PBS and the 100 μL sample was assayed for particle size by dynamic light scattering (DLS) measured on a Brookhaven Instruments Corp ZetaPALS instrument. Particle sizing measurements were performed at a wavelength of 659.0 nm with a detection angle of 90° at RT.
DNA Condensation
The gWiz-Luc plasmid was mixed with the bis-intercalating dye YOYO-1 iodide (Invitrogen/Molecular Probes) at a dye:base pair (D:BP) ratio of 1:25 and incubated at 50°C for 2 hours to equilibrate the dye complexes with DNA (19). Polyplexes were formed at various N/P ratios from 0 to 5 by complexing YOYO-labeled DNA with the polymers or peptides in HBG as described above. 20 μL of the polyplex solution and 80 μL of HBG were added to each well of a 96-well plate and the fluorescence from each well was measured on a Tecan Safire2 plate reader with excitation at 491 nm and emission at 513 nm. Fluorescence signal from polyplexes with N/P > 0 was normalized to N/P = 0 (DNA only) signal.
Polyplex Unpackaging Studies
The gWiz-Luc plasmid was labeled with YOYO and poly-L-lysine (PLL, 12–24 kDa, from Sigma), K11-DNP peptide, and copolymers B1, C1, and D1 were used to form polyplexes at N/P = 5 in HBG as described above. 20 μL of polyplex solution were mixed with 80 μL of HBG, HBG containing 0.1 mg/mL heparan sulfate (HS, from Sigma), or HBG containing 0.5 mg/mL HS in each well of a 96-well plate. Additionally, 20 μL of polyplexes formed with copolymer B1 or D1 were mixed with 80 μL of HBG containing DTT and/or HS at specific concentrations in each well of a 96-well plate. The polyplexes mixed with 10 mM DTT were allowed to incubate for 30 min at RT and the polyplexes mixed with 25 mM DTT were allowed to incubate for 1 h at RT. The fluorescence from each well was measured on a Tecan Safire2 plate reader with excitation at 491 nm and emission at 513 nm. Fluorescence signal from polyplexes with N/P > 0 was normalized to N/P = 0 (DNA only) signal.
IN VITRO Transfection Efficiency
Human cervix epithelial adenocarcinoma cells (HeLa, passage 25, ATCC # CCL-2) were seeded in complete cell culture medium (MEM + 10% FBS + 1% antibiotic/antimicrobial) at a density of 5 × 104 cells/well in each well of a 24-well plate. The 24-well plates were placed in a 37°C incubator with 5% CO2 for 24 hours to allow the HeLa cells to attach. Polyplexes were formed at N/P = 5 using 1 μg of gWiz-Luc plasmid DNA in 20 μL total volume. Each sample was brought up to 200 μL with OptiMEM I (Invitrogen). The cells were washed twice with PBS and the transfection solutions were added. The 24-well plates were returned to the incubator for four hours, at which time the polyplex solution was replaced with complete cell culture media after washing the cells with PBS. The cells were returned to the incubator for a further 44 hours. After a total incubation of 48 hours, the luciferase expression was quantified with a luciferase assay kit (Promega Corp.) according to the manufacturer’s instructions, except that a freeze-thaw cycle at −20°C was included after the addition of the lysis buffer to ensure complete cell lysis. Luminescence intensity was measured on a Tecan Safire2 plate reader with integration for 1 s. The total protein content in each well was measured by a BCA Protein Assay Kit (Pierce) according to the manufacturer’s instructions so that the luciferase activity measured in each well could be normalized by the total protein content in each well. Each sample was tested in replicates with n = 8, but one outlier was identified in the copolymer D1 data set (using the interquartile method of identifying outliers). The statistical significance of differences in the samples was determined using Student’s t-test, where a p-value of less than 0.05 was considered significant.
Cellular Toxicity
HeLa cells were seeded in complete cell culture medium at a density of 1 × 104 cells/well in each well of a 96-well plate. The 96-well plates were placed in a 37°C incubator with 5% CO2 for 24 hours to allow the HeLa cells to attach. The cells were washed with PBS, the polymer and peptide samples were diluted to the indicated concentrations with OptiMEM, and 100 μL of polymer or peptide sample was added to the cells. The 96-well plates were returned to the incubator for four hours, at which time the polymer or peptide solution was replaced with complete cell culture media after washing the cells with PBS. The cells were returned to the incubator for a further 40 hours. Finally, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS reagent, from Promega Corp.) was added to the cells, the cells were returned to the incubator for four hours (for a total incubation of 48 h), and the number of viable cells in each well was quantified by measuring the absorbance at 490 nm on a Tecan Safire2 plate reader.
Results and Discussion
Synthesis of Fmoc-AEDP and Peptide Monomers
Oligolysine peptides containing 11 Lysine residues and a Lysine-DNP residue (K11-DNP) were synthesized by SPPS. DNP was included as a label for the peptide; antibodies to DNP can be used in future studies to track the polymer. An Fmoc-protected, nonreducible or reducible linker (Ahx or AEDP, respectively) was then conjugated to the N-terminus of the peptides using the same SPPS chemistry. Finally, the deprotected N-termini of the linker-modified peptides 1 were reacted with MC 2 in order to introduce a reactive vinyl group for radical polymerization (Scheme 1). The masses of K11-DNP, linker-modified and MC-modified peptides agreed very well with the calculated masses, as shown in Table 2, and the purity of each peptide was found to be greater than 90% by HPLC (Supporting Information).
Table 2.
Calculated weights of the various peptide monomers and peptide monomer precursors compared to the weights determined by ESI-MS
| Peptide | Calculated (g/mol) | Found (g/mol)a |
|---|---|---|
| K11 | 1722.3 | 1722.6 |
| K11-Ahx | 1835.3 | 1835.7 |
| K11-Ahx-MC | 1903.3 | 1903.8 |
| K11-AEDP | 1885.3 | 1885.7 |
| K11-AEDP-MC | 1953.3 | 1953.7 |
All weights found by mass spectrometry correlate within 0.03% of the actual mass.
Copolymerization of HPMA with Peptide Monomer
The vinyl-functionalized peptides 4 were copolymerized with HPMA by radical polymerization to generate copolymers 5 (Scheme 1). Poly(N-(2-hydroxypropyl)methacrylamide) (HPMA), originally synthesized by Kopeček and coworkers (20–21), has been incorporated in polymer-drug conjugates that have entered phase I/II clinical trials (22–26). In a landmark clinical study, it was found that cumulative doses of HPMA copolymer in excess of 20 g/m2 could be administered to patients without any polymer-related toxicity or immunogenicity (22). In addition, HPMA has been grafted onto quaternary amine-containing polymers and used to sterically stabilize polyplexes formed with these graft copolymers (27–29). HPMA was therefore chosen for copolymerization with peptide monomers due to its ease of incorporation into copolymers, ability to sterically stabilize colloids, and lack of toxicity when injected into human patients.
Three series of nonreducible poly(HPMA-stat-K11) copolymers were synthesized in order to determine the effect of polymerization conditions on polymer properties and composition (Table 1). In series A, the amount of VA044 initiator used during polymerization was varied while keeping the total monomer concentration and ratios of comonomers (5 mol% peptide, 95 mol% HPMA) fixed. For uncontrolled radical polymerizations, increasing the radical to monomer ratio increases the number of chains initiated, thereby reducing the molecular weight of the resulting polymers. As expected, the copolymers synthesized in series A follow the trend of decreasing molecular weight with increasing amount of initiator (Table 1). HPLC analysis revealed that increasing the amount of initiator increased the incorporation of peptide into the copolymer but also increased the amount of HPMA homopolymer without any peptide. Thus, 5 wt% VA044 initiator was used for the subsequent polymerizations.
In series B and C, the amount of peptide monomer added in the feed was changed to 1 and 10 mol%, respectively. Between copolymers A3, B1, and C1, the wt% initiator and total monomer concentration are constant and the only difference is the mol% of peptide monomer in the feed. A comparison of these copolymers shows that increasing the amount of peptide monomer in the feed leads to increased wt% of peptide in the final copolymer, but also a reduced overall molecular weight (Table 1). It is very likely that this reduction in overall molecular weight is caused by a lower reactivity of the peptide macromonomer, as it has been shown that macromonomers often have lower reactivities than smaller monomers (30–31). Radke and Müller investigated copolymerization of macromonomers with methyl methacrylate and showed that increasing the macromonomer concentration leads to an increase in viscosity of the polymerization solution and a lower reactivity of the macromonomer due to decreased mobility of the larger macromonomer compared to the smaller comonomer (30). The authors also demonstrated that the average segment densities surrounding the propagating radical site decrease with increasing distance between the reactive center and the nearest macromonomer (30). The macromonomer is therefore most reactive when it is at its lowest mol% in the feed, resulting in long stretches of comonomer that extend the distance between macromonomers and reduce the steric hindrance of high segment densities that prevents the addition of the next macromonomer.
The incorporation efficiency of the peptide macromonomer decreases with increasing mol% of peptide macromonomer in the feed (63% of feed for B1, 60% of feed for A3, and 44% of feed for C1), which is consistent with lower reactivity of the peptide monomer at higher mol percents in the feed. Furthermore, as the mol% of peptide macromonomer is increased, the formation of long polymers becomes more dependent on the incorporation of the macromonomer to continue the polymerization. Thus, the decreased reactivity of the peptide macromonomer most likely plays a significant role in the reduction in total copolymer size seen with increasing mol% of peptide monomer in the feed. Similar behavior was reported in another system where increasing the feed mol% of a comonomer with lower reactivity decreased the overall molecular weight of the resulting copolymer (32). The decrease in molecular weight with increasing mol% of peptide may also be attributed to the DNP label on the peptide, because it is known that DNP can act as a chain transfer agent during radical polymerization (33–34). The ability of DNP to accept radicals retards the polymerization, extending the time required for polymerization in the presence of DNP compared to a polymerization in the same conditions except without DNP (33). Thus, the copolymers synthesized at the higher mol% of peptide will be retarded the most due to the presence of the highest concentration of DNP and 48 hr may not be long enough to extend the chains fully, resulting in lower molecular weights.
Copolymers B2 and C2 were synthesized to determine the effect of changing the total monomer concentration in the polymerization solution. First, it should be noted that changing total monomer concentration had only a minor effect on the wt% of peptide in the final copolymers. Also, the inverse relationship between peptide mol% in the feed and resulting copolymer MW still held. A comparison of B1 vs. B2 and C1 vs. C2 shows that polymerizations at lower monomer concentrations led to copolymers with lower total molecular weights. It has been shown that the degree of polymerization (DP) of a polymer generated from macromonomers during uncontrolled radical polymerization increases with increasing macromonomer concentration, mainly due to the increase in viscosity at higher macromonomer concentrations (35). Higher viscosity reduces the rate of termination, thus extending the lifetime of the polymerization and increasing the DP, due to a diffusion controlled limit on bimolecular termination (35). Thus, the viscosity of the reaction solution is an important factor for optimization: not only does a high viscosity decrease the reactivity of the macromonomers, which tends to lead to smaller copolymers, but a higher viscosity also decreases the rate of termination, which tends to allow for larger copolymers. Comparison within series B or series C in Table 1 shows that, for a given composition, increasing the monomer concentration leads to higher percent incorporation of the peptide in the final copolymer (44% for C1 compared to 39% for C2 and 63% for B1 compared to 40% for B2). Furthermore, when comparing between series, these studies demonstrate that the peptide monomer concentration has less of an effect on the percent peptide incorporation than does the mol% of peptide monomer in the feed. Thus, the maximum concentration of 85 mg/mL (above which gelation of the reaction mixture would occur during polymerization) was employed for all further polymerizations.
Copolymer D1 was synthesized to investigate the effect of a reducible AEDP linker incorporated in the peptide monomer. The polymerization conditions for D1 were the same as for A3. Copolymer D1 has a lower MW and a slightly higher peptide mol% incorporation in the final copolymer compared with A3. The difference in polymer properties might be due to the change in the hydrophobicity of the Ahx versus AEDP linkers in the peptide monomers especially since the linker group is close to the propagating radical site. It is known that radical attack on an alkene can lead to radical transfer to a nearby sulfur atom and the opening of a ring during polymerization (36). However, the increased distance of the disulfide bond in the AEDP to the alkene group in the MC, compared to the single carbon spacer in the ring opening polymerization (36), will greatly reduce the ability of the sulfur atom to accept radicals. Thus, only very minimal reversible chain transfer or chain extension through the sulfur atom is expected to occur in this system.
In summary, a library of p(HPMA-stat-K11) copolymers was synthesized so that materials with a range of properties could be tested to determine the optimal formulation for gene delivery. The successful synthesis of copolymers with different amounts of peptide, different molecular weights, and reducible versus non-reducible linkages between the peptides and the copolymer backbone has been demonstrated.
Demonstration of degradability of copolymer D1
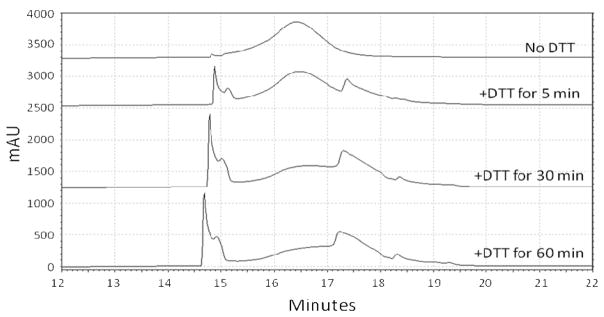
To test the degradation of copolymers synthesized with reducible (D1, with AEDP linker) and non-reducible (A4, with Ahx linker) peptides, D1 and A4 were incubated with the reducing agent DTT and polymer degradation was monitored by HPLC (Figure 1). D1 copolymer elutes between 15–17 minutes. Treatment with 10 mM DTT results in free peptide release by 5 min (~14.7 min elution time). The resulting HPMA homopolymer elutes at 17.4 min. Taken together, these results show that under reducing conditions, copolymer D1 is efficiently degraded into free peptide and the HPMA polymer backbone. Conversely, the DTT had no effect on the HPLC traces when incubated with non-reducible copolymer A4, indicating that the reducing agent was not able to cleave the peptide attached with a non-reducible linker (data not shown).
Figure 1.
Evaluation of copolymer D1 degradation under reducing conditions. HPLC elution traces of copolymer D1 with no DTT and after being treated with DTT for various times. DTT treatment results in liberation of K11-DNP peptide (14.5 min) and HPMA homopolymer (17–18 min).
Polyplex Formulation and Characterization
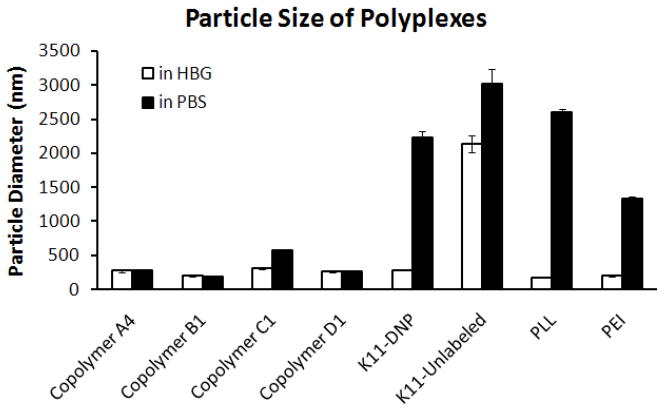
Polyplexes were first formed by mixing copolymers with plasmid DNA in HBG. HBG was selected because it has previously been shown to be an ideal buffer to improve polyplex formulation and reduce aggregation (37). The cationic polymers poly-L-lysine (PLL) and polyethylenimine (PEI) formed small polyplexes with sizes on the order of 200 nm when complexed with DNA at N/P = 5 in HBG (Figure 2). Similarly, all of the copolymers tested (A4, B1, C1, and D1) also formed small polyplexes with sizes around 200 nm when complexed with DNA at N/P = 5 in HBG. The size of polyplexes formed with unlabeled K11 peptide was very large (over a micron at N/P = 5 in HBG), consistent with the decreased ability of short cationic peptides to bind and condense DNA. Unexpectedly, the size of polyplexes formed with DNP-labeled K11 peptide at N/P = 5 in HBG was similar to the sizes of the polyplexes formed with the copolymers, PEI, and PLL. We hypothesize that the hydrophobic DNP label may increase the peptide’s affinity for DNA, thus increasing its ability to condense DNA into small particles.
Figure 2.
Polyplex size in HBG (20 mM salt) and 150 mM PBS. All peptides and polymers efficiently condensed plasmid in HBG to nanoparticles < 200 nm in diameter except unlabeled K11. HPMA copolymers formed stabilized nanoparticles even in PBS.
The salt stability of the polyplexes formed with the copolymers is an important characteristic because polyplexes prepared for in vivo use will need to be salt stable when administered intravenously in order to avoid aggregation that can lead to embolism. The polyplexes are able to maintain small sizes in the low-salt HBG buffer because of intermolecular repulsion due their positive surface charge at N/P = 5. However, when PBS is added, the higher amount of salt reduces the electrical double-layer thickness, rendering the particles susceptible to aggregation. As seen in Figure 2, PLL- and PEI-based polyplexes aggregate to sizes over one micron when diluted in PBS. Similar aggregation behavior is observed for the polyplexes formed with either unlabeled or DNP-labeled peptide. In contrast, polyplexes formed with all of the copolymers remain stable with a small size, even when diluted into PBS. It has been shown that cationic polymers modified with HPMA can be used to condense DNA and remain stable at high salt concentrations (27), where the HPMA is believed to take on a brush-like conformation on the surface and prevent aggregation by acting as a steric barrier. In a similar manner, a portion of the hydrophilic HPMA segments of the copolymer will be present on the surface of the polyplexes, and we hypothesize that they also adopt a brushlike conformation on the polyplex surface to resist aggregation. In support of this theory is the fact that copolymer C1, which has the lowest percentage of HPMA, almost doubles in size upon addition of PBS, indicating that it may be just outside the threshold level of HPMA required for complete salt stability in PBS.
Determination of Plasmid Packaging Efficiency
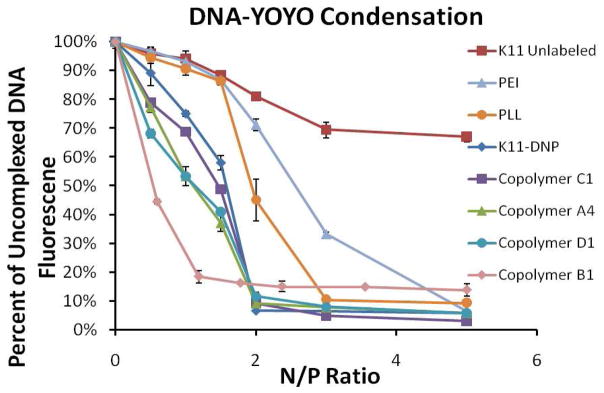
The efficiency of plasmid condensation by polycations can be monitored by a YOYO-1 quenching assay (19). The fluorescence of the YOYO-1 DNA intercalator is quenched when YOYO-labeled plasmid is packaged into a condensed state. Addition of PLL or PEI to YOYO-labeled plasmid results in decreasing fluorescence with increasing N/P ratio, with full condensation occurring at N/P = 3 for PLL and N/P = 5 for PEI (Figure 3). The unlabeled K11 peptide is unable to efficiently condense plasmid at the N/P ratios investigated. These results are consistent with the particle sizing results, where at N/P = 5 the unlabeled peptide was not able to form small particles. In contrast, the DNP-labeled K11 peptide fully condensed DNA at N/P = 2. Again, these results are fully consistent with the particle sizing results, which indicated that the K11-DNP peptide was able to condense DNA into small particles. The copolymers condensed DNA more efficiently than K11-DNP likely due to the higher cationic valency of the copolymers. However, this effect is minor compared to the benefit derived from having HPMA in the copolymer. As can be seen in Figure 3, the copolymer able to most effectively condense DNA is B1, which has the lowest mol% of peptide, and the copolymer to least effectively condense DNA is C1, which has the highest mol% of peptide. Copolymers A4 and D1, which contain an intermediate amount of peptide between that of B1 and C1, possess an intermediate ability to condense DNA. This data suggests that the presence of HPMA drastically increases the affinity of the copolymer for the DNA. A YOYO condensation assay was performed with HPMA homopolymer, and YOYO fluorescence was partially quenched, lending support to the hypothesis that HPMA itself is able to interact with the DNA.
Figure 3.
Extent of DNA condensation versus N/P ratio for HPMA copolymers, control polymers, and unpolymerized peptides. The fluorescence signal from DNA-YOYO was normalized to the fluorescence signal of DNA only (N/P = 0). Fluorescence signal decreases with increasing N/P ratio due to quenching of YOYO fluorescence resulting from DNA condensation.
Determination of Plasmid Unpackaging
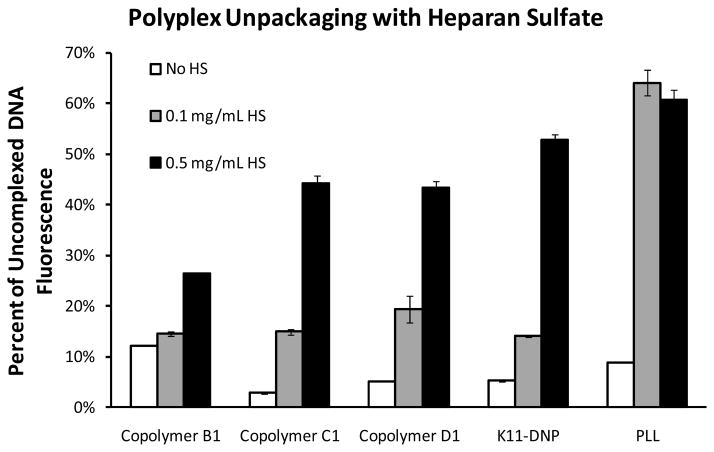
For systemic delivery applications, vector stability in the bloodstream and tissue is an important consideration. We and others have previously demonstrated that extracellular polyanions cause premature PEI polyplex unpackaging in vivo likely due to a polyelectrolyte exchange reaction (19, 38–39). Polyplex stability in the presence of destabilizing agents can also be evaluated using a YOYO-based fluorescence assay. As can be seen in Figure 4, polyplexes formed at N/P = 5 and diluted with HBG containing no HS are fully packaged, indicated by low YOYO fluorescence. Polyplexes formed with PLL or PEI are fully unpackaged after dilution into HBG containing HS at a concentration of 0.1 mg/mL (PEI data not shown). In contrast, polyplexes formed with copolymer B1 maintained DNA in a fully condensed state after incubation with 0.1 mg/mL HS. Polyplexes formed with copolymer C1 or D1 and polyplexes formed with K11-DNP were found to unpackage only slightly when incubated with 0.1 mg/mL HS. When the concentration of HS was increased to 0.5 mg/mL, polyplexes formed with K11-DNP peptide fully unpackaged, while polyplexes formed with copolymers B1, C1 and D1 remained partially packaged. These results are consistent with the affinities of the copolymers for DNA determined in the DNA-YOYO condensation assay. We hypothesize that both the DNP label and the HPMA can interact with the DNA, thus increasing the affinity of the binding of the copolymers and peptides to DNA and decreasing the susceptibility of the polyplexes to unpackaging.
Figure 4.
HS-mediated unpackaging of YOYO-labeled polyplexes. HS was diluted in HBG at various concentrations, and 80 μL of the indicated solution was added to 20 μL of polyplex (N/P = 5) containing 1 μg of DNA. Fluorescence signal increases with incubation with HS due to polyplex unpackaging and dequenching of YOYO fluorescence.
The optimal design of the polymeric in vivo gene delivery vehicle will need to allow for intracellular unpackaging, which is required for transcription (40). If the vectors are simply designed for maximum extracellular stability, they will be unable to unpackage intracellularly and the delivered cargo will be rendered useless. To overcome this challenge, copolymers were synthesized containing the DNA-condensing peptides linked to the polymer backbone through a reducible disulfide linkage. When the peptides are cleaved from the copolymer backbone, the affinity for DNA should be decreased and unpackaging should be more likely. The extracellular milieu is known to be an oxidizing environment; in contrast, the cytosol is a highly reducing environment due to the excess of reduced glutathione (GSH), thioredoxin, glutaredoxin, peroxiredoxins, and nicotinamide adenine dinucleotides (NADH and NADPH) over the oxidized analogues of these molecules (41–43).
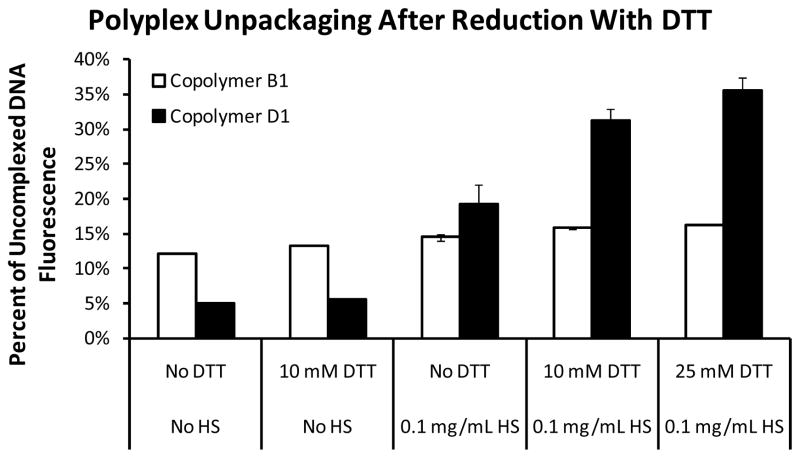
The extent of polyplex unpackaging due to competitive displacement of the polycation by HS was evaluated under reducing and non-reducing conditions for polyplexes formed with copolymer D1 (Figure 5). The polyplexes were first formed and then diluted in a buffer containing DTT, HS, or both. Reducing the linkage on copolymer D1 between peptide and polymer backbone without adding HS did not lead to any destabilization. Adding HS without DTT decreased the condensation state of the DNA-YOYO from 5% of uncomplexed DNA fluorescence to 19% of uncomplexed DNA fluorescence. Including 10 mM DTT or 25 mM DTT in the incubation with HS decreased the condensation state of the DNA-YOYO further, to 31% and 35% of uncomplexed DNA fluorescence, respectively. Reducing agents are present intracellularly at low millimolar concentrations (41), so these conditions were chosen to mimic the intracellular environment most closely. As can be seen from Figure 4, the maximum fluorescence recovery achieved with even 0.5 mg/mL HS was 60% of uncomplexed DNA fluorescence. There is a slight further increase in unpackaging when the DTT concentration and incubation time are increased. Thus, the polyplexes formed from copolymer D1 are more easily unpackaged under the reducing conditions of the cell cytosol. Polymers synthesized with peptides that do not contain the DNP label are expected to exhibit even more efficient DNA release under reducing conditions. In contrast, polyplexes formed from the non-reducible copolymer B1 were unaffected by any of the treatments.
Figure 5.
HS-mediated unpackaging of YOYO-labeled polyplexes formed with reducible (D1) and non-reducible (B1) copolymers before and after incubation with the reducing agent DTT. HS and DTT were diluted in HBG at various concentrations, and 80 μL of the indicated solution was added to 20 μL of polyplex (N/P = 5) containing 1 μg of DNA. The samples containing 10 mM DTT were incubated for 30 min and the samples containing 25 mM DTT were incubated for 1 h to ensure complete reduction of the disulfide linker.
IN VITRO Transfection Efficiency
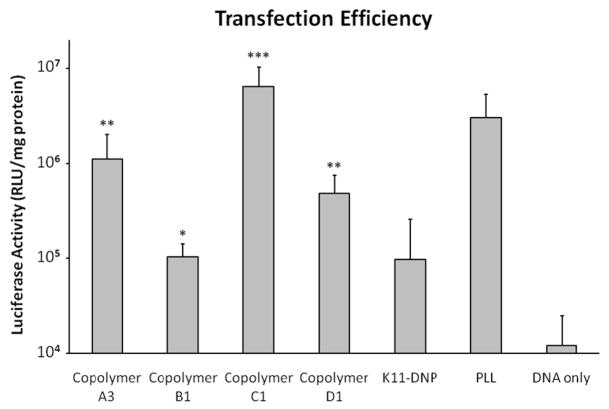
gWiz-Luc plasmid was delivered to HeLa cells using various delivery vehicles, and the efficiency of delivery was determined by the quantitative luciferase reporter system (Figure 6). Unlabeled K11 peptide did not mediate increased gene delivery over plasmid DNA only (data not shown) most likely due to its inability to form stable polyplexes; however, the DNP-labeled K11 peptide, which was able to form stable polyplexes, did mediate an increased gene delivery efficiency compared to DNA only, though this difference was not statistically significant due to high variability. All of the copolymers were found to mediate statistically significant (p < 0.01) increases in transgene expression compared to plasmid only. In addition, copolymers A3, C1 and D1 mediated statistically significantly (p < 0.01) increased transfection efficiency compared to unpolymerized K11-DNP peptide.
Figure 6.
Gene delivery efficiency of selected carriers (formulated at N/P = 5) to HeLa cells as measured by luciferase activity 48 h after transfection. Results are reported as the mean RLU/mg protein ± SD for replicate samples, with all samples having n > 7. The * denotes that copolymer B1 was statistically significantly different from DNA only (p < 0.01). The ** denotes that copolymer A3 and copolymer D1 were both statistically significantly different from copolymer B1 (p < 0.01), K11-DNP peptide (p < 0.01), and DNA only (p < 0.01). The *** denotes that copolymer C1 was statistically significantly different from copolymer A3 (p < 0.01), copolymer B1 (p < 0.01), copolymer D1 (p < 0.01), K11-DNP peptide (p < 0.01), PLL (p < 0.05), and DNA only (p < 0.01).
Since the delivery efficiency of oligolysine and PLL is known to depend on the molecular weight of the cationic delivery agent, it was expected that the copolymers that have more than one peptide incorporated and thus a higher effective molecular weight of lysine segments, would have higher delivery efficiency than peptide alone. Due to the same effect, it was also expected that the copolymers based on the non-reducible Ahx linker would show increasing efficiency with increasing amounts of peptide in the copolymer. This trend was observed, with C1 having 37.8 wt% peptide and the highest delivery efficiency, A3 having 30 wt% peptide and an intermediate delivery efficiency, and B1 having 7.7 wt% peptide and the lowest delivery efficiency. However, the average number of peptides per copolymer was inversely proportional to the copolymer transfection efficiency. This data indicates that the wt% of peptide in the copolymer but not the absolute number of peptide monomers in the copolymer is correlated with transfection efficiency.
One possible explanation is that the density of peptides in the copolymer could determine the cooperativity of the increase in effective polycation molecular weight. For example, copolymer B1 has an average of approximately 158 HPMA monomers per peptide monomer. In contrast, copolymer A3 has ~32 HPMA monomers per peptide monomer on average, and copolymer C1 has ~22 HPMA monomers per peptide monomer. Assuming even distribution of peptides in the polymer chain, copolymers with peptides that are more closely spaced could possess a higher effective polycation molecular weight and mediate more effective gene delivery, while the copolymers with peptides spaced further apart could have a lower effective polycation molecular weight and lower transfection efficiency.
Another possible explanation is that vector unpackaging affects transfection efficiency. Schaffer et al. have shown that the vectors with high intracellular stability mediated the lowest amounts of gene expression, while expression was greatly enhanced by using vehicles that easily dissociated from the DNA because the DNA could more easily be transcribed (40). From Figure 3, it can be seen that copolymer B1 condenses DNA most efficiently, while copolymer C1 condenses DNA least efficiently (among the Ahx-based copolymers). Analogously, from Figure 4, it can be seen that copolymer B1 is more resistant to unpackaging than copolymer C1.
A third possible explanation for the differences in transfection efficiencies is that polyplexes formed with the different copolymers may have unequal abilities to mediate cellular internalization. Polyplexes generated from covalently linked PLL-HPMA were found to have reduced cellular association and internalization compared to polyplexes formed with unmodified PLL (29). Conjugation of poly(ethylene glycol), another polar nonionic polymer, to PEI polyplexes causes a reduction in the amount of polyplex internalization that leads to a decreased transfection efficiency (44–45). It has been noted that increasing the amount of shielding polymer PEG in PEI-based polyplexes reduces transfection efficiency (45), which is similar to the observed trends of decreasing transfection efficiency for poly(HPMA-stat-K11) copolymers with increasing amounts of HPMA.
The AEDP-based, reducible copolymer D1 has a similar wt% of peptide and resistance to unpackaging under non-reducing conditions when compared to copolymer C1 (Figure 4). However, from Figure 6, it can be seen that the transfection efficiency of copolymer D1 is statistically significantly lower (p < 0.01) than that of copolymer C1. If copolymer D1 degradation and plasmid release occurs much faster than nuclear delivery of plasmid, the DNA may be degraded by nucleases before transcription can occur. Thus, the data suggests that there is an optimal intracellular stability of polyplexes for efficient gene expression.
It should be noted that copolymer C1 was found to mediate a higher transfection efficiency than PLL (p < 0.05). Copolymer C1 was able to more efficiently deliver DNA even though it only has about 8 kDa of mass from oligolysine in each copolymer. Copolymer C1 may have nearly optimal oligolysine content for DNA packaging without limiting intracellular DNA release. In this study, the copolymers were compared to PLL because, like PLL, they contain only primary amines on the lysine side chains to serve as cationic groups for DNA condensation. PEI, in contrast, has secondary and tertiary (in the case of branched PEI) amines that have lower pKa values. It is currently believed that PEI mediates endosomal escape through a proton sponge effect due to this buffering capacity (46–48), which leads to typically higher transfection efficiencies than those achieved by PLL. This modular copolymer system has the ability to incorporate other peptides that may include endosomal escape functionality to further increase the transfection efficiency to rival that of PEI.
Cellular Toxicity
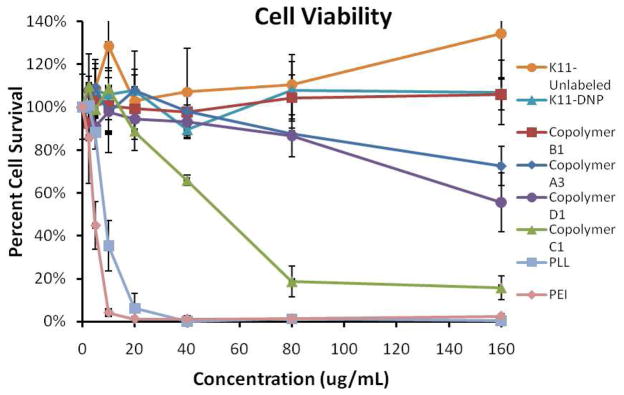
Both the efficacy and the toxicity of cationic polymer gene delivery vehicles are directly related to their molecular weight (9). The toxicity of the synthesized polymers, PLL and PEI were tested in HeLa cells. Cheng et al. recently demonstrated that the toxicity profiles of polycations against HEK293, HeLa and HepG2 are very similar (49); therefore HeLa cell lines were used as a representative cell line. All of the synthesized copolymers were found to be much less toxic than PLL (Figure 7). One factor very likely contributing to the lower toxicity is that the copolymers contain HPMA, and HPMA has been shown to be non-toxic up to very high concentrations (22). Even still, copolymer A3 has on average 15.7 kDa of oligolysine for each polymer strand, which is comparable to the 12–24 kDa PLL that was used. Copolymer A3 is found to have good biocompatibility, with greater than 70% cell viability at the highest concentration tested. The lower toxicity may be due to the spacing of HPMA between peptides that reduces the toxicity associated with the cationic lysines. Copolymer C1 contains on average 8 kDa of oligolysine per polymer strand, but has a high peptide wt% (and thus the fewest HPMA monomers spaced between its peptides) and was the most toxic of all the copolymers tested. It has been suggested that one mechanism for cationic polymer-induced cellular toxicity is for the polymers to form channels in the outer mitochondrial membrane, which allows the caspase-3 translocation and cytochrome c release that are hallmarks of apoptosis (50). We speculate that the HPMA between the peptides hinders the formation of these channels in the outer mitochondrial membrane, thus reducing the toxicity of the copolymers with large amounts of HPMA spaced between the peptides. The high toxicity of copolymer C1 may be due to it having an amount of HPMA between the peptides that is below the threshold that would be required to hinder the formation of the membrane channels.
Figure 7.
Cell viability of HeLa cells after incubation with free polymers, copolymers, and peptides at the indicated concentrations, as measured by an MTS assay.
Several investigators have crosslinked low molecular weight polycations using reducible disulfide bonds to generate high molecular weight conjugates that efficiently condense and deliver DNA, but which become less cytotoxic fragments upon intracellular reduction (9–15). Copolymer D1 was synthesized to allow reduction of the peptide linkers, and release of peptides from the polymer backbone. In composition, reducible copolymer D1 is very comparable to the non-reducible copolymer C1, with 10 kDa and 8 kDa from oligolysine in the copolymer, 35.5 and 37.8 wt% peptide, and 25 and 22 HPMA monomers between each peptide, respectively. Despite the similarities in composition, copolymer D1 is much less toxic than copolymer C1 (Figure 7).
Conclusions
We have demonstrated the synthesis of novel copolymers for gene delivery by radical copolymerization of a modified peptide monomer with an HPMA comonomer. These copolymers can condense DNA into small polyplexes that are stable at physiological salt concentrations and also resistant to unpackaging. The copolymers were also shown to deliver DNA into HeLa cells with an efficiency comparable to PLL while mediating much less cytotoxicity than PLL. This modular system has the capacity to incorporate other types of peptides into the polymer backbone, and these peptides may perform functions that increase transfection efficiency (such as endosomal escape) or specificity (such as targeting). Furthermore, the polymerization conditions could be further optimized to produce better copolymers. In contrast to traditional free radical polymerization that leads to sub-optimal incorporation of macromonomers, it has been shown that controlled polymerization schemes greatly improve the incorporation of macromonomers into copolymers by reducing the disparity in reactivities between macromonomers and the smaller comonomers (31). Moving to a controlled radical polymerization technique also has the added benefit of producing more well-defined copolymers of more consistent composition and with lower PDIs. The development of nearly monodisperse, targeted copolymers that mediate high levels of gene delivery can be possible in the near future using this system as a basis for further improvements.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Anthony Convertine for helpful discussions regarding the polymerization strategy. Mass spectrometry was performed in the Department of Chemistry Mass Spectrometry Facility, with special thanks to Dr. Martin Sadilek for his training and support on the ESI-MS instrument. GPC was performed with equipment in the laboratory of Dr. Patrick Stayton and Dr. Allan Hoffman, with special thanks to Dr. Scott Henry for his assistance. This work was supported by an NSF CAREER Award (CBET-0448547) to S.H.P., and NIH/NINDS grants 5R21NS052030 and 1R01NS064404. R.S.B. is grateful to the National Science Foundation for a graduate research fellowship.
Footnotes
Supporting Information Available: Figure S1 shows the mass spectrum of K11 peptide. Figure S2 shows the mass spectrum of K11-Ahx peptide. Figure S3 shows the mass spectrum of K11-Ahx-MC peptide. Figure S4 shows HPLC chromatograms for K11, K11-AEDP, and K11-AEDP-MC peptides. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bergen J, Pun S. Peptide-enhanced nucleic acid delivery. MRS Bulletin. 2005;30:663–667. [Google Scholar]
- 2.Kwon E, Bergen J, Park I, Pun S. Peptide-modified vectors for nucleic acid delivery to neurons. Journal of Controlled Release. 2008;132:230–235. doi: 10.1016/j.jconrel.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin ME, Rice KG. Peptide-guided gene delivery. Aaps Journal. 2007;9:E18–E29. doi: 10.1208/aapsj0901003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.OBrienSimpson NM, Ede NJ, Brown LE, Swan J, Jackson DC. Polymerization of unprotected synthetic peptides: A view toward synthetic peptide vaccines. Journal of the American Chemical Society. 1997;119:1183–1188. [Google Scholar]
- 5.Jackson DC, O’Brien-Simpson N, Ede NJ, Brown LE. Free radical induced polymerization of synthetic peptides into polymeric immunogens. Vaccine. 1997;15:1697–705. doi: 10.1016/s0264-410x(97)00085-6. [DOI] [PubMed] [Google Scholar]
- 6.Graham KL, Zeng WG, Takada Y, Jackson DC, Coulson BS. Effects on rotavirus cell binding and infection of monomeric and polymeric peptides containing alpha 2 beta 1 and alpha x beta 2 integrin ligand sequences. Journal of Virology. 2004;78:11786–11797. doi: 10.1128/JVI.78.21.11786-11797.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu ZH, Li MY, Cui DF, Fei J. Macro-branched cell-penetrating peptide design for gene delivery. Journal of Controlled Release. 2005;102:699–710. doi: 10.1016/j.jconrel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Ding H, Prodinger WM, Kopecek J. Identification of CD21-binding peptides with phage display and investigation of binding properties of HPMA copolymer-peptide conjugates. Bioconjugate Chemistry. 2006;17:514–523. doi: 10.1021/bc0503162. [DOI] [PubMed] [Google Scholar]
- 9.Breunig M, Lungwitz U, Liebl R, Goepferich A. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14454–14459. doi: 10.1073/pnas.0703882104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKenzie DL, Smiley E, Kwok KY, Rice KG. Low molecular weight disulfide cross-linking peptides as nonviral gene delivery carriers. Bioconjugate Chemistry. 2000;11:901–909. doi: 10.1021/bc000056i. [DOI] [PubMed] [Google Scholar]
- 11.Gosselin MA, Guo WJ, Lee RJ. Efficient gene transfer using reversibly cross-linked low molecular weight polyethylenimine. Bioconjugate Chemistry. 2001;12:989–994. doi: 10.1021/bc0100455. [DOI] [PubMed] [Google Scholar]
- 12.Siprashvili Z, Scholl FA, Oliver SF, Adams A, Contag CH, Wender PA, Khavari PA. Gene transfer via reversible plasmid condensation with cysteine-flanked, internally spaced arginine-rich peptides. Human Gene Therapy. 2003;14:1225–1233. doi: 10.1089/104303403767740768. [DOI] [PubMed] [Google Scholar]
- 13.Read ML, Singh S, Ahmed Z, Stevenson M, Briggs SS, Oupicky D, Barrett LB, Spice R, Kendall M, Berry M, Preece JA, Logan A, Seymour LW. A versatile reducible polycation-based system for efficient delivery of a broad range of nucleic acids. Nucleic Acids Research. 2005;33:e86. doi: 10.1093/nar/gni085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kloeckner J, Wagner E, Ogris M. Degradable gene carriers based on oligomerized polyamines. European Journal of Pharmaceutical Sciences. 2006;29:414–425. doi: 10.1016/j.ejps.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Peng Q, Zhong ZL, Zhuo RX. Disulfide cross-linked polyethylenimines (PEI) prepared via thiolation of low molecular weight PEI as highly efficient gene vectors. Bioconjugate Chemistry. 2008;19:499–506. doi: 10.1021/bc7003236. [DOI] [PubMed] [Google Scholar]
- 16.Wadhwa MS, Collard WT, Adami RC, McKenzie DL, Rice KG. Peptide-mediated gene delivery: influence of peptide structure on gene expression. Bioconjug Chem. 1997;8:81–8. doi: 10.1021/bc960079q. [DOI] [PubMed] [Google Scholar]
- 17.Fields CG, Fields GB, Noble RL, Cross TA. Solid-Phase Peptide-Synthesis of N-15-Gramicidin-a, N-15-Gramicidin-B, and N-15-Gramicidin-C and High-Performance Liquid-Chromatographic Purification. International Journal of Peptide and Protein Research. 1989;33:298–303. doi: 10.1111/j.1399-3011.1989.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 18.Jiang XL, van der Horst A, van Steenbergen MJ, Akeroyd N, van Nostrum CF, Schoenmakers PJ, Hennink WE. Molar-mass characterization of cationic polymers for gene delivery by aqueous size-exclusion chromatography. Pharmaceutical Research. 2006;23:595–603. doi: 10.1007/s11095-006-9574-4. [DOI] [PubMed] [Google Scholar]
- 19.Burke RS, Pun SH. Extracellular barriers to in Vivo PEI and PEGylated PEI polyplex-mediated gene delivery to the liver. Bioconjugate Chemistry. 2008;19:693–704. doi: 10.1021/bc700388u. [DOI] [PubMed] [Google Scholar]
- 20.Kopecek J, Bazilova H. Poly[N-(2-Hydroxypropyl)Methacrylamide] .1. Radical Polymerization and Copolymerization. European Polymer Journal. 1973;9:7–14. [Google Scholar]
- 21.Strohalm J, Kopecek J. Poly[N-(2-Hydroxypropyl)Methacrylamide] .4. Heterogeneous Polymerization. Angewandte Makromolekulare Chemie. 1978;70:109–118. [Google Scholar]
- 22.Vasey PA, Kaye SB, Morrison R, Twelves C, Wilson P, Duncan R, Thomson AH, Murray LS, Hilditch TE, Murray T, Burtles S, Fraier D, Frigerio E, Cassidy J. Phase I clinical and pharmacokinetic study of PK1 [N-(2-hydroxypropyl)methacrylamide copolymer doxorubicin]: first member of a new class of chemotherapeutic agents-drug-polymer conjugates. Cancer Research Campaign Phase I/II Committee. Clin Cancer Res. 1999;5:83–94. [PubMed] [Google Scholar]
- 23.Duncan R, Gac-Breton S, Keane R, Musila R, Sat YN, Satchi R, Searle F. Polymer-drug conjugates, PDEPT and PELT: basic principles for design and transfer from the laboratory to clinic. Journal of Controlled Release. 2001;74:135–146. doi: 10.1016/s0168-3659(01)00328-5. [DOI] [PubMed] [Google Scholar]
- 24.Duncan R. The dawning era of polymer therapeutics. Nature Reviews Drug Discovery. 2003;2:347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 25.Rihova B, Kubackova K. Clinical implications of N-(2-hydroxypropyl)methacrylamide copolymers. Curr Pharm Biotechnol. 2003;4:311–22. doi: 10.2174/1389201033489711. [DOI] [PubMed] [Google Scholar]
- 26.Haag R, Kratz F. Polymer therapeutics: concepts and applications. Angew Chem Int Ed Engl. 2006;45:1198–215. doi: 10.1002/anie.200502113. [DOI] [PubMed] [Google Scholar]
- 27.Oupicky D, Konak C, Ulbrich K. DNA complexes with block and graft copolymers of N-(2-hydroxypropyl)methacrylamide and 2-(trimethylammonio)ethyl methacrylate. J Biomater Sci Polym Ed. 1999;10:573–90. doi: 10.1163/156856299x00496. [DOI] [PubMed] [Google Scholar]
- 28.Oupicky D, Konak C, Dash PR, Seymour LW, Ulbrich K. Effect of albumin and polyanion on the structure of DNA complexes with polycation containing hydrophilic nonionic block. Bioconjug Chem. 1999;10:764–72. doi: 10.1021/bc990007+. [DOI] [PubMed] [Google Scholar]
- 29.Oupicky D, Konak C, Ulbrich K, Wolfert MA, Seymour LW. DNA delivery systems based on complexes of DNA with synthetic polycations and their copolymers. J Control Release. 2000;65:149–71. doi: 10.1016/s0168-3659(99)00249-7. [DOI] [PubMed] [Google Scholar]
- 30.Radke W, Muller AHE. Copolymerization of Methacryloyl-Terminated Pmma Macromonomers with Methyl-Methacrylate. Makromolekulare Chemie-Macromolecular Symposia. 1992;54–5:583–594. [Google Scholar]
- 31.Shinoda H, Miller PJ, Matyjaszewski K. Improving the structural control of graft copolymers by combining ATRP with the macromonomer method. Macromolecules. 2001;34:3186–3194. [Google Scholar]
- 32.Neugebauer D, Zhang Y, Pakula T. Gradient graft copolymers derived from PEO-based macromonomers. Journal of Polymer Science Part a-Polymer Chemistry. 2006;44:1347–1356. [Google Scholar]
- 33.Lang JL. Retardation of Polymerization in Substituted Styrenes by Chelated Nitrophenols. Journal of Chemical & Engineering Data. 1960;5:53–56. [Google Scholar]
- 34.Motyakin MV, Wasserman AM, Stott PE, Zaikov GE. Inhibitor radicals in styrene polymerization. Journal of Applied Polymer Science. 2004;91:1599–1603. [Google Scholar]
- 35.Tsutsumi K, Okamoto Y, Tsukahara Y. Radical Polymerization Behavior of Macromonomers .3. Effect of Macromonomer Concentration. Polymer. 1994;35:2205–2209. [Google Scholar]
- 36.Paulusse JMJ, Amir RJ, Evans RA, Hawker CJ. Free Radical Polymers with Tunable and Selective Bio- and Chemical Degradability. Journal of the American Chemical Society. 2009;131:9805–9812. doi: 10.1021/ja903245p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R, Wagner E. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Therapy. 1998;5:1425–1433. doi: 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- 38.Ruponen M, Yla-Herttuala S, Urtti A. Interactions of polymeric and liposomal gene delivery systems with extracellular glycosaminoglycans: physicochemical and transfection studies. Biochimica Et Biophysica Acta-Biomembranes. 1999;1415:331–341. doi: 10.1016/s0005-2736(98)00199-0. [DOI] [PubMed] [Google Scholar]
- 39.Moret I, Peris JE, Guillem VM, Benet M, Revert F, Dasi F, Crespo A, Alino SF. Stability of PEI-DNA and DOTAP-DNA complexes: effect of alkaline pH, heparin and serum. Journal of Controlled Release. 2001;76:169–181. doi: 10.1016/s0168-3659(01)00415-1. [DOI] [PubMed] [Google Scholar]
- 40.Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnology and Bioengineering. 2000;67:598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 41.Holmgren A. Thioredoxin and Glutaredoxin Systems. Journal of Biological Chemistry. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 42.Arunachalam B, Phan UT, Geuze HJ, Cresswell P. Enzymatic reduction of disulfide bonds in lysosomes: Characterization of a Gamma-interferon-inducible lysosomal thiol reductase (GILT) Proceedings of the National Academy of Sciences of the United States of America. 2000;97:745–750. doi: 10.1073/pnas.97.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J, Chen H, Vlahov IR, Cheng JX, Low PS. Evaluation of disulfide reduction during receptor-mediated endocytosis by using FRET imaging. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13872–13877. doi: 10.1073/pnas.0601455103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mishra S, Webster P, Davis ME. PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. European Journal of Cell Biology. 2004;83:97–111. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 45.Kursa M, Walker GF, Roessler V, Ogris M, Roedl W, Kircheis R, Wagner E. Novel shielded transferrin-polyethylene glycol-polyethylenimine/DNA complexes for systemic tumor-targeted gene transfer. Bioconjugate Chemistry. 2003;14:222–231. doi: 10.1021/bc0256087. [DOI] [PubMed] [Google Scholar]
- 46.Boussif O, Lezoualch F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and In-vivo - Polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonawane ND, Szoka FC, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. Journal of Biological Chemistry. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 48.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. Journal of Gene Medicine. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 49.Cheng H, Zhu JL, Zeng X, Jing Y, Zhang XZ, Zhuo RX. Targeted Gene Delivery Mediated by Folate-polyethylenimine-block-poly(ethylene glycol) with Receptor Selectivity. Bioconjug Chem. 2009;20:481–487. doi: 10.1021/bc8004057. [DOI] [PubMed] [Google Scholar]
- 50.Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Adv Drug Deliv Rev. 2006;58:1523–31. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.