Abstract
Current therapeutics and prophylactics for malaria are under severe challenge as a result of the rapid emergence of drug-resistant parasites. The human malaria parasite Plasmodium falciparum expresses two neutral aminopeptidases, PfA-M1 and PfA-M17, which function in regulating the intracellular pool of amino acids required for growth and development inside the red blood cell. These enzymes are essential for parasite viability and are validated therapeutic targets. We previously reported the x-ray crystal structure of the monomeric PfA-M1 and proposed a mechanism for substrate entry and free amino acid release from the active site. Here, we present the x-ray crystal structure of the hexameric leucine aminopeptidase, PfA-M17, alone and in complex with two inhibitors with antimalarial activity. The six active sites of the PfA-M17 hexamer are arranged in a disc-like fashion so that they are orientated inwards to form a central catalytic cavity; flexible loops that sit at each of the six entrances to the catalytic cavern function to regulate substrate access. In stark contrast to PfA-M1, PfA-M17 has a narrow and hydrophobic primary specificity pocket which accounts for its highly restricted substrate specificity. We also explicate the essential roles for the metal-binding centers in these enzymes (two in PfA-M17 and one in PfA-M1) in both substrate and drug binding. Our detailed understanding of the PfA-M1 and PfA-M17 active sites now permits a rational approach in the development of a unique class of two-target and/or combination antimalarial therapy.
Keywords: drug design, malaria, protease, structural biology, neutral aminopeptidases
An estimated 1–2 million people die each year as a consequence of infection with apicomplexan parasites of the genus Plasmodium (1). The widespread appearance of drug-resistant malaria parasites, even to newly developed second and third generation therapeutics such artemisinin and its derivatives, makes the development of unique antimalarial drug treatments all the more urgent (2).
Neutral aminopeptidases are ubiquitous in mammalian cells and are generally involved in protein processing by clipping N-terminal amino acids, such as the trimming of peptides to achieve optimal binding to Major Histocompatibility Complex class I molecules (3), or in the final stages of protein degradation. Malaria parasites express two such enzymes, a 122 kDa M1-family alanyl aminopeptidase and a 67.8 kDa M17-family leucyl aminopeptidase, termed PfA-M1 (MAL13P1.56, plasmodb.org) and PfA-M17 (Pf14_0439, plasmodb.org), respectively (4 –8). The concerted functions of these enzymes are essential for parasite viability as targeted gene-knock-outs have been unsuccessful (4, 7, 8). Both enzymes function in the blood stage of the infection and are responsible for regulating the intracellular amino acid pool within malaria cells, most likely by releasing amino acids from host-derived haemoglobin. In addition to providing essential nutrients for parasite growth and development, digestion of haemoglobin is important for maintaining the osmotic integrity of infected red blood cells (9). PfA-M1 and PfA-M17 are validated therapeutic targets for a unique class of antimalarial drugs. Blocking the activity of these aminopeptidases by dipeptide analogs prevents growth of P. falciparum in culture and kills P. c. chabaudi malaria in vivo (4 –7, 10).
Both PfA-M17 and PfA-M1 are able to efficiently cleave N-terminal amino acids from peptide substrates; however their specificities differ dramatically. PfA-M1 exhibits a far broader substrate specificity than PfA-M17 and rapidly hydrolyses substrates containing Leu, Ala, Arg, and Lys, but can also cleave substrates containing Phe, Tyr, Ser, and Asn (but not Glu or Asp) (4, 11, 12). In contrast, PfA-M17 possesses a severely restricted specificity for N terminally exposed leucine; alanine substrates are cleaved relatively poorly (∼850 fold lower than Leu) and substrates containing an Arg, Val, Pro, Gly, Ile, Glu, or Asp in the P1 position are not cleaved at all (7). These insights into to the differing specificities of the two enzymes have already permitted development of an P1-Arg-specific enzymatic assay for detecting the activity of PfA-M1 in malaria cytosolic extracts (4).
The two enzymes also have distinct differences in the arrangement of divalent metal cations in their active site. PfA-M1 binds a single, tightly-bound Zn2+ metal ion whereas PfA-M17 contains two metal-binding sites, a readily exchangeable site (site 1) and a tight-binding site (site 2) (13). Whereas PfA-M17 retains residual catalytic activity when the metal ion from site 1 is removed, the removal of metal ions from both sites results in an inactive apoenzyme that cannot be reactivated by addition of divalent metal cations (13). However, the divalent metal cation binding at site 1 of PfA-M17 can be functionally exchanged for other metal cations, the enzyme displaying a preference in the order Zn2+ > Mn2+ > Co2+ > Mg2+ (13). The type of metal cation in the active site of these enzymes can therefore influence the catalytic efficiency against peptide substrates as well as the binding of inhibitors. Together, these data highlight the importance of understanding the catalytic role of metal atoms for the future optimization of lead inhibitory compounds.
In this study we present the structure of the hexameric PfA-M17 enzyme, with one ( ) and two (
) and two ( ) metal ions bound in the active site. We also have determined the structure of PfA-M17 in the complex with the inhibitors bestatin (PfA-M17-BES) and phosphinic dipeptide analogue hPheP[CH2]Phe (termed compound 4, Co4; PfA-M17-Co4). We compare and contrast these data with our previously determined structure of PfA-M1 in complex with the same inhibitors and reveal dramatic differences in the manner in which the two enzymes receive, bind, and cleave N-terminal amino acid substrates. These studies will lead a rational approach in the development of inhibitors that have dual and selective activity for PfA-M17 and PfA-M1. Such reagents will permit key insights into the precise role of each enzyme in the parasite lifecycle and guide the structure-based design of unique classes of antimalarial therapeutics.
) metal ions bound in the active site. We also have determined the structure of PfA-M17 in the complex with the inhibitors bestatin (PfA-M17-BES) and phosphinic dipeptide analogue hPheP[CH2]Phe (termed compound 4, Co4; PfA-M17-Co4). We compare and contrast these data with our previously determined structure of PfA-M1 in complex with the same inhibitors and reveal dramatic differences in the manner in which the two enzymes receive, bind, and cleave N-terminal amino acid substrates. These studies will lead a rational approach in the development of inhibitors that have dual and selective activity for PfA-M17 and PfA-M1. Such reagents will permit key insights into the precise role of each enzyme in the parasite lifecycle and guide the structure-based design of unique classes of antimalarial therapeutics.
Results
The Overall Structure of PfA-M17.
The 2.0 Å x-ray crystal structure of the  aminopeptidase revealed two hexamers in the asymmetric unit (SI Text Table S1). Each PfA-M17 monomer contains an N-terminal regulatory domain and a C-terminal catalytic domain that together adopt the leucine aminopeptidase (LAP) fold (14). Further, the general hexameric arrangement of monomers is consistent with the physiological assembly described for PfA-M17 and other LAP family members (14
–16). Structural comparison (17) of the twelve monomers reveals only minor differences are apparent in loop regions, particularly in the N-terminal domain (average r.m.s.d = 0.3 Å over 555 Cα atoms). Unless indicated otherwise we refer to monomer A (Fig. 1
A) and/or the functional hexamer (Fig. 1
B, C) formed by molecules A–F. Numbering is for PfA-M17.
aminopeptidase revealed two hexamers in the asymmetric unit (SI Text Table S1). Each PfA-M17 monomer contains an N-terminal regulatory domain and a C-terminal catalytic domain that together adopt the leucine aminopeptidase (LAP) fold (14). Further, the general hexameric arrangement of monomers is consistent with the physiological assembly described for PfA-M17 and other LAP family members (14
–16). Structural comparison (17) of the twelve monomers reveals only minor differences are apparent in loop regions, particularly in the N-terminal domain (average r.m.s.d = 0.3 Å over 555 Cα atoms). Unless indicated otherwise we refer to monomer A (Fig. 1
A) and/or the functional hexamer (Fig. 1
B, C) formed by molecules A–F. Numbering is for PfA-M17.
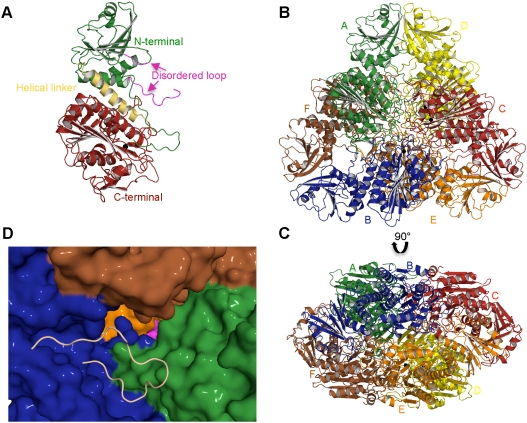
Fig. 1.
The structure of PfA-M17. (A) Diagram of the unbound PfA-M17 monomer colored by domain: N-terminal domain (Green), helical region linking the two domains (Yellow) and C-terminal catalytic domain (Red). The partially disordered loop that sits at the entrance to the active site chamber in the hexameric assembly is shown in magenta. The single zinc atom present in the site 2 position (the site 1 position is unoccupied in this structure,  ) is shown as a black sphere and is labeled. (B, C) Cartoon diagrams of the biologically functional PfA-M17 hexamer colored by chain: A (Green); B (Blue); C (Red); D (Yellow) E (Orange) F (Brown). The six active sites line an interior cavity. (D) The molecular surface of
) is shown as a black sphere and is labeled. (B, C) Cartoon diagrams of the biologically functional PfA-M17 hexamer colored by chain: A (Green); B (Blue); C (Red); D (Yellow) E (Orange) F (Brown). The six active sites line an interior cavity. (D) The molecular surface of  by chain (as colored in (B, C). The active site zinc and carbonate of chain B are visible (Purple spheres). Chains C & D are occluded in this view. The position of the loop (with the molecular surface omitted) in chain B that sits at the entrance to the catalytic cavity is shown by yellow coil (residues 246-265). This region is disordered in the other chains.
by chain (as colored in (B, C). The active site zinc and carbonate of chain B are visible (Purple spheres). Chains C & D are occluded in this view. The position of the loop (with the molecular surface omitted) in chain B that sits at the entrance to the catalytic cavity is shown by yellow coil (residues 246-265). This region is disordered in the other chains.
For each monomer, the N-terminal domain (85-276) comprises a central six-stranded β-sheet and three α-helices (Fig. 1 A). A ∼30 Å helix (277-300) connects the N-terminal domain with the C-terminal catalytic domain (301-605) through the center of the molecule (Fig. 1 A).
The catalytic domain comprises a central eight-stranded β-sheet flanked on both sides by nine α-helices and with a two-stranded β-sheet capping one end (Fig. 1 A). The active site is entirely contained within the catalytic domain (Fig. 1 A) and is located near the edge of the central β-sheet. The active site cleft measures 18 Å by 18 Å and contains a cluster of negatively charged residues that function to coordinate metal cations.
Whereas the crystal trials were established on monomeric material, the structure of PfA-M17 revealed the classic LAP hexameric assembly within the asymmetric unit and, in keeping with other enzymes in this superfamily, we suggest this represents the biologically relevant active form.
The six active sites are located in the interior of the molecule, where they line a disk-shaped cavity of diameter of ∼35 Å and height ∼15 Å. Access to the cavity occurs by channels formed by the N-terminal domains of each monomer. Most importantly, a ∼20 Å loop between helix 3 and strand 6 which is partially disordered in 10 of the 12 monomers, sits at the entrance to the catalytic cavity (Fig. 1 A, D). The complete loop is visible in electron density in chains C and I where it occludes the entrance to the enzyme interior (Fig 1 D). We suggest that flexibility of this loop may regulate substrate entrance and product exit from the buried aminopeptidase active sites (Fig. 1 D).
Binding of Metal Ions in the Active Site of PfA-M17.
The LAP structures described to date contain two metal ions in the active site. Biophysical studies on this enzyme family reveal that the first site is readily exchangeable and is thus termed the “loose” or regulatory site 1 (18). In contrast, the second metal ion is “tight-binding” and is known as the catalytic site 2 (18). Metal replacement studies on PfA-M17 show that the enzyme retains activity when only the tight-binding catalytic site 2 is occupied with a metal ion; removal of both metal ions lead to complete and irreversible inactivity (7, 13). Consistent with these ideas, in the presence of increasing concentrations of EDTA, loss of M17 activity follows a biphasic pattern as each metal ion is sequentially chelated (13) (SI Text Fig. S1). In contrast, studies on PfA-M1 reveal that this enzyme contains a single, essential, tightly-bound metal ion. Accordingly, increasing concentrations of EDTA eventually remove this metal ion in a monophasic manner, resulting in a sudden and irreversible loss of enzyme activity (SI Text Fig. S1).
Interestingly, the structure of 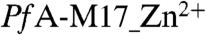 revealed that only the catalytic site 2 was occupied by a zinc ion and no electron density was visible for the site 1 metal ion. It is therefore suggested that the loosely bound metal site was lost during purification. High B-factors were also noted for the single zinc ion—suggesting local disorder of this atom.
revealed that only the catalytic site 2 was occupied by a zinc ion and no electron density was visible for the site 1 metal ion. It is therefore suggested that the loosely bound metal site was lost during purification. High B-factors were also noted for the single zinc ion—suggesting local disorder of this atom.
In 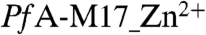 , the site 2 zinc is coordinated by Lys374, Asp379, Asp399, and Glu461 and two water molecules, creating a hexameric coordination of this ion (SI Text Fig. S2). For this, and all the other structures we report, we also observed strong planar density consistent with the presence of a carbonate ion in the PfA-M17 active site (previously noted in all other LAP crystal structures (19)) which forms hydrogen bonds to residues Lys374, Ala460, Gly462, Arg463, and Leu487 (SI Text Fig. S2). The carbonate ion has been proposed to act as a general base, accepting the proton and activating a (nucleophilic) metal-bridging water (19).
, the site 2 zinc is coordinated by Lys374, Asp379, Asp399, and Glu461 and two water molecules, creating a hexameric coordination of this ion (SI Text Fig. S2). For this, and all the other structures we report, we also observed strong planar density consistent with the presence of a carbonate ion in the PfA-M17 active site (previously noted in all other LAP crystal structures (19)) which forms hydrogen bonds to residues Lys374, Ala460, Gly462, Arg463, and Leu487 (SI Text Fig. S2). The carbonate ion has been proposed to act as a general base, accepting the proton and activating a (nucleophilic) metal-bridging water (19).
To understand the position and role of the metal atom in the site 1 position, we conducted soaking experiments on crystals and determined the 2.4 Å structure of  (SI Text Table S1). These data revealed a di-zinc center in the active site with strong Fo-Fc density for both zinc positions. The two metal ions are 2.9 Å apart (SI Text Fig. S2), however, in comparison to
(SI Text Table S1). These data revealed a di-zinc center in the active site with strong Fo-Fc density for both zinc positions. The two metal ions are 2.9 Å apart (SI Text Fig. S2), however, in comparison to  , the position of the modeled and refined Zn in catalytic site 2 is slightly shifted by ∼0.4 Å. In
, the position of the modeled and refined Zn in catalytic site 2 is slightly shifted by ∼0.4 Å. In 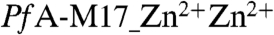 , the catalytic site 1 Zn2+ is coordinated by Asp379, Asp459, and Glu461, whereas coordination of the catalytic site 2 Zn is similar to
, the catalytic site 1 Zn2+ is coordinated by Asp379, Asp459, and Glu461, whereas coordination of the catalytic site 2 Zn is similar to 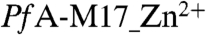 (SI Text Fig. S2).
(SI Text Fig. S2).
Our biochemical studies suggest that the ability of PfA-M17 to rapidly gain or lose a divalent cation, most likely at the regulatory site 1, may represent an important mechanism of regulation of catalytic activity not only for the binding of substrates but also inhibitors (13). Consistent with this idea, we found that in the absence of environmental metal, no metal ion was observed at the catalytic site 1 position (SI Text Fig. S2). Both metal atoms are also visible in electron density in the inhibitor-bound forms (discussed in the next section).
Interaction of the Antimalarial Inhibitors Bestatin and Co4 with PfA-M17 and Comparison with PfA-M1.
Two inhibitors of PfA-M1 and PfA-M17 enzyme have been well characterized, bestatin and compound 4 (Co4; phosphinic dipeptide analog) (5). We determined each inhibitor/enzyme complex structures to 2.0 and 2.6 Å resolution, respectively (SI Text Table S1). The bestatin-bound (PfA-M17-BES) and Co4-bound (PfA-M17-Co4) enzyme structures were obtained from crystallization experiments conducted in the presence of Mg2+ and Zn2+, respectively.
The electron density of the modeled ligands within the active site was well defined (Fig. 2). Both ligands form extensive interactions with the active site and the carbonate ion, however, the presence of the inhibitor did not result in any major conformational change or movement of active site side-chains (Overall r.m.s.d. of representative monomer chain was 0.2 Å for PfA-M17-BES and 0.3 Å for PfA-M17-Co4 over 522 cα atoms).
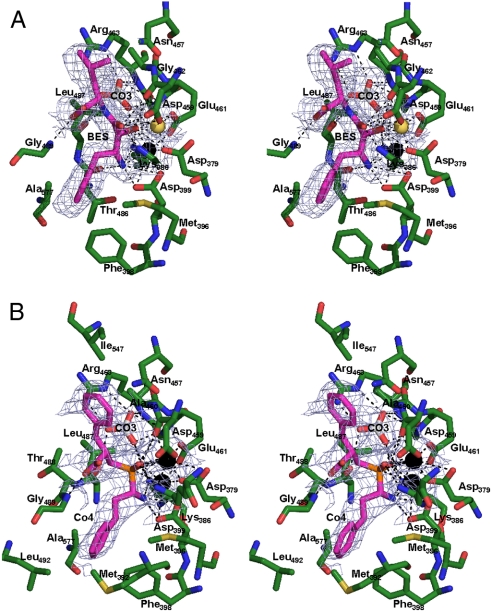
Fig. 2.
Stereo diagram of inhibitors binding to active site of PfA-M17. (A) 2.0 Å PfA-M17-BES and (B) 2.6 Å PfA-M17-Co4 active site showing inhibitors bound in the active site. Inhibitors (BES/Co4) are colored in magenta. Carbon atoms of residues are colored green. Zinc is shown as black sphere; Magnesium as yellow sphere and water molecules are light-blue spheres. Hydrogen and metallo-bonds are indicated (dashed lines). Electron density is a composite omit map showing averaged density of 12 protein chains contoured at 3.0 σ calculated by using a model containing only PfA-M17 atoms (no metal, ligand, or water was included in calculation).
The PfA-M17-BES structure revealed pentavalent coordination of the site 1 Mg2+ ion (Fig. 2 A). The bestatin molecule buried a surface area of 44.2 Å 2 and fitted neatly into each active site with the hydroxyl group (O2) and central nitrogen (N2) coordinating the active site metals (Fig. 2 A). Bestatin forms hydrogen bonds with 12 residues in the active site (predominantly with metal coordinating residues, SI Text Table S2) and in addition makes eight van der Waal interactions with other residues lining the pocket. The P1 Phe side-chain of bestatin is packed into the S1 pocket and forms interactions with the hydrophobic residues Met392, Met396, Phe398, Gly489, and Ala577. The P1′ position makes only two interactions with Asn457 and Ile547.
The PfA-M17-Co4 structure revealed that the buried surface area of the Co4 inhibitor (195.1 Å 2) was significantly larger than that occluded by bestatin. Strong Fo-Fc density was observed for both zinc ions in the active site. As a consequence of its central phosphate group, and in contrast to bestatin, Co4 is able to coordinate the catalytic site 1 Zn2+ in a hexavalent fashion (Fig. 2 B). Indeed, the central phosphate group of the ligand dominates interactions with the active site metals with both phosphoryl oxygens (O3/O4) interacting with the catalytic site 1Zn2+, whereas the catalytic site 2 Zn2+ is bound by the O4 atom as well as Co4 N atom. The differences in how Co4 and bestatin coordinate the site 1 Zn2+ (Fig. 2 and SI Text Fig. S3) may explain the linear inhibitory kinetics exhibited by Co4 versus the slow/tight-binding kinetics exhibited by bestatin (5, 7).
Aside from the differences in coordination of metal atoms, Co4 and bestatin interact with the active site in a similar fashion (Fig. 2), although Co4 forms only eight hydrogen bonds and one extra van der Waal interaction (at the P1 position with Leu492; SI Text Table S2).
Both bestatin and Co4 are far superior inhibitors of PfA-M17 than PfA-M1 (the K i for bestatin is 25 and 478 and for Co4 are 13 and 79 nM, respectively, SI Text Table S4). Consistent with this observation, a comparison of the four inhibitor-bound structures reveal that each inhibitor makes far more extensive interactions with the PfA-M17 active site than with PfA-M1 (SI Text Table S4). Furthermore, in contrast to PfA-M17, in PfA-M1 both inhibitors coordinate the single active site Zn2+ atom (equivalent to the site 1 metal in PfA-M17; SI Text Fig. S3) in the same (pentavalent) fashion. We also compared the inhibitor conformation in the active site of PfA-M17 with that seen in PfA-M1 (4). These data reveal that the general conformation of the inhibitor scaffold in both enzymes is quite similar, however, variation in the side-chain rotamers of the P1 and P1′ positions are observed (SI Text Fig. S3).
Structural Basis for the Differing Substrate Specificity of PfA-M1 and PfA-M17.
Although the two Plasmodium neutral aminopeptidases exhibit some overlap in the amino acids that can be accommodated into their active sites (in particular Leu and Phe (4, 7); SI Text Table S3), each protease has distinct substrate profiles that can be explained by key features within their active sites.
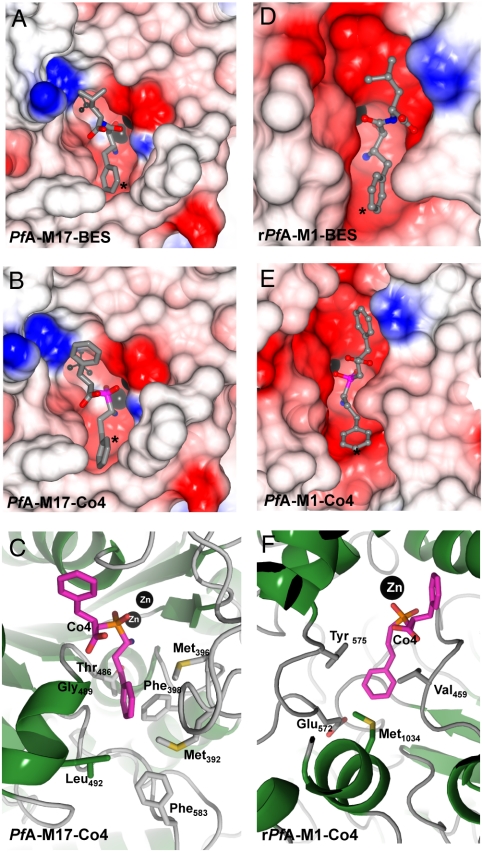
The PfA-M17 S1 pocket that interacts with the substrate P1 residue is a narrow and substantially hydrophobic pocket lined at the entrance by Met392, Met396, Phe398, Thr486, Gly489, Leu492, and Phe583 (Fig. 3 A–C). In both inhibitor-bound structures the P1 Phe-like moieties are tightly packed into this S1 pocket, in particular stacking against the aromatic ring of Phe398 (Fig. 3 A–C). The hydrophobic nature of this narrow pocket underlies the restricted substrate profile of PfA-M17, and explains the inability of this enzyme to cleave peptides/proteins after polar residues such as Arg, Glu, or Asp residues. Indeed, inspection of PfA-M17 structure reveals no suitable polar hydrogen-bonding partners at the base of the S1 pocket that could interact with a charged P1 side-chain (Fig. 3 C). With respect to smaller P1 residues, we suggest that amino acids such as Ala and Gly would fail to reach far enough into the S1 pocket to bind properly.
Fig. 3.
Comparison of the active site cleft and S1 binding pockets of PfA-M17 and PfA-M1. (A), (B) CCP4MG electrostatic potential surface of the active sites of (A) PfA-M17-BES and (B) PfA-M17-Co4. (C) Cartoon of PfA-M17-Co4 showing the residues lining the S1 pocket (labeled and in stick). (D), (E) show a CCP4MG electrostatic potential surface of the active site of PfA-M1-BES (D) and PfA-M1-Co4 (E). (F) shows a cartoon depicting the residues lining the S1 pocket (labeled and in stick) for PfA-M1-Co4. Throughout this figure, electrostatic surfaces are color-coded according to electrostatic potential calculated by the Poisson-Boltzmann solver within CCPMG. Metal ions are shown as black spheres. The carbonate ion in PfA-M17 (A), (B) is shown as ball and stick (Black). The S1 pocket of active sites are indicated (*).
In contrast, the molecular basis for PfA-M1 substrate promiscuity is apparent upon inspection of its S1 pocket which is much larger than that of PfA-M17 (Fig. 3 D–F). Comparison of inhibitor-bound and unbound PfA-M1 structures also revealed that the side-chain of Met1034 is able to move upon inhibitor binding; this residue may provide further flexibility in regards to the shape of the S1 pocket. Most notably, whereas the entrance to the specificity pocket is lined by hydrophobic residues (Val459, Tyr575, and Met1034), a polar glutamic acid (Glu572) residue is located at the base of the pocket where it would be available to form an ionic interaction with the side-chain of a P1 Arg (Fig. 3 F).
Discussion
Malaria parasites utilize proteases in many aspects of their intraerythrocytic cycle, including employing the serine protease subtilisin 1 and a dipeptidyl peptidase 3 to effect egress from the cell (20). Numerous proteases also degrade host haemoglobin in the digestive food vacuole as intraerythrocytic parasites (21). Although initial studies suggested that these digestive vacuole enzymes might represent attractive drug targets, additional gene targeting and inhibitory studies have now revealed that this is not the case with many of these enzymes having redundant functions (9).
In malaria parasites we, and others, have identified two essential netural aminopeptidases, PfA-M1 and PfA-M17, each of which is encoded by a single-copy gene. Gene targeting and in vivo animal experiments have demonstrated that parasites are unable to tolerate inhibition or inactivation of either enzyme, validating these molecules as therapeutic targets (4 –7, 10).
Whereas the precise functions of PfA-M1 and PfA-M17 are still speculative it is suggested that these enzymes function in the final stages of haemoglobin digestion and degrade the dipeptides produced by the action of various enzymes within the parasite’s acidic digestive vacuole (4, 5, 7, 8). This process is essential for the provision of free amino acids for parasite protein synthesis. However, where in the malaria parasite and how these enzymes may perform this process (or any other function) is under debate largely because of reports that present conflicting data depicting the cellular location of the PfA-M1 enzyme (22, 23). The consensus is that the PfA-M17 is located within the cytosol of the parasite. However, while we (4) and others (11, 12) have also located the PfA-M1 within the cytosolic compartment by immunocytochemistry, Dalal and Klemba (2007), using immunocytochemistry, light microscopy and YFP-tagging, observed the enzyme in the acidic digestive vacuole and nucleus (8).
Our biochemical data show that the two enzymes exhibit optimal activity at neutral pH and little or no activity at the resident pH of the digestive vacuole (pH 5.2) suggesting that their primary function is cytosolic. By extension, we had proposed that small peptides and dipeptides derived from haemoglobin digestion were transported from the digestive vacuole to the cytosol for final break-down by these enzymes to free amino acids (4, 22). Consistent with this idea, our studies reveal that both enzymes contain buried active sites that would most likely be inaccessible to all but small peptides. However, our structural data reveal that such peptides enter the two enzymes in remarkably different ways. The present study reveals that the six active sites of the PfA-M17 hexamer are arranged in a disc-like fashion so that they are orientated inwards to form a central catalytic cavity. Whereas this large space itself does not present any obstacle to substrate ingress and amino acid egress, it suggests that flexible loops sit at each of the six entrances to the catalytic cavern and function to regulate peptide access (Fig. 1 D). In contrast, our previous analysis of the three-dimensional structure of the PfA-M1 showed how four distinct domains within the protein combine to form two channels that connect the exterior environment to the deeply buried active site. The configuration of these channels led us to propose that peptide substrates reach the PfA-M1 active site through a large (13 Å in diameter and 30 Å in length) entrance tunnel and that amino acids are released through the smaller opening (8 Å) located on the opposite side of the enzyme (4). Taken together, however, both strategies would effectively limit the size of substrates that could access the active site, thus preventing unwanted and potentially deleterious PfA-M1 and PfA-M17 activity against larger protein substrates.
A number of interesting and important questions remain. Most notably, given the broad specificity of the PfA-M1, including its ability to efficiently cleave Leu and Phe, it is unclear why this enzyme would need to function alongside the Leu-specific PfA-M17. One suggestion is that the role of PfA-M17 may be linked to the inability of malaria parasites to synthesize isoleucine. Whereas Ile is one of the most abundant amino acids in malaria proteins it is nevertheless absent from host haemoglobin and therefore the parasite must source this amino acid from the extracellular milieu (24). The parasite achieves this by exchanging intracellular leucine by a two-step process that involves an ATP-independent transporter capable of exchanging intracellular leucine for isoleucine, followed by the ATP-dependent storage or accumulation of isoleucine within the parasite (25). These data lead us to speculate that the essential nature of the Leu-specific PfA-M17 may relate to the requirement for an ample supply of free leucine within the malaria cytosol to exchange for isoleucine.
The structural information obtained in this study, together with our previously published work on PfA-M1, will facilitate a rational approach in the design of unique PfA-M1 and PfA-M17 inhibitors and, furthermore will readily permit fragment-based screening and crystallographic studies on new leads. It is also notable that our studies reveal that PfA-M17 is able to exist with only a single metal ion bound to catalytic site 2, and that inhibitors, and perhaps substrates, themselves play an important role in the capture and/or coordination of the site 1 (loose) divalent cation. This feature, together with inhibitor interactions with the catalytic site 2 cation and other key differences between the active sites may be utilized for the development of specific compounds that target PfA-M17 but are unable to inhibit PfA-M1, and vice versa. Such reagents will help identify the relative efficacy (in terms of killing parasites) of inhibiting each enzyme alone and in combination as well as permitting a greater understanding of how malaria parasites acquire nutrients from inside and outside the erythrocyte. Together, such biological insights will be imperative for driving the overall drug development strategy for this enzyme system towards either a conventional approach that targets one or the other enzyme or a single molecule (or combination therapy) that inhibits both PfA-M1 and PfA-M17. The latter strategy in particular may reduce the likelihood of parasites being able to rapidly evolve resistance.
Materials and Methods
Purification and Molecular Analysis of Recombinant M17 Leucyl Aminopeptidase.
A truncated form of PfA-M17 lacking the N-terminal Asn-rich repeat region was prepared by PCR amplification using the synthesized gene (7) as a template followed by directional cloning into the Escherichia coli expression vector pTrcHis2B (Invitrogen). Previous studies have shown that the N-terminal Asn-rich region is highly variable between species and that its removal does not affect proper protein folding or catalytic activity (7). This construct was used to overexpress the truncated form of PfA-M17 in Rosetta2 BL21 cells grown in AutoInduction Media. Overexpressed cells were lysed in 50 mM Hepes pH 8.0; 300 mM NaCl. Hexa-Histidine tagged PfA-M17 was purified using a two-step purification process of Ni-NTA-agarose column followed by size exclusion chromatography on a Superdex 200 16/60 using a AKTAxpress high throughput chromatography system. Biochemical analysis indicated that kinetic parameters (k cat, K m, k cat/K m) of material purified and used in subsequent crystallisation trials were the same as published (7).
Enzymatic Analysis.
Aminopeptidase activity was determined as previously described (7) by measuring the release of the fluorogenic leaving group, 7-amino-4-methyl-coumarin (NHMec) from the fluorogenic peptide substrates H-Leu-NHMec.
Crystallization and X-ray Data Collection.
Purified PfA-M17 enzyme was concentrated to ∼13 mg/mL and crystals grown using the hanging drop vapour diffusion method, with 1∶1 (v/v) ratio of protein to mother liquor (0.5 mL well volume). The crystals appeared in 40% (v/v) polyethylene glycol 400, 0.1 M Tris (pH 8.5), 0.2 M Lithium sulphate and 1 mM TCEP. Crystal morphology were typically highly stacked thin plates (length typically 0.8–1.0 mm) that generally diffracted to only low-resolution.  crystals were obtained by soaking crystals in 1 mM ZnCl2 before data collection. Crystals of the PfA-M17-Co4 and PfA-M17-bestatin complex were obtained by cocrystallising in the presence of 1 mM ligand before soaking crystals overnight in mother liquor containing 1 mM metal chloride (MgCl2-bestatin and ZnCl2-Co4) and 1 mM ligand. Soaks in the presence of different divalent cations were undertaken to try and improve crystal morphology and diffraction data quality. The final crystal for the M17-bestatin structure that data was collected for was from a MgCl2 soak whereas the M17-Co4 data was collected from a ZnCl2 soak. Data were collected at 100 K using synchrotron radiation at the Australian synchrotron micro crystallography beamline 3ID1. The diffraction data for the ligand-free, ligand-free zinc-soak, bestatin-bound, and Co4-bound protease were collected to 2.0, 2.4, 2.0, and 2.6 Å resolution, respectively. Fluorescence scans of each crystal were collected and indicated the presence of zinc in all crystals (peaks detected at 8.63 and 9.58 against theoretical zinc absorbance of 8.6389, 8.6158, and 9.5720).
crystals were obtained by soaking crystals in 1 mM ZnCl2 before data collection. Crystals of the PfA-M17-Co4 and PfA-M17-bestatin complex were obtained by cocrystallising in the presence of 1 mM ligand before soaking crystals overnight in mother liquor containing 1 mM metal chloride (MgCl2-bestatin and ZnCl2-Co4) and 1 mM ligand. Soaks in the presence of different divalent cations were undertaken to try and improve crystal morphology and diffraction data quality. The final crystal for the M17-bestatin structure that data was collected for was from a MgCl2 soak whereas the M17-Co4 data was collected from a ZnCl2 soak. Data were collected at 100 K using synchrotron radiation at the Australian synchrotron micro crystallography beamline 3ID1. The diffraction data for the ligand-free, ligand-free zinc-soak, bestatin-bound, and Co4-bound protease were collected to 2.0, 2.4, 2.0, and 2.6 Å resolution, respectively. Fluorescence scans of each crystal were collected and indicated the presence of zinc in all crystals (peaks detected at 8.63 and 9.58 against theoretical zinc absorbance of 8.6389, 8.6158, and 9.5720).
Diffraction images were processed using MOSFLM (26), pointless (27) and SCALA (27) from the CCP4 suite (28). 5% of each dataset was flagged for calculation of RFree (29) with neither a sigma nor a low-resolution cut-off applied to the data. A summary of statistics is provided in SI Text Table S1. Subsequent crystallographic and structural analysis was performed using the CCP4i interface (30) to the CCP4 suite (28), unless stated otherwise.
Structure Determination and Refinement.
Structure determination proceeded using the molecular replacement method and the program MOLREP (31). A pseudotranslation vector (0.5 Å) was present on the c-axis. A search model was constructed from the crystal structure of aminopeptidase P from E. coli (PDB ID 1GYT) (probe identified using the FFAS server (32)). A “poly serine” model consisting of only C-terminal domain of 1GYT was constructed with residues other than alanine/glycine residues truncated at Cγ atom. 12 molecules, arranged as 2 hexamers, were present in the asymmetric unit and packed well within the unit cell. Strong unbiased features were observed for the N-terminal region of each PfA-M17 monomer in the initial electron density maps, these data supporting the correctness of the molecular replacement solution was confirmed.
Initial structure refinement and model building proceeded using one molecule in the asymmetric unit (with the other noncrystallographic symmetry (NCS)-related molecules generated using NCS operators). Maximum likelihood refinement using REFMAC (33), incorporating translation, liberation and screw-rotation displacement refinement was carried out, using a bulk solvent correction (Babinet model with mask). After initial C-α backbone trace was completed for one chain, NCS restraints were reduced to identical molecules in each hexamer and retained throughout to improve observation to parameter ratio. Imposed restraints were guided by manual inspection of the model and Rfree. Simulated annealing composite omit maps were generated using Crystallography and NMR System (34) omitting 5 % of the model. All model building and structural validation was done using Crystallography Object-Oriented Toolkit (35). Water molecules were added to the model using ARP/wARP (36) when the Rfree reached 25%. Solvent molecules were retained only if they had acceptable hydrogen-bonding geometry contacts of 2.5 to 3.5 Åwith protein atoms or with existing solvent and were in good 2Fo-Fc and Fo-Fc electron density. When restraints were relaxed, no change in electron density was noted.
Each inhibitor complex was initially solved and refined against the unbound PfA-M17 structure (protein atoms only) and clearly showed unbiased features in the active site for both structures. Positive electron density was apparent in the M17 - BES and M17 - Co4 structure in the position of the loose metal site 1, creating a dimetal center for the active site.
Pymol was used to produce all structural representations. Hydrogen bonds (excluding water-mediated bonds), were calculated using Lig_contact and CONTACT (28). Superpositions were conducted using MUSTANG (17). CCP4MG was used to produce electrostatic diagrams in (Fig. 3) (37). The coordinates and structure factors are available from the Protein Data Bank (3KQX; 3KQZ; 3KR4; 3KR5). Raw data and images are available from TARDIS (www.tardis.edu.au)(38).
Supplementary Material
Acknowledgments.
We thank the Monash University Protein Production Unit, Noelene Quinsey and Nikolaos Sotirellis for technical assistance with the production of the PfA-M17 enzyme. J.C.W. is an Australian Research Council Federation Fellow and a National Health and Medical Research Council (NHMRC) Honorary Principal Research Fellow. A.M.B. is an NHMRC Senior Research Fellow. D.L.G. is an NHMRC Career Development Award recipient. We thank the National Health and Medical Research Council and the Australian Research Council for funding support. We thank the Australian synchrotron for beamtime and for technical assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911813107/DCSupplemental.
References
- 1.Enserink M. Malaria. Signs of drug resistance rattle experts, trigger bold plan. Science. 2008;322(5909):1776. doi: 10.1126/science.322.5909.1776. [DOI] [PubMed] [Google Scholar]
- 2.D’Alessandro U. Existing antimalarial agents and malaria-treatment strategies. Expert Opin Pharmaco. 2009;10(8):1291–1306. doi: 10.1517/14656560902942319. [DOI] [PubMed] [Google Scholar]
- 3.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419(6906):480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 4.McGowan S, et al. Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase. Proc Natl Acad Sci USA. 2009;106(8):2537–2542. doi: 10.1073/pnas.0807398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skinner-Adams TS, et al. Identification of phosphinate dipeptide analog inhibitors directed against the Plasmodium falciparum M17 leucine aminopeptidase as lead antimalarial compounds. J Med Chem. 2007;50(24):6024–6031. doi: 10.1021/jm070733v. [DOI] [PubMed] [Google Scholar]
- 6.Skinner-Adams TS, et al. Plasmodium falciparum neutral aminopeptidases: New targets for anti-malarials. Trends Biochem Sci. 2009 doi: 10.1016/j.tibs.2009.08.004. http://dx.doi.org/10.1016/j.tibs.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Stack CM, et al. Characterization of the Plasmodium falciparum M17 leucyl aminopeptidase A protease involved in amino acid regulation with potential for antimalarial drug development. J Biol Chem. 282(3):2069–2080. doi: 10.1074/jbc.M609251200. [DOI] [PubMed] [Google Scholar]
- 8.Dalal S, Klemba M. Roles for two aminopeptidases in vacuolar hemoglobin catabolism in Plasmodium falciparum . J Biol Chem. 2007;282(49):35978–35987. doi: 10.1074/jbc.M703643200. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Istvan ES, Gluzman IY, Gross J, Goldberg DE. Plasmodium falciparum ensures its amino acid supply with multiple acquisition pathways and redundant proteolytic enzyme systems. Proc Natl Acad Sci USA. 2006;103(23):8840–8845. doi: 10.1073/pnas.0601876103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flipo M, et al. Novel selective inhibitors of the zinc plasmodial aminopeptidase PfA-M1 as potential antimalarial agents. J Med Chem. 2007;50(6):1322–1334. doi: 10.1021/jm061169b. [DOI] [PubMed] [Google Scholar]
- 11.Allary M, Schrevel J, Florent I. Properties, stage-dependent expression and localization of Plasmodium falciparum M1 family zinc-aminopeptidase. Parasitology. 2002;125(Pt 1):1–10. doi: 10.1017/s0031182002001828. [DOI] [PubMed] [Google Scholar]
- 12.Florent I, et al. A Plasmodium falciparum aminopeptidase gene belonging to the M1 family of zinc-metallopeptidases is expressed in erythrocytic stages. Mol Biochem Parasitol. 1998;97(1–2):149–160. doi: 10.1016/s0166-6851(98)00143-1. [DOI] [PubMed] [Google Scholar]
- 13.Maric S, et al. The M17 leucine aminopeptidase of the malaria parasite Plasmodium falciparum: Importance of active site metal ions in the binding of substrates and inhibitors. Biochemistry. 2009;48(23):5435–5439. doi: 10.1021/bi9003638. [DOI] [PubMed] [Google Scholar]
- 14.Burley SK, David PR, Taylor A, Lipscomb WN. Molecular structure of leucine aminopeptidase at 2.7-A resolution. Proc Natl Acad Sci USA. 1990;87(17):6878–6882. doi: 10.1073/pnas.87.17.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsui M, Fowler JH, Walling LL. Leucine aminopeptidases: Diversity in structure and function. Biol Chem. 2006;387(12):1535–1544. doi: 10.1515/BC.2006.191. [DOI] [PubMed] [Google Scholar]
- 16.Kim H, Lipscomb WN. X-ray crystallographic determination of the structure of bovine lens leucine aminopeptidase complexed with amastatin: Formulation of a catalytic mechanism featuring a gem-diolate transition state. Biochemistry. 1993;32(33):8465–8478. doi: 10.1021/bi00084a011. [DOI] [PubMed] [Google Scholar]
- 17.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: A multiple structural alignment algorithm. Proteins. 2006;64(3):559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 18.Carpenter FH, Vahl JM. Leucine aminopeptidase (Bovine lens) Mechanism of activation by Mg2+ and Mn2+ of the zinc metalloenzyme, amino acid composition, and sulfhydryl content. J Biol Chem. 248(1):294–304. [PubMed] [Google Scholar]
- 19.Strater N, Lipscomb WN. Two-metal ion mechanism of bovine lens leucine aminopeptidase: Active site solvent structure and binding mode of L-leucinal, a gem-diolate transition state analogue, by x-ray crystallography. Biochemistry. 1995;34(45):14792–14800. doi: 10.1021/bi00045a021. [DOI] [PubMed] [Google Scholar]
- 20.Blackman MJ. Malarial proteases and host cell egress: An ‘emerging’ cascade. Cell Microbiol. 2008;10(10):1925–1934. doi: 10.1111/j.1462-5822.2008.01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sijwali PS, Rosenthal PJ. Gene disruption confirms a critical role for the cysteine protease falcipain-2 in hemoglobin hydrolysis by Plasmodium falciparum . Proc Natl Acad Sci USA. 2004;101(13):4384–4389. doi: 10.1073/pnas.0307720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whisstock JC, McGowan S, Trenholme KR, Gardiner DL, Dalton JP. Reply to Klemba: Intracellular processing of the membrane-bound PfA-M17 neutral aminopeptidase, a target for new antimalarials. Proc Natl Acad Sci USA. 2009;106(22):E56. [Google Scholar]
- 23.Klemba M. On the location of the aminopeptidase N homolog PfA-M17 in Plasmodium falciparum . Proc Natl Acad Sci USA. 2009;106(22):E55. doi: 10.1073/pnas.0903493106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldberg DE, Slater AF, Cerami A, Henderson GB. Hemoglobin degradation in the malaria parasite Plasmodium falciparum: An ordered process in a unique organelle. Proc Natl Acad Sci USA. 1990;87(8):2931–2935. doi: 10.1073/pnas.87.8.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin RE, Kirk K. Transport of the essential nutrient isoleucine in human erythrocytes infected with the malaria parasite Plasmodium falciparum . Blood. 2007;109(5):2217–2224. doi: 10.1182/blood-2005-11-026963. [DOI] [PubMed] [Google Scholar]
- 26.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D. 2006;62(1):48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 27.Evans P. Scaling and assessment of data quality. Acta Crystallogr D. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 28.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT. Assessment of phase accuracy by cross validation: The free R value methods and applications. Acta Crystallogr D. 1993;49(Pt 1):24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- 30.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D. 2003;59(Pt 7):1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 31.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 32.Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: A server for profile-profile sequence alignments. Nucleic Acids Res. 2005;33:W284–288. doi: 10.1093/nar/gki418. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60((Part 12, number 1)):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Cohen SX, et al. ARP/wARP and molecular replacement: The next generation. Acta Crystallogr D. 2008;64(Pt 1):49–60. doi: 10.1107/S0907444907047580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potterton L, et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D. 2004;60((Part 12, number 1)):2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 38.Androulakis S, et al. Federated repositories of x-ray diffraction images. Acta Crystallogr D. 2008;D64(Pt 7):810–814. doi: 10.1107/S0907444908015540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.