Abstract
Disruption of mitotic events contributes greatly to genomic instability and results in mutator phenotypes. Indeed, abnormalities of mitotic components are closely associated with malignant transformation and tumorigenesis. Here we show that ninein-like protein (Nlp), a recently identified BRCA1-associated centrosomal protein involved in microtubule nucleation and spindle formation, is an oncogenic protein. Nlp was found to be overexpressed in approximately 80% of human breast and lung carcinomas analyzed. In human lung cancers, this deregulated expression was associated with NLP gene amplification. Further analysis revealed that Nlp exhibited strong oncogenic properties; for example, it conferred to NIH3T3 rodent fibroblasts the capacity for anchorage-independent growth in vitro and tumor formation in nude mice. Consistent with these data, transgenic mice overexpressing Nlp displayed spontaneous tumorigenesis in the breast, ovary, and testicle within 60 weeks. In addition, Nlp overexpression induced more rapid onset of radiation-induced lymphoma. Furthermore, mouse embryonic fibroblasts (MEFs) derived from Nlp transgenic mice showed centrosome amplification, suggesting that Nlp overexpression mimics BRCA1 loss. These findings demonstrate that Nlp abnormalities may contribute to genomic instability and tumorigenesis and suggest that Nlp might serve as a potential biomarker for clinical diagnosis and therapeutic target.
Introduction
The centrosome is a small cytoplasmic nonmembranous organelle involved in the nucleation and organization of microtubules and critical for proper cell cycle progression. It establishes polarity and orientation of microtubule during interphase and contributes to assembly of spindle and chromosomal segregation. The centrosome may also serve as a central site that anchors important molecules and regulates fundamental cellular function, such as cell motility and adhesion. Multiple studies show that centrosome activity is also important to the completion of cytokinesis and progression from the G1 phase to the S phase of the cell cycle (1, 2). Therefore, centrosome aberrations may result in disruption of cell cycle progression, including chromosomal missegregation and aneuploidy, which often lead to cell transformation, tumorigenesis, and the development of malignancies (3–5). Consistently, centrosome abnormalities have been frequently observed in different types of clinical human tumors and are often correlated with poor clinical prognosis. (6–8).
Centrosome instability has been linked to certain genetic aberrations (3, 9), including the loss of the tumor suppressor proteins p53 (10), BRCA1 (11, 12), and BRCA2 (13). Mouse embryonic fibroblasts (MEFs) carrying targeted deletion of p53 or Brca1 genes exhibit centrosome amplification and abnormalities of spindle formation, which are coupled with spontaneous tumorigenesis (10, 14, 15). Mammalian cells harboring p53 mutations or infected with HPVE6, a potent p53 inhibitor, often display centrosome abnormalities (16). It has been shown that the p53 target genes p21waf1 (17) and Gadd45 (18) regulate centrosome stability. The p21/Waf1 protein regulates centrosome replication in the S phase by inhibiting Cdk2/cyclin E kinase (17, 19), and p53- and BRCA1-regulated Gadd45 exert their role on maintaining centrosome stability by inhibiting Cdc2/cyclin B1 and Aurora-A (20, 21). Consistently, disruption of p21waf1 (17) or Gadd45 (18, 21) results in centrosome amplification and aneuploidy. In addition, several mitotic protein kinases have been implicated in centrosome amplification, such as Aurora-A (22), Plk1 (23, 24), Nek2 (25), and Cdc2 (26, 27). As a result, gene amplification or deregulated expression of those mitotic proteins is associated with centrosome abnormalities in human tumors. Introduction of exogenous Aurora-A or Plk1 expression vectors into cells leads to centrosome amplification and genomic instability (21–24).
Centrosomal protein ninein-like protein (Nlp) was initially characterized as an important molecule involved in centrosome maturation (28) and has most recently been defined as the tumor suppressor BRCA1-associated protein (29). Nlp physically interacts with γ-tubulin ring complex and stimulates microtubule nucleation in the interphase. Upon the G2/M transition, Nlp is subjected to phosphorylation by Plk1 and Nek2 (28, 30), leading to its dissociation from the centrosome. Therefore, the Nlp association with centrosome and its interaction with γ-tubulin are regulated by Plk1 and Nek2 activity. Nlp expression is cell cycle dependent and regulated by anaphase-promoting complex (APC/c) for its degradation (31). Inhibition of endogenous Nlp has been shown to interfere with MT nucleation. Overexpression of Nlp also results in aberrant mitotic spindle formation (28) and chromosomal missegregation as manifested by the presence of multiple nuclei in cells (31). It is thus thought that the fine level of Nlp is required for proper centrosome maturation and spindle assembly, and that disrupted expression of Nlp might lead to genomic instability and tumorigenesis.
In spite of genomic instability caused by abnormal expression of Nlp in cell models (28, 31), deregulated expression and genetic alterations of Nlp in human clinical tumors remain unclear. In addition, the oncogenic nature of this centrosomal protein needs to be defined. In this report, we demonstrate that levels of Nlp were upregulated in human breast cancers and lung carcinomas. The deregulated expression of Nlp in human tumors was associated with its gene amplification. Additionally, we show that Nlp is an oncogenic protein, the overexpression of which allowed anchorage-independent growth and induced cell malignant transformation. In parallel, we also used the transgenic model in which Nlp was overexpressed and found that elevated expression of Nlp resulted in centrosome amplification and spontaneous tumors. The Nlp transgenic animals were also susceptible to DNA damage–induced tumorigenesis.
Results
Nlp is overexpressed in human breast and lung carcinomas.
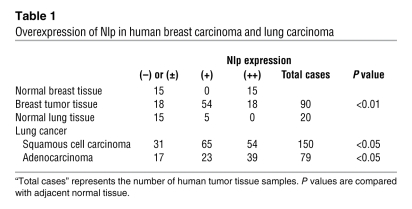
As demonstrated by others and our group, disruptions of centrosomal protein Nlp cause defects of spindle formation and chromosomal segregation (28, 31). We therefore first examined whether Nlp expression is deregulated in human tumors. Using an immunohistochemical approach, we found that Nlp was overexpressed in both breast and lung cancers compared with their normal (non-cancerous) tissues (Figure 1, A and B). The comparative results of immunohistochemistry are summarized in Table 1. Strong cytoplasmic staining of Nlp protein was detected in 72 of 90 (80%) human breast cancer samples. Weak or negative staining was detected in 18 tumors (20%). In contrast, none of 15 normal breast tissue samples exhibited strong staining for Nlp. We also analyzed 150 lung squamous cell carcinoma samples and 79 lung adenocarcinomas samples. Strong positive staining of Nlp was observed in 79% (119 of 150) of squamous cell carcinomas and 78% (62 of 79) of adenocarcinomas, but only 5 of 20 non-cancerous samples (25%) showed positive Nlp staining, and the remaining 15 normal lung tissues showed weak or negative Nlp staining. To further confirm the overexpression of Nlp in human tumors, we used Western blots to analyze 30 tumor samples and their paired normal adjacent tissues. In agreement with the observations in the immunohistochemical assays, Nlp overexpression was detected in 21 tumor samples and 5 representative samples (nos. 1, 8, 9, 11, and 14; shown in Figure 1C). In addition, Nlp overexpression and positive staining were also found in other types of tumors, including liver cancer, cervical cancer, and bladder carcinomas (data not shown). Collectively, these observations indicate that expression of the centrosomal protein Nlp is deregulated in human tumors, suggesting that abnormal levels of Nlp are correlated to clinical tumor development.
Figure 1. Deregulated expression Nlp in human lung and breast carcinomas.
(A) Normal (non-cancerous) human breast tissue and breast carcinoma tissue samples were collected and subjected to immunohistochemical staining with antibody to Nlp. Original magnification, ×100 (left) and ×200 (right). (B) Immunohistochemical assays using Nlp antibody were conducted in normal (non-cancerous) lung tissue and tumor samples. Original magnification, ×100 (left) and ×200 (right). (C) Cell lysates were prepared from 30 paired clinical lung tumor tissues (T) and normal adjacent tissues (N) and analyzed with antibodies to Nlp (top) and actin (bottom). The experiments were repeated 3 times, and only the representative results of 5 paired samples were shown.
Table 1 .
Overexpression of Nlp in human breast carcinoma and lung carcinoma
NLP gene is amplified in human tumors.
To determine the mechanism by which Nlp protein is overexpressed in human tumors, we measured Nlp mRNA levels by RT-PCR analysis in 30 paired clinical lung tumor and normal adjacent tissues. Consistent with the findings that Nlp protein is overexpressed in different types of human tumors, higher levels (greater than 2-fold) of NLP mRNA were observed in 18 of 30 primary lung tumor specimens, and 5 representative samples (nos. 1, 8, 9, 11, and 14) are shown in Figure 2A. Additionally, NLP mRNA expression in most human cancer lines (such as HeLa, A549, MCF-7, H157) were also higher than that seen in non-cancerous HEK293T cells (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI39447DS1).
Figure 2. NLP gene amplification in human tumor.
(A) Total RNA (0.5 μg) isolated from 30 paired lung tumor tissues and normal adjacent tissues were used for semi-quantitative RT-PCR assays. (B) DNA was extracted from the lung cancer samples and their normal adjacent tissues (See Figure 1C). DNA (20 μg per specimen) was digested with EcoRI and hybridized with NLP probe. The estimated size of the hybridized NLP band was approximately 7.7 kb. In addition, the digested DNA was also hybridized with GAPDH probe, which was included as an internal control. (C) Human lung carcinomas and their normal adjacent tissues from A were used for FISH analysis. In addition, multiple human cancer cell lines (lung cancer, cervical cancer, and esophageal cancer cells) were included for analysis of NLP gene copy numbers. NLP-hybridizing loci were visualized as red spots, and the hybridizing signals for chromosome 20–specific centromeric probe were green. Original magnification, ×400.
Next, genomic Southern blot analyses were used to examine the amplification of the NLP gene in the same 30 paired clinical lung tumor tissues and normal adjacent tissues mentioned above, which exhibited elevated levels of both Nlp protein and NLP mRNA (Figure 1C and Figure 2A). Consistently, NLP gene amplifications were detected in 11 lung carcinomas, and 5 representative samples (nos. 1, 8, 9, 11, and 14) are presented in Figure 2B. In addition, genomic PCR Southern blot assays also demonstrated that the NLP gene is amplified in human lung cancer samples (Supplemental Figure 1B).
To further confirm NLP amplification in lung carcinomas, FISH was used to measure NLP copy numbers in primary lung tumors. Amplifications of NLP were visualized as an increased number of loci hybridizing to an NLP BAC probe. Fifteen paired lung carcinoma and normal adjacent lung tissues were tested. Among them, 6 tumors were found to have NLP amplifications, but none of the normal adjacent lung tissues showed NLP amplifications (Figure 2C). Hybridization of a centromere of chromosome 20 (green spots) to the same tumor samples indicated that the increase in NLP hybridizing loci was not due to polyploidy of chromosome 20. Additionally, human lung cancer cells A549 and human breast cancer cells MCF-7 also showed NLP amplifications, but not in human renal cells HEK293T (Figure 2D). These findings indicate that there are genetic alterations of the NLP gene during tumorigenesis. It should be noted here that NLP gene amplifications are correlated to increased expression of Nlp mRNA and protein. Representative lung cancer samples with identified numbers (labeled 1, 8, 9, 11, and 14) that showed NLP gene amplification, elevated mRNA, and protein expression are presented in Figure 1C, Figure 2, A–C, and Supplemental Figure 1B, respectively.
Nlp induces cell transformation and tumorigenesis.
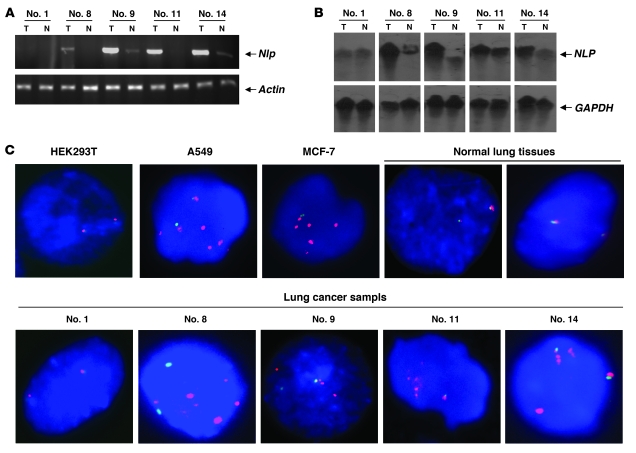
Further efforts were made to determine whether Nlp has an oncogenic property. We stably transfected the pEGFP-Nlp expression vector into NIH3T3 rodent fibroblast cells and obtained isogenic cells with high expression of Nlp. We used pooled clones (3T3-Pool) and 3 independent clones (3T3-53, 3T3-58, and 3T3-78) for tumorigenic assays (Figure 3A). These isogenic cell lines displayed fast-growing capabilities compared with the parental NIH3T3 cells (3T3) or cells transfected with empty pEGFP vector (3T3-GFP) (Figure 3B), indicating that Nlp may confer to the cell strong growing capability. To evaluate whether the Nlp-expressing NIH3T3 cells were transformed, their ability to form colonies in semi-solid medium (soft agar) was tested. Pooled clones and all 3 isogenic lines expressing Nlp formed significant numbers of large colonies when grown in soft agar. However, the parental NIH3T3 and cells transfected with empty vector only formed a few tiny colonies (Figure 3, C and D), which suggested that Nlp confers anchorage-independent growth to cells.
Figure 3. Anchorage-independent growth induced by Nlp expression.
(A) NIH3T3 cells were transfected with pEGFP-Nlp or empty vectors. Following G418 selection for 14 days, Nlp-expressing lines were established and analyzed by immunoblot assays. In the tumorigenic assays, pooled clone and 3 independent clones of NIH3T3 cells were used. (B) Cells (104) from Nlp-expressing cell lines (3T3-Pool, 3T3-53, 3T3-58, and 3T3-78) and the parental line (3T3) or 3T3-GFP were seeded in 100-mm dishes and grown in DMEM supplemented with 10% FBS. Cells were collected each day to count cell numbers. Data are presented as mean ± SEM; n = 9–12. (C) NIH3T3 or its isogenic cells (103) were seeded in 0.3% top agar and incubate at 37°C in a humidified incubator for 14 days. Nlp-expressing cells formed larger colonies in soft agar. Original magnification, ×20. (D) The quantitative results of colony formation were obtained. Data are presented as mean ± SEM; n = 9.
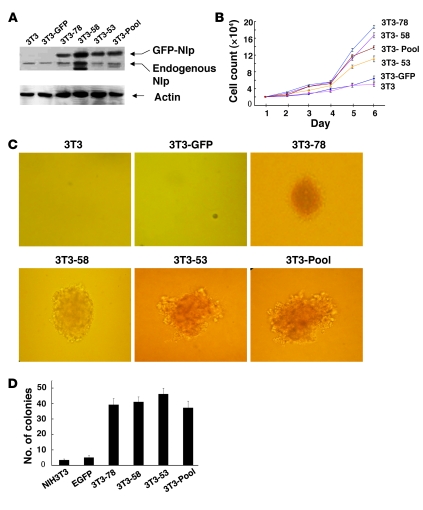
Next, Nlp-expressing NIH3T3 cells were subcutaneously injected into nude mice. Of the 60 animals injected with NIH3T3 cells expressing Nlp (3T3-Pool, 3T3-78, 3T3-58, and 3T3-53), 58 displayed tumors, but none of the 10 mice injected with control cells (3T3-GFP) generated measurable tumors (Figure 4A). After the animals were sacrificed, the xenografts were removed and collected for further analysis. H&E staining showed that the tumors were histologically sarcomas with medium differentiation, and immunohistochemistry demonstrated that tumors were strongly positive for Nlp staining (Figure 4B). In addition, we isolated genomic DNA from tumors and measured both mouse Nlp and human NLP genes using PCR analysis. As the positive controls, mouse Nlp gene was detected in MEFs and the human NLP gene was detected in HeLa cells. However, both mouse Nlp and human NLP genes were found in all xenografts (Figure 4C). These results further confirmed that the xenografts were derived from Nlp-expressing NIH3T3 rodent fibroblast cells. Taken together, these findings indicate that Nlp is oncogenic and able to induce cell transformation.
Figure 4. Oncogenic properties of Nlp in a nude mouse model.
(A) Nlp-expressing NIH3T3 cell lines (3T3-Pool, 3T3-78, 3T3-58, and 3T3-53), parental 3T3, and 3T3-GFP cells were subcutaneously injected into anesthetized 4-week-old nude mice. After 6 weeks, mice were sacrificed when tumors were visible and tumor tissues were removed for histological and molecular analysis. (B) H&E and Nlp staining analyses in tumors derived from the Nlp-expressing cells. Original magnification, ×100 (top) and ×200 (bottom). (C) Genomic DNA analysis in the Nlp-induced tumor samples. The primers were designed to specifically define either mouse Nlp DNA or human NLP DNA. As controls, MEFs and human HeLa cells were included.
Nlp transgenic mice develop spontaneous tumors and are susceptible to radiation-induced lymphoma.
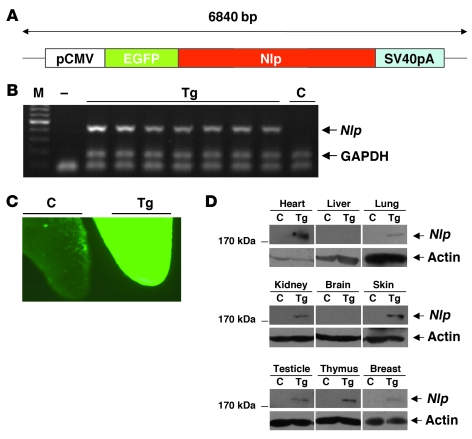
To investigate the oncogenic potentials of Nlp in vivo, transgenic mice overexpressing human Nlp were generated (Figure 5A). Mice overexpressing Nlp were viable and fertile, and development appeared normal. The expressions of Nlp in transgenic animals were determined by fluorescent examination and Northern and Western blotting assays (Figure 5, B–D). Expression of exogenous Nlp was detected in most tissues, including lung, kidney, breast, heart, thymus, testicle, and skin (Figure 5D).
Figure 5. Characterization of Nlp transgenic mice.
(A). NLP construct for generation of transgenic mice. (B) PCR analysis of tail DNA of NLP transgenic founders (Tg) and wild type (C) mice. M, DNA marker; —, negative control. (C) Immunofluorescent examination of newborn tail tips. The tail tips were obtained from 2-day-old mice and were observed by fluorescent microscopy. Original magnification, ×10. (D) Western blot analysis of different organs of NLP transgenic mice. Nlp expression was detected in skin, thymus, heart, testicle, breast, lung, and kidney of transgenic mice.
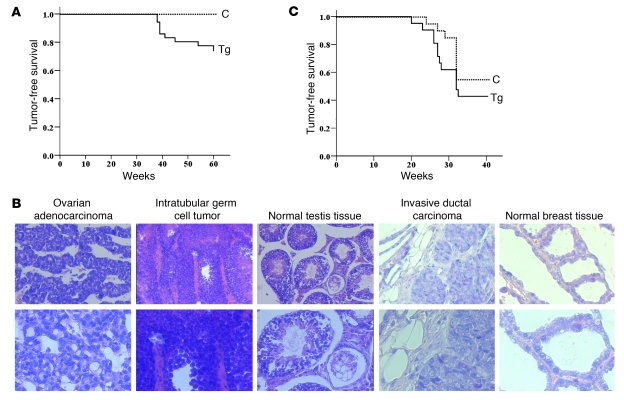
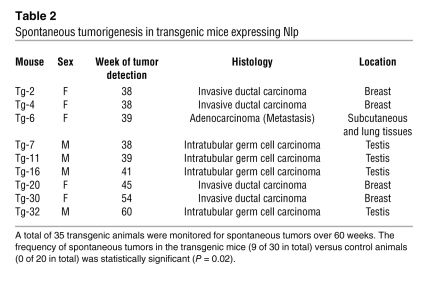
Nlp transgenic mice and normal littermates were monitored for spontaneous tumorigenesis. Normal animals (20 mice) did not develop any tumor by the age of 60 weeks, but Nlp-overexpressing mice (35 animals) exhibited spontaneous tumors (Figure 6A). In the group of transgenic mice, 1 female (at age of 39 weeks) was found to have a metastatic tumor in the right hind leg and lung tissue. This carcinoma was thought to come from ovarian gland epithelia (Figure 6B). Four male mice (38–60 weeks old) developed primary tumors in the testis (Figure 6B). Four female animals (38–54 weeks old) developed breast ductal cancers (Figure 6B), and the tumors were poorly differentiated with invasive features. The normal breast tissue and testis tissue were included for comparison (Figure 6B). A summary of those spontaneous tumors is shown in Table 2, and the frequency of spontaneous tumors in the transgenic mice versus control animals was statistically significant (P = 0.02). Thus, mice overexpressing Nlp are able to develop spontaneous tumorigenesis.
Figure 6. Spontaneous tumorigenesis in Nlp transgenic mice.
(A) Both normal and Nlp transgenic mice were maintained in normal conditions and monitored for spontaneous tumorigenesis. During 60 weeks, none of 20 normal mice exhibited spontaneous tumors, but 9 of 30 transgenic mice showed spontaneous tumors. (B) Transgenic animals with tumors were sacrificed for histological examinations. Ovarian adenocarcinoma was poorly differentiated and exhibited abnormal glandular structures. Intractable germ cell carcinomas were poorly differentiated with irregularly arrayed anaplastic cells, abundant cytoplasm, and irregular nuclei. Spermatogenesis was absent. Invasive breast ducal carcinoma was poorly differentiated without visible adenoid structures in the cancer nests. Nuclei of malignant cells were large and irregular, and hemorrhagic necrosis was observed. (C) Animals were treated with a total dose of 9 Gy (exposure to IR at 3 doses of 3 Gy each). Mice monitored for tumorigenesis for 32 weeks. Original magnification, ×100 (top) and × 200 (bottom).
Table 2 .
Spontaneous tumorigenesis in transgenic mice expressing Nlp
We also treated both normal mice and Nlp transgenic animals with ionizing radiation (IR) at a total dose of 9 Gy (Figure 6C and Table 3). For the Nlp transgenic mice treated with IR, the first lymphoma was detected at the 20th week, and 8 mice developed lymphomas over the period of 28 weeks, but in the group of normal animals, the first tumor appeared at 24th week and 2 mice developed lymphomas within the period of 28 weeks. The occurrence of IR-induced lymphomas in the transgenic mice compared with normal animals is statistically significant (P = 0.03). However, by 32 weeks, 12 of 20 transgenic animals and 8 of 20 control animals were found to have lymphomas. In addition, the lymphomas generated in Nlp transgenic mice appeared more malignant with strong invasive features. The lymphomas in normal mice treated by IR were often confined to the thymus and rarely invaded the neighbor organs, while most lymphomas that appeared in the transgenic mice occurred not only in the thymus but frequently invaded to the tissues of heart and lung. As shown in Supplemental Figure 2, C–F, lymphoma cells invaded the pericardium, a large number of lymphoma cells invaded the bronchial tissues, and some cells also invaded the alveolar interstitium. In some cases, lymphomas developed to an advanced stage and striated muscles of the chest wall were also invaded. At the same time, thymus adhered to the chest wall (Supplemental Figure 2, G and H). During the invasion of liver and kidney, lymphoma cells invaded interstitial tissues (Supplemental Figure 2, I–L). Therefore, Nlp transgenic mice are more susceptible to radiation-induced carcinogenesis.
Table 3 .
Tumorigenesis in transgenic mice expressing Nlp following IR treatment
MEFs derived from Nlp transgenic mice display centrosome amplification and resistance to DNA-induced apoptosis.
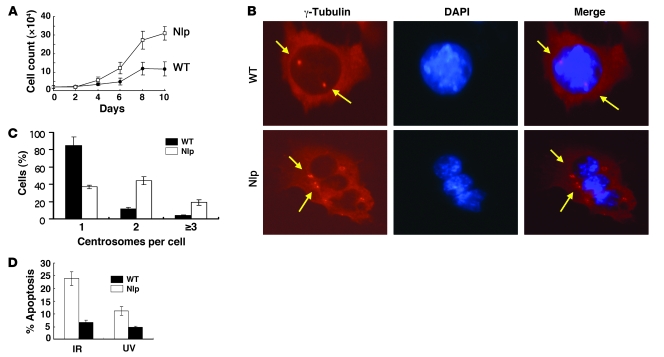
We isolated MEFs from Nlp transgenic mice and found that cells overexpressing Nlp exhibited faster growing compared with MEFs from normal animals (Figure 7A). Interestingly, approximately 20% of Nlp-overexpressing MEFs displayed centrosome amplification (defined as at least 3 centrosome copies per cell) (Figure 7, B and C). The centrosome amplification was also observed in about 22% of glomerular cells (data not shown). Additionally, most MEFs or glomerular cells with centrosome amplifications appeared to be multinucleate.
Figure 7. Analysis of MEFs derived from transgenic mice.
(A) MEFs isolated from both normal and Nlp transgenic mice were plated in 30-mm tissue culture dishes, and cells were collected for counting each day. Three separate experiments were conducted. Data are presented as mean ± SEM; n = 6–9. (B) MEFs isolated from normal or Nlp transgenic mice were stained for centrosomes with γ-tubulin antibody and counterstained with DAPI. Arrows indicate the positions of centrosomes in MEFs derived from either normal (WT) or Nlp transgenic animals. Original magnification, ×400. (C) Quantitative results of centrosome analysis in MEFs were obtained from 3 separate experiments, and in each experiment, more than 400 cells were examined. Data represent percentages of cells with different centrosome copies and are presented as mean ± SEM. (D) Expression of Nlp-attenuated apoptosis induced by IR and UV radiation. MEFs from normal and Nlp transgenic mice were treated with IR at a dose of 15 Gy and UV radiation at a dose of 30 J/m2. Cells were collected 16 hours later for examination of apoptosis. Data are presented as mean ± SEM; n = 9–12.
Next, the MEFs isolated from both Nlp transgenic and normal mice were treated with IR at a dose of 15 Gy or UV radiation at 30 J/m2 and subjected to analysis of apoptosis. Following IR, 22% of MEFs derived from normal mice underwent apoptosis, but 7% MEFs from Nlp transgenic mice showed apoptosis. Similarly, 12% of UV radiation–induced apoptosis in normal MEFs was observed, but 5% of Nlp-expressing MEFs displayed apoptosis after UV radiation (Figure 7D). Thus, overexpression of Nlp substantially attenuated DNA damage–induced apoptosis. Moreover, we also found that overexpression of Nlp disrupted apoptosis induced by some chemotherapeutic agents such as paclitaxel in human breast cancer cells (data not shown).
Discussion
A number of investigations have demonstrated that disruption of mitotic events greatly contribute to cell transformation. Abnormalities of mitotic components such as Plk1 (32), Aurora-A (22, 33), survivin (34), cyclin B1 (35), and Nek2 (36) are closely associated with tumorigenesis and development of malignancy. These proteins often exhibit gene amplification or deregulated expression in various types of human tumors. Interestingly, mitotic defects are closely associated with centrosome abnormalities, including structural alterations, such as an increase in centrosome number and size, excess of pericentriolar material, supernumerary centrioles, and functional abnormalities, such as increased microtubule nucleation activity (37). It has been well accepted that centrosome abnormalities may be an early event in tumorigenesis and sometime can serve as the cause rather than consequence of genomic instability (3, 5, 16, 38). In addition, the centrosome abnormalities often contribute to malignant tumor progression (6–8). However, the underlying mechanism by which centrosome aberrations cause tumorigenesis and malignant progression remains to be further defined.
In this study, we have shown that expression of the centrosomal protein Nlp, as a critical mitotic component, is deregulated in human lung and breast carcinomas. Using an immunochemical staining technique, we found that about 80% of breast tumor tissues and 78% of lung cancer tissues exhibit overexpression of Nlp (Figure 1 and Table 1). The fact that Nlp protein is overexpressed in human tumors is consistent with the observations that expression of NLP mRNA was abnormally upregulated in about 60% of lung cancer specimens. Importantly, abnormal expressions of Nlp protein and mRNA appear to be associated with NLP gene amplification in human tumors. Using FISH and Southern blot analysis, we demonstrated amplified signals of NLP in primary lung cancer samples and in lung carcinoma cell lines as well (Figure 2). These observations indicate that there are genetic alterations of the NLP gene in human tumors. More informatively, NLP gene amplifications were closely related to abnormal expression of Nlp mRNA and protein. Five representative lung cancer samples with identified numbers showed consistent patterns for NLP gene amplification and elevated mRNA and protein. In addition, several reports show amplifications of chromosomal region 20p11, where NLP gene is located, in human breast and esophageal tumors (39, 40). However, it is noted here that NLP amplification may not count for all deregulated expression of Nlp protein in human tumors, and elevated levels of Nlp in human tumors might also be caused by the defects in the processes of Nlp degradation.
Interestingly, Nlp exhibits a potent transforming capability when it is overexpressed, which is demonstrated by the fact that NIH3T3 cells expressing exogenous Nlp reveal anchorage-independent growth and tumor formation in nude mice. Pooled clone and 3 independent clones of NIH3T3 expressing Nlp exhibited the ability to grow quickly and formed colonies in soft agar. Following subcutaneous injection of NIH3T3 expressing Nlp into nude mice, tumors formed in almost all animals tested within 6 weeks (Figures 3 and 4). Consistently, the Nlp transgenic animals showed spontaneous tumorigenesis. In 35 mice aged 38–60 weeks, 9 animals developed cancers, including both primary and metastatic tumors (Figure 6 and Table 2). In addition, the Nlp transgenic mice were more susceptible to IR-induced lymphoma. The frequency of spontaneous and IR-induced tumors in the transgenic mice versus control animals is statistically significant. All those findings demonstrate that Nlp is an oncogenic protein. As discussed above, fine levels of Nlp are required for centrosome maturation and stability, and thus deregulated expression (both overexpression and downregulation) of Nlp may result in aberrant spindle formation and disruption of chromosomal segregation, which leads to aneuploidy and chromosomal aberration (28, 31). It is likely that when Nlp is deregulated, it causes centrosome aberrations and genomic instability, which in turn lead to tumorigenesis. In addition, since Nlp is involved in organization of the microtubule network that supports many important cellular functions including cell adhesion and migration, disruption of Nlp may cause defects in those biological aspects and contribute to the development of cell malignancy.
It is worth noting that we have recently characterized Nlp as a BRCA1-interacting protein and Nlp might mediate the role of BRCA1 in the control of mitotic progression (29). Indeed, there were similarities between the overexpression of Nlp and the loss of BRCA1. It has been shown that deletion of BRCA1 exon 11 in knockout mice results in centrosome amplification and abnormalities of spindle formation (14). The observations from our group and others have shown that abnormal expression of Nlp causes disrupted spindle assembly, chromosomal missegregation, and aneuploidy (28, 29, 31). In the model of transgenic mice, both MEFs and glomerular cells derived from Nlp transgenic mice exhibited centrosome amplification. About 20% of MEFs and glomerular cells displayed 3 or more copies of centrosomes (Figure 7), indicating that deregulated expression of Nlp disrupts the normal machinery of controlling centrosome stability. Thus, overexpression of Nlp mimics the phenotypes of disrupted BRCA1, including both centrosome amplification and in vivo oncogenic potentials.
In spite of its deregulated expression in human tumors and oncogenic potentials, overexpression of Nlp in cancer cells has been shown to affect clinical therapeutic sensitivity and to enhance cancer cell survival. Following introduction of exogenous Nlp into ovarian cancer SKOV3 cells, the cell death induced by the chemotherapeutic agent Taxol, which is thought to induce apoptosis by stabilizing the spindle microtubules and causing mitotic arrests at the metaphase stage, is substantially inhibited (41). In support of these findings, we have shown that MEFs derived from Nlp transgenic mice exhibit resistance to apoptosis activated by IR and UV radiation (Figure 7D). Collectively, the findings presented in this report imply that Nlp is an oncoprotein whose abnormal expression is correlated with human breast and lung cancers and that induces carcinogenesis. These findings thus provide what we believe are novel insights into our understanding of the role of disrupted regulatory machinery of mitotic progression in the development of human cancers. It is also possible that Nlp can be used as a potential molecular target for the development of a novel therapeutic drug and serve as a diagnostic marker in the clinical cancer treatment.
Methods
Clinical tissues samples collections.
Fresh tumor tissues and normal adjacent tissues were collected from patients with pathologically and clinically confirmed carcinomas. In some cases, non-cancerous tissues were also collected to serve as controls in the experiments. All human tumor tissues were obtained with written informed consent from patients or their guardians at Dalian Medical University prior to participation in the study. The Institutional Review Board of the Cancer Institute, Chinese Academy of Medical Sciences, approved use of the tumor specimens and animal models in this study. Most of tissue samples were fixed in formalin, embedded in paraffin, and sectioned at 5 μm thick. One section was stained with H&E for histological examination, and the others were used for immunohistochemistry or immunofluorescence staining. A portion of tissue specimens were kept in liquid nitrogen and sectioned for protein or mRNA extraction.
Cell culture and transfection.
Human lung cancer line A549, human breast carcinoma line MCF7, and NIH3T3 line were grown in DMEM supplemented with 10% FBS. Cell transfection was carried out as described previously (42). Briefly, 4 × 105 NIH3T3 cells were seeded onto 100-mm plates 1 day prior to transfection. Plasmid DNA (5 μg) and 15 μl of lipofectamine (Gibco-BRL) were used for each transfection, and cells were selected in the medium containing G418 (400 μg/ml). The G418-resistant colonies were collected and examined for Nlp expression.
Plasmid clones.
HJ07083, a KIAA0980/NLP cDNA clone, was generously provided by T. Nagase (KAZUSA DNA Research Institute, Kisarazu, Japan). pEGFP-Nlp was constructed by cloning KIAA0980 fragment (75-4848) into the BamHI and NotI sites of pGEX-5X-1 vector.
Cellular protein preparation, Western blotting analysis, and antibodies.
Cellular proteins were prepared and assayed as described previously (42). Nlp polyclonal antibody was produced using Ac-CQEKVDKLKEQFEKNTKSD-amide short peptide (putative KIAA0980 protein 1310–1327 aa; BioSource). This polyclonal antibody was demonstrated to be specific against human KIAA0980 protein (29).
Southern blot assays.
The Southern blot was performed by standard methods. Briefly, DNA was extracted from the lung tumor and paired with normal adjacent tissue samples, followed by phenol-chloroform purification. Approximately 20 μg of DNA per specimen was digested with EcoRI overnight and electrophoresed on 0.8% agarose gels. The DNA was transferred to a HyBond N+ Nylon filter (Pharmacia USA) and fixed for 2 min under UV light. Then the filters were hybridized with DIG-labeled NLP probes under stringent conditions using the DIG DNA labeling kit (Mylab). After hybridization with the NLP probe overnight at 65°C, the filters were washed at room temperature. An anti-digoxigenin–alkaline phosphatase conjugate was used for visualization with the DIG DNA detection kit (Mylab). The size of the hybridization band for NLP was approximately 7.7 kb.
FISH analysis.
The slide samples from tumor cell lines or tumor tissue were hybridized by 2-color FISH with an NLP-specific BAC probe RP11-96L6 (a gift from the Wellcome Trust Sanger Institute) and a chromosome 20–specific centromeric probe (Vysis). The NLP and centromere 20 probes were labeled with Cy3-dUTP (red) and FITC-dUTP (green), respectively, using the Bioprime DNA labeling system (Invitrogen) and hybridized with nuclei from cell lines or tumor tissue samples. We counterstained slides with DAPI and used the Olympus BX51 fluorescence microscope to capture FISH images. Results were expressed as a ratio of the number of copies of the NLP gene to the number of chromosome 20–centromeric markers. A ratio of greater than 2 indicated gene amplification.
Immunohistochemical analyses and semiquantitative evaluation.
The sample sections were analyzed using immunohistochemistry as described previously (43). For evaluation, the visible brown granules in cytoplasm were determined to indicate positive staining. Specimens were reviewed with staining intensity and staining extent (43). The relationship between Nlp expression and pathological parameters was analyzed using the χ2 test with SPSS software (SPSS Inc.). Statistical significance was reached at P < 0.05.
RT-PCR assay.
Total RNA was extracted using TRIzol reagent according to the manufacturer’s instructions. Total RNA (0.5 μg) in 1 μl of RNase-free water was used in 20 μl of room-temperature mixtures and subjected to DNA amplification using GeneAmp PCR System 9600 (PerkinElmer). PCR products were loaded on 1% agarose gel for analysis.
Soft agar assay.
Equal volumes of 1% agar and 2× DMEM medium were mixed and placed onto 60-mm dishes to generate 0.5% base agar. Nlp-expressing NIH3T3 cells were seeded in 0.3% top agar (103 cells per plate) and incubated at 37°C in humidified atmosphere for 10–14 days. Colonies were analyzed using a microscope.
Tumorigenicity assay.
Parental NIH3T3 cells or Nlp-expressing NIH3T3 cells were harvested by trypsinization, washed in PBS, counted, and collected by centrifugation. Cells (5 × 106) were suspended in 0.2 ml of PBS and injected subcutaneously into 4-week-old nude mice. After 6 weeks, mice were sacrificed, and tumor tissues were removed for further analysis. The experiments were performed in accordance with relevant institutional and national guidelines and regulations.
Generation of transgenic mice.
To generate the pEGFP-C3-NLP expression plasmid, full-length human NLP cDNA was cloned into the pEGFP-C3 vector. The complete insert, which was isolated as an ApaLI-SspI (6.8-kb) fragment, contained the CMV promoter, the EGFP gene, NLP, and the simian virus 40 poly(A). It was microinjected into the pronuclei of fertilized zygotes under sterile conditions, and subsequently the injected eggs were transferred into pseudopregnant recipients to generate transgenic mice. At 3 weeks after birth, potential founders were analyzed by PCR of tail DNA for the presence of transgenes. Such transgenic mice were generated in the inbred FVB/N genetic background and were housed in a controlled, specific pathogen–free environment. Nlp transgenic mice grew to adulthood without obvious differences from their wild-type littermates. The animals survived without development malformation and had no problems in procreation.
Preparation of MEFs.
MEFs were prepared from E13.5 embryos, which were isolated under sterile conditions. After rinsing twice with HBSS, these tissues were cut up and digested with 0.25% trypsin at 37°C. Single cells were collected by centrifugation at a speed of 75 g for 5 minutes. Collected cells were washed twice in HBSS and subsequently resuspended and maintained in DMEM supplemented with 10% FBS.
IR-induced carcinogenesis.
Transgenic and control mice 6–8 weeks old were irradiated 3 times with a dose of 3 Gy, for a total dose of 9 Gy. The animals were maintained in normal conditions for observation of carcinogenesis. Usually, the mice were observed 3 times each week for development of tumors and illness. At 9 months after irradiation, all animals were sacrificed for pathological examinations.
Statistics.
Results are expressed as mean ± SEM values, and P values less than 0.05 were considered statistically significant. Data processing was performed using the software package SPSS for Windows (SPSS Inc.).
Supplementary Material
Acknowledgments
We thank T. Nagase (Kazusa DNA Research Institute, Kisarazu, Japan) for the KIAA0980 (NLP) cDNA clone and the Wellcome Trust Sanger Institute for the RP11-96L6 probe. This work is supported by funding from the 973 National Key Fundamental Research Program of China (grant 2009CB521801) and the National Natural Science Foundation of China (grants 30730046 and 30721001).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(2):498–507. doi:10.1172/JCI39447.
References
- 1.Doxsey SJ. Centrosomes as command centres for cellular control. Nat Cell Biol. 2001;3(5):E105–E108. doi: 10.1038/35074618. [DOI] [PubMed] [Google Scholar]
- 2.Raff JW. Centrosomes: Central no more? Curr Biol. 2001;11(5):R159–R161. doi: 10.1016/S0960-9822(01)00082-3. [DOI] [PubMed] [Google Scholar]
- 3.Salisbury JL, Whitehead CM, Lingle WL, Barrett SL. Centrosomes and cancer. Biol Cell. 1999;91(6):451–460. [PubMed] [Google Scholar]
- 4.Marx J. Cell biology. Do centrosome abnormalities lead to cancer? Science. 2001;292(5516):426–429. doi: 10.1126/science.292.5516.426. [DOI] [PubMed] [Google Scholar]
- 5.Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2(11):815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 6.Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc Natl Acad Sci U S A. 1998;95(6):2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kramer A, Neben K, Ho AD. Centrosome aberrations in hematological malignancies. Cell Biol Int. 2005;29(5):375–383. doi: 10.1016/j.cellbi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119(12):2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 9.Duensing S, Munger K. Centrosome abnormalities, genomic instability and carcinogenic progression. Biochim Biophys Acta. 2001;1471(2):M81–M88. doi: 10.1016/s0304-419x(00)00025-1. [DOI] [PubMed] [Google Scholar]
- 10.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271(5256):1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 11.Deng CX. Roles of BRCA1 in centrosome duplication. Oncogene. 2002;21(40):6222–6227. doi: 10.1038/sj.onc.1205713. [DOI] [PubMed] [Google Scholar]
- 12.Hsu LC, White RL. BRCA1 is associated with the centrosome during mitosis. Proc Natl Acad Sci U S A. 1998;95(22):12983–12988. doi: 10.1073/pnas.95.22.12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tutt A, et al. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol. 1999;9(19):1107–1110. doi: 10.1016/S0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- 14.Xu X, et al. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3(3):389–395. doi: 10.1016/S1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 15.Tarapore P, Fukasawa K. Loss of p53 and centrosome hyperamplification. Oncogene. 2002;21(40):6234–6240. doi: 10.1038/sj.onc.1205707. [DOI] [PubMed] [Google Scholar]
- 16.Duensing S, et al. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc Natl Acad Sci U S A. 2000;97(18):10002–10007. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarapore P, Horn HF, Tokuyama Y, Fukasawa K. Direct regulation of the centrosome duplication cycle by the p53- p21Waf1/Cip1 pathway. Oncogene. 2001;20(25):3173–3184. doi: 10.1038/sj.onc.1204424. [DOI] [PubMed] [Google Scholar]
- 18.Hollander MC, et al. Genomic instability in Gadd45a-deficient mice. Nat Genet. 1999;23(2):176–184. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- 19.Ciciarello M, et al. p53 displacement from centrosomes and p53-mediated G1 arrest following transient inhibition of the mitotic spindle. J Biol Chem. 2001;276(22):19205–19213. doi: 10.1074/jbc.M009528200. [DOI] [PubMed] [Google Scholar]
- 20.Zhan Q, et al. Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated protein Gadd45. Oncogene. 1999;18(18):2892–2900. doi: 10.1038/sj.onc.1202667. [DOI] [PubMed] [Google Scholar]
- 21.Shao S, et al. Gadd45a interacts with aurora-A and inhibits its kinase activity. J Biol Chem. 2006;281(39):28943–28950. doi: 10.1074/jbc.M600235200. [DOI] [PubMed] [Google Scholar]
- 22.Zhou H, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20(2):189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 23.do Carmo Avides M, Tavares A, Glover DM. Polo kinase and Asp are needed to promote the mitotic organizing activity of centrosomes. Nat Cell Biol. 2001;3(4):421–424. doi: 10.1038/35070110. [DOI] [PubMed] [Google Scholar]
- 24.Donaldson MM, Tavares AA, Hagan IM, Nigg EA, Glover DM. The mitotic roles of Polo-like kinase. . J Cell Sci. 2001;114(Pt 13):2357–2358. doi: 10.1242/jcs.114.13.2357. [DOI] [PubMed] [Google Scholar]
- 25.Fry AM, Meraldi P, Nigg EA. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998;17(2):470–481. doi: 10.1093/emboj/17.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayor T, Meraldi P, Stierhof YD, Nigg EA, Fry AM. Protein kinases in control of the centrosome cycle. FEBS Lett. 1999;452(1–2):92–95. doi: 10.1016/S0014-5793(99)00534-7. [DOI] [PubMed] [Google Scholar]
- 27.Hinchcliffe EH, Sluder G. Two for two: Cdk2 and its role in centrosome doubling. Oncogene. 2002;21(40):6154–6160. doi: 10.1038/sj.onc.1205826. [DOI] [PubMed] [Google Scholar]
- 28.Casenghi M, Meraldi P, Weinhart U, Duncan PI, Korner R, Nigg EA. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev Cell. 2003;5(1):113–125. doi: 10.1016/S1534-5807(03)00193-X. [DOI] [PubMed] [Google Scholar]
- 29.Jin S, et al. BRCA1 interaction of centrosomal protein Nlp is required for successful mitotic progression. J Biol Chem. 2009;284(34):22970–22977. doi: 10.1074/jbc.M109.009134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rapley J, et al. Coordinate regulation of the mother centriole component nlp by nek2 and plk1 protein kinases. Mol Cell Biol. 2005;25(4):1309–1324. doi: 10.1128/MCB.25.4.1309-1324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Zhan Q. Cell cycle-dependent expression of centrosomal ninein-like protein in human cells is regulated by the anaphase-promoting complex. . J Biol Chem. 2007;282(24):17712–17719. doi: 10.1074/jbc.M701350200. [DOI] [PubMed] [Google Scholar]
- 32.Eckerdt F, Yuan J, Strebhardt K. Polo-like kinases and oncogenesis. Oncogene. 2005;24(2):267–276. doi: 10.1038/sj.onc.1208273. [DOI] [PubMed] [Google Scholar]
- 33.Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53–/– cells. . EMBO J. 2002;21(4):483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu B, et al. Nuclear survivin as a biomarker for non-small-cell lung cancer. Br J Cancer. 2004;91(3):537–540. doi: 10.1038/sj.bjc.6602027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan W, et al. BRCA1 regulates GADD45 through its interactions with the OCT-1 and CAAT motifs. J Biol Chem. 2002;277(10):8061–8067. doi: 10.1074/jbc.M110225200. [DOI] [PubMed] [Google Scholar]
- 36.Hayward DG, Clarke RB, Faragher AJ, Pillai MR, Hagan IM, Fry AM. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Res. 2004;64(20):7370–7376. doi: 10.1158/0008-5472.CAN-04-0960. [DOI] [PubMed] [Google Scholar]
- 37.Duensing S. A tentative classification of centrosome abnormalities in cancer. Cell Biol Int. 2005;29(5):352–359. doi: 10.1016/j.cellbi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 38.Duensing S, Munger K. Human papillomavirus type 16 E7 oncoprotein can induce abnormal centrosome duplication through a mechanism independent of inactivation of retinoblastoma protein family members. J Virol. 2003;77(22):12331–12335. doi: 10.1128/JVI.77.22.12331-12335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larramendy ML, et al. Comparative genomic hybridization reveals complex genetic changes in primary breast cancer tumors and their cell lines. Cancer Genet Cytogenet. 2000;119(2):132–138. doi: 10.1016/S0165-4608(99)00226-5. [DOI] [PubMed] [Google Scholar]
- 40.Pimkhaokham A, et al. Nonrandom chromosomal imbalances in esophageal squamous cell carcinoma cell lines: possible involvement of the ATF3 and CENPF genes in the 1q32 amplicon. Jpn J Cancer Res. 2000;91(11):1126–1133. doi: 10.1111/j.1349-7006.2000.tb00895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu D, et al. Increased expression of Nlp, a potential oncogene in ovarian cancer, and its implication in carcinogenesis. Gynecol Oncol. 2008;110(2):230–236. doi: 10.1016/j.ygyno.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 42.Fan W, et al. BRCA1 regulates GADD45 through its interactions with the OCT-1 and CAAT motifs. Journal of Biological Chemistry. 2002;277(10):8061–8067. doi: 10.1074/jbc.M110225200. [DOI] [PubMed] [Google Scholar]
- 43.Tong T, et al. Overexpression of Aurora-A contributes to malignant development of human esophageal squamous cell carcinoma. Clin Cancer Res. 200410217304–7310. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.