Abstract
Human telomeres contain numerous copies of (TTAGGG)n•(AATCCC)n repeated sequence with multiple TTAGGG repeats in 3’ single-stranded overhangs. Single-stranded oligonucleotides consisting of four TTAGGG repeats can fold into various intramolecular quadruplexes structures stabilized by quartets of guanines. The quadruplex structures are believed to play role in telomere functions and considered as targets for anticancer drugs design. In an effort to create a more realistic model of telomeric DNA we designed oligonucleotides containing a duplex region at the 5’ end and four telomeric repeats in the 3’ overhang. We applied CD spectroscopy and 125I radioprobing to determine the conformation of the quadruplexes formed in the 3’ overhangs. We found that in the presence of NaCl the conformation of the quadruplex changes with formation of the 5’ duplex, and depends on the position of the interface between the duplex and the 3’ telomeric sequence. When the duplex region extended to the first T of the first TTAGGG repeat both CD and radioprobing data are consistent with parallel propeller conformation of the overhang. In the presence of the KCl, formation of the duplex at the 5’ end of DNA molecules did not change the fold of the quadruplex in the overhang which was interpreted as a mixture of two the isomers of 3+1 conformation regardless of the duplex/overhang interface position. Our results demonstrate that the interface between the duplex and single-stranded overhang can affect the conformation of the telomeric quadruplex.
At the ends of eukaryotic chromosomes are often found repeated sequences containing stretches of guanines; for example, all mammalian telomeres contain numerous (TTAGGG)n•(AATCCC)n repeats with multiple TTAGGG repeats in the 3’-single-stranded overhangs (1, 2). The exact mechanism of function of these sequences is still unclear, but they all have a common feature: a single-stranded G-rich strand can form a quadruplex structure that is stabilized by hydrogen bonds between four guanines forming a G-tetrad (3-6). To form an intramolecular quadruplex a human telomeric sequence must contain at least four runs of guanines, i.e. AGGG(TTAGGG)3. Most of the data on the structure of telomeric DNA were obtained by studying conformations of such “minimal” G-quadruplex forming telomeric oligonucleotides (7-10). It was found that these oligonucleotides could adopt a variety of quadruplex conformations that can be broadly divided into parallel, antiparallel and mixed types depending on the relative orientation of the neighboring runs of G’s. Within these broad classes there is a variety of folds differing by the conformations of the TTA connecting loops. Some of the quadruplex conformations are shown in Figure 1.
Figure 1.
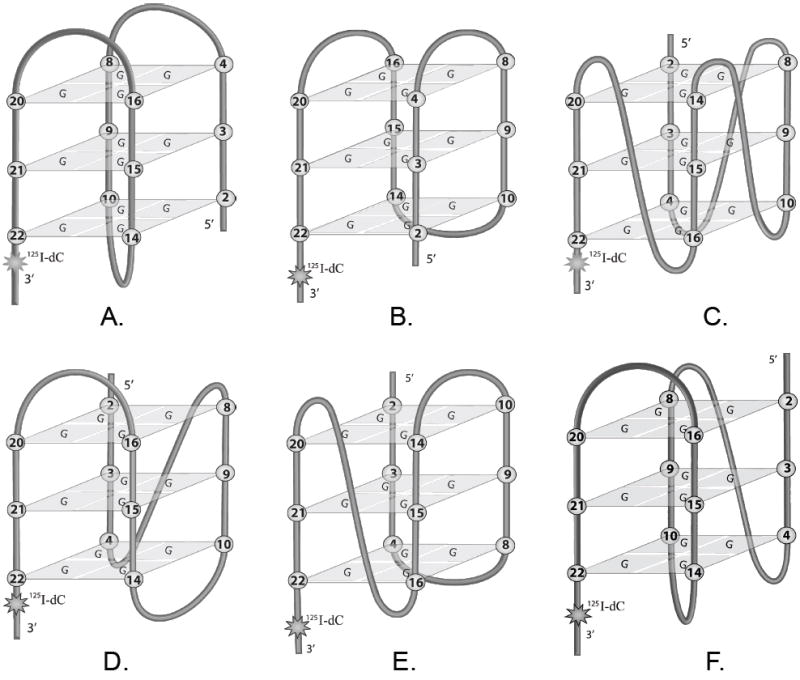
The schematic diagrams of possible intramolecular conformations of human telomeric quadruplexes. Antiparallel: (A) Basket (G22↑G20, G16↓G14, G10↑G8, G4↓G2) and (B) Chair (G22↑G20, G16↓G14, G10↑G8, G4↓G2); parallel: (C) Propeller (G22↑G20, G16↑G14, G10↑G8, G4↑G2); mixed 3+1: (D) Type I (G22↑G20, G16↓G14, G10↑G8, G4↑G2); (E) Type II (G22↑G20, G16↑G14, G10↓G8, G4↑G2) and (F) 3+1 basket (G22↑G20, G16↓G14, G10↑G8, G4↑G2).
The fold of the telomeric oligonucleotide depends on the type of stabilizing monovalent ions (9, 11-13), and on the flanking nucleotides that can interact with the loops of the quadruplex, thus stabilizing a particular conformation (9, 10, 14-16). Some of the conformations were implicated in compaction of the telomeric DNA (8, 9) and, thus, in protecting of the ends of the chromosomes. For example, in all-parallel, so-called propeller conformation (8) and in so-called 3+1 conformation (9, 10), neighboring quadruplexes could stack to each other forming a well-compacted structure.
It is not clear if the structural polymorphism of the telomeric DNA has a biological role. To address this question it is important to find out what conformation(s) the telomeric repeats adopt inside cells. This information would also help in the design of G-quadruplex binding drugs developed for anticancer activities by virtue of inhibiting the function of telomeres (reviewed in (17, 18)). In an attempt to create a more realistic model of the telomeric DNA, we designed DNA molecules containing a 5’ duplex region and G-quadruplex forming telomeric sequence at the 3’ end. We first characterized the general type of G-quadruplex conformation of the molecules by CD spectroscopy. We then applied 125I radioprobing to determine the exact folding of the quadruplexes in these molecules. [125I]-IdC (125I-dC) was incorporated at the 3’ end of the telomeric repeats. Decay of 125I produced DNA strand breaks in a distance-dependent manner, allowing us to determine the changes in folding of G-quadruplex. We found that in the presence of Na+ the conformation of quadruplex dramatically changed upon 5’ duplex formation, and depended on the position of the duplex/overhang interface. In contrast, in the presence of K+ the folding of the quadruplex was not affected by either the addition of the duplex or the position of the duplex/overhang interface.
Materials and Methods
Oligodeoxyribonucleotides and reagents
All oligodeoxyribonucleotides (ODNs, Table 1) were synthesized on an ABI394 DNA synthesizer (Applied Biosystems, Foster City, CA), and purified by denaturating polyacrylamide gel electrophoresis (PAGE) as described in detail in (11). The concentration of single-stranded ODN was measured at 260 nm on an Agilent 8453 diode array spectrophotometer,and was calculated with the extinction coefficient calculator software (http://www.basic.northwestern.edu/biotools/oligocalc.html).
Table 1.
Oligonucleotides used in this study
| T1 | 5’-32P-GAGAACAGTCAACATACAGTGTT-1A1G2G3G4T5T6A7G8G9G10T11T12A13G14G15G16T17T18A19G20G21G22C*23T24 | |
| Oligonucleotides used for 125I-labeling of T1 | ||
| T2 | 5’-GAGAACAGTCAACATACAGTGTTAGGGTTAGGGTTAGGGTTAGGG | Primer |
| T3 | 3’-TGTATGTCACAATCCCAATCCCAATCCCAATCCCGA | Complementary template |
| Complementary oligonucleotide complexes | ||
| TD1 | 5’-32P-GAGAACAGTCAACATACAGTGTTAGGGTTAGGGTTAGGGTTAGGGC*T | T1 |
| 3’-CTCTTGTCAGTTGTATGTCACAAT | T4 | |
| TD2 | 5’-32P-GAGAACAGTCAACATACAGTGTTAGGGTTAGGGTTAGGGTTAGGGC*T | T1 |
| 3’-CTCTTGTCAGTTGTATGTCACAA | T5 | |
| TD3 | 5’-32P-GAGAACAGTCAACATACAGTGTTAGGGTTAGGGTTAGGGTTAGGGC*T | T1 |
| 3’-CTCTTGTCAGTTGTATGTCACA | T6 | |
| T7 | 5’-GAGAACAGTCAACATACAGTGTTA | |
C* = [125I]-IdC
Labeling and purification of oligodeoxyribonucleotides
The telomeric ODNs were labeled with 125I using [125I]-IdCTP (Perkin Elmer, Waltham, MA) and Klenow fragment of DNA polymerase I (Fermentas, Hanover, MD) by the primer extension reaction (19). Primer and template oligonucleotides (T2 and T3, Table 1) were annealed by incubation for 40 min at 37 °C in Klenow buffer (Fermentas), in total amount of 20 pmol each in 10 μl final volume. Then the whole sample was transferred to the tube containing 77 pmol (170 μCi) of lyophilized [125I]-IdCTP. The labeling reaction was initiated by addition 0.5 μl (5 units) of Klenow fragment of DNA polymerase I. After 5 min at room temperature 1 μl of 1mM dTTP solution was added and the reaction continued for 2 more minutes. The reaction was stopped by addition of 20 μL of 10 mM EDTA, extracted once with phenol/chloroform mixture, and the product was purified by gel filtration on MicroSpin G50 TE equilibrated column (GE Healthcare, Piscataway, NJ) and vacuum dried. The resulting 125I labeled oligonucleotide was dissolved in 1X Reaction Buffer A (Fermentas) in the presence 30 pmol (150 μCi) of [γ-32P]-ATP (Perkin Elmer), 10 units of T4 PNK (Fermentas) and incubated at 37 °C for 40 minutes. The resulting 5’-32P labeled oligonucleotide was extracted with phenol/chloroform, purified by gel filtration on MicroSpin G25 water equilibrated column (GE Healthcare), lyophilized, dissolved in 5 μl of formamide stop solution (USB Corporation, Cleveland, OH) and loaded onto 6% denaturing PAGE. The band corresponding to the 125I and 32P labeled telomeric oligonucleotide was excised and eluted from crushed gel with 100 μl of 1x TBS for 1 hour on shaking platform at room temperature. Then sample was purified from acrylamide debris on 0.22 μm cellulose acetate X-spin filter (Corning Inc., Corning, NY) and oligonucleotide was precipitated with ethanol. The pellet was washed 3 times with 70% ethanol, vacuum dried and dissolved in water.
Preparation of G-4 quadruplexes and radioprobing
Telomeric oligonucleotide labeled with 125I and 32P (1 pmol) was annealed with complementary T4, T5 and T6 oligonucleotides (1 pmol) in 44 mM Tris/HCl buffer pH 7.4. Neither NaCl nor KCl were added at this point because we noticed that the presence of salt prevents complete annealing of the complementary oligonucleotides. Annealing was tested by a band-shift assay in 4-20% gradient pre-cast TBE PAGE (Invitrogen, Carlsbad, CA). After annealing, NaCl and KCl were added to the samples to final concentration 100 mM. The samples were incubated for 1 hr at 22 °C, flash frozen in liquid nitrogen and stored at -80 °C for DNA breaks accumulation. After 30 days the samples were thawed, the integrity of the duplexes was tested once again in 4-20% gradient pre-cast TBE PAGE. Strand breaks in the telomeric oligonucleotide were analyzed in 12% denaturing PAGE. Probabilities of breaks were calculated based on the measurement of the intensity of the bands by densitometry and peak deconvolution as was described in detail in (15).
CD spectroscopy
CD spectra were recorded on a Jasco J-810 spectropolarimeter in a 2 mm pathlength cuvette and a wavelength scanning speed of 100 nm/min. The spectra are the averages of three scans between 200 and 320 nm at 25 °C. Telomeric oligonucleotide samples were prepared substantially the same as for radioprobing assay: 200 μl of 2 μM solution of oligonucleotide T1 (not labeled with any radioisotope) was annealed in 20 mM sodium or potassium phosphate pH 7.4 with complementary oligonucleotides (2 μM) to yield TD1, TD2 and TD3 duplexes. After annealing, NaCl and KCl were added to the samples to final concentration 100 mM (Na+ or K+), and samples were incubated for 1 hr at 22 °C. All spectra were baseline subtracted and curves were smoothed using a three-point moving average. Spectra of TD1-TD3 complexes were additionally corrected by subtraction of spectra of corresponding short duplexes.
Calculation of distances
Interatomic distances were calculated using the coordinates from Protein Data Bank (PDB) files 143D (7) and 1KF1 (8), 2GKU (10) and 2JPZ (20). Guanine cores of the structures were aligned using MatchMaker function of UCSF Chimera (21), except 143D that was aligned manually. Distances to the guanines of the aligned structures were measured from atom O4 of residue T25 of the 2JPZ structure.
Results
Gel-shift assay – demonstration of double stranded complex formation
Oligonucleotides, TD1, TD2 and TD3, containing 5’ duplex and 3’ telomeric overhangs are shown in Table 1. They are different in length of the duplex region; in TD1 it covers the first TTA triad of the first telomeric repeat while in TD3 it is two nucleotides shorter, and extends only to the first T on the TTA triad (underlined in Table 1). To obtain TD1, TD2 and TD3 the 5’-32P and 125I labeled T1 (Table 1) was annealed with 5’-complementary oligonucleotides. We noticed that annealing was not 100% (data not shown) if carried out in the presence of 100 mM of KCl or NaCl, probably due to the interaction of the single-stranded 5’-end of T1 with G-quadruplex structure at the 3’-end (14). Therefore, the annealing was carried out in Tris/HCl buffer followed by addition of KCl or NaCl to allow G-quadruplex formation in the 3’ overhang at the specified conditions. The extent of annealing was verified by the gel-shift assay (Figure 2). Results shown in Figure 2 demonstrate duplex formation between the 5’-complementary oligonucleotides T4, T5 and T6 and complementary portion of T1. Mobility of TD1, TD2 and TD3 duplexes (lanes 3-5) is slower then that of the single stranded T1 (lane 2) and comparable with the mobility of the long duplex T1+T3 (lane 1); thus confirming almost complete formation of the desired DNA constructs.
Figure 2.
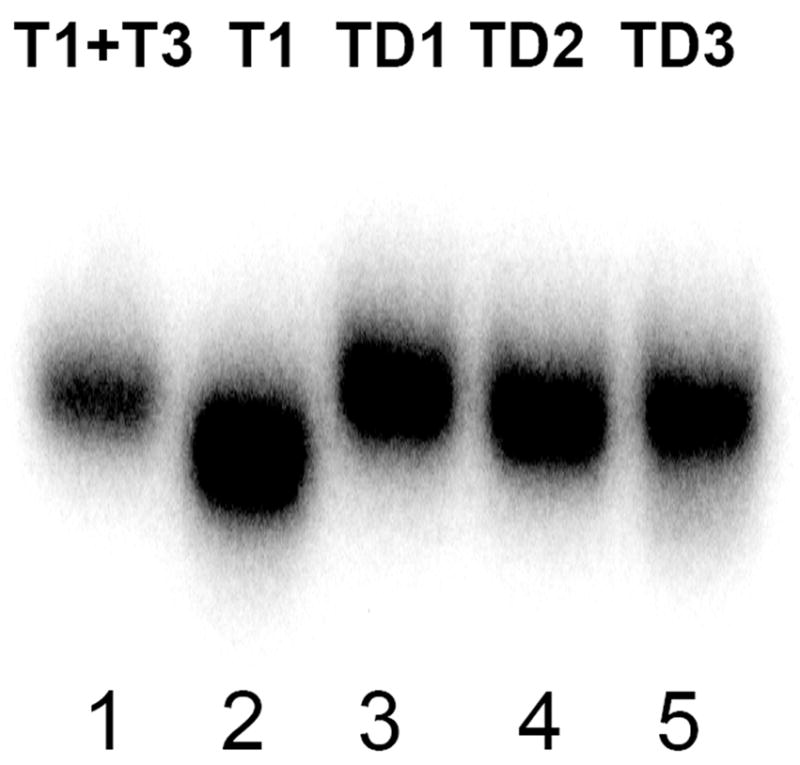
Gel-shift assay for double stranded complex formation. Autoradiograph of native 6% PAGE run at room temperature in 1x TBE. Lane 1 - telomeric oligonucleotide T1 + complementary template T3; Lane 2 - telomeric oligonucleotide T1; Lanes 3, 4 and 5 -telomeric duplexes TD1, TD2 and TD3 correspondingly.
CD spectroscopy profiles
We first applied the analysis of circular dichroism (CD) spectra to determine the general type of G-quadruplex conformation in our DNA constructs. CD spectroscopy has been widely used for the characterization of G-quadruplex-forming oligonucleotides (9, 22-28). CD spectra are generally affected by the orientation of guanine residues relative to the sugar moieties, i.e. anti or syn, and by sugar pucker. Therefore, antiparallel and mixed-type quadruplexes containing both anti and syn conformations of Gs and parallel quadruplexes with all Gs in anti conformation can be distinguished by their CD spectra. Nucleotides of the loops, and that in the 5’ and 3’ extensions, also contribute to CD spectra, complicating their interpretation. Strictly speaking, it is impossible to predict the exact fold of a quadruplex based solely on its CD spectra. Nevertheless, reference spectra have been determined for different G-quadruplex conformations. Thus, CD spectra of antiparallel basket and chair conformations of telomeric DNA have characteristic strong positive peak at 295 nm and a smaller negative peak at 265 nm and a positive peak at 245 nm (29). Mixed 3+1 conformations exhibit a strong peak at 290 nm, a shoulder peak at 268 nm and a negative peak at 240 nm (9). CD spectra of the parallel conformation of telomeric DNA are expected to have a positive peak near 260 nm and a negative peak near 240 nm based on the spectra of short G-rich telomeric oligonucleotides that formed tetramolecular parallel structures (29). Intramolecular structures with such CD spectra were described as parallel quadruplexes (25, 27), however, parallel fold of these intramolecular quadruplexes has not been confirmed by other methods in solution. Also, based on CD spectra of GGGTTAGGG oligonucleotide forming a dimeric all-parallel quadruplex a positive band at 290 nm was assigned to the contribution of external loop residues (25).
The duplex regions of TD1, TD2 and TD3 significantly contribute to the overall CD spectra of these molecules. To extract these contributions, oligonucleotides forming just the duplex parts of TD1, TD2 and TD3 were synthesized, annealed, and their CD spectra were subtracted from the overall spectra of TD1, TD2 and TD3. CD spectra of T1, and the “duplex-subtracted” spectra of TD1, TD2 and TD3 in the presence of Na+ or K+ are shown in Figure 3. The raw CD spectra before subtraction are available in supplemental material (Supplemental Figure 1).
Figure 3.
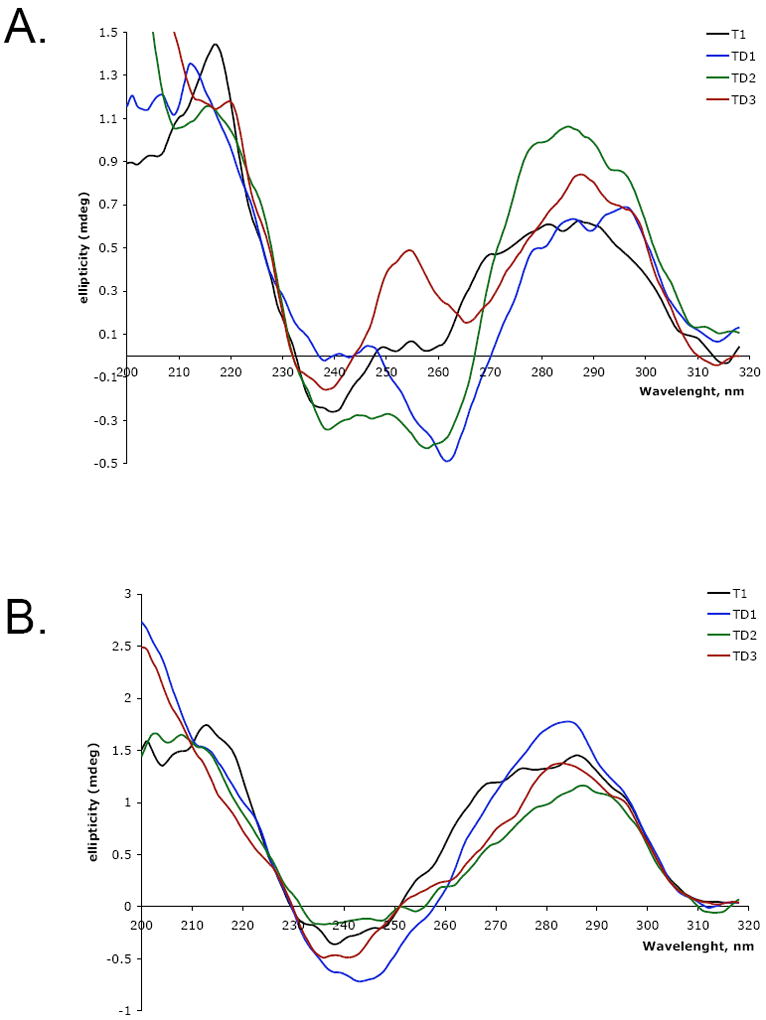
Duplex subtracted CD spectra of telomeric oligonucleotide T1 and complexes TD1, TD2 and TD3 measured in 100 mM Na+ (A) or in 100 mM K+ (B) solutions at 25 °C.
In the presence of Na+ CD spectra are clearly affected by the duplex formation at the 5’ end of T1. The spectra of TD1, TD2 and TD3 are quite different from that of T1. At the same time while CD spectra of TD1 and TD2 are similar to each other they are completely different from CD spectrum of TD3 that exhibit a strong positive peak at about 255 nm. CD spectrum of T1 with a positive peak at 290 nm, a shoulder peak at 268 nm and a negative peak at 240 nm is consistent with the signature of a mixed 3+1 quadruplex conformation. CD spectrum of TD1 shows general features of an antiparallel quadruplex - a positive peak at 295 nm and a negative around 260 nm, but the lack of a positive peak at 240 nm and a positive peak at 280 nm indicate the presence of other quadruplex conformations. CD spectrum of TD2 does not correspond to a single quadruplex conformation and can be interpreted as a spectrum of a mixture of antiparallel and mixed conformations. TD3 is the only conformation that has a distinct positive peak at 255 nm; that, along with the negative peak at 240 nm, is consistent with the presence of the parallel quadruplex conformation.
In the presence of K+ CD spectra of all tested complexes (Figure 3B) are consistent with the signature of a mixed 3+1 quadruplex conformation. With slight variations, they all have a maximum at ~285-295 nm, a shoulder between 260 and 270 nm and a minimum at ~240-245.
Radioprobing of telomeric quadruplexes
To assess the exact fold of intramolecular quadruplexes in TD1, TD2 and TD3 we applied 125I-radioprobing. TD1, TD2 and TD3 were allowed to fold in the specified conditions and then samples were frozen in liquid nitrogen and stored for two weeks to accumulate 125I-induced DNA strand breaks. Then the samples were analyzed for strand breaks by sequencing PAGE (Figure 4). Figure 5 represents probabilities of breaks at the individual nucleotides based on the measurement of the intensity of the bands by densitometry and peak deconvolution (for details see (15)).
Figure 4.
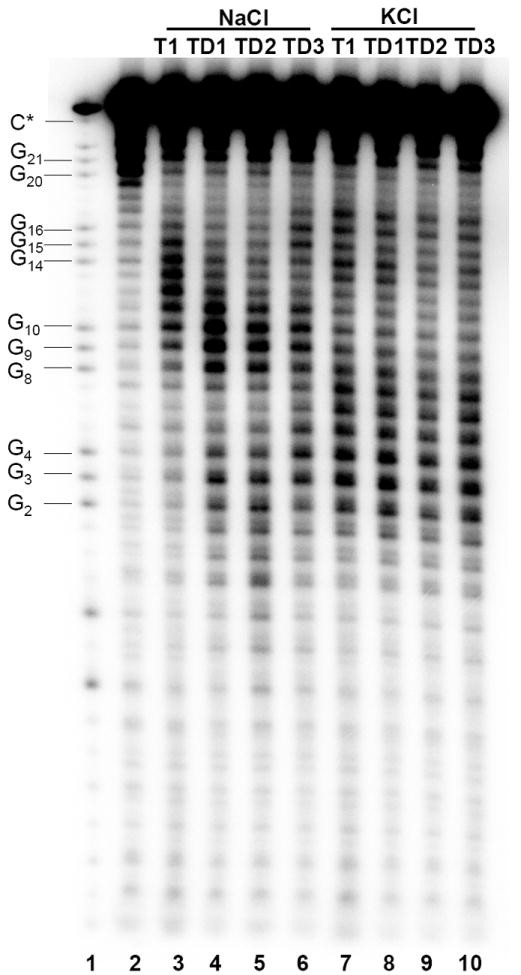
Analysis of DNA breaks in telomeric oligonucleotides by 12% denaturing PAGE. Lanes: 1 – G sequencing ladder; 2 – Duplex of T1 with complementary template T3; 3-6 – oligonucleotide T1 and complexes TD1, TD2 and TD3 folded in 100 mM NaCl solution; 7-10 – oligonucleotide T1 and complexes TD1, TD2 and TD3 folded in 100 mM KCl solution.
Figure 5.
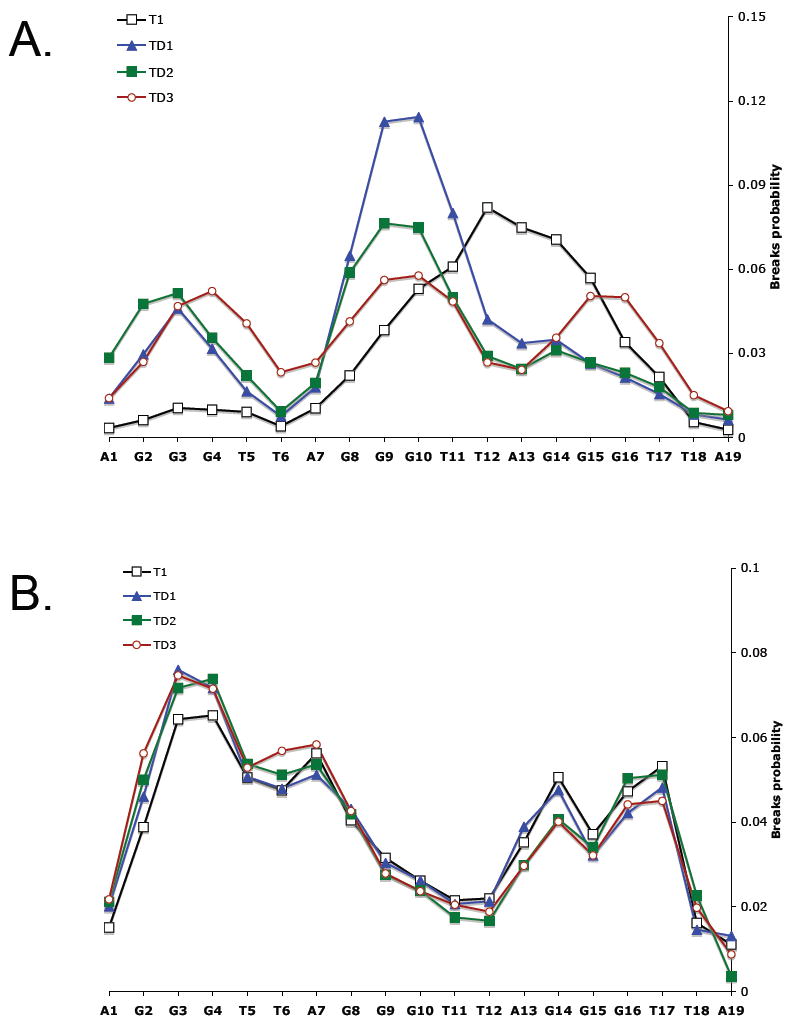
Distribution of 125I induced strand break probability in the telomeric oligonucleotide T1 and complexes TD1, TD2 and TD3 in 100 mM NaCl (A) or in 100 mM KCl (B).
For analysis of the radioprobing results we used the same considerations and rules as we described in our previous paper (15). We focused our attention on G’s in the core of the G-quadruplex. We showed that the geometry of the cores is very similar for all quadruplex conformations; disregarding, of course, that different guanines are in different positions in these conformations. Our interpretation of radioprobing data was based on a simple geometric consideration; if I-125 is at the “bottom” of the core then the G’s at the “top” of the core are the most distant from it and, consequently, would have the least probability of breaks. To prove this we now calculated distances from the 3’ T25(O4) located at the “bottom” of the 3+1 type 2 NMR structure (2JPZ (20)) to the sugars of the core G’s. In addition, using UCSF Chimera (21) we align cores of other quadruplexes (1KF1, 143D, 2GKU (7, 8, 10)) with that of 2JPZ. Then we measured distances from T25(O4) of 2JPZ to the sugars of G’s of the other aligned structures. The results are presented in Figure 6. As expected, for all tested quadruplex conformations the distances increase as we move from the position in the bottom of the core to the top (as they are oriented in Figure 1). Therefore, if the probability of breaks decreases along a run of three G’s then it means that the distance increases from the first to the last G in the run. If “↑” indicates the increase in distances and “↓” the decrease, then, for example, the basket conformation can be described as G22↑G20, G16↓G14, G10↑G8, G4↓G2, “3+1” type 1 as G22↑G20, G16↓G14, G10↑G8, G4↑G2, and propeller as G22↑G20, G16↑G14, G10↑G8, G4↑G2. Obviously the distances always increase along G22-G20 side; therefore, we will omit it in our descriptions.
Figure 6.
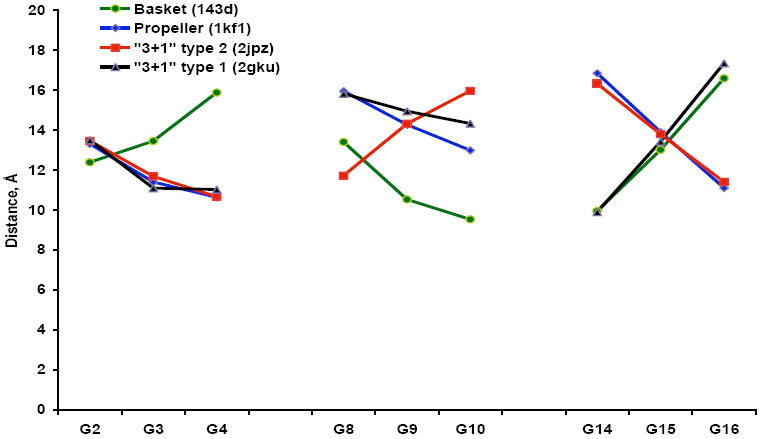
Average distances (in angstroms) from the O4 of the T25 of the quadruplex structure from PDB file 2JPZ to the sugars of the guanines in the core of 2JPZ and three other quadruplex conformations from PDB files 1KF1, 143D, and 2GKU that were aligned with 2JPZ as described in the Materials and Methods. Different conformations of the G-quadruplex; green – basket, PDB ID 143D; blue - propeller, PDB ID 1KF1; red – “3+1” type 2, PDB ID 2JPZ; black – “3+1” type 1, PDB ID 2GKU.
Conformation of TD complexes in Na+
Figure 5A shows the results of radioprobing experiment of single-stranded T1 and TD1, TD2 and TD3 complexes in the presence of Na+. The higher the probability of breaks the closer the nucleotide to the 125I-dC at the 3’ end of the structure. Accordingly, for T1 the drop of the breaks probability from G14 to G16 shows that the former is closer to the bottom of the structure than the latter. Likewise, the G10 is closer than G8. The probabilities of breaks are the lowest for the G2-G4 side indicating that it is diagonal to the G22 position. Overall, the distribution of breaks for T1 in Na+ can be described as G16↓G14, G10↑G8, G4↑G2.
Duplex formation at the 5’ end of T1 (TD1, TD2 and TD3) drastically changes distributions of breaks. For TD3 there is a clear pattern of increase of breaks probability along all G-sides. The structure can be described as G22↑G20, G16↑G14, G10↑G8, G4↑G2 and corresponds to all-parallel propeller conformation of quadruplex; consistent with the CD spectra data. For TD1 and TD2 distributions of breaks are similar to each other; both have a pronounced maximum at G10 and G9. TD1 conformation can be formally described as G16↓G14, G10↑G8, G4↓↑G2 and TD2 as G16↓G14, G10↓↑G8, G4↓↑G2 where “↓↑” indicates the lack of a clear increase or decrease in breaks probability along G2-G4 and G8-G10 sides. This uncertainty does not allow unambiguously ascribing the observed distribution of breaks to any single G-quadruplex conformation. Therefore, we conclude that in Na+ TD1 and TD2 are present as a mixture of different isomers of G-quadruplex. This conclusion is in accord with the CD spectroscopy data (Figure 3).
Conformation of TD complexes in K+
Figure 5B shows the results of radioprobing experiment of T1, TD1, TD2 and TD3 complexes in the presence of KCl. In contract to the NaCl data and in agreement with CD spectra, there is no significant difference in distribution of breaks probabilities between all tested oligonucleotides. Probabilities of breaks are almost the same for G16 and G14. They are lowest for the G8-G10 with a weak increase in the breaks probabilities along the G8-G10 side. At the same time there is a clear drop in breaks probability for G4-G2 and the nucleotides at the 5’ from them indicating that this side and the 5’ region going away from the 125I that is at the 3’ end (bottom in Figure 1) of the molecules. Quadruplex conformations can be formally described as G16↓↑G14, G10↓G8, G4↑G2 where “↓↑” indicates the lack of a clear increase or decrease in breaks probability along G16-G14 side. As it was in our previous study, in the presence of K+ (15) it is impossible to clearly attribute this breaks distribution to a single G-quadruplex conformation. Hence, the radioprobing results point to a mixture of quadruplex conformations in the presence of K+ for all tested oligonucleotides.
Discussion
The simultaneous presence of several quadruplex conformations in solution poses a challenge to any structural method and complicates interpretation of experimental results. Next we will try to deduce structural information from the results of our radioprobing experiments in the light of our CD spectra data. CD spectra data indicate that in K+ the quadruplexes are in a mixed 3+1 conformation. From the NMR studies (9, 10, 20, 30) we know that telomeric oligonucleotides in the presence of K+ can adopt two isomers of mixed 3+1 conformation, type1 and type 2 (Figure 1). The main ambiguity in our radioprobing data in K+ (Figure 5B) is the lack of increase or decrease in breaks probability along G16-G14 side. We believe that this ambiguity is consistent with the presence of the mixture of type 1 and type 2 isomers of mixed 3+1 conformation. Indeed, in type 1 and type 2 isomers (Figure 1) G16-G14 side points in different directions, therefore, in type 1 isomer maximum of probability of breaks along this side is at G14, while in type 2 isomer it is at G16. Superposition of these two distributions would result in the pattern of breaks observed in Figure 5B, i.e. equal at G14 and G16 and somewhat lower at G15. Guanines along G10-G8 side have the lowest breaks probability consistent with this side being diagonal (i.e. most distant) from the 125I position in both type 1 and type 2 isomers of mixed 3+1 conformation. Although G10-G8 side is in different orientations in type 1 and type 2 isomers, we observe steady decrease in breaks probability from G8 to G10. This discrepancy could be due, for example, to the non-linear relation between the distance and probability of breaks. Thus, in type 2 isomer the distance from G8 to 125I is considerably shorter than the distance from G10 to 125I in type 1 isomer (Figure 6). This could explain why the overall breaks probability at G8 is higher than that at G10 in the mixture of the isomers.
The distribution of breaks for T1 in Na+ was G16↓G14, G10↑G8, G4↑G2. This distribution is consistent with both type 1 and basket mixed 3+1 conformations (Figure 1). The probabilities of breaks are the lowest for the G2-G4 side indicating that it is the most distant from (i.e. diagonal to) the 125I position. Therefore, T1 is most likely in the mixed 3+1 basket conformation that we proposed in our previous study for a similar telomeric oligonucleotide with a 5’ extension (15). The CD spectrum of T1 in Na+ is also consistent with a mixed 3+1 conformation.
CD spectra of TD1 and TD2 in Na+ showed certain features of both antiparallel and 3+1 conformations. The main ambiguity in their radioprobing profiles (Figure 5A, blue and green curves) is the lack of any clear increase or decrease in breaks probability along the G4-G2 side. This is consistent with a mixture of the antiparallel basket and 3+1 basket conformations. In these two conformations only the G4-G2 side points in different directions relative the 125I position. In the antiparallel basket G2 is the closest to 125I, while in the 3+1 basket it is G4. Superposition of the break distributions characteristic to these two basket conformations is consistent with that observed for TD1 and TD2. The difference in the break distributions for TD1 and TD2 could be attributed to various proportions of these conformers in the mixtures.
The above considerations define major quadruplex conformations present simultaneously in K+ and Na+ solutions; i.e. 3+1 type 1 and type 2 for all studied molecules in K+, and antiparallel and 3+1 basket for TD1 and TD2 in Na+. However, not all the features of break distribution profiles can be explained by these considerations. For example, we cannot explain considerably higher breaks probability at G10 and G9 compared to that at G14 and G15 for TD1 in Na+. As we previously discussed (15), radioprobing, (i.e. distribution of DNA breaks probability), in principle, should provide a true information about internal distances in a DNA molecule. However, in the labeling scheme used in our study the exact position of the 125I was not fixed, thus, causing some degree of uncertainty in the interpretation of the results.
We should note here that the order of the distances to the sugars of the core G’s depends considerably on the position of 125I at the bottom of the core. For example, the distance from the C1’ of G22 that is in the corner of the core is actually shorter to G2 at the top than to G4 at the bottom of the core (Figure 2 in (15)). Therefore, in our previous paper (15) interpretation of radioprobing data was actually based on simple geometrical considerations rather than on the actual distances. The distances were presented to prove that the cores of all quadruplexes had similar geometries and G22 was chosen simply because the “extra-core” nucleotides are different in different structures (some of the structures do not have extra nucleotides at the 3’ end at all). Herein, by aligning cores of different G-quadruplexes we actually showed that the distances from a reference position at the bottom of the core follow the predicted pattern, i.e. G’s at the top of the core are more distant than that at the bottom. At the same time we noticed that if the reference position was moved to the periphery of the core and closer to the G-tetrad at the bottom the order of the G’s in terms of distances could be altered (IP, unpublished observation), like it was in the extreme case of the C1’ position of G22 (see above). Therefore, formation of the duplex at the 5’ end of the molecules could affect the position of the 125I-dC especially in antiparallel conformations driving it closer to one or another G-side. This could be an alternative to the presented above explanation for the ambiguities in radioprobing data observed for TD1 and TD2 in Na+. At the same time we could not exclude a minor presence of other quadruplex conformations in solutions that would affect fine details of breaks distribution profiles.
Nevertheless, combination of radioprobing data and CD spectra strongly indicate the presence of all-parallel propeller quadruplex conformation in TD3 in Na+. This quadruplex conformation of telomeric DNA was first observed by X-ray crystallography. There were also several reports describing this conformation in some special conditions in solution (25, 31). It is possible that folding of telomeric overhangs into this conformation requires stabilizing interactions with similar neighboring quadruplexes as it is in crystal (8) or in the case of multiple telomeric repeats (27). In our study the duplex region could provide such stabilization. Thus folding of the first (from the duplex) four repeats into the parallel quadruplex may trigger folding of the rest of the repeats in the single-stranded overhang into the same conformation. This would result in contraction of the overhangs normally prevented by telomeric proteins such as POT1, and may play a role in the functioning of telomeres as was previously discussed (8).
Recently, a new basket conformation of telomeric quadruplex stabilized by two G-tetrads was described by NMR in K+ (16). CD spectrum of this conformation is somewhat similar to that observed in our study for TD3 in Na+ (Figure 2A). However, our radiorobing data for TD3 are inconsistent with the presence of this conformation. For example, in this new basket conformation G14 would be the closest to 125I at the 3’ end, while radioprobing profile (Figure 5A) shows that G16 is the closest to 125I. Radioprobing profiles of other molecules described here do not support the presence of this new basket conformation either. Nevertheless, as it was discussed above, we cannot exclude a minor presence of this form of quadruplex.
Previous studies utilizing methods of NMR and X-ray crystallography determined molecular structures of telomeric quadruplexes in single-stranded G-rich oligonucleotides, laying down a framework for further structure-function relation studies in telomeric DNA. Their application to more realistic models of telomeres containing multiple repeats in both single- and double-stranded form is currently complicated by the size of such DNA molecules. In our study, by combining radioprobing and CD spectroscopy, we were able to gain detailed information regarding conformation of telomeric quadruplexes in DNA molecules containing both duplex and quadruplex domains. We showed that in the presence of Na+ one of these molecules adopts the all-parallel propeller quadruplex conformation. We also concluded that in Na+ two other molecules were present as a mixture of antiparallel and 3+1 basket conformations, while in K+ all the molecules were present as a mixture of 3+1 type 1 and type 2 conformations.
Supplementary Material
Acknowledgments
We would like to thank Dr. P. Schuck for the help with CD measurements. This research was supported by the Intramural Research Program of the NIH, Clinical Center, and in part by the National Institute of Biomedical Imaging and Bioengineering.
Abbreviations
- ODN
oligodeoxyribonucleotide
- T4 PNK
T4 Polynucleotide Kinase
- PDB
Protein Data Bank
- CD
circular dichroism
- PAGE
polyacrylamide gel electrophoresis
Footnotes
This research was supported by the Intramural Research Program of the NIH, Clinical Center and National Institute of Biomedical Imaging and Bioengineering.
References
- 1.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sfeir AJ, Chai W, Shay JW, Wright WE. Telomere-end processing the terminal nucleotides of human chromosomes. Mol Cell. 2005;18:131–138. doi: 10.1016/j.molcel.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 3.Gellert M, Lipsett MN, Davies DR. Helix Formation by Guanylic Acid. Proc Natl Acad Sci U S A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 5.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 6.Panyutin IG, Kovalsky OI, Budowsky EI, Dickerson RE, Rikhirev ME, Lipanov AA. G-DNA: a twice-folded DNA structure adopted by single-stranded oligo(dG) and its implications for telomeres. Proc Natl Acad Sci U S A. 1990;87:867–870. doi: 10.1073/pnas.87.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 8.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 9.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J Am Chem Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y, Neumann RD, Panyutin IG. Intramolecular quadruplex conformation of human telomeric DNA assessed with 125I-radioprobing. Nucleic Acids Res. 2004;32:5359–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wlodarczyk A, Grzybowski P, Patkowski A, Dobek A. Effect of ions on the polymorphism, effective charge, and stability of human telomeric DNA. Photon correlation spectroscopy and circular dichroism studies. J Phys Chem B. 2005;109:3594–3605. doi: 10.1021/jp045274d. [DOI] [PubMed] [Google Scholar]
- 13.Redon S, Bombard S, Elizondo-Riojas MA, Chottard JC. Platinum cross-linking of adenines and guanines on the quadruplex structures of the AG3(T2AG3)3 and (T2AG3)4 human telomere sequences in Na+ and K+ solutions. Nucleic Acids Res. 2003;31:1605–1613. doi: 10.1093/nar/gkg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaynutdinov TI, Neumann RD, Panyutin IG. Structural polymorphism of intramolecular quadruplex of human telomeric DNA: effect of cations, quadruplex-binding drugs and flanking sequences. Nucleic Acids Res. 2008;36:4079–4087. doi: 10.1093/nar/gkn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim KW, Amrane S, Bouaziz S, Xu W, Mu Y, Patel DJ, Luu KN, Phan AT. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J Am Chem Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neidle S, Parkinson GN. Quadruplex DNA crystal structures and drug design. Biochimie. 2008;90:1184–1196. doi: 10.1016/j.biochi.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Rezler EM, Bearss DJ, Hurley LH. Telomere inhibition and telomere disruption as processes for drug targeting. Annu Rev Pharmacol Toxicol. 2003;43:359–379. doi: 10.1146/annurev.pharmtox.43.100901.135733. [DOI] [PubMed] [Google Scholar]
- 19.Panyutin IG, Neumann RD. Radioprobing of DNA: distribution of DNA breaks produced by decay of 125I incorporated into a triplex-forming oligonucleotide correlates with geometry of the triplex. Nucleic Acids Res. 1997;25:883–887. doi: 10.1093/nar/25.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai J, Carver M, Punchihewa C, Jones RA, Yang D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 22.Rezler EM, Seenisamy J, Bashyam S, Kim MY, White E, Wilson WD, Hurley LH. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J Am Chem Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 23.Giraldo R, Suzuki M, Chapman L, Rhodes D. Promotion of parallel DNA quadruplexes by a yeast telomere binding protein: a circular dichroism study. Proc Natl Acad Sci U S A. 1994;91:7658–7662. doi: 10.1073/pnas.91.16.7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardin CC, Watson T, Corregan M, Bailey C. Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of d(CGCG3GCG) Biochemistry. 1992;31:833–841. doi: 10.1021/bi00118a028. [DOI] [PubMed] [Google Scholar]
- 25.Rujan IN, Meleney JC, Bolton PH. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray RD, Li J, Chaires JB. Energetics and Kinetics of a Conformational Switch in G-Quadruplex DNA. J Phys Chem B. 2009 doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vorlickova M, Chladkova J, Kejnovska I, Fialova M, Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dapic V, Abdomerovic V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue Y, Kan ZY, Wang Q, Yao Y, Liu J, Hao YH, Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J Am Chem Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


