Abstract
Sp1 transcription factor regulates the expression of multiple genes, including the Sp1 gene itself. We analyzed the ability of different cell cycle regulatory proteins to interact with Sp1 and to affect Sp1 promoter activity. Using an antibody array, we observed that CDK4, SKP2, Rad51, BRCA2 and p21 could interact with Sp1 and we confirmed these interactions by co-immunoprecipitation. CDK4, SKP2, Rad51, BRCA2 and p21 also activated the Sp1 promoter. Among the known Sp1-interacting proteins, E2F-DP1, Cyclin D1, Stat3 and Rb activated the Sp1 promoter, whereas p53 and NFκB inhibited it. The proteins that regulated Sp1 gene expression were shown by positive chromatin immunoprecipitation to be bound to the Sp1 promoter. Moreover, SKP2, BRCA2, p21, E2F-DP1, Stat3, Rb, p53 and NFκB had similar effects on an artificial promoter containing only Sp1 binding sites. Transient transfections of CDK4, Rad51, E2F-DP1, p21 and Stat3 increased mRNA expression from the endogenous Sp1 gene in HeLa cells whereas overexpression of NFκB, and p53 decreased Sp1 mRNA levels. p21 expression from a stably integrated inducible promoter in HT1080 cells activated Sp1 expression at the promoter and mRNA levels, but at the same time it decreased Sp1 protein levels due to the activation of Sp1 degradation. The observed multiple effects of cell cycle regulators on Sp1 suggest that Sp1 may be a key mediator of cell cycle associated changes in gene expression.
Keywords: Sp1, protein-protein interactions, cell cycle, gene regulation, transcription, p21
Introduction
Sp1 transcription factor regulates a wide range of cellular processes,1 including cell cycle regulation, hormonal activation, apoptosis and angiogenesis.2 It belongs to a family of nuclear proteins, called Sp/KLF (specificity protein/Krüppel-like factor) that recognizes the GC-rich DNA-binding core sequences GC-(GGGGCGGGG) and GT-(GGTGTGGGG) boxes due to the presence of three conserved Cys2His2 zinc fingers that form the DNA-binding domain.3
Regulation of Sp1-dependent transcription can be affected by changes in Sp1 abundance, as it occurs during the cell cycle with an increase during G1 phase; DNA binding activity or transactivation activity, and it can involve direct protein-protein interactions with other nuclear factors in which the other factor is not necessarily bound to promoter DNA. Several protein-binding sites have been identified throughout Sp1.4 For example, Sp1-interacting proteins include several viral proteins; members of the basal transcription machinery, several transcription activators and cell cycle regulators.4 In the latter case, the retinoblastoma protein has been described to interact physically with Sp1 in a complex that enhances the transcriptional activation by Sp1.5 In addition, Sp1 can also interact with the E2F-family of transcription factors which is believed to integrate cell cycle progression with transcription through its cyclical interactions with important cell cycle regulators, such as Rb, cyclins and cyclin-dependent kinases.6 Moreover, Sp factors have been proposed to be essential for the transactivation of the p21Cip1 promoter by members of the p53 family of proteins.7
Posttranslational modifications also regulate Sp1 activity, including glycosylation, acetylation, phosphorylation and sumoylation.4,8 Many kinases are able to phosphorylate Sp1 and its phosphorylation state could also be affected by protein phosphatases.9 CyclinE-CDK2 and CyclinA-CDK2 complexes which are important regulators of the mammalian cell cycle, phosphorylate Sp1 forming stable complexes with this transcription factor bound to the DNA.10
We previously described the cloning of the 5′-flanking region of the human Sp1 gene, where a number of putative binding sites for transcription factors were found. Within the minimal promoter sequence, expanding 217 bp from the transcription start site, two Sp1 binding sites were described. Sp1 promoter was mainly regulated by Sp1, Sp3 and NF-Y, although E2F was also found to bind and activate the Sp1 promoter.11,12
In the present study we performed a more detailed analysis of the Sp1 promoter taking into account its regulation by protein-protein interactions. Using a proteomic array system, we first screened for new Sp1-interacting proteins among cell cycle regulators. We confirmed by coimmunoprecipitation the interaction of Sp1 with Rad51, CDK4, SKP2, BRCA2 and p21 and we studied the effect of the overexpression of these proteins both on the Sp1 promoter and on the endogenous Sp1 mRNA levels. Other known Sp1-interacting proteins were also tested for their effect on Sp1 regulation. We analyzed the regulation of the Sp1 promoter in situ by chromatin immunoprecipitation using specific antibodies against the different proteins studied. Additionally, an artificial Sp1-dependent promoter was used in order to ascertain if the effects observed were unique to the Sp1 promoter or a general mechanism regulating Sp1-dependent transcription. Interaction of Sp1 with p21 was further studied using HT1080 p21-9 cells with inducible p21 expression.13 Although p21 activated Sp1 transcription both at the promoter and mRNA levels, it did not increase Sp1 protein levels due to the activation of Sp1 protein degradation. The observed multiple effects of cell cycle regulators on Sp1 suggest that Sp1 may be a key mediator of cell cycle associated changes in gene expression.
Results
Sp1 interacts with different cell cycle regulators
To identify proteins that might interact with transcription factor Sp1, whole cell extracts from HeLa cells were screened for Sp1 interactors using an antibody array (Hypromatrix). Among all the Sp1-interacting proteins identified in the Antibody Array, the following were selected for further study: CDK4 and p21, that play a pivotal role in the control of eukaryotic cell cycle; SKP2 that binds to different substrates for ubiquitination by the SCF complex; Rad51 and BRCA2 that participate in DNA-damage responses; and Stat3, p53, NFκB, E2F and Rb for which the interaction with Sp1 had been previously described and that were used as positive controls.
The interactions between Sp1 and several selected proteins identified in the antibody array were confirmed by coimmunoprecipitation. Sp1, CDK4, SKP2, Rad51 and BRCA2 were immunoprecipitated from HeLa whole-cell extracts using specific antibodies and the presence of Sp1 in the immunoprecipitates was determined by western blot analysis. Sp1 was detected in the immunoprecipitates of antibodies against these proteins, demonstrating that all the selected proteins interacted with Sp1 (Fig. 1A). No signal corresponding to Sp1 was detected when the immunoprecipitation was performed with unspecific IgGs. The interaction between Sp1 and p21 was established by immunoprecipitation using a p21-specific antibody and HeLa cells overexpressing p21 (Fig. 1B).
Figure 1.
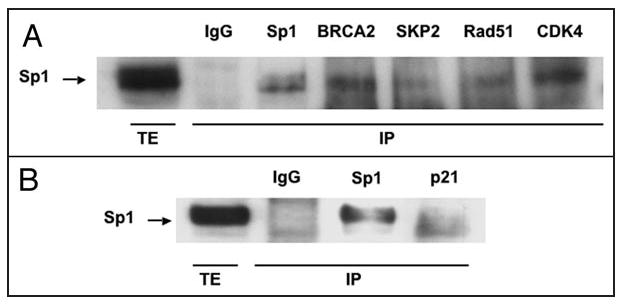
Coimmunoprecipitation of Sp1 with BRCA2, SKP2, Rad51, CDK4 and p21. (A) Immunoprecipitates were obtained from 1 mg of HeLa total cell extract by incubation with 5 μg of the indicated specific antibodies. After washing, the bound proteins were resolubilized and Sp1 protein was detected by Western analysis. The first lane (TE) shows the signal corresponding to Sp1 from 200 μg of HeLa total extract. (B) Coimmunoprecipitation using p21-antibody. 5 μg of p21 expression vector were transfected into HeLa cells using Fugene 6. Total extracts were obtained and 1 mg of each extract was used for coimmunoprecipitation with p21 antibody. Other conditions were as in Figure 1A.
Effects of Sp1-interacting proteins on Sp1-promoter
We used the minimal promoter of the Sp1 gene (pGL3FOR2), which is mainly regulated through two Sp1 sites,12 to test the effect of the Sp1-interacting proteins. SKP2, Rad51, CDK4, BRCA2 and p21 showed significant transcriptional activation of the Sp1 promoter (Fig. 2A). In addition, Rb, Stat3 and CyclinD1 activated the Sp1 promoter, whereas p53 and NFκB decreased Sp1 promoter activity by more than 50%. E2F/DP1 was an activator of Sp1 transcription as previously described.12
Figure 2.
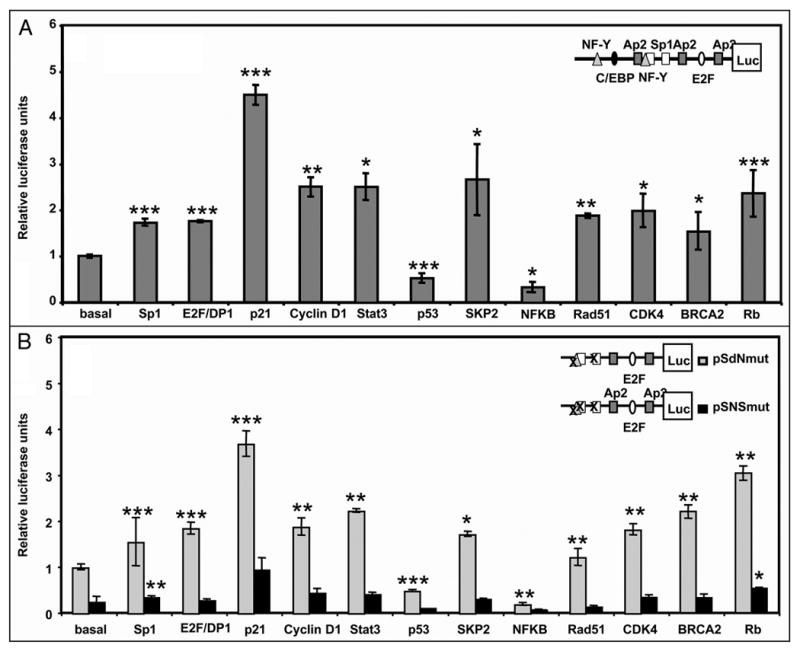
Regulation of Sp1 promoter by overexression of Sp1-interacting proteins. (A) HeLa cells were cotransfected with 250 ng of pGL3FOR2 (scheme at upright A) and 500 ng of the indicated expression vectors. After 30 h, cell lysates were assayed for luciferase activity. Firefly luciferase activity was normalized to the renilla luciferase activity for each sample. Results are expressed relative to the activity obtained upon transfection with pGL3FOR2 alone. (B) HeLa cells were cotransfected with 250 ng of pSdNmut or pSNSmut (scheme at upright B) together with 500 ng of the indicated expression vectors. Results are expressed relative to the activity obtained upon transfection with pSdNmut alone. Other conditions were as in Figure 3A. Results represent the mean ± SE of 3 different experiments. *p < 0.05, **p < 0.01 and ***p < 0.001 Results are expressed relative to the pGL3FOR2 or pSdNmut constructs alone.
To better characterize the effect of Sp1-interacting proteins on Sp1-dependent regulation, we used two Sp1 promoter constructs containing different point mutations: pSdNmut, bearing mutations at the NF-Y site and one of the Sp1 boxes, and pSNSmut, with the NF-Y and both Sp1 boxes mutated (scheme in Fig. 2B). All the Sp1-interacting proteins had the same effect on pSdNmut as on the wild type promoter (Fig. 2B). In contrast, a decrease in promoter activity was observed when the two Sp1 boxes were mutated, demonstrating the relevance of the Sp1 binding site for the effect of the interacting proteins.
Overexpression of IκB and a dominant negative mutant of p53 caused activation of the Sp1 promoter whereas dominant negatives of Rb and Stat3 decreased the activity of the construct (data not shown). The combination of BRCA2 and Rad51 produced a slight further increase in SP1 promoter-luciferase activity compared to the effect caused by overexpression of each protein alone (Fig. 3A).
Figure 3.
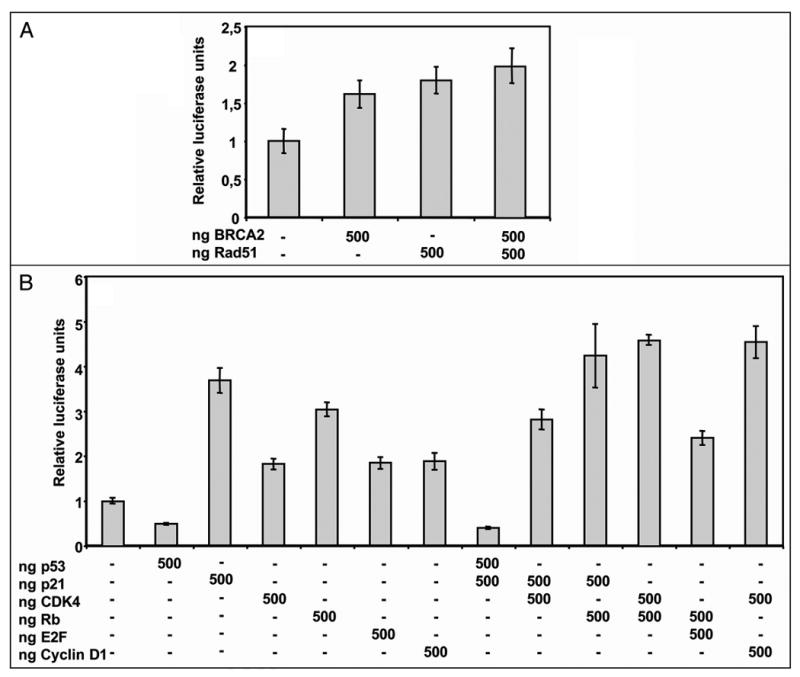
Effect of overexpression of different combinations of Sp1-interacting proteins on Sp1 promoter activity. (A) Combination of the DNA-repair proteins Rad51 and BRCA2. Cells were co-transfected with 250 ng of pSdNmut and the indicated amounts of BRCA2 and/or Rad51. After 30 h, cell lysates were assayed for luciferase activity. Other conditions were as in Figure 2B. (B) Different combinations of cell cycle regulators p53, p21, CDK4, Rb, E2F and cyclinD1. HeLa cells were co-transfected with 250 ng pSdNmut construct and the indicated amounts of the corresponding expression vectors. Other conditions were as in Figure 2B.
Different combinations of cell cycle regulators p53, p21, CDK4, CyclinD1, Rb and E2F1 were also tested for their effect on the Sp1 promoter. As shown in Figure 3B, p53 was able to counteract p21-induced activation of Sp1 promoter. Moreover, the combination of p21 and CDK4 activated Sp1 promoter but to a lower extend than p21 alone. The combination of p21 and Rb caused an increase in Sp1 promoter, to a higher level than p21 or Rb alone. Additionally, the combination of Rb and CDK4 also increased Sp1 promoter activity and this effect was additive compared to the activation caused by either Rb or CDK4 alone. Finally, the combination of Rb with E2F activated Sp1 promoter to an intermediated level, higher than E2F alone but lower than Rb alone.
Regulation of the endogenous Sp1 gene
To study the effect of the interacting proteins on the endogenous levels of Sp1, we performed RT- Real Time PCR to quantify Sp1 mRNA levels in HeLa cells after the overexpression of these proteins. As shown in Figure 4, overexpression of p53 and NFκB decreased Sp1 mRNA levels more than 50%, whereas E2F1, p21, Cyclin D1, Stat3, SKP2, Rad51, CDK4, BRCA2 and Rb, increased the Sp1 mRNA levels significantly, in accordance with the results on Sp1 promoter activity. All the changes were in keeping with the changes in the transcriptional activity of the Sp1 promoter measured in the luciferase assays (Figs. 2 and 3).
Figure 4.
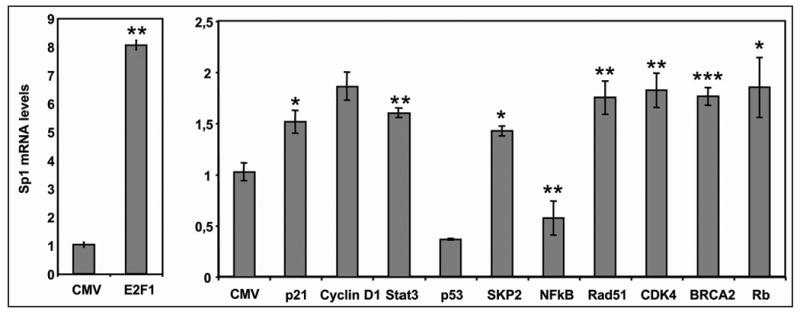
Effect of Sp1-interacting proteins on Sp1 endogenous levels. HeLa cells were transfected with 1 μg of the indicated expression vectors using 3 μl of FUGENE™ 6. After 72 h RNA was extracted and RT-Real Time PCR was performed as described. Results are expressed relative to the Sp1 mRNA levels obtained upon transfection with the empty vector pCMV. Results represent the mean ± SE of 3 different experiments and are expressed relative to the control using a CMV vector without insert. *p < 0.05, **p < 0.01 and ***p < 0.001.
Sp1-interacting proteins are bound to the Sp1 promoter in vivo
To examine whether the effects of the exogenous Sp1-interacting proteins on Sp1 transcription reflected the interaction of the corresponding endogenous proteins with the native Sp1 promoter, we performed ChIP analysis of chromatin from HeLa cells using specific antibodies against different proteins. As shown in Figure 5, amplification of the Sp1 promoter was observed from chromatin immunoprecipitated using antibodies against Sp1, Stat3, SKP2, NFκB, p21, p53, BRCA2, CDK4 and E2F, demonstrating that both Sp1 and the endogenous Sp1-interactive proteins are bound at the Sp1 promoter containing Sp1-binding sites present in the Sp1 promoter. In the Real Time results, a control region was used for normalization.
Figure 5.
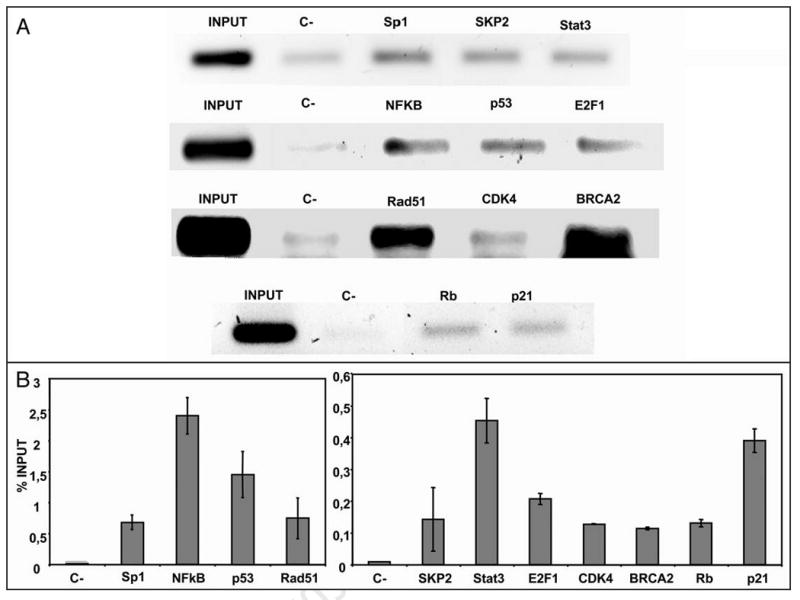
ChIP analysis of the Sp1 promoter. ChIP assays for the Sp1 promoter region were performed using HeLa cells. DNA bound to the immunoprecipitated Sp1, SKP2, Stat3, NFκB, p53, E2F1, Rad51, CDK4, BRCA2, Rb and p21 using specific antibodies, was amplified by PCR. Rabbit IgG was used as negative control (marked as C-). (A) Representative images of the PCR products corresponding to the amplification for each antibody using Sp1- specific primers are shown. (B) Quantification of ChIP analysis was performed by Real Time PCR and results are expressed as percent of the input, taking into account that ΔCt (Sp1) = Ct Sp1 promoter - Ct genomic DNA, ΔΔCt = ΔCt (Sp1) - ΔCt (IgG) and then referred to the input, considering the dilution factor.
Effect of Sp1-interacting proteins on an artificial Sp1-dependent promoter
To test if the effects observed with the Sp1 gene promoter were a general mechanism that could affect other Sp1-regulated promoters, we used a luciferase construct with a promoter that contained five tandem Sp1 binding sites (pG5-5x(GC)-Luc).14 As shown in Figure 6A, p53 and NFκB were able to significantly decrease promoter activity whereas Rb, SKP2, E2F1/DP1, Stat3, BRCA2 and p21 increased promoter activity by 2-fold. CDK4, Rad51, CyclinD1 had no effect on the artificial promoter controlled by Sp1.
Figure 6.
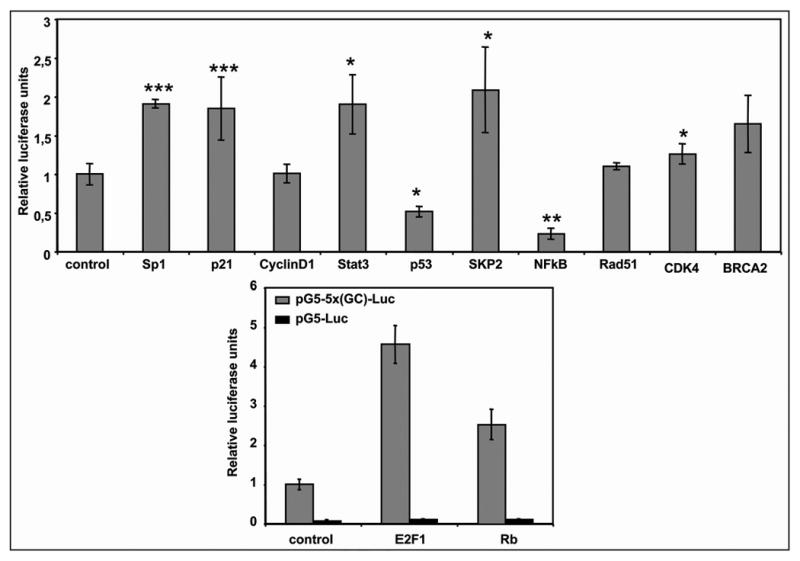
Effect of Sp1-interacting proteins on an artificial promoter containing only Sp1-binding sites. (A) HeLa cells were cotransfected with 250 ng of pG5-5x(GC)-Luc and 500 ng of the indicated expression vectors. (B) HeLa cells were cotransfected with 250 ng of pG5-5x(GC)-Luc or pG5-Luc and 500 ng of the indicated expression vectors. In all cases, after 30 h, cell lysates were assayed for luciferase activity. Firefly luciferase activity was normalized to the renilla luciferase activity for each sample. Results are expressed relative to the activity obtained upon transfection with pG5-5x(GC)-Luc and pCMV. Results represent the mean ± SE of 3 different experiments and are expressed compared to the pG5-5x(GC)-Luc construct cotransfected with 500 ng of an empty CMV vector. *p < 0.05, **p < 0.01 and ***p < 0.001 Results are expressed relative to the pGL3FOR2 or pSdNmut constructs alone.
The effects of E2F1 and Rb overexpression were also analyzed using a construct in which the Sp1 sites were deleted. As shown in Figure 6B, the activity of this construct was almost abolished with the elimination of the Sp1 sites, and the addition of either E2F or Rb did not increase significantly the promoter activity. Altogether these results indicated that the effects of E2F and Rb are due to a direct interaction with Sp1.
Effect of p21 on Sp1-dependent transcription
Given that p21 caused the higher effects on the Sp1 promoter and that p21 expression has been shown to result in multiple changes in gene expression in human HT1080 fibrosarcoma and other cell lines,15,16 we further characterized the effect of p21 on Sp1. Transient transfection used in the other experiments leads to very high and often supraphysiological expression levels of the transfected protein, and therefore we used HT1080 p21-9 cells with stably integrated p21 which is inducible by IPTG (isopropyl-β-thio-galactoside) to the levels that are similar to those observed in response to DNA damage or at the onset of senescence.15 First, we confirmed by coimmunoprecipitation that the interaction between Sp1 and p21 takes place in HT1080 p21-9 cells (Fig. 7A). Then, we transfected HT1080 p21-9 with the Sp1 promoter construct pSdNmut. p21 induction by IPTG increased Sp1 promoter activity in a dose-dependent manner (Fig. 7B), thus confirming our previous results with transiently transfected p21. Moreover, endogenous Sp1 mRNA levels increased up to 2-fold after 72 h of IPTG addition (Fig. 7C). Finally, the artificial promoter controlled exclusively by Sp1 was activated by 2-fold after IPTG addition (Fig. 7D).
Figure 7.
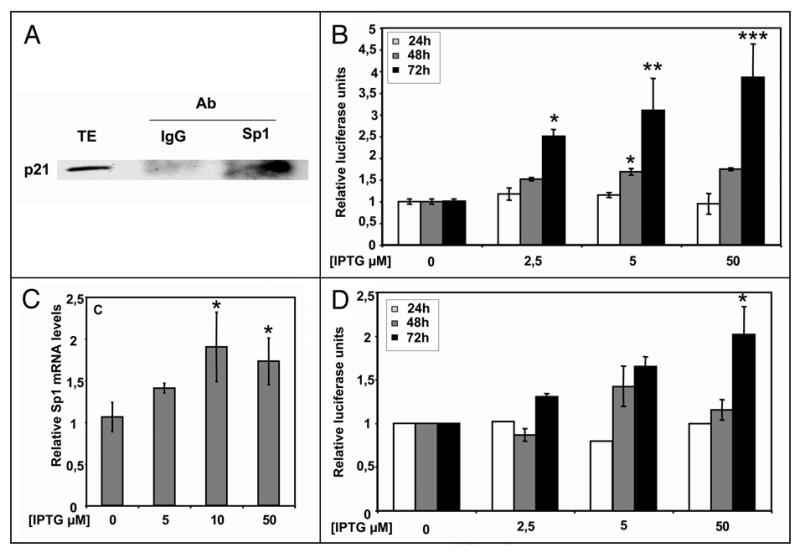
Effect of inducible p21 on Sp1-dependent function and expression. (A) Coimmunoprecipitation of p21 with Sp1. Immunoprecipitates were obtained from 1 mg of total cell extracts from HT1080 p21-9 treated with 50 μM IPTG, by incubation with 5 μg of the indicated specific antibodies. After washing, the bound proteins were resolubilized and p21 protein was detected by Western analysis. The first lane (TE) shows the signal corresponding to p21 from 50 μg of induced HT1080 p21-9 cells total extract. (B) Effect of p21 induction on Sp1 promoter. HT1080 p21-9 cells were cotransfected with 500 ng of pSdN construct before induction with increasing amounts of IPTG. At indicated times, cells were assayed for luciferase activity. Firefly luciferase activity was normalized to renilla luciferase activity as described in Materials and Methods. Results are expressed relative to the activity obtained upon transfection with pSdN without IPTG addition. (C) Effect of p21 induction on Sp1 mRNA levels. HT1080 p21-9 cells were treated with the indicated amounts of IPTG. After 72 h RNA was extracted and RT-Real Time PCR was performed as described. Results are expressed relative to the Sp1 mRNA levels obtained without adding IPTG. (D) Effect of p21 induction on a promoter containing only Sp1-binding sites. HT1080 p21-9 cells were transfected with the pG5-5x(GC)-Luc vector and IPTG was added to the medium at the concentrations indicated. Results are expressed relative to the activity obtained upon transfection with pG5-5x(GC)-Luc without IPTG addition. Results represent the mean ± SE of 3 different experiments. *p < 0.05, **p < 0.01 and ***p < 0.001 compared to the pSp3-FOR4 construct alone.
To confirm Sp1 induction by p21 as observed at the RNA level through protein analysis, Sp1 protein levels were analyzed after IPTG addition. Surprisingly, p21 induction caused a decrease rather than an increase in Sp1 protein levels (Fig. 8A). To determine whether this effect on Sp1 protein levels was due to an increase in Sp1 degradation we tested the effect of the calpain inhibitor E64d on p21-induced Sp1 depletion. Incubation of p21-9 induced cells with E64d for 24 hours not only prevented the decrease in the Sp1 protein but also increased its levels up to 190% of the control (Fig. 8C). This result is consistent with previously observed induction of Sp1 mRNA by p21 and suggests that Sp1 depletion was due to protein degradation. Incubation of p21-9 cells with E64d for longer periods of time also caused the inhibition of p21 degradation and its accumulation (data not shown). Incubation of HT1080 p21-9 cells with IPTG in the presence of E64d for 24 hours after transfection with either Sp1 promoter or Sp1-dependent promoter constructs caused an increase in the activity of both constructs, as compared to control cells or to cells incubated with IPTG alone (Fig. 8D).
Figure 8.
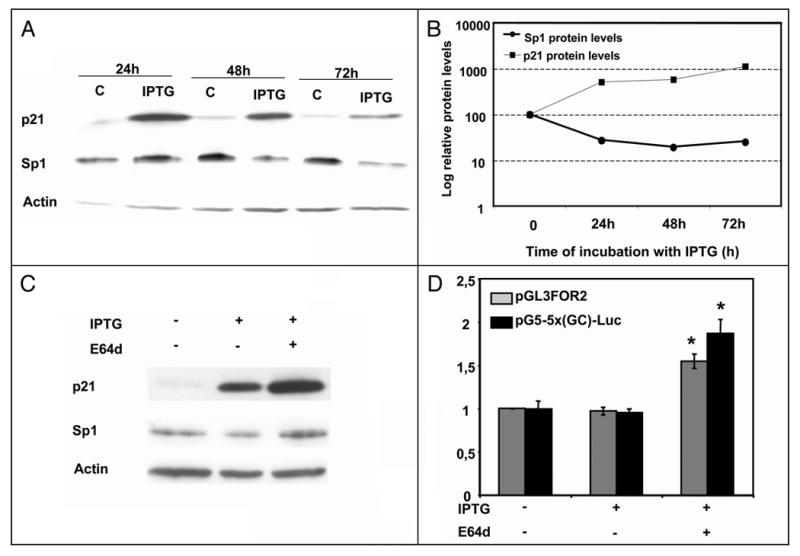
Effect of p21 induction on Sp1 protein levels. (A) Sp1, p21 and Actin protein levels were determined by Western Blot in HT1080 p21-9 cells after the addition of 50 μM IPTG at different times. (B) Quantification of the results presented in (A). Quantification was performed using ImageQuant software (Molecular Dynamics). The representation is in a semilogarithmic scale. (C) Immunoblotting of Sp1, p21 and Actin in HT1080 p21-9 cells treated for 24 hours with 50 μM IPTG and 3 μg/ml of the inhibitor E64d (Sigma). (D) Effect of E64d on Sp1 and Sp1-regulated promoters. HT1080 p21-9 cells were cotransfected with 250 ng of pGL3FOR2 or pG5-5x(GC)-Luc constructs before the addition of 50 μM of IPTG and 3 μg/ml of E64d. Results represent the mean ± SE of 3 different experiments. *p < 0.05.
Discussion
Protein-protein interactions among sequence-specific DNA-binding transcription factors suggest novel modes of regulation. One DNA-bound transcription factor can recruit a second factor to the promoter where binding sites for the latter are absent. The aim of this work was to identify new Sp1-interacting proteins and to study the regulation of the Sp1 promoter by such proteins. Interactions between Sp1 and cell cycle regulators have been well documented and some of the proteins that interact with Sp1 are able to bind among themselves, suggesting that Sp1 could be part of different protein complexes. The results of the present study expand the network of SP1 interactions and suggest new links of SP1 to cell cycle controlling proteins (Fig. 9).
Figure 9.
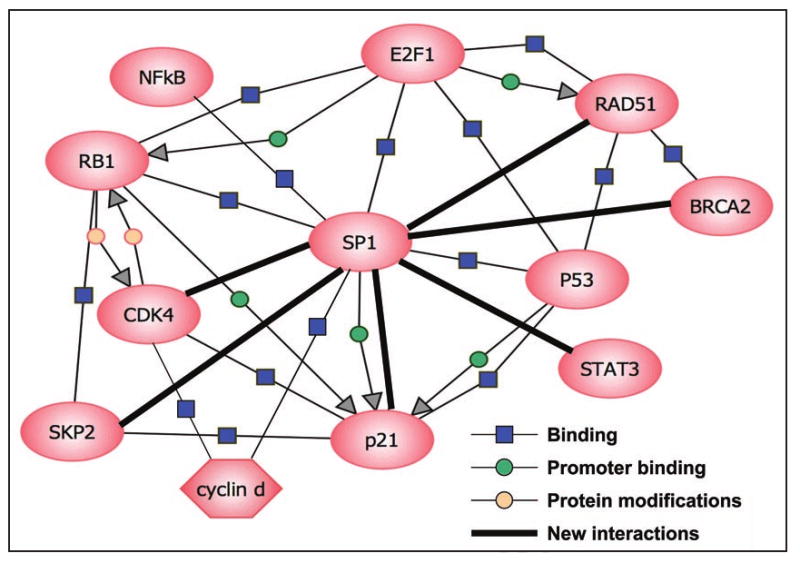
Network diagram of Sp1-interacting proteins. Different Sp1-interacting proteins are presented. This interaction network was created with the aid of software Pathway Architect 2.01 (Stratagene), and represents both the previously known and the newly discovered binding associations among all of these proteins.
We previously described the interaction between Sp1 and the retinoblastoma gene product Rb.5 Rb is a target of the cyclin/CDK complexes, mainly CDK4/cyclinD, and its function depends, at least in part, on the interactions with the E2F family of DNA-binding transcription factors.17 E2F1 and CDK2 also interact with Sp1 forming stable complexes.5,10,18 In the present work we demonstrate that Sp1 is able to bind to CDK4 and a CDK inhibitor p21. Overexpression of all these cell cycle regulators (CDK4, Rb, cyclinD1, E2F and p21) increased Sp1 promoter activity and Sp1 mRNA levels. Although it could had been expected that p21 decreased CDK4 activity, the combined overexpression of p21 and CDK4 results in a higher transcription than in CDK4 alone, although lower than with p21. This is in agreement with the fact that p21 plays a permissive role in cell cycle progression as a consequence of its ability to cooperate with CDK4, and to function as an assembly factor for CyclinD-CDK4 complex, as reported in.19-21 One might expect that the combination of Rb and CDK4 would lead to a change in the phosphorylation state of Rb. However, since the interaction between Sp1 and Rb is independent of Rb phosphorylation state,5 the combination of Rb and CDK4 produces an additive effect in Sp1 transcription. In keeping with this observation and taking into account that p21 activates Rb dephosphorylation,22 the combination of p21 and Rb caused the activation of the Sp1 promoter. In the case of combining E2F and Rb, the resulting effect on Sp1 transcription is mainly due to Rb, present in its hypophosphorylated form, which is able to repress E2F, either by sequestering this factor or by turning E2F into an active repressor.17 E2F and Rb were able to activate the artificial promoter containing only Sp1 sites demonstrating that their effect on the Sp1 gene promoter was mediated directly by Sp1-interaction. On the contrary, overexpression of Cyclin D1 and CDK4 had no effect on this promoter suggesting that they might need the presence of additional factors rather than Sp1 alone to activate Sp1 transcription.
Another complex interacting with Sp1 is SCFskp2 that plays an important role in the ubiquitin-mediated degradation of some cell cycle proteins such as p27,23 and some transcription factors such as E2F1.24 The SCFskp2 complex is composed of four subunits: cullin, SKP1, RBX1 and the F-box protein SKP2.25 The first three proteins form a common scaffold onto which different F-box proteins can be assembled, conferring specificity to the complex. Cdc34 is also included in this complex and enables transfer of the ubiquitin molecule to specific targets.26 Cdc34, p19Skp1, RbX1&2 and p45SKP2 are present in the antibody array and gave positive signals for Sp1 interaction. Given that SKP2 is the F-box of the complex and recognizes the substrate, we validated its interaction with Sp1 by co-immunoprecipitation. Different studies have suggested a close link between the activator function of many transcription factors and their ubiquitylation/degradation.27 Our data demonstrate that SKP2, instead of degrading Sp1, activates the Sp1 promoter and increases Sp1 mRNA levels and protein levels up to 2-fold (data not shown). This effect has also been observed for the interaction between c-myc and SKP2.28 It is conceivable that Sp1 could be recruiting SCFSKP2 to the promoter to degrade negative regulators of transcription at the promoter level. This hypothesis is supported by the results of the ChIP assays that were positive for SKP2 at the Sp1 promoter level and with the result that SKP2 also activates the artificial promoter containing only Sp1 boxes.
BRCAs are tumor suppressor genes involved in multiple pivotal cellular processes. These proteins contribute to DNA repair, transcriptional regulation in response to DNA damage and maintenance of chromosomal stability, thereby protecting the genome from damage. Many of these functions are mediated by a large number of cellular proteins that interact with BRCAs.29 The interaction between BRCA1 and Sp1 has been described for the regulation of IGF-IR gene expression.30 Here we show that Sp1 is able to bind to BRCA2 in addition to BRCA1 and that this interaction increases Sp1 promoter activity and Sp1 mRNA endogenous levels. Moreover, BRCA2 is able to activate a promoter driven only by Sp1 indicating that BRCA2 interacts with Sp1 and that additional factors present in the Sp1 promoter are not necessary for the resulting activation. Rad51 and RBBP are additional components of the repair machinery present in the antibody array that originated positive spots for Sp1 interaction. The interaction of Sp1 with Rad51 was also confirmed by co-immunoprecipitation. Rad51 was able to activate Sp1 transcription at the promoter and mRNA levels. However, Rad51 did not affect Sp1-dependent activation of the promoter containing only Sp1 binding sites. Combination of BRCA2 and Rad51 increased Sp1 promoter activity to a moderate degree, suggesting that they are forming a part of a complex in which more proteins are needed for the maximum increase in Sp1 transcription. The interaction between BRCA2 and Rad51 has been reported31 and there is genetic evidence that it is fundamental for the maintenance of cell division and chromosome structure.29
Sp1 interactions with other proteins also involve several transcription factors: Stat1, Stat3, p53 and NFκB. Tumor suppressor p53 is involved in transcriptional activation of important genes, such as p21, in the regulation of cell cycle and apoptosis32 and it is involved both in DNA damage and in cell cycle control. In our experiments, p53 overexpression decreased the activity of the Sp1 promoter. Transcriptional repression of other genes by p53 revealed that Sp1 was prevented from binding to the promoter region by a p53-Sp1 protein complex.33-35 Our results are in keeping with these observations; p53 counteracted the positive effect of p21 on Sp1 transcription, probably because Sp1 is prevented by p53 from binding to its binding site. NFκB is a nuclear effector in a signaling pathway responsive to a large number of extracellular stimuli in many cells.36 NFκB is capable of participating in the transcriptional regulation of target genes, independently of its DNA binding site, through cooperation with other transcription factors. NFκB and Sp1 bind cooperatively and activate transcription of the human immunodeficiency virus synergistically.37 Additionally, NFκB has been described as a transcriptional inhibitor for the gluconeogenic enzyme PEPCK by sequestration of a co-activator protein such as CBP.38 Accordingly, in our model NFκB behaved as a repressor of Sp1-dependent activation of both the Sp1 promoter and the artificial promoter.
Stats are a family of transcription factors involved in ligand-dependent growth stimulation or differentiation as well as in antiproliferative effects.39,40 Both Stat1 and Stat3 have been reported to interact with Sp1.41,42 Stat3 activates transcription of VEGF through its interaction with a Sp1/DNA complex and not by its direct binding to a palindromic enhancer element.42 In agreement with these observations, in our conditions, Stat3 is an activator of both Sp1 promoter and the artificial promoter, and it binds to the Sp1 promoter.
p21 was studied in further detail as it was able to strongly activate the Sp1 promoter and there are several reports showing that p21 affects transcription of many different genes.15,43,44 We tested the effects of p21 on Sp1 promoter and Sp1 mRNA levels in HT1080 p21-9 cells, which carry a stably integrated p21 transgene inducible by IPTG addition to physiologically meaningful levels. We confirmed that p21 was able to activate transcription form the Sp1 promoter and to increase Sp1 mRNA expression in this system, as previously observed in HeLa cells transiently transfected with p21. The activation of Sp1 at the promoter and mRNA levels after p21 induction is in accordance with previous observations using CDK inhibitors.45 This effect could be caused by dephosphorylation of Sp1 at concrete sites that could increase its transcriptional activity directly or increase the degradation of its inhibitory domain.8 An increase in Sp1 transcriptional activity would increase Sp1 transcription as Sp1 is positively autoregulated.
To further characterize the effects of p21 on Sp1, we analyzed the effect of p21 on Sp1 protein levels by Western blot. Surprisingly, p21 induction did not increase but rather reduced Sp1 protein levels by 80%. The decrease in Sp1 protein levels observed after 24 hours of p21 induction was reverted by the protease inhibitor E64d, indicating that p21 induced Sp1 protein degradation. Both Sp1 full length and the cleaved form were able to activate either the Sp1 promoter or an Sp1-dependent promoter construct as determined in luciferase assays in HT1080 p21-9 cells incubated with IPTG in the absence or in the presence of E64d.
The effect of p21 on Sp1 protein levels was in accordance with Sp1 degradation in aged animal tissues as well as in cells undergoing replicative senescence,3,46,47 where p21 induction plays a pivotal role.22 p21 was previously found to promote the degradation of key cell cycle regulatory proteins, including p53,48 and Rb;49 the latter protein can be degraded through the same pathway as Sp1.50 Aside from recruiting Sp1 into this proteolytic pathway, p21, as mentioned above, could potentially cause dephosphorylation of Sp1 increasing the degradation of its inhibitory domain, converting Sp1 into a more unstable protein.8 The dual effect of p21 on Sp is similar to its effect on Rb, where p21 activates Rb by dephosphorylation and concomitantly inactivates it by triggering its degradation.49
In summary, Sp1 function and transcription are regulated by different cell cycle related proteins identified by their interaction with Sp1 protein. Multiple effects of cell cycle regulators on Sp1, observed in the present study, suggest that Sp1 may be a key mediator of cell cycle associated changes in gene expression.
Materials and Methods
Plasmid constructs
pGL3FOR2, containing the minimal promoter of Sp1, and pSdNmut and pSNSmut, containing point mutations for the Sp1 and NF-Y boxes on the Sp1 promoter, have been previously described in detail11 and in a scheme presented in Figure 2.
pG5-5x(GC)-Luc contains the luciferase gene controlled by five tandem Sp binding sites and was a generous gift from Dr. Man-Wook Hur. pG5-Luc was engineered by deletion of the five tandem Sp binding sites.
The expression vectors for the different proteins used in this study were obtained from the following investigators: Sp1 (Dr. R. Tjian), E2F1 and DP1 (Dr. T. Kouzarides), pCMV-Cip1 (p21) (Dr. S. Elledge), Stat3 (Dr. J.C. Lacal), PC53-SN3 (Dr. B. Vogelstein), p45SKP2 (Dr. C. Serra-Pagès), Rad51 (Dr. M. Defais), BRCA2 (Dr. J. Bueren), NFκB (Dr. C. Caelles), CyclinD1 (Dr. N. Agell), CDK4 (Dr. M. Barbacid) and Rb (Dr. W.G. Kaelin).
Cell culture
HeLa human cervical carcinoma cells were grown in Ham's F-12 medium supplemented with 5% foetal bovine serum (Invitrogen). p21-9 cells were grown in DMEM/GlutaMAX supplemented with 10% foetal calf serum (Invitrogen). Cultures were maintained at 37°C in a humidified 5% CO2-containing atmosphere. The derivation, maintenance and p21 induction by IPTG in HT1080 p21-9 cells has been previously described.13
Screening of the antibodyarray
HeLa total extracts were prepared in extraction solution at 4°C (120 mm NaCl, 25 mm KCl, 2 mm EGTA, 1 mm EDTA, 0.1 mm DTT, 15 mm Tris-HCl, pH 7.5, 0.5% Triton X-100, 10 mg/mL leupeptin, 0.5 mm phenylmethylsulfonyl fluoride). The debris was removed by centrifugation (10,000 g, 15 min).
HeLa total extracts were diluted at a concentration of 2 mg/ml in extraction solution containing 1% dry milk. Subsequently, the total extract was incubated with the AntibodyArray™ (Cell Cycle array, Hypromatrix) containing 60 antibodies, as indicated by the manufacturer (see list at http://www.hypromatrix.com). Briefly, the AntibodyArray™ was blocked with 5% dry milk in TBST (150 mm NaCl, 25 mm Tris-HCl, 0.05% Tween-20, pH 7.5) for 1h at room temperature. Then, the membrane was incubated for 2 h at room temperature with the total extract, and washed in TBST (3 × 15 min). The membrane was incubated with biotinylated anti-Sp1 antibody (PEP 2, Santa Cruz) and then with streptavidin peroxidase. Binding of the Sp1 antibody was detected by enhanced chemiluminiscence, as recommended by the manufacturer (GE Healthcare).
Co-immunoprecipitations
HeLa total extracts were prepared by incubation in extraction solution for 30 minutes on ice (50 mM Tris-HCl pH 8, 50 mM KCl, 5% Glicerol, 0.1 mM EDTA, 0.5% NP-40, 1 mM PMSF, 10 mg/ml Leupeptin). Cell debris was removed by centrifugation (10,000 g, 10 min). In the case of p21 immunoprecipitation, 5 × 105 HeLa cells were transfected with 5 μg of p21 expression vector using FUGENE™ 6 (Roche Applied Science) 48 h before preparation of the extracts. Total extracts from HeLa cells were immunoprecipiated using specific antibodies (Santa Cruz) for Sp1 (PEP 2), BRCA2 (H-300), SKP2 (H-435), Rad51 (H-92), CDK4 (H-22), p21 (C-19) or unspecific IgGs (Sigma) with the aid of Protein G-Sepharose (GE Healthcare). Then, the pellets were washed three times in extraction solution, resolubilized in loading buffer, boiled for 10 minutes and subjected to Western blot analysis using anti-Sp1 antibody (PEP 2, Santa Cruz) as described in.5
For the immunoprecipitation using HT1080 p21-9 cells, cells were treated with 50 μM IPTG and 24 hours later, total extracts were prepared as described above. These cell extracts were immunoprecipitated using specific antibodies for Sp1 (PEP 2) or IgGs as described above and subjected to Western blot analysis using anti-p21 antibody (C-19, Santa Cruz).
Cotransfections and luciferase assays
HeLa cells were seeded into 6-well plates the day before transfection at a density of 2 × 105 cells/well in Ham's F12 medium containing 5% foetal bovine serum. The medium was renewed before transfection, which was performed using FUGENE™ 6 (Roche Applied Science). For each well, 3 μl of FUGENE™ 6 in 100 μl of serum-free F-12 medium was incubated at room temperature for 5 min. The mixture was added to the vectors used. In cotransfections, 250 ng of the constructs pGL3FOR2, pSdNmut or pSNSmut, described in11 or pG5-5x(GC)-Luc or pG5-Luc, were mixed with the indicated amounts of Sp1, E2F, DP1, p21, CyclinD1, Stat3, p53, SKP2, NFκB, p21, CDK4, BRCA2 and Rb expression vectors before the addition of FUGENE™ 6 in serum-free F-12 medium.
Luciferase activity was assayed 30 h after transfection. Cell extracts were prepared by lysing the cells with 200 μl of freshly diluted 1× Reporter Lysis Buffer (Promega). The lysate was centrifuged at 12,000 g for 2 min to pellet the cell debris. The supernatants were transferred to a fresh tube and a 30 μl-aliquot of the extract was added to 30 μl of the luciferase assay substrate (Promega). The luminiscence of the samples was read after 10 min on a Glomax™ 20/20 Luminometer, in which the light production (relative luminiscence units) was measured for 10 seconds. Each transfection was performed in triplicate.
Transfection efficiency was corrected by by normalizing the firefly luciferase activity expressed from tested promoter by the control renilla luciferase resulting from cotransfection with the pCMV renilla luciferase vector. 30 μl of the Stop & Glo solution were added to the luciferase mix after reading luciferase activity and were incubated for 10 min before reading the luminescence. Alternatively, when overexpression of the plasmid increased the renilla activity, luciferase results were corrected by protein concentration using the Bio-Rad protein assay reagent according to the manufacturer's protocol.
For luciferase assays with HT1080 p21-9 cells, cells were seeded into 6-well plates the day before transfection at a density of 2 × 105 cells/well in DMEM/Glutamax medium containing 10% foetal calf serum. The medium was renewed before transfection, which was performed using FUGENE™ 6 (Roche Applied Science). For each well, 3 μl of FUGENE™ 6 in 100 μl of serum-free F12 medium was incubated at room temperature for 5 min. The mixture was added to the vectors used. 24 hours after transfection, cells were split and IPTG was added to the medium at the indicated concentrations in one of the two wells corresponding to one transfection. Cells were collected at the indicated times and luciferase activity was measured using the Dual Luciferase Reporter Kit (Promega) according to the manufacturer's instructions. Renilla activity determined for cells in the absence of IPTG was used to normalize the results. For the E64d treatments, 3 μg/ml of E64d were added to the medium together with IPTG and luciferase was measured after 24 hours.
ChIP analysis
Formaldehyde cross-linking and ChIP were performed as described in.51,52 Briefly, HeLa cells (semiconfluent 100 mm dish) were washed with phosphate-buffered saline and incubated for 10 minutes with 1% formaldehyde. The reaction was quenched with 0.1 M glycine and cells were sonicated to obtain chromatin fragments of an average length of 500 to 800 bp. Immunoprecipitation was performed with protein G-Sepharose (GE Healthcare) and 5 μg of antibodies (Santa Cruz) specific for Sp1 (PEP 2), BRCA2 (H-300), SKP2 (H-435), Rad51 (H-92), CDK4 (H-22), Stat3 (H-190), E2F-1 (C-20), p53 (FL-393) and NFκB p65 (A) or 10 μg of antibodies Rb (C-15) and p21 (C-19). Non-specific IgG (Sigma) was used as a negative control. The chromatin solution was pre-cleared by incubation with protein G-sepharose for 2 h at 4°C, divided into aliquots, and incubated overnight with different antibodies. Before use, protein G-Sepharose was blocked with herring sperm DNA (1 μg/μl) (120–3000 nucleotides length) and BSA (1 μg/μl) for 2 h and then overnight, all at 4°C.
Sp1 promoter was amplified by PCR using the following specific primers:
Fwd 5′- GCAAGCGAGTCTTGCCATTGG -3′
Rev 5′- CGCTCATGGTGGCAGCTGAGG -3′
PCR was performed in a final volume of 30 μl for at least 28 cycles. Then, 10 μl samples of the PCR products were electrophoresed in a 2% agarose gel and the amplified fragments were visualized after EtBr staining.
For quantification, amplification was performed by Real Time-PCR in an ABI Prism 7000 Sequence Detection System (Applied Biosystems) with 2 μl of immunoprecipitated DNA and the primers indicated above using the Fast SYBR Green Master Mix (Applied Biosystems). The reaction was performed following the manufacturers recommendations. The percent of input was calculated by the standard ΔΔCt method. As a negative control, the following primers were also used:
Fwd: 5′-ATGGTTGCCACTGGGGATCT-3′
Rev: 5′-TGCCAAAGCCTAGGGGAAGA-3′
These negative control primers flank a region of genomic DNA between the GAPDH gene and the chromosome condensation-related SMC-associated protein (CNAP1) gene.
Sp1 mRNA endogenous levels
1 μg of expression vectors for E2F, DP1, p21, CyclinD1, Stat3, p53, SKP2, NFκB, p21, CDK4, BRCA2 and Rb was transfected using Fugene 6.
Total RNA was extracted from HeLa cells using the Ultraspec™ RNA reagent (Biotecx) in accordance with the manufacturer's instructions.
cDNA was synthesized in a total volume of 20 μl from RNA samples by mixing 1 μg of total RNA, 125 ng of random hexamers (Roche Molecular Biochemicals), in the presence of 75 mM KCl, 3 mM MgCl2,10 mM dithiothreitol, 20 units of RNAsin (Promega), 0.5 mM dNTPs (AppliChem), 200 units of M-MLV reverse transcriptase (Invitrogen) and 50 mM Tris-HCl buffer, pH 8.3. The reaction mixture was incubated at 37°C for 60 min. The cDNA product was used for subsequent amplification by Real Time-PCR.
Sp1 mRNA levels were determined in an ABI Prism 7000 Sequence Detection System (Applied Biosystems) using 3 μl of the cDNA mixture and the Assays-on-demand Hs00412720_m1 for Sp1 and Hs99999901_s1 for 18S RNA (both from Applied Biosystems). 18S RNA was used as an endogenous control. The reaction was performed following the manufacturers recommendations. Fold-changes in gene expression were calculated using the standard ΔΔCt method.
Sp1 protein levels measurement
HT1080 p21-9 total extracts were prepared in extraction solution for 1 hour at 4°C (50 mM Hepes, 0.5 M NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% Glicerol (V/V), 1% Triton X-100, Protease inhibitor cocktail). Cell debris was removed by centrifugation (10,000 g, 10 min).
Cells extracts were resolved on SDS-7% or 12% polyacrylamide gels,53 and transferred to a PVDF membrane (Immobilon P, Millipore) using a semidry electroblotter. The membranes were probed with antibodies against Sp1, p21 (both from Santa Cruz) or Actin (Sigma). Signals were detected by secondary horseradish peroxidase-conjugated antibody (1:2500 dilution) and enhanced chemiluminiscence, as recommended by the manufacturer (GE Healthcare). Quantification of the bands was performed using the ImageQuant software (Molecular Dynamics).
Statistical analysis
Data are presented as mean ± SEM. Statistical analysis was performed using Student's t test using SPSS 13 software for Macintosh. p values of less than 0.05 were considered statistically significant.
Acknowledgments
This work was supported by grants SAF05-0247 and SAF08-00043 from “Plan Nacional de Investigación Científica” and ISCIII-RETIC RD06/0020 (Spain) (to C.J.C. and V.N), and grants R01 AG17921 and R01 AG028687 from the U.S. National Institute of Aging (to I.B.R.). The Barcelona research group holds the Quality mention from the “Generalitat de Catalunya” (SGR0883). A.T. is the recipient of a predoctoral fellowship from the University of Barcelona.
Abbreviations
- CDK
cyclin dependent kinase
- ChIP
chromatin immunoprecipitation
- dNTPs
deoxynucleosides triphosphate
- DTT
dithiothreitol
- EDTA
ethylene diamine tetraacetic acid
- EGTA
ethylene glycol tetraacetic acid
- EtBr
ethidium bromide
- IgG
immunoglobulin G
- IPTG
isopropyl-beta-D-thiogalactopyranoside
- SCF
Skp, cullin, F-box
- PMSF
phenylmethanesulphonylfluoride
- SEM
standard error of the mean
- TBST
tris buffered saline tween
References
- 1.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–48. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 4.Li L, He S, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochem Cell Biol. 2004;82:460–71. doi: 10.1139/o04-045. [DOI] [PubMed] [Google Scholar]
- 5.Noe V, Alemany C, Chasin LA, Ciudad CJ. Retinoblastoma protein associates with SP1 and activates the hamster dihydrofolate reductase promoter. Oncogene. 1998;16:1931–8. doi: 10.1038/sj.onc.1201718. [DOI] [PubMed] [Google Scholar]
- 6.Slansky JE, Farnham PJ. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 7.Koutsodontis G, Vasilaki E, Chou WC, Papakosta P, Kardassis D. Physical and functional interactions between members of the tumour suppressor p53 and the Sp families of transcription factors: importance for the regulation of genes involved in cell cycle arrest and apoptosis. Biochem J. 2005;389:443–55. doi: 10.1042/BJ20041980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spengler ML, Brattain MG. Sumoylation inhibits cleavage of Sp1 N-terminal negative regulatory domain and inhibits Sp1-dependent transcription. J Biol Chem. 2006;281:5567–74. doi: 10.1074/jbc.M600035200. [DOI] [PubMed] [Google Scholar]
- 9.Chu S, Ferro TJ. Sp1: regulation of gene expression by phosphorylation. Gene. 2005;348:1–11. doi: 10.1016/j.gene.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Banchio C, Schang LM, Vance DE. Phosphorylation of Sp1 by cyclin-dependent kinase 2 modulates the role of Sp1 in CTP: phosphocholine cytidylyltransferase alpha regulation during the S phase of the cell cycle. J Biol Chem. 2004;279:40220–6. doi: 10.1074/jbc.M406468200. [DOI] [PubMed] [Google Scholar]
- 11.Nicolas M, Noe V, Jensen KB, Ciudad CJ. Cloning and characterization of the 5′-flanking region of the human transcription factor Sp1 gene. J Biol Chem. 2001;276:22126–32. doi: 10.1074/jbc.M010740200. [DOI] [PubMed] [Google Scholar]
- 12.Nicolas M, Noe V, Ciudad CJ. Transcriptional regulation of the human Sp1 gene promoter by the specificity protein (Sp) family members nuclear factor Y (NF-Y) and E2F. Biochem J. 2003;371:265–75. doi: 10.1042/BJ20021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang BD, Xuan Y, Broude EV, Zhu H, Schott B, Fang J, Roninson IB. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene. 1999;18:4808–18. doi: 10.1038/sj.onc.1203078. [DOI] [PubMed] [Google Scholar]
- 14.Lee JA, Suh DC, Kang JE, Kim MH, Park H, Lee MN, Kim JM, Jeon BN, Roh HE, Yu MY, Choi KY, Kim KY, Hur MW. Transcriptional activity of Sp1 is regulated by molecular interactions between the zinc finger DNA binding domain and the inhibitory domain with corepressors, and this interaction is modulated by MEK. J Biol Chem. 2005;280:28061–71. doi: 10.1074/jbc.M414134200. [DOI] [PubMed] [Google Scholar]
- 15.Chang BD, Watanabe K, Broude EV, Fang J, Poole JC, Kalinichenko TV, Roninson IB. Effects of p21Waf1/Cip1/Sdi1 on cellular gene expression: implications for carcinogenesis, senescence and age-related diseases. Proc Natl Acad Sci USA. 2000;97:4291–6. doi: 10.1073/pnas.97.8.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roninson IB. Tumor cell senescence in cancer treatment. Cancer Res. 2003;63:2705–15. [PubMed] [Google Scholar]
- 17.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 18.Rotheneder H, Geymayer S, Haidweger E. Transcription factors of the Sp1 family: interaction with E2F and regulation of the murine thymidine kinase promoter. J Mol Biol. 1999;293:1005–15. doi: 10.1006/jmbi.1999.3213. [DOI] [PubMed] [Google Scholar]
- 19.Cheng M, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM, Sherr CJ. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. Embo J. 1999;18:1571–83. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Saha P, Kornbluth S, Dynlacht BD, Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Mol Cell Biol. 1996;16:4673–82. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–62. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 22.Roninson IB. Oncogenic functions of tumour suppressor p21(Waf1/Cip1/Sdi1): association with cell senescence and tumour-promoting activities of stromal fibroblasts. Cancer Lett. 2002;179:1–14. doi: 10.1016/s0304-3835(01)00847-3. [DOI] [PubMed] [Google Scholar]
- 23.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–9. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 24.Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol. 1999;1:14–9. doi: 10.1038/8984. [DOI] [PubMed] [Google Scholar]
- 25.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–78. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 26.Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 1998;14:236–43. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 27.Conaway RC, Brower CS, Conaway JW. Emerging roles of ubiquitin in transcription regulation. Science. 2002;296:1254–8. doi: 10.1126/science.1067466. [DOI] [PubMed] [Google Scholar]
- 28.von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, Cetinkaya C, Hydbring P, Weidung I, Nakayama K, Nakayama KI, Soderberg O, Kerppola TK, Larsson LG. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida K, Miki Y. Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription and cell cycle in response to DNA damage. Cancer Sci. 2004;95:866–71. doi: 10.1111/j.1349-7006.2004.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abramovitch S, Glaser T, Ouchi T, Werner H. BRCA1-Sp1 interactions in transcriptional regulation of the IGF-IR gene. FEBS Lett. 2003;541:149–54. doi: 10.1016/s0014-5793(03)00315-6. [DOI] [PubMed] [Google Scholar]
- 31.Marmorstein LY, Ouchi T, Aaronson SA. The BRCA2 gene product functionally interacts with p53 and RAD51. Proc Natl Acad Sci USA. 1998;95:13869–74. doi: 10.1073/pnas.95.23.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–31. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 33.Yamabe Y, Shimamoto A, Goto M, Yokota J, Sugawara M, Furuichi Y. Sp1-mediated transcription of the Werner helicase gene is modulated by Rb and p53. Mol Cell Biol. 1998;18:6191–200. doi: 10.1128/mcb.18.11.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borellini F, Glazer RI. Induction of Sp1-p53 DNA-binding heterocomplexes during granulocyte/macrophage colony-stimulating factor-dependent proliferation in human erythroleukemia cell line TF-1. J Biol Chem. 1993;268:7923–8. [PubMed] [Google Scholar]
- 35.Ohlsson C, Kley N, Werner H, LeRoith D. p53 regulates insulin-like growth factor-I (IGF-I) receptor expression and IGF-I-induced tyrosine phosphorylation in an osteosarcoma cell line: interaction between p53 and Sp1. Endocrinology. 1998;139:1101–7. doi: 10.1210/endo.139.3.5832. [DOI] [PubMed] [Google Scholar]
- 36.Gerondakis S, Grumont R, Gugasyan R, Wong L, Isomura I, Ho W, Banerjee A. Unravelling the complexities of the NFkappaB signalling pathway using mouse knockout and transgenic models. Oncogene. 2006;25:6781–99. doi: 10.1038/sj.onc.1209944. [DOI] [PubMed] [Google Scholar]
- 37.Perkins ND, Agranoff AB, Pascal E, Nabel GJ. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol Cell Biol. 1994;14:6570–83. doi: 10.1128/mcb.14.10.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao W. Advances in NFkappaB signaling transduction and transcription. Cell Mol Immunol. 2004;1:425–35. [PubMed] [Google Scholar]
- 39.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 40.Schindler C. STATs as activators of apoptosis. Trends Cell Biol. 1998;8:97–8. doi: 10.1016/s0962-8924(98)01233-1. [DOI] [PubMed] [Google Scholar]
- 41.Look DC, Pelletier MR, Tidwell RM, Roswit WT, Holtzman MJ. Stat1 depends on transcriptional synergy with Sp1. J Biol Chem. 1995;270:30264–7. doi: 10.1074/jbc.270.51.30264. [DOI] [PubMed] [Google Scholar]
- 42.Loeffler S, Fayard B, Weis J, Weissenberger J. Interleukin-6 induces transcriptional activation of vascular endothelial growth factor (VEGF) in astrocytes in vivo and regulates VEGF promoter activity in glioblastoma cells via direct interaction between STAT3 and Sp1. Int J Cancer. 2005;115:202–13. doi: 10.1002/ijc.20871. [DOI] [PubMed] [Google Scholar]
- 43.Poole JC, Thain A, Perkins ND, Roninson IB. Induction of transcription by p21Waf1/Cip1/Sdi1: role of NFkappaB and effect of non-steroidal anti-inflammatory drugs. Cell Cycle. 2004;3:931–40. [PubMed] [Google Scholar]
- 44.Gregory DJ, Garcia-Wilson E, Poole JC, Snowden AW, Roninson IB, Perkins ND. Induction of transcription through the p300 CRD1 motif by p21WAF1/CIP1 is core promoter specific and cyclin dependent kinase independent. Cell Cycle. 2002;1:343–50. [PubMed] [Google Scholar]
- 45.Penuelas S, Alemany C, Noe V, Ciudad CJ. The expression of retinoblastoma and Sp1 is increased by low concentrations of cyclin-dependent kinase inhibitors. Eur J Biochem. 2003;270:4809–22. doi: 10.1046/j.1432-1033.2003.03874.x. [DOI] [PubMed] [Google Scholar]
- 46.Oh JE, Han JA, Hwang ES. Downregulation of transcription factor, Sp1, during cellular senescence. Biochem Biophys Res Commun. 2007;353:86–91. doi: 10.1016/j.bbrc.2006.11.118. [DOI] [PubMed] [Google Scholar]
- 47.Ammendola R, Mesuraca M, Russo T, Cimino F. Sp1 DNA binding efficiency is highly reduced in nuclear extracts from aged rat tissues. J Biol Chem. 1992;267:17944–8. [PubMed] [Google Scholar]
- 48.Broude EV, Demidenko ZN, Vivo C, Swift ME, Davis BM, Blagosklonny MV, Roninson IB. p21 (CDKN1A) is a negative regulator of p53 stability. Cell Cycle. 2007;6:1468–71. [PubMed] [Google Scholar]
- 49.Broude EV, Swift ME, Vivo C, Chang BD, Davis BM, Kalurupalle S, Blagosklonny MV, Roninson IB. p21(Waf1/Cip1/Sdi1) mediates retinoblastoma protein degradation. Oncogene. 2007;26:6954–8. doi: 10.1038/sj.onc.1210516. [DOI] [PubMed] [Google Scholar]
- 50.Nishinaka T, Fu YH, Chen LI, Yokoyama K, Chiu R. A unique cathepsin-like protease isolated from CV-1 cells is involved in rapid degradation of retinoblastoma susceptibility gene product, RB and transcription factor SP1. Biochim Biophys Acta. 1997;1351:274–86. doi: 10.1016/s0167-4781(96)00210-2. [DOI] [PubMed] [Google Scholar]
- 51.Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M, Dobbelstein M, Del Sal G, Piaggio G, Mantovani R. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol Cell Biol. 2005;25:3737–51. doi: 10.1128/MCB.25.9.3737-3751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tapias A, Ciudad CJ, Noe V. Transcriptional regulation of the 5′-flanking region of the human transcription factor Sp3 gene by NF-1, c-Myb, B-Myb, AP-1 and E2F. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbagrm.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 53.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]


