Abstract
Pyrrole-imidazole (Py-Im) polyamides are a group of chemicals that are able to bind specifically to DNA sequences in vitro and in mammalian cells. Using a cell based reporter assay, we investigated the size and linker affects on the cellular permeability of polyamides. We found that the conventional β-alanine – 3,3’-diamino-N-methyldipropylamine (βDa) linker strongly limited the cellular permeability. We discovered that a short ethylene diamine (Et) linker displayed high cellular permeability. With the improved Et linker, we found that the cellular permeability of polyamides was size-dependent.
Based on the general concept of the mechanisms of natural transcription activators in eukaryotic cells,1 it was proposed that a synthetic chemical containing a DNA-binding moiety and a protein-binding moiety that interacts with transcriptional coactivators should mimic the biological functions of natural transcription activators.2 Since then, significant progress has been made toward validating this idea.3–5 For the DNA-binding moiety, pyrrole-imidazole (Py-Im) polyamides, a group of compounds developed by Dervan and colleagues6, have shown considerable promise. Replacing N-methylpyrrole rings in the natural products distamycin and netropsin with N-methylimidazole rings by single atom changes creates a group of molecules capable of both A•T and G•C DNA base pair recognitions with 2:1 stoichiometry.7, 8 Connecting two Py-Im polyamides via a 4-amino butyric acid generates “hairpin” Py-Im polyamides that exhibit high affinity binding to duplex DNA in vitro.9 Remarkably, there exists a simple “code” that relates the structures of Py-Im polyamides and specific DNA sequences to which they bind.10. The hairpin Py-Im polymides have been shown to be capable of antagonizing specific protein-DNA interactions in living cells as well, demonstrating the feasibility of using them to deliver artificial activation domains to specific genetic loci in cells.11 However, despite these impressive advances, more information about the properties of these compounds in living cells, such as cellular permeability, the ability to localize in the nucleus and the kinetics and dynamics of binding to chromosomal DNA, will be critical if artificial activators are to become reliable tools for biological research, let alone drug candidates. For example, results from fluorescence microscopy experiments indicate that the cellular permeability of Py-Im polyamides varies considerably with relatively minor changes in the molecules.12, 13
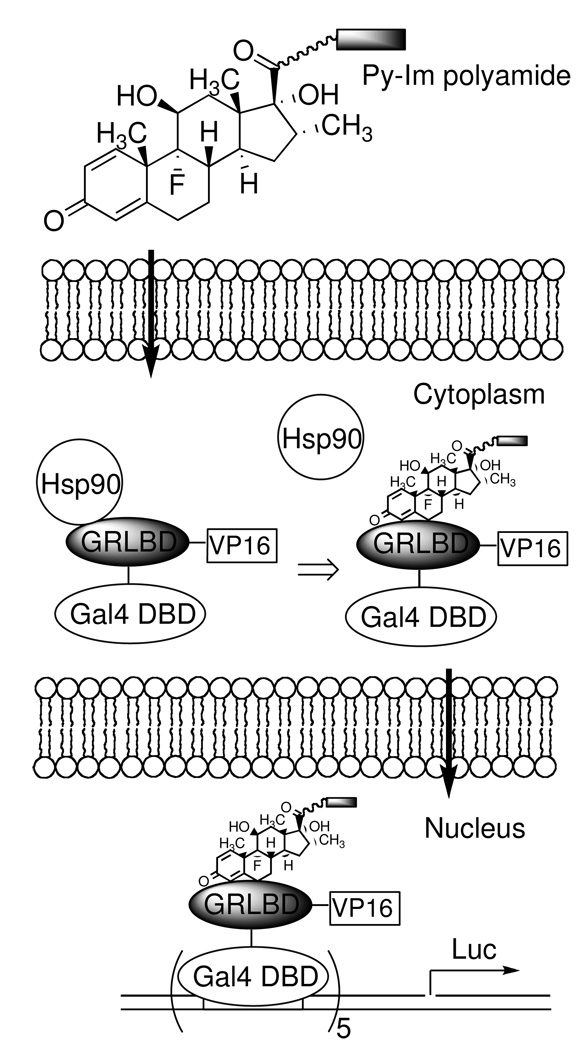
Given the importance of cellular permeability in the design of hairpin polyamide conjugates, we have focused on the development of a more quantitative assay. The use of fluorescence microscopy provides only a qualitative estimate of permeability and, because of the requirement for confocal fluorescence microscopy, has relatively low throughput.14 We recently reported a more convenient and quantitative assay that allows the relative permeability of a series of related molecules to be examined.15 In this scheme, a dexamethasone (dex) conjugate of the molecule of interest is incubated with cells that express a fusion protein that includes the yeast Gal4 DNA-binding domain (Gal4DBD), human glucocorticoid receptor ligand-binding domain (GRLBD), and the C-terminal activation domain of the herpes simplex virus (HSV) transcriptional activator VP16 (VP16). In the apo form, this fusion protein is trapped in the cytoplasm through interaction with heat shock protein 90 (Hsp90).16 When the dex conjugate permeates the cell membrane, the fusion protein (Gal4DBD-GRLBD-VP16) is released from Hsp90 due to dex-GRLBD interaction, and the complex is allowed to translocate into the nucleus where it activates the transcription of activate a Gal4-reponsive luciferase (Luc) reporter gene. The dose-dependent Luc activity readouts provide a measure of the relative permeability of the test molecule relative to some other dex conjugate that is employed as a standard. Here we report a study on Py-Im polyamide cellular permeability using this assay. We find that, not surprisingly, the permeability of Py-Im polyamides is size-dependent. Moreover, we show that linkers used commonly to conjugate molecules to Py-Im polyamides can severely retard the conjugate from entering the cell. The implications of our results for design of synthetic gene expression regulators are discussed.
Results
Dex derivatives with different sensitivities in functional cellular permeability assay
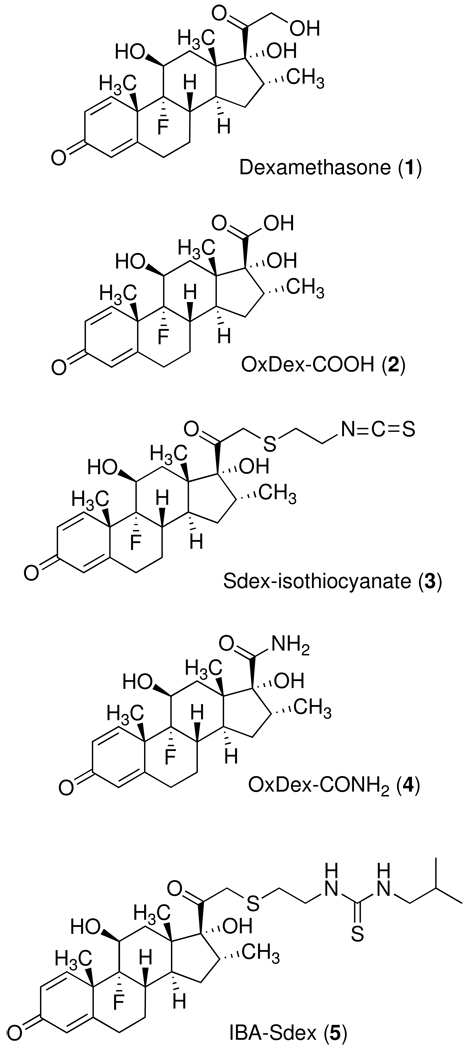
Two dex derivatives Dex-17β-carboxylic acid (Oxdex-COOH, 2)17 and Dex-21-S(CH2)2NCS (Sdex-isothiocyanate, 3)18, 19 were prepared as described previously (Figure 1). Py-Im polyamide amines were conjugated to Oxdex-COOH via amide bond formation15 and to Sdex-isothiocyanate via thiourea formation.20 The GR-binding affinities of Oxdex and Sdex conjugates were evaluated by a GR competitive binding assay15, in which the dex conjugates competed with a fluorescein-labeled dex derivative called Fluormone (Invitrogen). The displacement of Fluormone from GR was monitored by a decrease in fluorescence polarization. It has been shown that Sdex conjugates (IC50 = 6 ~ 30 nM) are better GR binders than Oxdex conjugates (IC50 = 1 ~ 5 µM).20 Because of the high GR-binding affinity of Sdex derivatives, only low concentrations of Sdex conjugates inside living cells were required to induce Luc activity (see Scheme 1). Therefore, Sdex-containing conjugates serve as a high sensitivity tool for permeability measurements with sparingly permeable molecules. In contrast, a much higher concentration of Oxdex conjugates is needed for significant Luc induction due to the lower affinity of the OxDex-GRLBD complex, making these derivatives ideal for the comparison of more highly permeable compounds.
Figure 1.
Structures of dexamethasone derivatives employed in this study.
Scheme 1.
Py-Im polyamide permeability is affected by different linkers and sizes
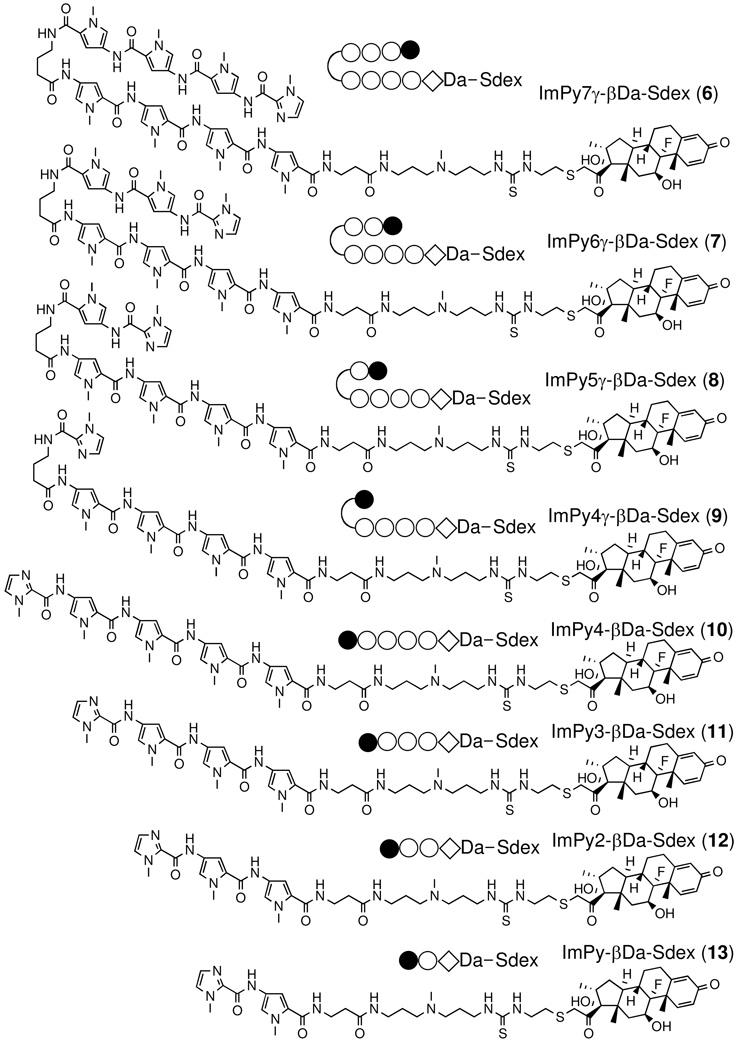
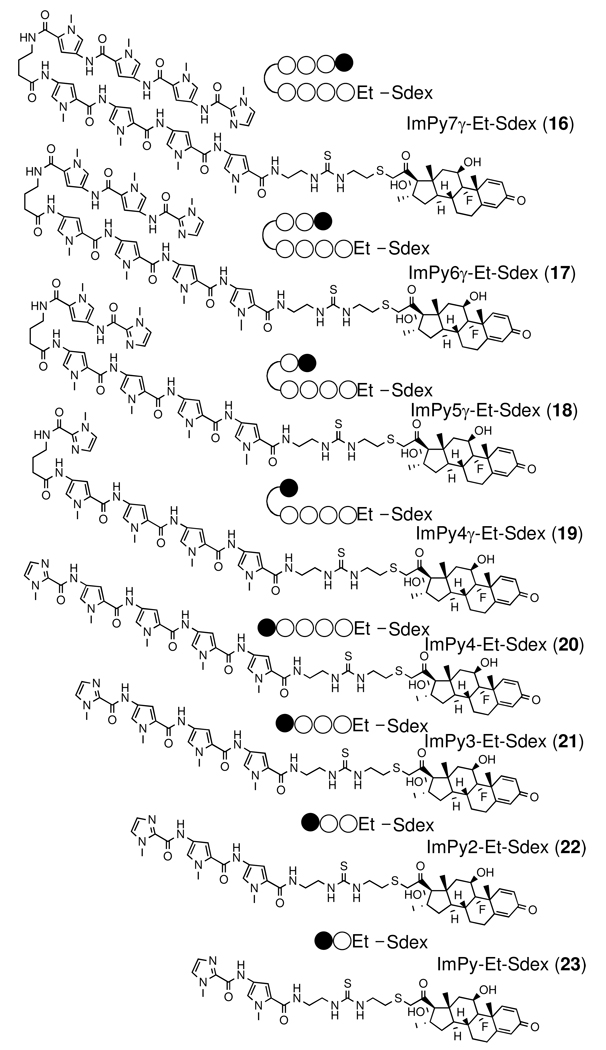
Previously we made the Py-Im – dex conjugate ImPy7γ-βDa-Sdex (6) in an effort to construct a synthetic transcription activator.20 This conjugate recognizes the DNA sequence 5’- TGTTAT-3’ with high affinity (KD = 5 ~ 50 nM). The GR competitive binding assay showed that ImPy7γ-βDa-Sdex (6) binds to GR with an affinity (IC50 = nM) close to that of dex, demonstrating that the polyamide does not inhibit steroid-GRLBD interactions. 20 These in vitro data suggested that ImPy7γ-βDa-Sdex (6) would be a good starting compound to test the biological activity of synthetic transcription activators in living mammalian cells. Note that ImPy7γ-βDa was used successfully as the DNA-binding moiety of the first synthetic transcription activator with activity in vitro.21 In the cell-based functional assay (Scheme 1) the ImPy7γ-βDa-SDex conjugate induced modest Luc activity, in stark contrast to the control compound isobutyl-Sdex (IBA-Sdex (5)).20 We concluded that, not unexpectedly, ImPy7γ-βDa is far less cell permeable than the small isobutyl group. We reasoned that the low permeability of ImPy7γ-βDa might be due to both the relative large size of the Py-Im polyamide (MW = 1265) and the floppy βDa linker. The β-alanine – 3,3’-diamino-N-methyl dipropylamine (ββDa) linker is convenient for conjugation of chemical agents to Py-Im polyamides, since it is produced efficiently by aminolytic cleavage of Py-Im polyamide after solid-phase synthesis.21, 22 To test our hypotheses, we made a series of compounds by gradually reducing the number of Py in ImPy7γ-βDa and conjugated the polyamides to Sdex (6–13)(Figure 2). Another series of compounds were made by substituting the βDa linker in the previous series with a shorter ethylene diamine (Et) linker (16–23)(Figure 2). All compounds were purified and tested for their binding to GR using the Fluormone competitive binding assay and subjected to the cell based functional assay. The GR competitive binding assay showed that all compounds in both series have similar affinities for GR (IC50 = 6 –30 nM, Table 2, Figure. S1). Thus, the dose-dependent Luc activity readouts were taken as measures of the relative cellular permeability of the Py-Im – Sdex conjugates.
Figure 2.
Structures of Py-Im polyamide – Sdex conjugates with β-Da and Et linkers.
Table 2.
IC50 values of Sdex and Oxdex Py-Im polyamide conjugates from in vitro GR competition binding assays.
| Compound | IC50 (nM) | Compound | IC50 (nM) |
|---|---|---|---|
| ImPy7γ̃βDa-Sdex (6) | 14 ± 0.87 | ImPy7γ-Et-Sdex (16) | 27 ± 2.1 |
| ImPy6γ̃βDa-Sdex (7) | 20 ± 1.1 | ImPy6γ-Et-Sdex (17) | 21 ± 1.7 |
| ImPy5γ̃βDa-Sdex (8) | 22 ± 2.9 | ImPy5γ-Et-Sdex (18) | 21 ± 2.3 |
| ImPy4γ̃βDa-Sdex (9) | 16 ± 1.1 | ImPy4γ-Et-Sdex (19) | 23 ± 2.6 |
| ImPy4-βDa-Sdex (10) | 16 ± 2.0 | ImPy4-Et-Sdex (20) | 34 ± 2.5 |
| ImPy3-βDa-Sdex (11) | 20 ± 1.1 | ImPy3-Et-Sdex (21) | 26 ± 1.8 |
| ImPy2-βDa-Sdex (12) | 12 ± 0.82 | ImPy2-Et-Sdex (22) | 14 ± 0.80 |
| ImPy-βDa-Sdex (13) | 8.5 ±0.43 | ImPy-Et-Sdex (23) | 5.7 ± 0.30 |
| ImPy7γ-Et-Oxdex (24) | 840 ± 130 | Ac-Et-Sdex (15) | 9.8 ± 0.88 |
| ImPy3-Et-Oxdex (25) | 550 ± 57 | Ac-Da-Sdex (28) | 8.3 ± 0.61 |
| Impy2-Et-Oxdex (26) | 480 ± 69 | Ac-βDa-Sdex (14) | 9.3 ± 0.48 |
| ImPy-Et-Oxdex (27) | 460 ± 42 | dexamethasonea (1) | 5 ± 1 |
| Oxdex-CONH2 c (4) | 600 ± 100 | IBA-Sdexb (5) | 15 ± 1.5 |
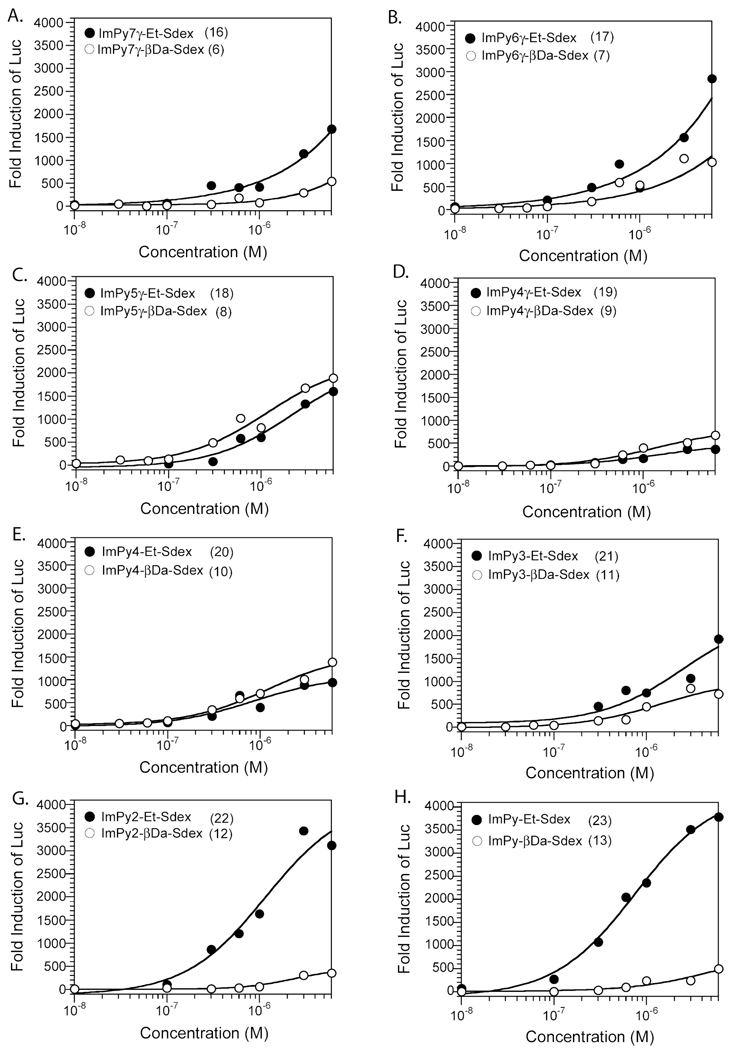
As shown in Figure 3, no significant difference in Luc induction by the βDa series (6–13) was found. Noticeably, the smallest compounds ImPy-βDa-Sdex (13) and ImPy2-βDa-Sdex (12) induced the similar low Luc activities as the largest compound ImPy7-βDa-Sdex (6). These data, in combination with the finding that all of the compounds had similar GR-binding affinities in vitro, lead us to conclude that the cellular permeability of Py-Im polyamides with the βDa linker was size-independent, and all of the βDa series of Py-Im – Sdex conjugates had similarly limited cellular permeability. This was unexpected and suggested that a part of the molecule other than the polyamide was limiting cellular permeability.
Figure 3.
Dose dependent Luc activities induced by Py-Im polyamide –β-Da – Sdex conjugates (○) and Py-Im polyamide – Et – Sdex conjugates (●) in HeLa cells. HeLa cells were transfected with a plasmid encoding Gal4DBD-GRLBD-VP16 protein and a Gal4-reponsive luciferase reporter.
In stark contrast to ImPy-βDa-Sdex (13), ImPy-Et-Sdex (23) induced robust Luc activity with a maximum induction of 3700-fold and an EC50 value of 0.75 µM (Table 1, Figure 3H). Both values were close to that of IBA-Sdex (5) (EC50 = 0.35 µM). With a slightly increased size, ImPy2-Et-Sdex still achieved a maximum Luc induction (3400 fold) at 6 µM concentration and gave a slightly higher EC50 value (EC50 = 1.2 µM, Table 1, Figure 3G), again, a drastic improvement relative to its counterpart ImPy2-βDa-Sdex (12). These data show that Et is a much more permeable linker than βDa.
Table 1.
EC50 values of Luc induction in HeLa cells by selected Py-Im polyamides dexamethasone conjugates and dexamethasone derivatives.
When the length of the polyamide chain was increased, reduced cell permeability was observed, as originally expected. ImPy3-Et-Sdex (21) and ImPy4-Et-Sdex (20) induced lower Luc activity and did not achieve the maximum induction at 6 µM (Figure 3E, F). ImPy4γ-Et-Sdex (19) gave the lowest Luc activity among all Sdex conjugates (Figure 3D). However, the larger molecules ImPy5γ-Et-Sdex (18), ImPy6γ-Et-Sdex (17), and ImPy7γ-Et-Sdex (16) induced Luc activities at higher levels compared to ImPy4γ-Et-Sdex (19) (Figure 3C-3A). The high cell permeability engineered by the Et linker was also observed in the longer polyamides. For example, ImPy7γ-Et-Sdex (16) showed modestly improved Luc activity induction (1,500 fold at 6 µM) compared to ImPy7γ-βDa-Sdex (500 fold at 6 µM). However, the differential between the cell permeabilities of the longer polyamides containing the Et and βDa linkers was far more modest than the large difference between these pairs observed with the lower molecular mass compounds (compare Figure 3A to Figure 3H, for example). This suggests that as the size of the polyamide increases, it begins to limit the cell permeability of the chimera.
The βDa linker has much lower intrinsic cell permeability than the Et linker
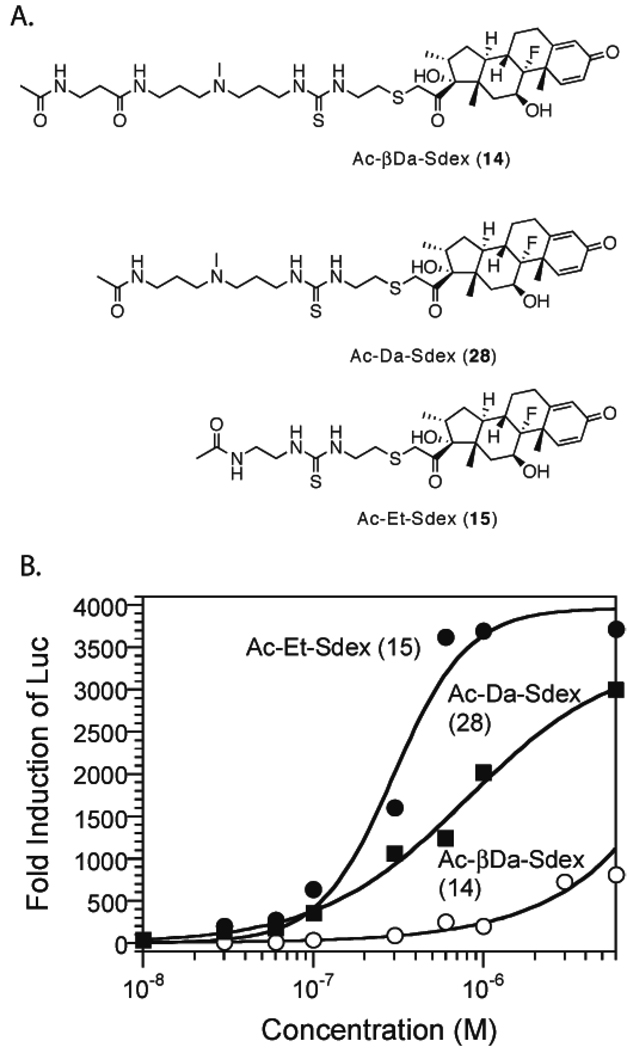
The difference in cellular permeability between the βDa and the Et series Py-Im polyamides lead us to investigate the cellular permeability of the linkers alone. Ac-βDa-Sdex and Ac-Et-Sdex (Figure 4A) were made with the terminal amines capped with acetyl groups. Both conjugates had similar GR-binding affinities (Table 2, Fig. S1Q, R), so the Luc induction levels can be taken as a direct indication of their relative cellular permeability. As shown in Figure 4B, Ac-Et-Sdex induced robust Luc activity, with a maximum induction of 3600-fold achieved at 0.6 µM and an EC50 value of 0.29 µM (Table 1). These data suggested that Ac-Et was highly permeable, similar to the isobutyl group (in IBA-Sdex (5), Table 1). In contrast to Ac-Et-Sdex (15), Ac-βDa-Sdex (14) induced very modest Luc induction (600 fold at 6 µM), similar to ImPy-βDa-Sdex (13), ImPy2-βDa-Sdex (12), and ImPy7γ-βDa-Sdex (6). These data show conclusively that the βDa linker was the limiting factor for Py-Im polyamide cellular permeability, at least for the lower molecular mass polyamides. To further probe which component in the βDa linker caused low cellular uptake, we prepared Ac-Da-Sdex (28, Figure 4A). Compound 28 induced better Luc activity than Ac-βDa-Sdex (14), with a maximum induction of 3,000 fold achieved at 6 µM and an EC50 value of 0.8 µM (Figure 4B). The EC50 value of Ac-Da-Sdex (28) was 2.5 times higher than that of Ac-Et-Sdex, indicating that Ac-Da was less permeable than Ac-Et.
Figure 4.
Measurement of the cellular permeability of the linkers. A. Structures of linker – Sdex conjugates. B. Dose-dependent Luc activities induced by Ac-βDa-Sdex (○), Ac-Da-Sdex (■), and Ac-Et-Sdex (●).
Cellular permeability of Py-Im polyamides using Oxdex conjugates
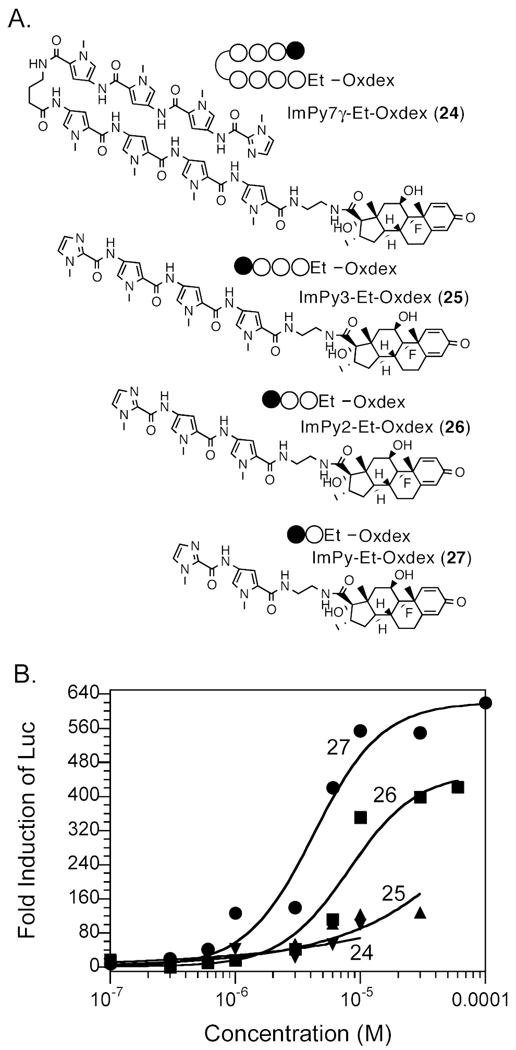
To confirm the conclusions drawn from the Sdex conjugates, we repeated the experiments using Oxdex conjugates and Et as the linker. Oxdex is a modest affinity GR ligand compared to Sdex, with an IC50 value about 40 times higher than that of Sdex. Due to the low GR-binding affinity, more steroid conjugate must permeate the cell membrane in order to trigger a reporter gene response. We focused on ImPy, ImPy2, ImPy3, and ImPy7 (Fig. 5A). All four polyamide-Et - Oxdex conjugates showed GRLBD-binding affinities similar to that of the control compound Oxdex-CONH2 (460 – 840 nM, Table 2, Figure S1S-V), so the permeability of polyamides could be compared directly by their ability to induce Luc activity. As shown in Figure 5B, ImPy-Et-Oxdex (27) and ImPy2-Et-Oxdex (26) achieved maximum Luc induction at about a 20 µM concentration, with EC50 values of 4.2 and 8.1 µM, respectively (Table 1). In comparison, the EC50 value of the control compound Oxdex-CONH2 (4) was 35 µM. These results were consistent with our conclusion that ImPy and ImPy2 are highly permeable. On the other hand, ImPy3-Et-Oxdex (25) and ImPy7γ-Et-Oxdex (24) exhibited similar low Luc induction at 10 µM concentration, 7 and 4 times lower than that of ImPy-Et-Oxdex and ImPy2-Et-Oxdex. Unfortunately, both compounds were insoluble at concentrations higher than 10 µM, so the maximum Luc induction could not be detected.
Figure 5.
Measurement of the cell permeability of Py-Im polyamide – Oxdex conjugates. A. Structures of Py-Im polyamide – Oxdex conjugates with the Et linker. B. Dose dependent Luc activities induced by Py-Im polyamide – Et – Oxdex conjugates in HeLa cells. Circles, ImPy-Et-Oxdex; squares, ImPy2-Et-Oxdex; triangle, ImPy3-Et-Oxdex; inverted triangle, ImPy7γ-Et-Oxdex.
Discussion
In conclusion, we have employed a novel system for the quantitative comparison of the relative cell permeability of steroid conjugates to probe the cell permeability of polyamides. The dose-dependent Luc activities directly reflect the permeability of the polyamide-dex conjugates that are explored as potential artificial transcriptional activators. The limited cellular permeability of ImPy7-βDa-Sdex led us to investigate the size and linker affects on the permeability of the chimera. For a series of related molecules with a similar dexamethasone derivative, the results of the functional assay may also indicate the relative permeability of Py-Im polyamides alone. However, this newly developed functional assay needs to be further validated as a general method for Py-Im polyamide permeability evaluation.
An interesting finding was that the βDa linker, used in the construction of artificial transcription activators for historical reasons21, 22, has limited cellular permeability. Indeed, the poor permeability of this linker largely restricts polyamides of almost any size from entering cells efficiently, explaining the initially surprising finding that polyamide-Sdex conjugates of different sizes containing this linker exhibit similar (and poor) cell permeability. A further study showed that β-alanine played an important role in affecting the permeability of Sdex conjugates. By eliminating β-alanine, Ac-Da-Sdex displayed improved cellular uptake than Ac-βDa-Sdex. Our results are consistent with the initial findings of Dervan and colleagues.12, 13 Through a systematic study, Edelson et al. and Best et al. concluded that in general, hairpin Py-Im polyamides with the Da linker conjugated to fluorescent dyes had better cellular uptake than that with the βDa linker. Furthermore, hairpin Py-Im polyamides with a C-terminal tail consisting of Da and 1,3-benzene-dicarboxylic acid showed improved efficacy in living mammalian cells.23
An important finding of this study is that the Et linker is highly permeable. When fused to the steroid alone or to small polyamides containing two or three units, a large difference in the relative permeability of the molecules containing the βDa and Et linkers was observed. This argues that polyamide conjugates of the type being developed as artificial activators24, 25 should employ the more permeable Et linker and avoid the βDa linker. Recently Nickols et al. reported a reporter assay that could be used to evaluate linker affect on hairpin polyamide cellular permeability.26 Various linkers were conjugated to the C-terminal end of a polyamide that targeted the hypoxia response element (HRE). The inhibition of HRE driven vascular endothelial growth factor (VEGF) expression was taken as a biological read-out for linker affect on cellular uptake and nuclear localization.
The favorable permeability properties of the Et linker were also observed in comparisons of larger polyamide-Sdex conjugates such as ImPy7-Et (or βDa)-Sdex, but the difference was much more modest (≈ 3-fold), arguing that at this point, the size of the polyamide itself begins to be responsible for the limited permeability of the steroid conjugate. These results are consistent with the previous findings that large hairpins (10 ring) had poor cellular uptake while small compounds (6 ring) had excellent cellular uptake.13 A molecule like ImPy7 is probably the smallest polyamide unit that would be of interest for the design of bioactive molecules, since it recognizes a DNA sequence of six base pairs. Thus, while cell culture experiments are clearly feasible with conjugates of this type of molecule, particularly using the Et linker, it remains to be determined whether polyamide-based artificial activators will be sufficiently bioavailable to be employed routinely in animal models. Although small Py-Im – Sdex conjugates ImPy-Et-Sdex and ImPy2-Et-Sdex showed excellent cellular uptake, they could not be used as artificial transcription activators due to low DNA-binding affinities and specificities of ImPy and ImPy2.27 Dervan and colleagues have shown that in 8-ring Py-Im hairpin polyamides, the sequences of polyamides were important on cellular uptake and nuclear localization.12, 13 The sequence affect on permeability should be considered in the design of artificial transcription activators in the future.
To further confirm our finding that the Et linker enhanced the cellular uptake of Py-Im – Sdex conjugates compared to the βDa linker, we conducted our assay with breast cancer cell line MCF7. The results are summarized in Figure S2 (supporting information). Ac-βDa-Sdex (14), ImPy-βDa-Sdex (13), and ImPy7-βDa-Sdex (6) induced similar low Luc activities. These results were consistent with the results in HeLa cells that the βDa linker was the limiting factor of cellular uptake. In stark contrast to Ac-βDa-Sdex (14), Ac-Et-Sdex (15) induced robust Luc activity in MCF7 cells with a maximum induction of 1,200 fold achieved at 1 µM and an EC50 value of 0.25 µM. Similar to the Luc activities observed in HeLa cells, ImPy-Et-Sdex (23) displayed improved cellular permeability compared to ImPy-βDa-Sdex (13) with an EC50 value of 0.94 µM. ImPy7-Et-Sdex (16) showed modest improvement of cellular uptake compared to ImPy7-βDa-Sdex (26), indicating that as size of the polyamide increased, it began to limit the cellular permeability of the chimera.
The better cellular uptake of Py-Im – Sdex conjugates with the Et linker than those with the βDa linker may be due to the higher lipophilicity of the Et linker conjugates. To probe this issue, we evaluated the n-octanol – water partition coefficient LogP values of both Et and βDa linker conjugates. Based on a computational program from Molinspiration,28 compounds 15, 23, and 16 gave LogP values of 1.66, 1.53, and 1.42, respectively, while 14, 13, and 6 gave LogP values of 1.60, 1.48, and 1.37, respectively. The Et linker conjugates gave higher logP values than their βDa linker counterparts, indicating that higher lipophilicity of the Et linker conjugates might be responsible for the higher cellular uptake.
Experimental Section
General
4-tert-Butoxycarbonylamino-1-methyl-1H-pyrrole-2-carboxylic acid benzotriazol-1yl ester (t-Boc-Py-Obt) was purchased from Oakwood Products, Inc. 1-Methyl-1H-imidazole-2-carboxylic acid (Im-COOH) was purchased from BACHEM. 4-tert-Butoxycarbonylamino-butanoic acid (t-Boc-γ-COOH) was purchased from NOVAbiochem. O-(7-azabenzo-triaol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU) was purchased from Applied Biosystems. Thiophenol (PhSH) was purchased from ACROS Organics. 3, 3’-Diamino-N-methyl dipropylamine, triethylamine, ethylene diamine, acetic anhydride, and anhydrous dichloromethane (CH2Cl2) were from Sigma. N-Acetylethylenediamine (technical grade) and N-hydroxysuccinimide (NHS), and anhydrous tetrahydrofuran (THF) were from Sigma. N, N’-Dicyclohexylcarbodiimide (DCC) was from Fluka. N-acetyl-β-alanine (Ac-β-Ala-OH) was from NOVAbiochem. Boc-β-alanine-(4-carbonylaminomethyl)-benzyl-ester-copoly(styrene-divinylbenzene) resin (Boc-β-Pam resin, 0.26 mmole / g) was from Peptide International. 4-Nitro-benzophenone-oxime-copoly(styrene-divinylbenzene) resin (Oxime resin LL, 0.85 mmole / g) was from NOVAbiochem. All chemical reagents were of analytical grade unless were otherwise stated.
Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), Dulbecco’s phosphate buffered saline, modified Eagle’s medium (MEM) non-essential amino acids, penicillin-streptomycin, reduced-serum medium (Opti-MEM 1), and transfection reagent Lipofectamine Plus were from Invitrogen. Dimethyl sulphoxide (DMSO, HYBRI-MAX) was from Sigma. Hela cells were from the American Type Culture Collection (ATCC, CCL-2). Cell densities were determined on a Hausser Scientific hemacytometer. Glucocorticoid receptor (GR) competition assay kit was purchased from Invitrogen.
Thin layer chromatography (TLC) was performed on Sigma-Aldrich silica gel 60 F254 pre-coated plates. Compounds were visualized under 254 nm ultraviolet lights. 1H 13C NMR spectra were recorded on a Varian Inova 400 or 500 spectrometer as noted. UV spectra were measured on a Beckman Coulter DU640 spectrophotometer. High-resolution electrospray ionization mass spectrometry (HRESI-MS) was performed at Resource for Biomedical and Bio-organic Mass Spectrometry at Washington University, St Louis. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) was carried out on a Voyager-DE PRO mass spectrometer from PerSeptive Biosystems. High performance liquid chromatograph (HPLC) was performed on a Waters HPLC system. Preparatory HPLC was carried out using a Vydac 22 × 250 mm, 300 Å, 10 µm C18 column with a flow rate of 10 mL / min. Semi-preparatory HPLC was carried out using a Vydac 10 × 250 mm, 300 Å, 5 µm C18 column with a flow rate of 5 mL / min. Analytical HPLC was carried out using a Vydac 4.6 × 250 mm, 300 Å, 5 µm C18 column with a flow rate of 5 mL / min. The elution solutions are H2O (0.1 % trifluoroacetic acid (TFA), v/v) and acetonitrile (0.1 % TFA, v/v). HPLC was monitored at 254 nm wavelength. Each product was analyzed by analytical HPLC. The purity of products was evaluated as > 95% by comparing the integration of the product peak and the integrations of all peaks in each chromatogram. Luciferase (Luc) activity was measured on a Berthold Sirius single-tube luminometer. Fluorescence polarization was measured on a Panvera Beacon 2000 fluorometer.
Synthesis. Sdex-isothiocyanate (3, Dex-21-S(CH2)2NCS)
Synthesis was done as described previously.19 18 TLC (20% ether in CHCl3) Rf = 0.2. mp = 86 – 88 °C. 1H NMR (400 MHz, DMSO-d6): δ7.27 (d, J = 10.3 Hz, 1H), 6.20 (d, J = 10.3 Hz, 1H), 5.98 (s, 1H), 5.28 (s, 1H), 5.09 (s, 1H), 4.11 (bs, 1H), 3.89 (d, J = 16.8 Hz, 1H), 3.81–3.84 (m, 2H), 3.47 (d, J = 16.8, 1H), 2.87–2.97 (m, 1H), 2.81–2.87 (m, 2H), 2.55–2.65 (m, 1H), 2.30–2.40 (m, 1H), 2.30 (d, J = 11 Hz, 1H), 2.17 (d, J = 11 Hz, 1H), 2.08 (dd, J = 11, 6.5 Hz, 1H), 1.70–1.80 (m, 1H), 1.58 (dd, J = 11, 6.5 Hz, 1H), 1.47 (1H), 1.44 (s, 3H), 1.25–1.40 (m, 1H), 0.98–1.07 (m, 1H), 0.84 (s, 3H), 0.76 (d, J = 6.9 Hz, 3H). 13C NMR (125 MHz, DMSO-d6): δ15.2, 16.7, 22.8, 27.1, 30.1, 31.6, 31.8, 33.4, 34.5, 35.9, 43.0, 44.5, 47.3, 47.8, 70.3, 70.6, 90.8, 100.4, 101.8, 124.0, 128.9, 152.5, 167.1, 185.2, 207.1. HRESI-MS: calcd for C25H32FNNaO4S2 (MNa+) 516.1654, found 516.1656.
ImPy7γ̃βDa-NH2, ImPy6γ̃βDa-NH2, ImPy5γ̃βDa-NH2, ImPy4γ̃βDa-NH2, ImPy4-βDa-NH2, ImPy3-βDa-NH2, ImPy2-βDa-NH2, and ImPy-βDa-NH2 were synthesized as described before22 on Boc-β-Pam resin. ImPy7γ̃βDa-NH2, ImPy6γ̃βDa-NH2, ImPy5γ̃βDa-NH2, ImPy4γ̃βDa-NH2, ImPy4-βDa-NH2, ImPy3-βDa-NH2 were purified by reversed-phase HPLC on a preparatory C18 column (start at 15% acetonitrile, 0.25% acetonitrile / min). The elution gradient for ImPy2-βDa-NH2 and ImPy-βDa-NH2 HPLC purification was 0.5% acetonitrile / min, starting at 0% acetonitrile. The collected fractions were lyophilized. The freeze-dried samples were dissolved in H2O and purified again by reversed-phase HPLC (2 % acetonitrile / min) and lyophilized. MALDI-TOF-MS (MH+): calcd for ImPy7γ̃βDa-NH2 1264.62, found 1265.22; calcd for ImPy6γ̃βDa-NH2 1142.58, found 1142.89; calcd for ImPy5γ̃βDa-NH2 1020.53, found 1020.88; calcd for ImPy4γ̃βDa-NH2 898.48, found 899.05; calcd for ImPy4̃βDa-NH2 813.43, found 813.58; calcd for ImPy3-βDa-NH2 691.38, found 691.68; calcd for ImPy2-βDa-NH2 569.33, found 569.85; calcd for ImPy-βDa-NH2 447.28, found 447.70.
ImPy7γ̃βDa-Sdex (6), ImPy6γ̃βDa-Sdex (7), ImPy5γ̃βDaSdex (8), ImPy4γ̃βDa-Sdex (9), ImPy4-βDa-Sdex (10), ImPy3-βDa-Sdex (11), ImPy2-βDa-Sdex (12), and ImPy-βDa-Sdex (13)
Individual Py-Im amine (1 mg), Sdex-isothiocyanate (2 mg), and triethylamine (7.5 µL) were added in anhydrous CH2Cl2 (500 µL). The mixture was vortexed at room temperature for 2 h, and CH2Cl2 was completely removed on a speedvac system. The residue was suspended in 50% acetonitrile and subjected to HPLC purification on a semi-preparatory C18 column, gradient elution 3.3% acetonitrile / min. The retention times of imidazole-pyrrole amines, the products, and Sdex-isothiocyanate were 11–14 min, 17 min, and 21 min, respectively. The fractions of the products were collected, freeze-dried, and stored in a desiccate chamber at room temperature. MALDI-TOF-MS (MH+): calcd for ImPy7γ̃βDa-Sdex 1757.80, found 1757.21; calcd for ImPy6γ̃βDa-Sdex 1635.75, found 1635.28; calcd for ImPy5γ̃βDaSdex 1513.70, found 1513.76; calcd for ImPy4γ̃βDa-Sdex 1391.66, found 1392.01; calcd for ImPy4-βDa-Sdex 1306.60, found 1306.40; calcd for ImPy3-βDa-Sdex 1184.55, found 1184.10; calcd for ImPy2-βDa-Sdex 1062.51, found 1062.95; calcd for ImPy-βDa-Sdex 940.46, found 940.91.
Ac-βDa-Sdex (14)
A mixture of Ac-β-Ala-OH (500 mg, 3.8 mmol), N-hydroxysuccinimide (434 mg, 3.8 mmol), and DCC (824 mg, 4 mmol) in anhydrous THF (10 mL) was stirred at 0 °C for 1h, and then at room temperature for 12 h. The solution was clarified by filtration and THF was removed under reduced pressure. The white gooey residue (Ac-β-Ala-CO-NHS ester) was used in the subsequent reaction without purification. Sdex-isothiocyanate (2 mg, 4 µmol), 3, 3’-Diamino-N-methyl dipropylamine (15 µL, 93 µmol), and triethylamine (7.5 µL) were added in anhydrous CH2Cl2 (500 µL). The mixture was vortexed at room temperature for 2 h. CH2Cl2 was completely removed on a speedvac system. The residue was suspended in 50% acetonitrile and subjected to HPLC purification on a semi-preparatory C18 column. Lyophilization yielded clear oil (NH2-Da-Sdex). MALDI-TOF-MS (MH+) calcd for NH2-Da-Sdex 639.34, found 639.52. All NH2-Da-Sdex was dissolved in anhydrous CH2Cl2 (500 µL). Triethylamine (7.5 µL) and Ac-β-Ala-CO-NHS (10 mg) were added, and the mixture was vortexed at room temperature for 2 h to yield Ac-βDa-Sdex. CH2Cl2 was completely removed on a speedvac system. The residue was suspended in 50% acetonitrile and purified by HPLC on a semi-preparatory C18 column. MALDI-TOF-MS (MH+) calcd for Ac-βDa-Sdex (14) 752.39, found 752.77.
Ac-Da-Sdex (28)
NH2-Da-Sdex was synthesized as described above and was dissolved in anhydrous CH2Cl2 (500 µL). Triethylamine (5 µL) and acetic anhydride (10 µL) were added, and the mixture was vortexed at room temperature for 2 h. CH2Cl2 was completely removed on a speedvac system. The residue was suspended in 50% acetonitrile and purified by HPLC on a semi-preparatory C18 column. MALDI-TOF-MS (MH+) calcd for Ac-Da-Sdex (28) 681.35, found 681.72.
Ac-Et-Sdex (15)
A mixture of N-Acetylethylenediamine (15 µL), triethylamine (7.5 µL), and Sdex-isothiocyanate (2 mg) in anhydrous CH2Cl2 (500 µL) was vortexed at room temperature for 2 h. Purification was done following the procedures described above. MALDI-TOF-MS (MH+) calcd for Ac-Et-Sdex (15) 596.26, found 596.46.
ImPy7γ-Et-NH2, ImPy6γ-Et-NH2, ImPy5γ-Et-NH2, ImPy4γ-Et-NH2, ImPy4-Et-NH2, ImPy3-Et-NH2, ImPy2-Et-NH2, and ImPy-Et-NH2 were synthesized according to published procedures29 on Oxime resin except that the Boc groups were removed with 25% TFA/CH2Cl2/0.5 M PhSH, instead of 80% TFA/CH2Cl2/0.5 M PhSH. After synthesis, the resin was stirred in ethylene diamine at 65 °C for 12 h to release the products. HPLC purification was carried out on a C18 preparatory column as described in the Im(Py)n(γ)nβ-Da-NH2 purification, 0.5% acetonitrile / min, starting 0% acetonitrile. MALDI-TOF-MS (MH+): calcd for ImPy7γ-Et-NH2 1108.50, found 1108.42; calcd for ImPy6γ-Et-NH2 986.45, found 986.39; calcd for ImPy5γ-Et-NH2 864.40, found 864.30; calcd for ImPy4γ-Et-NH2 742.35, found 742.31; calcd for ImPy4-Et-NH2 657.30, found 657.43; calcd for ImPy3-Et-NH2 541.30, found 541.90; calcd for ImPy2-Et-NH2 413.21, found 413.59; calcd for ImPy-Et-NH2 291.16, found 291.57.
ImPy7γ-Et-Sdex (16), ImPy6γ-Et-Sdex (17), ImPy5γ-Et-Sdex (18), ImPy4γ-Et-Sdex (19), ImPy4-Et-Sdex (20), ImPy3-Et-Sdex (21), ImPy2-Et-Sdex (22), and ImPy-Et-Sdex (23)
Synthesis was done as described in Im(Py)n(γ)m-βDa-Sdex synthesis with Im(Py)n(γ)m-Et-NH2. MALDI-TOF-MS (MH+): calcd for ImPy7γ-Et-Sdex 1601.67, found 1601.43; calcd for ImPy6γ-Et-Sdex 1479.63, found 1479.68; calcd for ImPy5γ-Et-Sdex 1357.58, found 1357.63; calcd for ImPy4γ-Et-Sdex 1235.53, found 1235.19; calcd for ImPy4-Et-Sdex 1150.48, found 1150.55; calcd for ImPy3-Et-Sdex 1028.43, found 1028.45; calcd for ImPy2-Et-Sdex 906.38, found 906.57; calcd for ImPy-Et-Sdex 784.33, found 784.76.
ImPy7γ-Et-Oxdex (24), ImPy3-Et-Oxdex (25), ImPy2-Et-Oxdex (26), and ImPy-Et-Oxdex (27)
ImPy7γ-Et-NH2, ImPy3-Et-NH2, ImPy2-Et-NH2, and ImPy-Et-NH2 were prepared as described above. ImPy7γ-Et-Oxdex, ImPy3-Et-Oxdex, ImPy2-Et-Oxdex, and ImPy-Et-Oxdex were synthesized as described in Im(Py)n(γ)m-Et-Sdex synthesis, except that Sdex-isothiocyanate was substituted with OxDex-CO-NHS ester (Dex-17β-carboxylic acid N-hydroxysuccinimide ester). MALDI-TOF-MS (MH+): calcd for ImPy7γ-Et-Oxdex 1468.67, found 1468.35; calcd for ImPy3-Et-Oxdex 895.43, found 895.77; calcd for ImPy2-Et-Oxdex 773.38, found 773.51; calcd for ImPy-Et-Oxdex 651.33, found 651.60.
Concentration determination
Stock solutions of all chemicals used in this study were prepared in DMSO. A DMSO stock solution (1 µL) was diluted in H2O (1 mL) for Sdex conjugates or 50 % methanol in water (1 mL) for Oxdex conjugates. The concentrations were determined by the distinct absorbance of Py-Im polyamides or dexamethasone derivatives. Extinction coefficients: ImPy7, ε310 nm=6.9 ×104 M−1 cm−1; ImPy6, ε310 nm = 6.0 ×104 M−1 cm−1; ImPy5, ε310 nm = 5.2 × 104 M−1 cm−1, ImPy4, ε310 nm = 4.3 × 104 M−1 cm−1; ImPy3, ε304 nm = 3.5 × 104 M−1 cm−1; ImPy2, ε304 nm = 2.6 × 104 M−1 cm−1; ImPy, ε304 nm = 1.7 × 104 M−1 cm−1; Ac-βDa-Sdex, Ac-Da-Sdex, and Ac-Et-Sdex, ε242 nm = 1.2 × 104 M−1 cm−1.
Cell culture, transfection, and luciferase assay
HeLa cells were grown in 100 mm tissue culture plates at 37 °C under 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) fetal bovine serum, 10 µg/mL penicillin, and 10 µg/mL streptomycin. Cells were passed every 3 days and never allowed to reach confluency. Cells (1.5 × 104 cells per well) were placed in 96-well plates and incubated in 100 µL of the previously prepared tissue culture media for 24 h before transfection by the Lipofectamine Plus method (Invitrogen). Cells in each well were transfected with 50.5 ng total DNA, including 25 ng pG5B reporter plasmid (Kodadek lab stock), 25 ng pEGal4DBD-GRLBD-VP16 (Kodadek lab stock), and 0.5 ng R. reniformis luciferase plasmid (pRL-SV40, Promega). After transfection, the transfection buffer was thoroughly removed and replaced by Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) fetal bovine serum. All chemicals were prepared in DMSO and added directly in each well after transfection with the final DMSO concentration under 1%. Luciferase assays were conducted 40 h after transfection and the addition of chemicals.
Binding competition assay
The binding affinities of Sdex and Oxdex conjugates for glucocorticoid receptor (GR) ligand-binding domain (GR LBD) were determined using GR Competitor Assay Kit from Invitrogen. Sdex and Oxdex conjugates with various concentrations in DMSO (0.5 µL) were mixed with Fluormone (a fluorescein labeled dexamethasone derivative provided by Invitrogen, 1 nM) and recombinant human GR (4 nM) in 100 µL of the binding buffer (10 mM potassium phosphate, pH 7.4, 10 mM Na2MoO4, 0.1 mM EDTA, 5 mM dithiothreitol). The mixtures were incubated in the dark at 25 °C for 2 h before measurement.
Supplementary Material
Acknowledgment
This work was supported by grants from the National Institute of Health (P01-DK58398) and the Welch Foundation (I-1299).
Abbreviations
- (Py-Im)
Pyrrole-Imidazole
- (βDa)
β-Alanine-3,3’-diamino-N-methyldipropylamine
- (Da)
3,3’-Diamino-N-methyldipropylamine
- (Et)
Ethylene diamine
- (IBA)
Isobutyl amine
- (Oxdex)
Dex-17β-carboxylic acid
- (dex)
Dexamethasone
- (Sdex)
Dex-21-S(CH2)2NCS
- (Gal4DBD)
Gal4 DNA-binding domain
- (GR)
Glucocorticoid receptor
- (GRLBD)
Glucocorticoid receptor ligand-binding domain
- (HSV)
Herpes simplex virus
- (Hsp90)
Heat shock protein 90
- (Luc)
Luciferase
Footnotes
Supporting Information Available: Competitive binding affinity assay data, Luc assay data with MCF7 cell line, and 1H and 13C NMR spectra of Sdex-isothiocyanate (3). This material is available free of charge via the Internet at http:://pubs.acs.org.
References
- 1.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 2.Denison C, Kodadek T. Small-molecule-based strategies for controlling gene expression. Chem. Biol. 1998;5:R129–R145. doi: 10.1016/s1074-5521(98)90167-3. [DOI] [PubMed] [Google Scholar]
- 3.Koh JT, Zheng J. The new biomimetic chemistry: artificial transcription factors. ACS Chem. Biol. 2007;2:599–601. doi: 10.1021/cb700183s. [DOI] [PubMed] [Google Scholar]
- 4.Majmudar CY, Mapp AK. Chemical approaches to transcriptional regulation. Curr. Opin. Chem. Biol. 2005;9:467–474. doi: 10.1016/j.cbpa.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Ansari AZ, Mapp AK. Modular design of artificial transcription factors. Curr. Opin. Chem. Biol. 2002;6:765–772. doi: 10.1016/s1367-5931(02)00377-0. [DOI] [PubMed] [Google Scholar]
- 6.Dervan PB. Molecular recognition of DNA by small molecules. Bioorg. Med. Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 7.Mrksich M, Wade WS, Dwyer TJ, Geierstanger BH, Wemmer DE, Dervan PB. Antiparallel side-by-side dimeric motif for sequence-specific recognition in the minor groove of DNA by the designed peptide 1-methylimidazole-2-carboxamide netropsin. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7586–7590. doi: 10.1073/pnas.89.16.7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wade WS, Mrksich M, Dervan PB. Design of peptides that bind in the minor groove of DNA at 5'-(a,T)G(a,T)C(a,T)-3' sequences by a dimeric side-by-side motif. J. Am. Chem. Soc. 1992;114:8783–8794. [Google Scholar]
- 9.Mrksich M, Parks ME, Dervan PB. Hairpin peptide motif. A new class of oligopeptides for sequence-specific recognition in the minor groove of double-helical DNA. J. Am. Chem. Soc. 1994;116:7983–7988. [Google Scholar]
- 10.White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature. 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 11.Melander C, Burnett R, Gottesfeld JM. Regulation of gene expression with pyrrole-imidazole polyamides. J. Biotechnol. 2004;112:195–220. doi: 10.1016/j.jbiotec.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Edelson BS, Best TP, Olenyuk B, Nickols NG, Doss RM, Foister S, Heckel A, Dervan PB. Influence of structural variation on nuclear localization of DNA-binding polyamide-fluorophore conjugates. Nucleic Acids Res. 2004;32:2802–2818. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Best TP, Edelson BS, Nickols NG, Dervan PB. Nuclear localization of pyrrole-imidazole polyamide-fluorescein conjugates in cell culture. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu CF, Dervan PB. Quantitating the concentration of Py-Im polyamide-fluorescein conjugates in live cells. Bioorg. Med. Chem. Lett. 2008;18:5851–5855. doi: 10.1016/j.bmcl.2008.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu P, Liu B, Kodadek T. A high-throughput assay for assessing the cell permeability of combinatorial libraries. Nat. Biotechnol. 2005;23:746–751. doi: 10.1038/nbt1099. [DOI] [PubMed] [Google Scholar]
- 16.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 17.Govindan MV, Manz B. Three-step purification of glucocorticoid receptors from rat liver. Eur. J. Biochem. 1980;108:47–53. doi: 10.1111/j.1432-1033.1980.tb04694.x. [DOI] [PubMed] [Google Scholar]
- 18.Lopez S, Simons J, Stoney S. Dexamethasone 21-(β-Isothiocyanatoethyl) thioether: A new affinity label for glucocorticoid receptors. J. Med. Chem. 1991;34:1762–1767. doi: 10.1021/jm00110a002. [DOI] [PubMed] [Google Scholar]
- 19.Simons J, Stoney S, Pons M, Johnson DF. α-Keto mesylate: A reactive, thiol-specific functional group. J. Org. Chem. 1980;45:3084–3088. [Google Scholar]
- 20.Liu B, Yu P, Alluri PG, Kodadek T. Simple reporter gene-based assays for hairpin poly(amide) conjugate permeability and DNA-binding activity in living cells. Mol. Biosyst. 2005;1:307–317. doi: 10.1039/b511514k. [DOI] [PubMed] [Google Scholar]
- 21.Mapp AK, Ansari AZ, Ptashne M, Dervan PB. Activation of gene expression by small molecule transcription factors. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3930–3935. doi: 10.1073/pnas.97.8.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baird EE, Dervan PB. Solid phase synthesis of polyamides containing imidazole and pyrrole amino acids. J. Am. Chem. Soc. 1996;118:6141–6146. [Google Scholar]
- 23.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Improved nuclear localization of DNA-binding polyamides. Nucleic Acids Res. 2007;35:363–370. doi: 10.1093/nar/gkl1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon Y, Arndt HD, Mao Q, Choi Y, Kawazoe Y, Dervan PB, Uesugi M. Small molecule transcription factor mimic. J. Am. Chem. Soc. 2004;126:15940–15941. doi: 10.1021/ja0445140. [DOI] [PubMed] [Google Scholar]
- 25.Xiao X, Yu P, Lim HS, Sikder D, Kodadek T. A cell-permeable synthetic transcription factor mimic. Angew. Chem. Int. Ed. Engl. 2007;46:2865–2868. doi: 10.1002/anie.200604485. [DOI] [PubMed] [Google Scholar]
- 26.Nickols NG, Dervan PB. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10418–10423. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly JJ, Baird EE, Dervan PB. Binding site size limit of the 2:1 pyrrole-imidazole polyamide-DNA motif. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6981–6985. doi: 10.1073/pnas.93.14.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aponte JC, Verastegui M, Malaga E, Zimic M, Quiliano M, Vaisberg AJ, Gilman RH, Hammond GB. Synthesis, cytotoxicity, and anti-Trypanosoma cruzi activity of new chalcones. J. Med. Chem. 2008;51:6230–6234. doi: 10.1021/jm800812k. [DOI] [PubMed] [Google Scholar]
- 29.Poulin-Kerstien AT, Dervan PB. DNA-templated dimerization of hairpin polyamides. J. Am. Chem. Soc. 2003;125:15811–15821. doi: 10.1021/ja030494a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.