Summary
Transforming growth factor-beta (TGFβ) is a secreted cytokine, which intricately controls a plethora of physiological and pathological processes during development and carcinogenesis. TGFβ exerts antiproliferative effects and functions as a tumor suppressor during early stages of tumorigenesis, whereas at later stages it functions as a tumor promoter aiding in metastatic progression through an autocrine TGFβ loop. Intricate knowledge of TGFβ signaling and its regulation are still evolving. In this review, we make an attempt to showcase the associated enigma of TGFβ signaling in its dual functional role as tumor suppressor and metastatic promoter during early and late stages of carcinogenesis, respectively.
Keywords: Transforming growth factor-beta (TGFβ), TGFβ receptors, Smads, canonical signaling, non-canonical signaling, carcinogenesis, cytostatic effects, epithelial to mesenchymal transition (EMT)
Introduction
TGFβ is a pleiotropic cytokine that is secreted by fibroblasts and epithelial cells in a tissue specific manner and functions in a context-dependent fashion. In the 1970s, a host of individual peptide growth factors that could confer a ’transformed’ phenotype on nonmalignant cells were identified (1). Repeated rounds of purification of extracts from virus transformed cells, which initially was used to identify sarcoma growth factor (2), identified two peptides responsible for growth of normal rat kidney epithelial (NRK) cells on soft agar (3-4). These peptides were christened as transforming growth factor-alpha (TGFα) and transforming growth factor-beta (TGFβ) (5). TGFβ was purified to homogeneity from human platelets, human placenta, and bovine kidney and characterized as a 25-kDa homodimer (6-8).
Structurally related peptides harboring a conserved set of cysteines characterize the TGFβ family members (9-11). Since the identification of TGFβ1 in 1980s, two other distinct isoforms of TGFβ have been identified in mammals, TGFβ2 and TGFβ3. Currently, this superfamily comprises 34 family members, inclusive of TGFβ, Activins, Bone Morphogenetic Proteins (BMP), Vg1, Mullerian Inhibiting Substance (MIS), Growth and Differentiation Factor (GDF) and Inhibin and is highly conserved in organisms ranging from Caenorhabditis elegans, Drosophila melanogaster, Xenopus laevis, and mammals (10).
Originally believed to stimulate cell proliferation and growth, TGFβ was subsequently shown to have the potential to inhibit cell growth (12-13). Specifically, TGFβ1 is involved in immune suppression, angiogenesis, apoptosis, cell growth, and epithelial to mesenchymal transitions (EMT) during development and metastatic cancer progressions (14-22).
TGFβ Signaling Cascade
Binding of TGFβ family ligands to the constitutively active TGFβ type II serine/threonine kinase receptor (TβRII) results in the recruitment of type I receptor (TβRI) and the formation of a stable oligomeric receptor complex (23). Formation of the complex results in the type II receptor to phosphorylate the type I receptor at the C-terminal GS domain, a highly conserved 30 amino acid sequence with a characteristic SGSGSG sequence directly upstream of the kinase domain (24-26). This phosphorylation leads to a conformational change resulting in type I receptor-kinase activation.
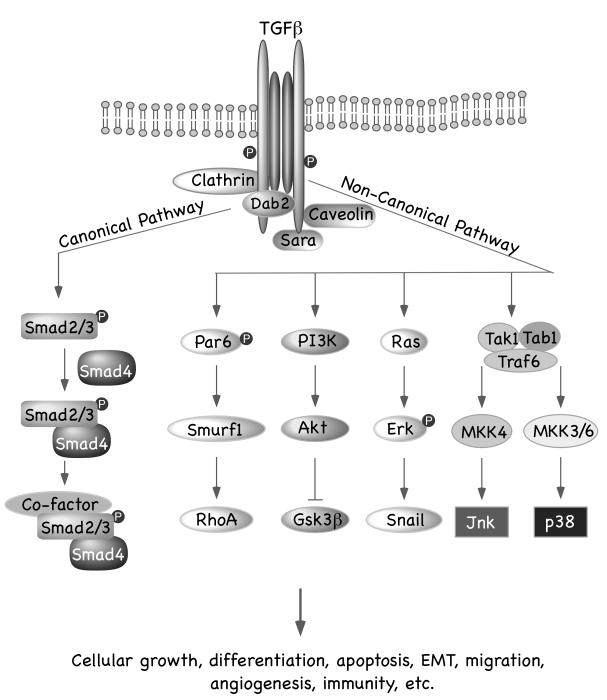
Recently, the structural basis for this two-step assembly process has been revealed (27). The extracellular domains of the TβRI and TβRII fit snugly around the dimeric TGFβ as a six-piece puzzle. It was earlier shown that TβRII binds the fingertip of the extended TGFβ3 ligand structure (28), with a conserved N-terminal extension in TβRII remaining disordered. In the active complex, seven residues of this N-terminal complex become ordered resulting in active heterotetrameric complex formation. Also a five-residue finger in TβRI was shown to hydrogen bond with an aspartate in TβRII, explaining the lack of avidity of TβRI to free TGFβ ligand (27, 29). The activated TβRI interacts with and phosphorylates a number of proteins, thereby activating multiple downstream signaling pathways. Downstream of this the signal is broadly transduced in either a Smad-dependent (canonical) or Smad-independent (non-canonical) signaling pathway (Figure 1).
Figure 1. Schematic Representation of Canonical and Non-canonical TGFβ Signaling Pathway.
Binding of TGFβ to its cognate receptor initiates the signaling pathway. In the canonical pathway, activated type I receptors phosphorylates R-Smads, which subsequently form a complex with the Co-Smad, Smad4. The resulting R-Smads/Co-Smad complex translocates to the nucleus and interacts with distinct transcription factors to turn on or off transcription of many TGFβ-responsive genes that regulate cell proliferation and differentiation. Additionally, TGFβ activates different non-Smad pathways, including PI3K, Ras, Par6, Jnk/p38/MAPK pathways, which cumulatively regulate TGFβ-mediated functions.
I. Canonical TGFβ Signaling Pathway
Smads are the central regulators
The Smads were identified as intermediates of the decapentaplegic (dpp) signaling pathway in Drosophila melanogaster (30-31). Loss of function mutations in Mothers against dpp (mad) in Drosophila melanogaster resulted in pupal lethality, gut defects, and other phenotypes similar to dpp mutant phenotypes. Genetic screens in Caenorhabditis elegans identified sma-2, sma-3, and sma-4 as genes that have mutant phenotypes similar to that observed for the TGFβ-like receptor gene, daf-4 (32). From these data, it was proposed that the mad and sma are homologous genes involved in TGFβ signaling cascade. Later, murine and human homologues to the mad and sma genes were identified and collectively called Smads (33-40).
To date eight mammalian Smad proteins have been characterized and are divided into three functional sub-groups: the receptor-activated Smads (R-Smads), common mediator Smad (Co-Smad), and the inhibitory Smads (I-Smads). Human Smad2, Smad4 and Smad7 map to chromosome 18q21-22, Smad3 and Smad6 map to chromosome 15q21-22, and Smad5, Smad1 and Smad8 map to chromosome 15q31, 15q4, and 15q13 respectively (34). The R-Smads are directly phosphorylated by the type I receptors on their carboxy terminal Ser-Ser-X-Ser (SSXS) motif and include Smad1, Smad2, Smad3, Smad5, and Smad8. Smad2 and Smad3 are phosphorylated in response to the TGFβs and activin, whereas Smad1, Smad5, and Smad8 are phosphorylated in response to BMP. It must be mentioned that in a recent study by Liu et al. (41), TGFβ has been shown to phosphorylate Smad1 and Smad5, which are normally activated by BMP. It was also observed that this phosphorylation is required for activation of TGFβ-mediated metastatic progression in breast cancer cells (41) This study challenges the paradigm that BMP and TGFβ signal through distinct pools of Smad proteins and is in fact suggestive of a more intricate regulation of ligand-dependent phosphorylation of Smads than currently understood. The only mammalian Co-Smad to be identified, thus far, is Smad4 and it mediates signals from both the activin/TGFβ and BMP signaling pathways. Smad4 functions to assist in the further transduction of the signaling pathways by oligomerizing with activated R-Smad(s). The I-Smads, Smad6 and Smad7, are induced by BMP and/or TGFβ/activin, respectively and act as negative feedback to inhibit activation of the R-Smads by inducing degradation of the receptors or by competing with the R-Smads for receptor binding (10).
The Smads are characterized by two conserved regions known as the amino terminal (N-terminal) Mad homology domain-1 (MH1) and C-terminal Mad homology domain-2 (MH2), which are joined by a short, poorly conserved linker region. The MH1 domain is highly conserved among the R-Smads and the Co-Smad, whereas the I-Smads lack a MH1 domain. The R-Smads and Smad4 have N-terminal nuclear localization signals (NLS) and Smad4 has a nuclear export signal (NES) in the MH1 domain (42-44). The MH1 domain plays a role in R- and Co-Smad nuclear import, cytoplasmic anchoring, DNA binding, and regulation of transcription. The MH2 domain is conserved among all of the Smad proteins and regulates Smad oligomerization, cytoplasmic anchoring, and transcription of target genes. The MH1 and MH2 domains bind to a number of proteins including ubiquitination adaptors and substrates, transcriptional co-activators and co-repressors, and a number of transcription factors (41). Furthermore, Smad3 has a transactivation domain in the linker region (45). The functional roles that are assigned to the linker region of the Smads are ubiquitination and transcriptional activation.
Receptor activation of Smad2 and Smad3: The role of adaptor proteins in TGFβ Signaling
The Smad signaling cascade is initiated by C-terminal phosphorylation of Smad2 and/or Smad3 by activated TβR1 (36). However, in order for Smad2 and Smad3 to be phosphorylated by TβRI, they must be recruited to the activated receptor complex. A number of proteins have been identified to interact with Smad2 and/or Smad3 to regulate R-Smad phosphorylation. Smad anchor for receptor activation (SARA) and hepatocytes growth factor-regulated tyrosine kinase substrate (Hrs/Hgs) are FYVE domain containing proteins that present Smad2 to TβRI (46-47). SARA is associated with the plasma membrane and can interact with both non-phosphorylated R-Smads and the TGFβ receptor complex (46). When the receptors become activated, and the R-Smads are phosphorylated, the R-Smads dissociate from SARA and the receptor complex, and bind to Smad4. SARA has a higher affinity for monomeric Smads; therefore it is thought that SARA may also act to regulate Smads by inhibiting aberrant R-Smad oligomerization (48). Hrs/Hgs is localized to early endosomes and synergizes with SARA to present Smad2 to the activated receptor complex (46, 49-51). We have earlier shown that Disabled-2 (Dab2) associates with TβRI and TβRII and functionally bridges the activated receptors to Smad2 and Smad3 through its N-terminal phosphotyrosine-binding (PTB)/-interacting (PID) domain (52). Subsequent work has shown that Dab2 acts as a critical switch of TGFβ-induced EMT (53). Additionally, TGFβ receptor-associated protein-1 (TRAP-1) (54), and the adaptor protein embryonic liver fodrin (ELF) (55) enable activation of R-Smads by the activated TGFβ receptor complex. Endocytosis of the active TGFβ receptor complex is another mechanism by which R-Smad activation is regulated. There is sufficient evidence supporting and arguing against the necessity for receptor endocytosis in R-Smad phosphorylation (56-57). The dependency on receptor endocytosis for R-Smad activation may be cell-type dependent.
The Smad Pathway
The activated TβRI phosphorylates R-Smads at its C-terminal SXSS motif. Phosphorylated R-Smads then form a complex with Smad4. The resulting complex of R-Smads/Co-Smads moves to the nucleus and functionally interacts with distinct transcription factors to turn on or off transcription of many TGFβ-responsive genes that regulate cell proliferation and differentiation (10). The L45 loop of activated type I receptor interacts with the L3 loop of the Smad proteins (10). The interaction plays an important role in determining the signaling specificity as the structure of the L45 loop differ between receptors and dictates which Smads will bind and be activated.
II. Non-canonical (Smad-independent) TGFβ signaling pathways
TGFβ signaling can activate the MAP kinases ERK, JNK, and p38 MAP kinase (58-62). Evidence for this activation came from studies with Smad4-deficient cells and cells overexpressing dominant negative Smad4 (63). In these cells, JNK/MAPK activation was shown to be adequate to elicit TGFβ regulated responses. Conversely, it was shown that TβRIs that were incapable of activating downstream Smads could still activate p38. Recently it was shown that activated TGFβ receptors directly induce polyubiquitination via a lysine at position 63 (K63) of TRAF6, which subsequently is required for activation of JNK and p38 (64). The consequences of MAPK activation by TGFβ remain unclear, however evidence suggests ERK activation is involved in TGFβ-mediated Ras signaling in epithelial cells. TβRs can also directly activate RhoA to induce actin stress fiber formation in fibroblasts, albeit evidences suggest a cooperative role of Smads (65-67). TGFβ–induced EMT integrates Smad as well as non-Smad signaling, and compulsorily requires signaling through PI3K/Protein kinase B (Akt) pathway (14, 68). Alternatively, phosphorylation of the polarity protein Par6 by the activated receptor complex has also been shown to be involved in EMT. This happens as a follow-up of Par6 induced ubiquitination and degradation of RhoA (69).
The Smad proteins can also serve as the platform for signaling crosstalk mechanisms. ERK has been shown to phosphorylate the linker region of Smad1, Smad2, and Smad3 through the Ras pathway (70-71). Phosphorylation of Smads by ERK prevents nuclear translocation of the Smad complex to the nucleus, as a result of which cells containing hyperactive Ras pathway become insensitive to TGFβ stimulation. Contrasting reports have noted nuclear translocation of Smad complex in Ras transformed cells and ERK-mediated Smad phosphorylation seems to increase the half-life of Smad, stabilize complex formation with Smad4, enhancing the overall transcriptional activity of Smad2 (72). Other kinases, like protein kinase C (PKC) can directly phosphorylate Smad3 to prevent its binding to DNA while NF-κβ and STAT signaling inhibit TGFβ signaling by increasing induction of Smad7 expression (73-75). Evidence also exists for the cooperation between the TGFβ and Wnt pathway as well as cooperation between p53 and Smads in modulating expression of TGFβ regulated genes (76-78). Recently, it has been shown that TGFβ acts in sync with Ras and mutant p53 to inhibit p63 and aid in metastatic progression of tumors (79). The multi-step crosstalk of Smad and non-Smad pathways affords a complex, yet meticulous regulation of TGFβ signaling and greater understanding of these crosstalk pathways in a cell type and context specific environment will elucidate the physiological and pathological relevance of this tight regulation (Figure 1).
Regulation and Signal Attenuation of TGFβ Response
Attenuation of TGFβ signaling is mediated either by the I-Smads or ubiquitination and proteosomal degradation of Smad2/3. The I-Smads antagonize TGFβ signaling by competitive inhibition of Smad2/3 binding to the activated TβRI. (80-83). Additionally, Smad7 dephosphorylates activated type I receptor by initiating interactions with a complex containing GADD34 and protein phosphatase 1 (84). Smad7 also contributes in signal attenuation by recruiting Smurf E3 ubiquitin ligase to type I receptor and initiating proteosomal degradation, thereby preventing sustained TGFβ signaling after endocytosis of the receptor complex into calveolar lipid rafts (85). Subsequent internalization of the Type II receptors occurs as a result of β-arrestin2 recruitment of TGFβ signalosome at the receptor level. The receptor internalization pathways dictate either downstream signal activation or attenuation. Whereas, clathrin-mediated internalization leads to downstream signaling, caveolin-mediated pathway leads to Smad7-Smurf2-dependent rapid receptor turnover (86). Smurf2and Skp1-Cul1F-box protein (SCF), the Smad2 and Smad3 E3 ubiquitin ligase, respectively, mediates the degradation of Smad2 and Smad3 (85). Smad3 is also degraded through the carboxy terminus of Hsp70 interacting protein (CHIP) dependent degradation (87). Smad3 can interact directly with Hsp70 resulting in TGF□ independent ubiquitination and degradation of Smad3 (88). This lends credence to the homeostatic regulation of TGF□-mediated signal amplification and subsequent attenuation.
Co-repressors, like Ski and SnoN render an additional level of regulation of TGFβ signaling. Within the nucleus, TGFβ stimulation potentiates Smad3 to bind SnoN and promote its subsequent degradation by the anaphase promoting complex (APC) or Smurf2 (89-90). TGFβ signaling transcriptionally induces SnoN, allowing for a negative feedback control of TGFβ signaling (91). SnoN and Ski function to inhibit TGFβ signaling by disrupting the Smad complex and by recruiting histone deacetylases such as the N-CoR complex, to the chromatin (92).
Finally, attenuation of the TGFβ signaling can also occur through dephosphorylation of the Smad/Co-Smad complex. PPM1A was identified as the phosphatase responsible for dephosphorylating the Smads and their subsequent release from the nucleus (93). The dephosphorylated Smads were shown to recycle back into the cytoplasm to await the next round of signaling (93). However, whether degradation of proteins or the modulation of the protein through post-translational modifications plays the dominant role in abrogating TGFβ signaling remains to be elucidated. It is likely that ubiquitin mediated proteosomal degradation and dephosphorylation events function cooperatively and in a redundant fashion to ensure rapid kinetics and tight control of TGFβ signal attenuation.
Paradoxical Role of TGFβ Signaling-The Yin and Yang of Carcinogenesis
Extensive evidences exist for deregulated TGFβ signaling pathway as a causative agent for tumor initiation and advanced stage disease progression. TGFβ exerts antiproliferative effects and functions as a tumor suppressor during early stages of tumorigenesis, whereas at later stages it functions as a tumor promoter aiding in metastatic progression through an autocrine TGFβ loop (94) (see Figure 2). Transgenic mice expressing a dominant negative TβRII in the epidermis and mammary glands show aggressive tumor formation and metastatic progression (95). Susceptibility of TGFβ-mediated antiproliferative effects is absent in lung cancer (96), head and neck squamous cell carcinoma (97), prostate cancer (98), gastric cancer (99-100), colon cancer (34, 101), pancreatic cancer (102), and breast cancers (103). Recently it has been shown that the tumor suppressor Merlin and a trans-acting negative regulator of signaling, Erbin fine regulates the context dependent response to TGFβ signaling (104). It was shown that in fibroblasts, Merlin is phosphorylated and subsequently inactivated by p21 activated kinase 2 (PAK2), inducing growth and proliferation. PAK2 is a serine/threonine protein kinase and is regulated by activated Rac and other GTPases. PAK2 activity in epithelial cells promotes apoptosis. To prevent antiproliferative effects in epithelial cells Merlin recruits Erbin and disrupts activation and function of PAK2 (104).
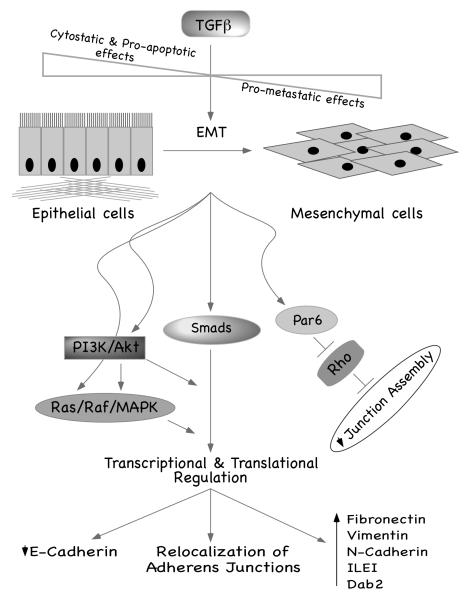
Figure 2. Paradoxical Effects of TGFβ Signaling.
In normal epithelium TGFβ functions as a tumor suppressor through its antiproliferative and pro-apoptotic effects. But with tumor progression, autocrine loops of TGFβ are activated and tumor cells become resistant to the antiproliferative effects of TGFβ. This change of sensitivity of tumor cells to TGFβ is accompanied by EMT with concomitant loss of adherens and tight junctions, downregulation of E-Cadherin expression, and increase in mesenchymal cell markers such as Dab2, N-Cadherin, and ILEI. EMT renders mobility to the tumor cells, which is a critical pre-requisite for metastatic progression of the tumor. Evidences exist for a role of both Smad-independent and Smad-dependent pathways in TGFβ-mediated EMT. It is important to note that both transcriptional and posttranscriptional regulatory pathways are involved in this process.
TGFβ as a tumor suppressor: cytostatic and pro-apoptotic effects
TGFβ functions as a tumor suppressor by mediating its antiproliferative effects in a large variety of cell types. During early stages of tumorigenesis, TGFβ inhibits cell cycle promotion and evasion of TGFβ-mediated antiproliferative effects is a prerequisite for advancement of tumor progression (15, 16, 105). TGFβ-mediated downregulation of c-Myc is a central event of antiproliferative regulatory effects (106-108). c-Myc functions as a transcriptional activator or inhibitor, depending on the target gene, thereby promoting cell growth through the G1 phase of the cell cycle (109-111). Ectopic overexpression of c-Myc results in insensitivity to the growth inhibitory effects of TGFβ (112). Defective repression of c-Myc and subsequent resistance to TGFβ is reported in a number of breast cancer cell lines. Repression of c-Myc by TGFβ has been shown to occur through the Smad pathway (113).
Additionally, TGFβ induces the cyclin-dependent kinase inhibitors (CKIs) p15 and p21 (It must be noted that cyclin-dependent kinase inhibitor, p21Cip1 is different from the serine/threonine kinase PAK2, discussed earlier) (112, 114-117). TGFβ transcriptionally upregulates p15 expression in a Smad-dependent fashion through inhibition of Cyclin D1/Cdk4 (118). TGFβ-dependent induction of p21 and/or p27 also regulates Cdk activity (114-117). p21 directly interacts with and inhibits Cyclin D-Cdk4/6, Cyclin E-Cdk2, and Cyclin A-Cdk2 complexes, therefore arresting progression of the cell cycle in the late G1 phase (119). But, perhaps the most direct way of TGFβ-mediated growth inhibition in epithelial cells involves the dephosphorylation and histone H1 kinase activity of p34cdc2 protein kinase at the G1/S transition (120). TGFβ regulation of the p21 promoter involves Sp1 and the Smads (117). But contrasting reports have shown that lymphocytes from p27 deficient mice remain sensitive to the growth inhibitory effect of TGFβ; therefore, suggesting that p27 may not be actually necessary for TGFβ-induced cell cycle arrest (121). Cell division cycle 25A (Cdc25A) mRNA and cyclin activating kinase (CAK) activity is downregulated by TGFβ (115, 122-123). Cdc25A is a Cdk tyrosine phosphatase that functions to inactivate Cdks by dephosphorylating threonine/tyrosine residues that are necessary for full activation of the Cdks. In contrast, CAK phosphorylates Cdks on a conserved threonine residue. Without this phosphorylation, the Cdks cannot be fully active. The decrease in Cdc25A expression, mediated by TGFβ, was observed in mammary gland epithelial cells (116, 123). As a result of TGFβ-mediated downregulation of Cdc25A and inactivation of CAK, the Cdks are not fully active and cell cycle progression stops during G1 phase.
Pro-apoptotic effects of TGFβ also contribute towards its cytostatic effects. TGFβ-induced apoptotic response has been seen in prostate epithelium, hepatocytes ad hepatoma cell lines, B-lymphocytes and B-cell lines (124). TRAIL and the AP-1/Smad pathway (125), Daxx and the JNK pathway (126), DAPK and the Smad pathway (127), GADD45b and the p38 pathway (128), and ARTS, a mitochondrial protein that aids in caspase activation (129) have all been indicated to be involved in TGFβ-mediated apoptotic events. Work from our lab has documented that TGFβ-induced expression of the proapoptotic protein, Bim, induces cell death in B-lymphocytes (130). It was also shown that stimulation of the pro-survival CD40 receptor inhibited TGFβ-mediated Bim expression and subsequent apoptosis in WEHI1231 B-lymphocytes (131). Smad3-dependent Bim induction has been shown in gastric epithelial cells undergoing TGFβ-induced apoptosis (132), in AML12 hepatocytes and NMuMG mammary epithelial cells (133). Data from our lab and others suggest that Smad3 and Bim are critical mediators of TGFβ-induced apoptosis (133).
TGFβ as a promoter of metastatic progression: TGFβ-Mediated Epithelial to Mesenchymal Transition (EMT)
During metastatic progression, TGFβ promotes epithelial to mesenchymal transition (EMT) (16, 18, 134), which is accompanied by a concomitant loss of cell-cell and cell-matrix adhesion and morphogenic changes from a polarized epithelial phenotype to an elongated fibroblastoid or mesenchymal phenotype (134-135) (see Figure 2). TGFβ-induced EMT is also indispensable during embryonic development for neural crest, heart, and craniofacial structures formation (21, 136). It is interesting to note that EMT during development is largely spatially and temporally regulated, whereas the EMT seen during advanced cancer progression may not reflect the order and timing of events observed during development (137). EMT has a critical role in cancer cell motility, invasion and metastasis. In order for cancer cells to invade surrounding tissues and metastasize to distant sites it is necessary for the cells to dissociate and penetrate the basement membrane, characteristics of developmental EMT.
Evidence suggests that non-Smad signaling pathways are primarily involved in the induction of EMT by TGFβ. Signaling through integrin β1 (138), p38MAPK (62), phosphoinositide 3-kinase (PI3K) (14, 68, 139), Ras homologous (Rho) A (65, 140), Jagged/Notch (141), nuclear factor κβ (NF-κβ) (142) have all been shown to be required for TGFβ-induced EMT. TGFβ treatment of non-transformed murine NMuMG cells and mouse cortical tubule (MCT) cells resulted in an induction of EMT and treatment of the cells with the MEK inhibitor U0126 blocked TGFβ-mediated induction of EMT (143).
Work in our lab has indicated induction of Disabled 2 (Dab2) as an important prerequisite for TGFβ-mediated EMT in NMuMG cells (53). TGFβ treatment of NMuMG cells harboring stable knockdown of Dab2 resulted in apoptosis rather than EMT (53). In an expression profiling analyses of polysome bound mRNA during TGFβ-mediated EMT in EpRas cells, interleukin like EMT inducer (ILEI) was shown to be translationally upregulated during EMT (144-146). Stable ILEI expression causes epithelial plasticity changes and/or tumor formation in NMuMG mammary epithelial cells (146). But the precise mechanism by which expression of Dab2 and ILEI is regulated by TGFβ remains elusive.
Activated Ras/Raf/Mitogen activated protein kinase (MAPK) pathway has been implicated for TGFβ-mediated EMT in human, rat, or mouse epidermal, pancreas, intestine, liver, prostate, and mammary epithelial cells (138-139, 147-149). In other models, TGFβ stimulates ERK, whose function is required for the relocalization of adherens junctions and cell motility induced by TGFβ. In addition, TGFβ activates both Snail and Slug, zinc-finger proteins that repress transcription of E-cadherin in certain cell culture models of EMT (150-152).
Even though it is widely believed that the non-Smad pathways are predominantly involved in TGFβ-mediated EMT, the Smad-dependent pathway has also been implicated in select cases. TGFβ signaling through TβRI and TβRII has also been implicated for TGFβ-mediated EMT and Smad overexpression has been shown to cause synergistic induction of EMT when combined with activated TGFβ receptors (153). Ectopic expression of Smad2 or Smad3, along with Smad4, in human and mouse non-transformed cell lines enhanced TGFβ-induced EMT, whereas the expression of a dominant negative Smad2, Smad3, or Smad4 blocked TGFβ-mediated EMT (154). In vivo evidence for Smad3 involvement in EMT result from experiments in Smad3 knockout mice where loss of Smad3 blocks injury-induced EMT in primary lens epithelial cells and fails to induce EMT in primary tubular epithelial cells derived from Smad3-/- mouse kidneys (151, 154). More recently it was shown that after deletion of Smad3 in mouse hepatocytes, TGFβ induced EMT only in control hepatocytes but not in Smad3-/- hepatocytes (155), suggesting involvement of Smad-dependent signaling in TGFβ-mediated EMT.
Concluding Remarks and Perspectives
Hence, TGFβ, which was initially discovered to be responsible for causing transformation and later shown to possess growth inhibitory properties is a unique cytokine functioning entirely paradoxically at early and late stages of tumorigenesis. Under normal physiological conditions or during onset of tumor formation, TGFβ exerts cytostatic effects and functions as a tumor suppressor whereas at advanced stages of cancer it functions as a tumor promoter aiding in metastatic progression. During metastatic progression, TGFβ promotes epithelial to mesenchymal transdifferentiation (EMT), accompanied by a concomitant loss of cell-cell and cell-matrix adhesion and morphogenic changes from a polarized epithelial phenotype to an elongated fibroblastoid or mesenchymal phenotype. Interestingly, TGFβ-induced EMT is also indispensable during embryonic development for neural crest, heart, and craniofacial structure formation (21). This again highlights deregulated developmental processes being involved in disease progression, underlying the importance of the regulation of these vastly interconnected pathways.
Even though research spanning over the last two decades has candidly outlined details of TGFβ signaling pathway in physiological and pathological conditions, much remains to be elucidated about the precise mechanisms of its deregulation in different forms and stages of cancer. Furthermore, detailed mechanistic knowledge is still unavailable for TGFβ-mediated EMT.Answers to these questions will undoubtedly lead not only to many interesting and surprising observations that reveal additional regulatory complexities, but hopefully lead to the development of TGFβ-dependent anti-cancer therapies. Right now we are at a very exciting phase having all the tools for this next phase of transition into translational research. The enigma continues.
Acknowledgements
This work was supported by Grants CA55536 and CA80095 from the National Cancer Institute to P.H.H. A.C. is supported by an American Heart Association (Ohio Valley Affiliate) Pre-doctoral Fellowship 075080B. We thank all past and present members of the lab for their contributions to the scientific work leading to knowledge advancement on TGFβ signaling pathway.
References
- 1.Sporn MB. TGF-beta: 20 years and counting. Microbes Infect. 1999;1(15):1251–3. doi: 10.1016/s1286-4579(99)00260-9. [DOI] [PubMed] [Google Scholar]
- 2.de Larco JE, Todaro GJ. Growth factors from murine sarcoma virus-transformed cells. Proc Natl Acad Sci USA. 1978;75(8):4001–5. doi: 10.1073/pnas.75.8.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts AB, Anzano MA, Lamb LC, Smith JM, Sporn MB. New class of transforming growth factors potentiated by epidermal growth factor: isolation from nonneoplastic tissues. Proc Natl Acad Sci USA. 1981;78(9):5339–43. doi: 10.1073/pnas.78.9.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Childs CB, Proper JA, Tucker RF, Moses HL. Serum contains a platelet-derived transforming growth factor. Proc Natl Acad Sci USA. 1982;79:5312–16. doi: 10.1073/pnas.79.17.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts AB, Frolik CA, Anzano MA, Sporn MB. Transforming growth factors from neoplastic and nonneoplastic tissues. Fed Proc. 1983;42(9):2621–6. [PubMed] [Google Scholar]
- 6.Assoian RK, Komoriya A, Meyers CA, Miller DM, Sporn MB. Transforming growth factor-beta in human platelets. Identification of a major storage site, purification, and characterization. J Biol Chem. 1983;258(11):7155–60. [PubMed] [Google Scholar]
- 7.Frolik CA, Dart LL, Meyers CA, Smith DM, Sporn MB. Purification and initial characterization of a type beta transforming growth factor from human placenta. Proc Natl Acad Sci USA. 1983;80(12):3676–80. doi: 10.1073/pnas.80.12.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts AB, Anzano MA, Meyers CA, Wideman J, Blacher R, Pan YC, Stein S, Lehrman SR, Smith JM, Lamb LC, et al. Purification and properties of a type beta transforming growth factor from bovine kidney. Biochemistry. 1983;22(25):5692–8. doi: 10.1021/bi00294a002. [DOI] [PubMed] [Google Scholar]
- 9.Kingsley DM. The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8(2):133–46. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 10.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–91. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113(6):685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 12.Tucker RF, Shipley GD, Moses HL, Holley RW. Growth inhibitor from BSC-1 cells closely related to platelet type beta transforming growth factor. Science. 1984;226(4675):705–7. doi: 10.1126/science.6093254. [DOI] [PubMed] [Google Scholar]
- 13.Moses HL, Tucker RF, Leof EB, Coffey RJ, Jr., Halper J, Shipley GD. Type β transforming growth factor is a growth stimulator and a growth inhibitor. In: Feramisco J, Ozanne b., Stiles C, editors. Growth Factors and Transformation. Cancer Cells. Vol. 3. Cold Spring Harbor Press; Cold Spring Harbpr, NY: 1985. pp. 65–71. [Google Scholar]
- 14.Pepper MS. Transforming growth factor-beta: vasculogeneis, angiogenesis, and vessel wall integrity. Cytokine Growth Factor Rev. 1997;8(1):21–43. doi: 10.1016/s1359-6101(96)00048-2. [DOI] [PubMed] [Google Scholar]
- 15.Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem. 2000;275(47):36803–10. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- 16.Akhurst RJ, Derynck R. TGF-beta signaling in cancer-a double-edged sword. Trends Cell Biol. 2001;11(11):S44–51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- 17.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29(2):117–29. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 18.Moustakas A, Pardali K, Gaal A, Heldin CH. Mechanisms of TGF-beta signaling in regulation of cell growth and differentiation. Immunol Lett. 2002;82(1-2):85–91. doi: 10.1016/s0165-2478(02)00023-8. [DOI] [PubMed] [Google Scholar]
- 19.Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci USA. 2003;100(15):8621–3. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007;178(3):437–51. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massague J. TGFbeta in Cancer. Cell. 2008;134(2):215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19(2):156–72. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamashita H, ten Dijke P, Franzen P, Miyazono K, Heldin CH. Formation of hetero-oligomeric complexes of type I and type II receptors for transforming growth factor-beta. J Biol Chem. 1994;269(31):20172–8. [PubMed] [Google Scholar]
- 24.Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370(6488):341–7. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 25.Wrana JL, Tran H, Attisano L, Arora K, Childs SR, Massague J, O’Connor MB. Two distinct transmembrane serine/threonine kinases from Drosophila melanogaster form an activin receptor complex. Mol Cell Biol. 1994;14(2):944–50. doi: 10.1128/mcb.14.2.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wieser R, Wrana JL, Massague J. GS domain mutations that constitutively activate T beta R-I, the downstream signaling component in the TGF-beta receptor complex. Embo J. 1995;14(10):2199–208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, Schwarz PM, Wrana JL, Hinck AP. Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell. 2008;29(2):157–68. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 28.Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP. Crystal structure of the human TbetaR2 ectodomain—TGF-beta3 complex. Nat Struct Biol. 2002;9(3):203–8. doi: 10.1038/nsb766. [DOI] [PubMed] [Google Scholar]
- 29.Massague J. A very private TGF-beta receptor embrace. Mol Cell. 2008;29(2):149–50. doi: 10.1016/j.molcel.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Raftery LA, Twombly V, Wharton K, Gelbart WM. Genetic screens to identify elements of the decapentaplegic signaling pathway in Drosophila. Genetics. 1995;139(1):241–54. doi: 10.1093/genetics/139.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics. 1995;139(3):1347–58. doi: 10.1093/genetics/139.3.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savage C, Das P, Finelli AL, Townsend SR, Sun CY, Baird SE, Padgett RW. Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor beta pathway components. Proc Natl Acad Sci USA. 1996;93(2):790–4. doi: 10.1073/pnas.93.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker JC, Harland RM. A novel mesoderm inducer, Madr2, functions in the activin signal transduction pathway. Genes Dev. 1996;10(15):1880–9. doi: 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- 34.Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, Thomsen GH, Wrana JL, Attisano L. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86(4):543–52. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 35.Derynck R, Gelbart WM, Harland RM, Heldin CH, Kern SE, Massague J, Melton DA, Mlodzik M, Padgett RW, Roberts AB, Smith J, Thomsen GH, Vogelstein B, Wang XF. Nomenclature: vertebrate mediators of TGFbeta family signals. Cell. 1996;87(2):173. doi: 10.1016/s0092-8674(00)81335-5. [DOI] [PubMed] [Google Scholar]
- 36.Macias-Silva M, Abdollah S, Hoodless PA, Pirone R, Attisano L, Wrana JL. MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87(7):1215–24. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 37.Riggins GJ, Thiagalingam S, Rozenblum E, Weinstein CL, Kern SE, Hamilton SR, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mad-related genes in the human. Nat Genet. 1996;13(3):347–9. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 38.Yingling JM, Das P, Savage C, Zhang M, Padgett RW, Wang XF. Mammalian dwarfins are phosphorylated in response to transforming growth factor beta and are implicated in control of cell growth. Proc Natl Acad Sci USA. 1996;93(17):8940–4. doi: 10.1073/pnas.93.17.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Feng X, We R, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-beta response. Nature. 1996;383(6596):168–72. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 40.Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin CH, Miyazono K, ten Dijke P. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. Embo J. 1997;16(17):5353–62. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu IM, Schilling SH, Knouse KA, Choy L, Derynck R, Wang X-F. TGFβ-stimulated Smad1/5 phosphorylation requires the ALK5 L45 loop and mediates the promigratory TGFβ switch. Embo J. 2008;28:88–98. doi: 10.1038/emboj.2008.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao Z, Liu X, Henis YI, Lodish HF. A distinct nuclear localization signal in the N terminus of Smad 3 determines its ligand-induced nuclear translocation. Proc Natl Acad Sci USA. 2000;97(14):7853–8. doi: 10.1073/pnas.97.14.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurisaki A, Kose S, Yoneda Y, Heldin CH, Moustakas A. Transforming growth factor-beta induces nuclear import of Smad3 in an importin-beta1 and Ran-dependent manner. Mol Biol Cell. 2001;12(4):1079–91. doi: 10.1091/mbc.12.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao Z, Brownawell AM, Macara IG, Lodish HF. A novel nuclear export signal in Smad1 is essential for its signaling activity. J Biol Chem. 2003;278(36):34245–52. doi: 10.1074/jbc.M301596200. [DOI] [PubMed] [Google Scholar]
- 45.Prokova V, Mavridou S, Papakosta P, Kardassis D. Characterization of a novel transcriptionally active domain in the transforming growth factor beta-regulated Smad3 protein. Nucleic Acids Res. 2005;33(12):3708–21. doi: 10.1093/nar/gki679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95(6):779–91. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- 47.Miura S, Takeshita T, Asao H, Kimura Y, Murata K, Sasaki Y, Hanai JI, Beppu H, Tsukazaki T, Wrana JL, Miyazono K, Sugamura K. Hgs (Hrs), a FYVE domain protein, is involved in Smad signaling through cooperation with SARA. Mol Cell Biol. 2000;20(24):9346–55. doi: 10.1128/mcb.20.24.9346-9355.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin L, Ding Y, Bromberg JS. Gene transfer of transforming growth factor-beta 1 prolongs murine cardiac allograft survival by inhibiting cell-mediated immunity. Hum Gene Ther. 1996;7(16):1981–8. doi: 10.1089/hum.1996.7.16-1981. [DOI] [PubMed] [Google Scholar]
- 49.Burd CG, Emr SD. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell. 1998;2(1):157–62. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 50.Gaullier JM, Simonsen A, D’Arrigo A, Bremnes B, Stenmark H, Aasland R. FYVE fingers bind PtdIns(3)P. Nature. 1998;394(6692):432–3. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- 51.Patki V, Lawe DC, Corvera S, Virbasius JV, Chawla A. A functional PtdIns(3)P-binding motif. Nature. 1998;394(6692):433–4. doi: 10.1038/28771. [DOI] [PubMed] [Google Scholar]
- 52.Hocevar BA, Smine A, Xu XX, Howe PH. The adaptor molecule Disabled-2 links the transforming growth factor beta receptors to the Smad pathway. Embo J. 2001;20(11):2789–801. doi: 10.1093/emboj/20.11.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prunier C, Howe PH. Disabled-2 (Dab2) is required for transforming growth factor beta-induced epithelial to mesenchymal transition (EMT) J Biol Chem. 2005;280(17):17540–8. doi: 10.1074/jbc.M500974200. [DOI] [PubMed] [Google Scholar]
- 54.Charng MJ, Zhang D, Kinnunen P, Schneider MD. A novel protein distinguishes between quiescent and activated forms of the type I transforming growth factor beta receptor. J Biol Chem. 1998;273(16):9365–8. doi: 10.1074/jbc.273.16.9365. [DOI] [PubMed] [Google Scholar]
- 55.Mishra B, Tang Y, Katuri V, Fleury T, Said AH, Rashid A, Jogunoori W, Mishra L. Loss of cooperative function of transforming growth factor-beta signaling proteins, smad3 with embryonic liver fodrin, a beta-spectrin, in primary biliary cirrhosis. Liver Int. 2004;24(6):637–45. doi: 10.1111/j.1478-3231.2004.0958.x. [DOI] [PubMed] [Google Scholar]
- 56.Hayes S, Chawla A, Corvera S. TGF beta receptor internalization into EEA1-enriched early endosomes: role in signaling to Smad2. J Cell Biol. 2002;158(7):1239–49. doi: 10.1083/jcb.200204088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Penheiter SG, Mitchell H, Garamszegi N, Edens M, Dore JJ, Jr., Leof EB. Internalization-dependent and -independent requirements for transforming growth factor beta receptor signaling via the Smad pathway. Mol Cell Biol. 2002;22(13):4750–9. doi: 10.1128/MCB.22.13.4750-4759.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hartsough MT, Mulder KM. Transforming growth factor beta activation of p44mapk in proliferating cultures of epithelial cells. J Biol Chem. 1995;270(13):7117–24. doi: 10.1074/jbc.270.13.7117. [DOI] [PubMed] [Google Scholar]
- 59.Atfi A, Djelloul S, Chastre E, Davis R, Gespach C. Evidence for a role of Rho-like GTPases and stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) in transforming growth factor beta-mediated signaling. J Biol Chem. 1997;272(3):1429–32. doi: 10.1074/jbc.272.3.1429. [DOI] [PubMed] [Google Scholar]
- 60.Engel ME, McDonnell MA, Law BK, Moses HL. Interdependent SMAD and JNK signaling in transforming growth factor-beta-mediated transcription. J Biol Chem. 1999;274(52):37413–20. doi: 10.1074/jbc.274.52.37413. [DOI] [PubMed] [Google Scholar]
- 61.Hocevar BA, Brown TL, Howe PH. TGF-beta induces fibronectin synthesis through a c-Jun N-terminal kinase-dependent, Smad4-independent pathway. Embo J. 1999;18(5):1345–56. doi: 10.1093/emboj/18.5.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bakin AV, Rinehart C, Tomlinson AK, Arteaga CL. p38 mitogen-activated protein kinase is required for TGFbeta-mediated fibroblastic transdifferentiation and cell migration. J Cell Sci. 2002;115(Pt 15):3193–206. doi: 10.1242/jcs.115.15.3193. [DOI] [PubMed] [Google Scholar]
- 63.Engel ME, Datta PK, Moses HL. Signal transduction by transforming growth factor-beta: a cooperative paradigm with extensive negative regulation. J Cell Biochem Suppl. 1998;30-31:111–22. [PubMed] [Google Scholar]
- 64.Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol Cell. 2008;31(6):918–24. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. Transforming growth factor-beta1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell. 2001;12(1):27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edlund S, Landstrom M, Heldin CH, Aspenstrom P. Transforming growth factor-beta-induced mobilization of actin cytoskeleton requires signaling by small GTPases Cdc42 and RhoA. Mol Biol Cell. 2002;13(3):902–14. doi: 10.1091/mbc.01-08-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vardouli L, Moustakas A, Stournaras C. LIM-kinase 2 and cofilin phosphorylation mediate actin cytoskeleton reorganization induced by transforming growth factor-beta. J Biol Chem. 2005;280(12):11448–57. doi: 10.1074/jbc.M402651200. [DOI] [PubMed] [Google Scholar]
- 68.Kattla JJ, Carew RM, Heljic M, Godson C, Brazil DP. Protein kinase B/Akt activity is involved in renal TGF-beta1-driven epithelial-mesenchymal transition in vitro and in vivo. Am J Physiol Renal Physiol. 2008;295(1):F215–25. doi: 10.1152/ajprenal.00548.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ozdamar B, Bose R, Barrios-Rodiles M, Wang HR, Zhang Y, Wrana JL. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307(5715):1603–9. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- 70.Kretzschmar M, Doody J, Massague J. Opposing BMP and EGF signalling pathways converge on the TGF-beta family mediator Smad1. Nature. 1997;389(6651):618–22. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- 71.Kretzschmar M, Doody J, Timokhina I, Massague J. A mechanism of repression of TGFbeta/ Smad signaling by oncogenic Ras. Genes Dev. 1999;13(7):804–16. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Caestecker MP, Parks WT, Frank CJ, Castagnino P, Bottaro DP, Roberts AB, Lechleider RJ. Smad2 transduces common signals from receptor serine-threonine and tyrosine kinases. Genes Dev. 1998;12(11):1587–92. doi: 10.1101/gad.12.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ulloa L, Doody J, Massague J. Inhibition of transforming growth factor-beta/SMAD signalling by the interferon-gamma/STAT pathway. Nature. 1999;397(6721):710–3. doi: 10.1038/17826. [DOI] [PubMed] [Google Scholar]
- 74.Yakymovych I, Ten Dijke P, Heldin CH, Souchelnytskyi S. Regulation of Smad signaling by protein kinase C. Faseb J. 2001;15(3):553–5. doi: 10.1096/fj.00-0474fje. [DOI] [PubMed] [Google Scholar]
- 75.Jenkins BJ, Grail D, Nheu T, Najdovska M, Wang B, Waring P, Inglese M, McLoughlin RM, Jones SA, Topley N, Baumann H, Judd LM, Giraud AS, Boussioutas A, Zhu HJ, Ernst M. Hyperactivation of Stat3 in gp130 mutant mice promotes gastric hyperproliferation and desensitizes TGF-beta signaling. Nat Med. 2005;11(8):845–52. doi: 10.1038/nm1282. [DOI] [PubMed] [Google Scholar]
- 76.Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell. 2003;113(3):301–14. doi: 10.1016/s0092-8674(03)00308-8. [DOI] [PubMed] [Google Scholar]
- 77.Wilkinson DS, Ogden SK, Stratton SA, Piechan JL, Nguyen TT, Smulian GA, Barton MC. A direct intersection between p53 and transforming growth factor beta pathways targets chromatin modification and transcription repression of the alpha-fetoprotein gene. Mol Cell Biol. 2005;25(3):1200–12. doi: 10.1128/MCB.25.3.1200-1212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang Y, Prunier C, Howe PH. The inhibitory effects of Disabled-2 (Dab2) on Wnt signaling are mediated through Axin. Oncogene. 2008;27(13):1865–75. doi: 10.1038/sj.onc.1210829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, Parenti AR, Rosato A, Bicciato S, Balmain A, Piccolo S. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137(1):87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 80.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389(6651):622–6. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 81.Hata A, Shi Y, Massague J. TGF-beta signaling and cancer: structural and functional consequences of mutations in Smads. Mol Med Today. 1998;4(6):257–62. doi: 10.1016/s1357-4310(98)01247-7. [DOI] [PubMed] [Google Scholar]
- 82.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389(6651):631–5. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 83.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276(16):12477–80. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 84.Shi W, Sun C, He B, Xiong W, Shi X, Yao D, Cao X. GADD34-PP1c recruited by Smad7 dephosphorylates TGFbeta type I receptor. J Cell Biol. 2004;164(2):291–300. doi: 10.1083/jcb.200307151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275(47):36818–22. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 86.Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signaling and turnover. Nat Cell Biol. 2003;5(5):410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- 87.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8(4):303–8. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xin H, Xu X, Li L, Ning H, Rong Y, Shang Y, Wang Y, Fu XY, Chang Z. CHIP controls the sensitivity of transforming growth factor-beta signaling by modulating the basal level of Smad3 through ubiquitin-mediated degradation. J Biol Chem. 2005;280(21):20842–50. doi: 10.1074/jbc.M412275200. [DOI] [PubMed] [Google Scholar]
- 89.Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol. 2001;3(6):587–95. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- 90.Stroschein SL, Bonni S, Wrana JL, Luo K. Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev. 2001;15(21):2822–36. doi: 10.1101/gad.912901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stroschein SL, Wang W, Zhou S, Zhou Q, Luo K. Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science. 1999;286(5440):771–4. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- 92.Wu JW, Krawitz AR, Chai J, Li W, Zhang F, Luo K, Shi Y. Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: insights on Ski-mediated repression of TGF-beta signaling. Cell. 2002;111(3):357–67. doi: 10.1016/s0092-8674(02)01006-1. [DOI] [PubMed] [Google Scholar]
- 93.Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, Shi Y, Chen YG, Meng A, Feng XH. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell. 2006;125(5):915–28. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6(7):506–20. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 95.Amendt C, Schirmacher P, Weber H, Blessing M. Expression of a dominant negative type II TGF-beta receptor in mouse skin results in an increase in carcinoma incidence and an acceleration of carcinoma development. Oncogene. 1998;17(1):25–34. doi: 10.1038/sj.onc.1202161. [DOI] [PubMed] [Google Scholar]
- 96.Yanagisawa K, Uchida K, Nagatake M, Masuda A, Sugiyama M, Saito T, Yamaki K, Takahashi T, Osada H. Heterogeneities in the biological and biochemical functions of Smad2 and Smad4 mutants naturally occurring in human lung cancers. Oncogene. 2000;19(19):2305–11. doi: 10.1038/sj.onc.1203591. [DOI] [PubMed] [Google Scholar]
- 97.Garrigue-Antar L, Munoz-Antonia T, Antonia SJ, Gesmonde J, Vellucci VF, Reiss M. Missense mutations of the transforming growth factor beta type II receptor in human head and neck squamous carcinoma cells. Cancer Res. 1995;55(18):3982–7. [PubMed] [Google Scholar]
- 98.Park BJ, Park JI, Byun DS, Park JH, Chi SG. Mitogenic conversion of transforming growth factor-beta1 effect by oncogenic Ha-Ras-induced activation of the mitogen-activated protein kinase signaling pathway in human prostate cancer. Cancer Res. 2000;60(11):3031–8. [PubMed] [Google Scholar]
- 99.Myeroff LL, Parsons R, Kim SJ, Hedrick L, Cho KR, Orth K, Mathis M, Kinzler KW, Lutterbaugh J, Park K, et al. A transforming growth factor beta receptor type II gene mutation common in colon and gastric but rare in endometrial cancers with microsatellite instability. Cancer Res. 1995;55(23):5545–7. [PubMed] [Google Scholar]
- 100.Kang SH, Bang YJ, Im YH, Yang HK, Lee DA, Lee HY, Lee HS, Kim NK, Kim SJ. Transcriptional repression of the transforming growth factor-beta type I receptor gene by DNA methylation results in the development of TGF-beta resistance in human gastric cancer. Oncogene. 1999;18(51):7280–6. doi: 10.1038/sj.onc.1203146. [DOI] [PubMed] [Google Scholar]
- 101.Markowitz S. TGF-beta receptors and DNA repair genes, coupled targets in a pathway of human colon carcinogenesis. Biochim Biophys Acta. 2000;1470(1):M13–20. doi: 10.1016/s0304-419x(99)00031-1. [DOI] [PubMed] [Google Scholar]
- 102.Goggins M, Shekher M, Turnacioglu K, Yeo CJ, Hruban RH, Kern SE. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58(23):5329–32. [PubMed] [Google Scholar]
- 103.Lucke CD, Philpott A, Metcalfe JC, Thompson AM, Hughes-Davies L, Kemp PR, Hesketh R. Inhibiting mutations in the transforming growth factor beta type 2 receptor in recurrent human breast cancer. Cancer Res. 2001;61(2):482–5. [PubMed] [Google Scholar]
- 104.Wilkes MC, Repellin CE, Hong M, Bracamonte M, Penheiter SG, Borg JP, Leof EB. Erbin and the NF2 tumor suppressor Merlin cooperatively regulate cell-type-specific activation of PAK2 by TGF-beta. Dev Cell. 2009;16(3):433–44. doi: 10.1016/j.devcel.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-beta signaling in cancer. J Natl Cancer Inst. 2000;92(17):1388–402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- 106.Mulder KM, Brattain MG. Alterations in c-myc expression in relation to maturational status of human colon carcinoma cells. Int J Cancer. 1988;42(1):64–70. doi: 10.1002/ijc.2910420113. [DOI] [PubMed] [Google Scholar]
- 107.Zentella A, Massague J. Transforming growth factor beta induces myoblast differentiation in the presence of mitogens. Proc Natl Acad Sci USA. 1992;89(11):5176–80. doi: 10.1073/pnas.89.11.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mulder KM, Levine AE, Hernandez X, McKnight MK, Brattain DE, Brattain MG. Modulation of c-myc by transforming growth factor-beta in human colon carcinoma cells. Biochem Biophys Res Commun. 1988;150(2):711–6. doi: 10.1016/0006-291x(88)90449-4. [DOI] [PubMed] [Google Scholar]
- 109.Alexandrow MG, Moses HL. Transforming growth factor beta 1 inhibits mouse keratinocytes late in G1 independent of effects on gene transcription. Cancer Res. 1995;55(17):3928–32. [PubMed] [Google Scholar]
- 110.Facchini LM, Penn LZ. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. Faseb J. 1998;12(9):633–51. [PubMed] [Google Scholar]
- 111.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19(1):1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Alexandrow MG, Moses HL. Transforming growth factor beta and cell cycle regulation. Cancer Res. 1995;55(7):1452–7. [PubMed] [Google Scholar]
- 113.Yagi K, Furuhashi M, Aoki H, Goto D, Kuwano H, Sugamura K, Miyazono K, Kato M. c-myc is a downstream target of the Smad pathway. J Biol Chem. 2002;277(1):854–61. doi: 10.1074/jbc.M104170200. [DOI] [PubMed] [Google Scholar]
- 114.Datto MB, Li Y, Panus JF, Howe DJ, Xiong Y, Wang XF. Transforming growth factor beta induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci USA. 1995;92(12):5545–9. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reynisdottir I, Massague J. The subcellular locations of p15(Ink4b) and p27(Kip1) coordinate their inhibitory interactions with cdk4 and cdk2. Genes Dev. 1997;11(4):492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- 116.Iavarone A, Massague J. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-beta in cells lacking the CDK inhibitor p15. Nature. 1997;387(6631):417–22. doi: 10.1038/387417a0. [DOI] [PubMed] [Google Scholar]
- 117.Moustakas A, Kardassis D. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc Natl Acad Sci USA. 1998;95(12):6733–8. doi: 10.1073/pnas.95.12.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sandhu C, Garbe J, Bhattacharya N, Daksis J, Pan CH, Yaswen P, Koh J, Slingerland JM, Stampfer MR. Transforming growth factor beta stabilizes p15INK4B protein, increases p15INK4B-cdk4 complexes, and inhibits cyclin D1-cdk4 association in human mammary epithelial cells. Mol Cell Biol. 1997;17(5):2458–67. doi: 10.1128/mcb.17.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–16. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 120.Howe PH, Draetta G, Leof EB. Transforming growth factor beta 1 inhibition of p34cdc2 phosphorylation and histone H1 kinase activity is associated with G1/S-phase growth arrest. 1991;11(3):1185–94. doi: 10.1128/mcb.11.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh DY, Nakayama K. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85(5):707–20. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 122.Nagahara H, Ezhevsky SA, Vocero-Akbani AM, Kaldis P, Solomon MJ, Dowdy SF. Transforming growth factor beta targeted inactivation of cyclin E:cyclin-dependent kinase 2 (Cdk2) complexes by inhibition of Cdk2 activating kinase activity. Proc Natl Acad Sci USA. 1999;96(26):14961–6. doi: 10.1073/pnas.96.26.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bhowmick NA, Ghiassi M, Aakre M, Brown K, Singh V, Moses HL. TGF-beta-induced RhoA and p160ROCK activation is involved in the inhibition of Cdc25A with resultant cell-cycle arrest. Proc Natl Acad Sci USA. 2003;100(26):15548–53. doi: 10.1073/pnas.2536483100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sanchez-Capelo A. Dual role for TGF-beta1 in apoptosis. Cytokine Growth Factor Rev. 2005;16(1):15–34. doi: 10.1016/j.cytogfr.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 125.Herzer K, Grosse-Wilde A, Krammer PH, Galle PR, Kanzler S. Transforming growth factor-beta-mediated tumor necrosis factor-related apoptosis-inducing ligand expression and apoptosis in hepatoma cells requires functional cooperation between Smad proteins and activator protein-1. Mol Cancer Res. 2008;6(7):1169–77. doi: 10.1158/1541-7786.MCR-08-0073. [DOI] [PubMed] [Google Scholar]
- 126.Perlman R, Schiemann WP, Brooks MW, Lodish HF, Weinberg RA. TGF-beta-induced apoptosis is mediated by the adapter protein Daxx that facilitates JNK activation. Nat Cell Biol. 2001;3(8):708–14. doi: 10.1038/35087019. [DOI] [PubMed] [Google Scholar]
- 127.Jang CW, Chen CH, Chen CC, Chen JY, Su YH, Chen RH. TGF-beta induces apoptosis through Smad-mediated expression of DAP-kinase. Nat Cell Biol. 2002;4(1):51–8. doi: 10.1038/ncb731. [DOI] [PubMed] [Google Scholar]
- 128.Yoo J, Ghiassi M, Jirmanova L, Balliet AG, Hoffman B, Fornace AJ, Jr., Liebermann DA, Bottinger EP, Roberts AB. Transforming growth factor-beta-induced apoptosis is mediated by Smad-dependent expression of GADD45b through p38 activation. J Biol Chem. 2003;278(44):43001–7. doi: 10.1074/jbc.M307869200. [DOI] [PubMed] [Google Scholar]
- 129.Gottfried Y, Voldavsky E, Yodko L, Sabo E, Ben-Itzhak O, Larisch S. Expression of the pro-apoptotic protein ARTS in astrocytic tumors: correlation with malignancy grade and survival rate. Cancer. 2004;101(11):2614–21. doi: 10.1002/cncr.20675. [DOI] [PubMed] [Google Scholar]
- 130.Wildey GM, Patil S, Howe PH. Smad3 potentiates transforming growth factor beta (TGFbeta )-induced apoptosis and expression of the BH3-only protein Bim in WEHI 231 B lymphocytes. J Biol Chem. 2003;278(20):18069–77. doi: 10.1074/jbc.M211958200. [DOI] [PubMed] [Google Scholar]
- 131.Patil S, Wildey GM, Brown TL, Choy L, Derynck R, Howe PH. Smad7 is induced by CD40 and protects WEHI 231 B-lymphocytes from transforming growth factor-beta - induced growth inhibition and apoptosis. J Biol Chem. 2000;275(49):38363–70. doi: 10.1074/jbc.M004861200. [DOI] [PubMed] [Google Scholar]
- 132.Ohgushi M, Kuroki S, Fukamachi H, O’Reilly LA, Kuida K, Strasser A, Yonehara S. Transforming growth factor beta-dependent sequential activation of Smad, Bim, and caspase-9 mediates physiological apoptosis in gastric epithelial cells. Mol Cell Biol. 2005;25(22):10017–28. doi: 10.1128/MCB.25.22.10017-10028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ramesh S, Wildey GM, Howe PH. Transforming growth factor beta (TGFbeta)-induced apoptosis: the rise & fall of Bim. Cell Cycle. 2009;8(1):11–7. doi: 10.4161/cc.8.1.7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24(37):5764–74. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 135.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7(2):131–42. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 136.Trelstad RL, Hay ED, Revel JD. Cell contact during early morphogenesis in the chick embryo. Dev Biol. 1967;16(1):78–106. doi: 10.1016/0012-1606(67)90018-8. [DOI] [PubMed] [Google Scholar]
- 137.Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene. 2005;24(50):7443–54. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 138.Bhowmick NA, Zent R, Ghiassi M, McDonnell M, Moses HL. Integrin beta 1 signaling is necessary for transforming growth factor-beta activation of p38MAPK and epithelial plasticity. J Biol Chem. 2001;276(50):46707–13. doi: 10.1074/jbc.M106176200. [DOI] [PubMed] [Google Scholar]
- 139.Gotzmann J, Huber H, Thallinger C, Wolschek M, Jansen B, Schulte-Hermann R, Beug H, Mikulits W. Hepatocytes convert to a fibroblastoid phenotype through the cooperation of TGF-beta1 and Ha-Ras: steps towards invasiveness. J Cell Sci. 2002;115(Pt 6):1189–202. doi: 10.1242/jcs.115.6.1189. [DOI] [PubMed] [Google Scholar]
- 140.Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, Beug H, Grunert S. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J Cell Biol. 2002;156(2):299–313. doi: 10.1083/jcb.200109037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. Embo J. 2004;23(5):1155–65. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huber MA, Azoitei N, Baumann B, Grunert S, Sommer A, Pehamberger H, Kraut N, Beug H, Wirth T. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114(4):569–81. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xie L, Law BK, Chytil AM, Brown KA, Aakre ME, Moses HL. Activation of the Erk pathway is required for TGF-beta1-induced EMT in vitro. Neoplasia. 2004;6(5):603–10. doi: 10.1593/neo.04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pradet-Balade B, Boulme F, Beug H, Mullner EW, Garcia-Sanz JA. Translation control: bridging the gap between genomics and proteomics? Trends Biochem Sci. 2001;26(4):225–9. doi: 10.1016/s0968-0004(00)01776-x. [DOI] [PubMed] [Google Scholar]
- 145.Jechlinger M, Grunert S, Tamir IH, Janda E, Ludemann S, Waerner T, Seither P, Weith A, Beug H, Kraut N. Expression profiling of epithelial plasticity in tumor progression. Oncogene. 2003;22(46):7155–69. doi: 10.1038/sj.onc.1206887. [DOI] [PubMed] [Google Scholar]
- 146.Waerner T, Alacakaptan M, Tamir I, Oberauer R, Gal A, Brabletz T, Schreiber M, Jechlinger M, Beug H. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell. 2006;10(3):227–39. doi: 10.1016/j.ccr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 147.Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996;10(19):2462–77. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 148.Oft M, Heider KH, Beug H. TGFbeta signaling is necessary for carcinoma cell invasiveness and metastasis. Curr Biol. 1998;8(23):1243–52. doi: 10.1016/s0960-9822(07)00533-7. [DOI] [PubMed] [Google Scholar]
- 149.Davies M, Robinson M, Smith E, Huntley S, Prime S, Paterson I. Induction of an epithelial to mesenchymal transition in human immortal and malignant keratinocytes by TGF-beta1 involves MAPK, Smad and AP-1 signalling pathways. J Cell Biochem. 2005;95(5):918–31. doi: 10.1002/jcb.20458. [DOI] [PubMed] [Google Scholar]
- 150.Zavadil J, Bitzer M, Liang D, Yang YC, Massimi A, Kneitz S, Piek E, Bottinger EP. Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc Natl Acad Sci USA. 2001;98(12):6686–91. doi: 10.1073/pnas.111614398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Saika S, Ikeda K, Yamanaka O, Sato M, Muragaki Y, Ohnishi Y, Ooshima A, Nakajima Y, Namikawa K, Kiyama H, Flanders KC, Roberts AB. Transient adenoviral gene transfer of Smad7 prevents injury-induced epithelial-mesenchymal transition of lens epithelium in mice. Lab Invest. 2004;84(10):1259–70. doi: 10.1038/labinvest.3700151. [DOI] [PubMed] [Google Scholar]
- 152.Giannelli G, Bergamini C, Fransvea E, Sgarra C, Antonaci S. Laminin-5 with transforming growth factor-beta1 induces epithelial to mesenchymal transition in hepatocellular carcinoma. Gastroenterology. 2005;129(5):1375–83. doi: 10.1053/j.gastro.2005.09.055. [DOI] [PubMed] [Google Scholar]
- 153.Oft M, Akhurst RJ, Balmain A. Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat Cell Biol. 2002;4(7):487–94. doi: 10.1038/ncb807. [DOI] [PubMed] [Google Scholar]
- 154.Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A. TGF-beta and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell. 2005;16(4):1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ju W, Ogawa A, Heyer J, Nierhof D, Yu L, Kucherlapati R, Shafritz DA, Bottinger EP. Deletion of Smad2 in mouse liver reveals novel functions in hepatocyte growth and differentiation. Mol Cell Biol. 2006;26(2):654–67. doi: 10.1128/MCB.26.2.654-667.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]