Abstract
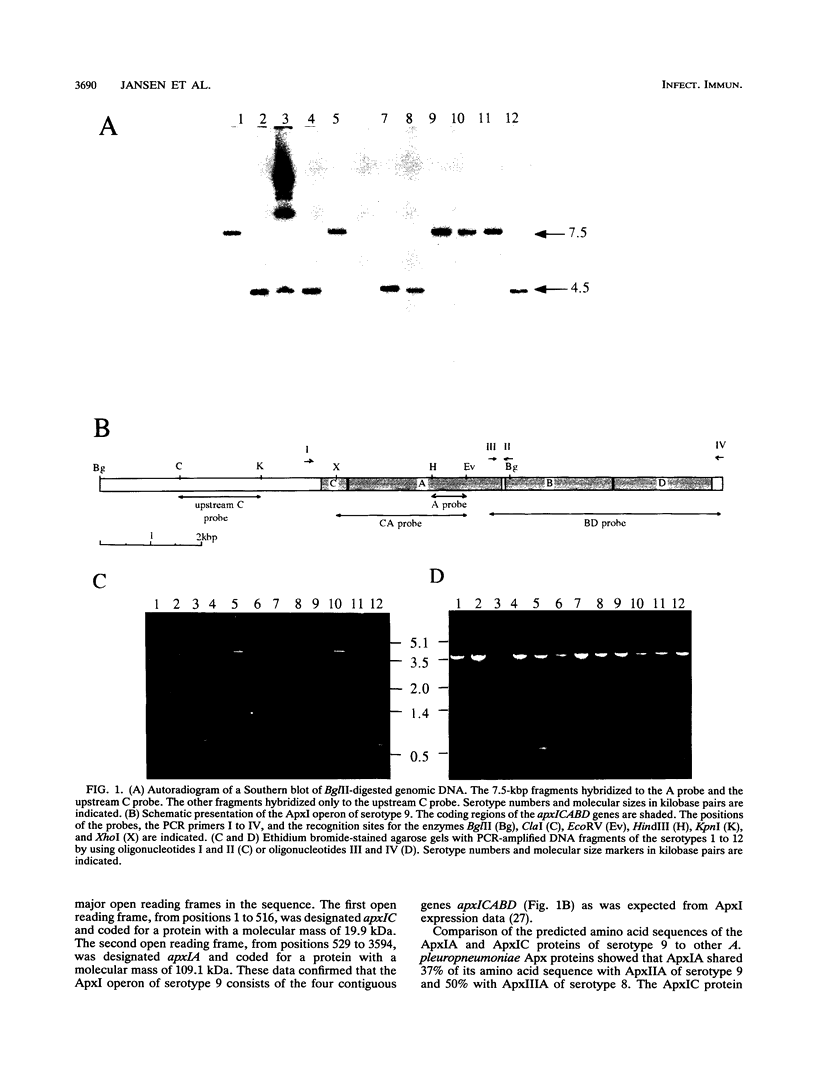
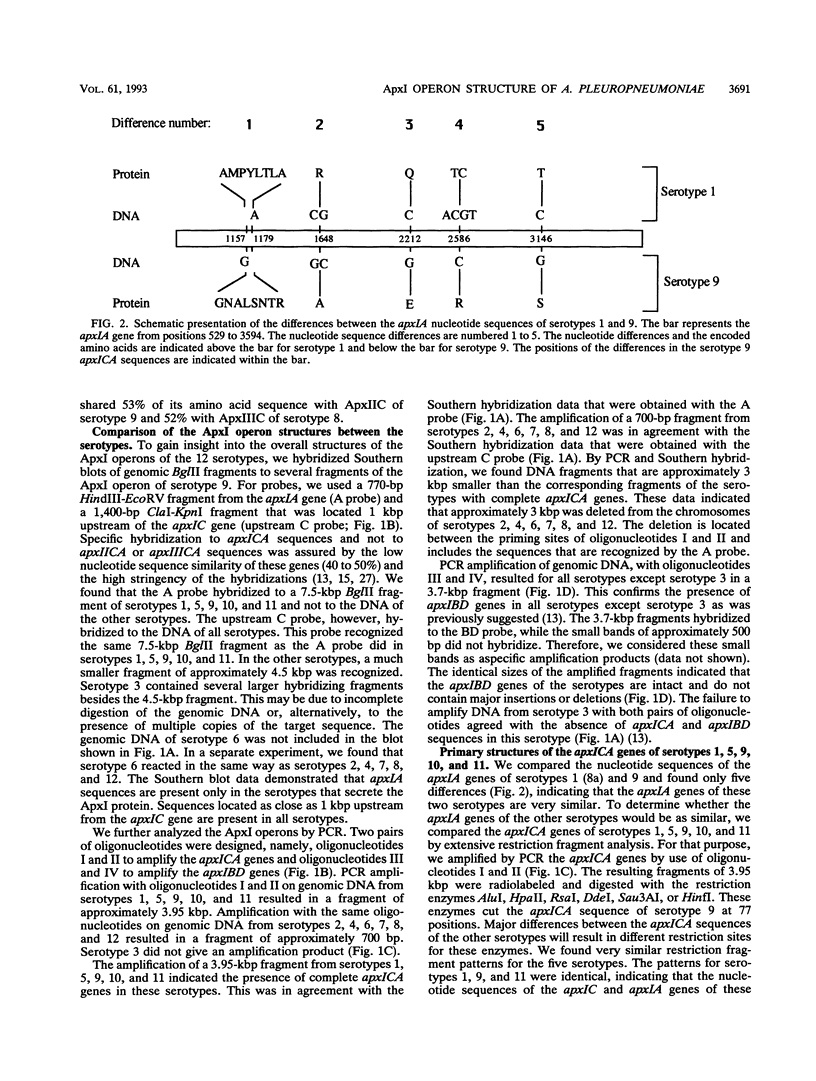
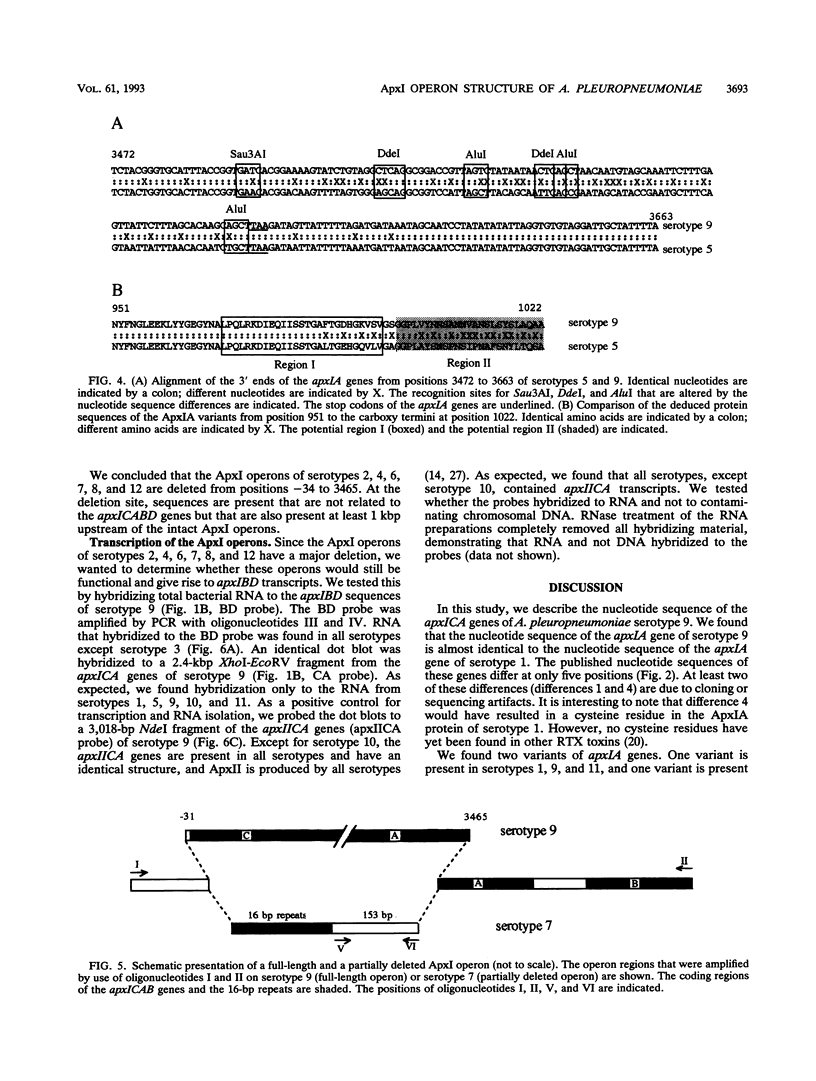
Actinobacillus pleuropneumoniae-RTX-toxin I (ApxI), an important virulence factor, is secreted by serotypes 1, 5, 9, 10, and 11 of A. pleuropneumoniae. However, sequences homologous to the secretion genes apxIBD of the ApxI operon are present in all 12 serotypes except serotype 3. The purpose of this study was to determine and compare the structures of the ApxI operons of the 12 A. pleuropneumoniae serotypes. We focused on the nucleotide sequence comparison of the ApxI-coding genes, the structures of the ApxI operons, and the transcription of the ApxI operons. We determined the nucleotide sequences of the toxin-encoding apxICA genes of serotype 9 and found that the gene for the structural toxin, apxIA, was almost identical to the apxIA gene of serotype 1. The toxin-encoding genes of the other serotypes are also similar for the main part; nevertheless, two variants were identified, one in serotypes 1, 9, and 11 and one in serotypes 5 and 10. The two apxIA variants differ mainly within the distal 110 nucleotides. Structural analysis demonstrated that intact ApxI operons, consisting of the four contiguous genes apxICABD, are present in serotypes 1, 5, 9, 10, and 11. ApxI operons with a major deletion in the apxICA genes are present in serotypes 2, 4, 6, 7, 8, and 12. Serotype 3 does not contain ApxI operon sequences. We found that all ApxI operons are transcriptionally active despite the partial deletion of the operon in some serotypes. The implications of these data for the expression and secretion of ApxI and the other Apx-toxins, ApxII and ApxIII, as well as for the development of a subunit vaccine against A. pleuropneumoniae will be discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C., Potter A. A., Gerlach G. F. Isolation and molecular characterization of spontaneously occurring cytolysin-negative mutants of Actinobacillus pleuropneumoniae serotype 7. Infect Immun. 1991 Nov;59(11):4110–4116. doi: 10.1128/iai.59.11.4110-4116.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram T. A. Actinobacillus pleuropneumoniae: molecular aspects of virulence and pulmonary injury. Can J Vet Res. 1990 Apr;54 (Suppl):S53–S56. [PubMed] [Google Scholar]
- Burrows L. L., Lo R. Y. Molecular characterization of an RTX toxin determinant from Actinobacillus suis. Infect Immun. 1992 Jun;60(6):2166–2173. doi: 10.1128/iai.60.6.2166-2173.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. F., Young R., Struck D. K. Cloning and characterization of a hemolysin gene from Actinobacillus (Haemophilus) pleuropneumoniae. DNA. 1989 Nov;8(9):635–647. doi: 10.1089/dna.1.1989.8.635. [DOI] [PubMed] [Google Scholar]
- Chang Y. F., Young R., Struck D. K. The Actinobacillus pleuropneumoniae hemolysin determinant: unlinked appCA and appBD loci flanked by pseudogenes. J Bacteriol. 1991 Aug;173(16):5151–5158. doi: 10.1128/jb.173.16.5151-5158.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felmlee T., Pellett S., Welch R. A. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985 Jul;163(1):94–105. doi: 10.1128/jb.163.1.94-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey J., Beck M., Stucki U., Nicolet J. Analysis of hemolysin operons in Actinobacillus pleuropneumoniae. Gene. 1993 Jan 15;123(1):51–58. doi: 10.1016/0378-1119(93)90538-e. [DOI] [PubMed] [Google Scholar]
- Frey J., Meier R., Gygi D., Nicolet J. Nucleotide sequence of the hemolysin I gene from Actinobacillus pleuropneumoniae. Infect Immun. 1991 Sep;59(9):3026–3032. doi: 10.1128/iai.59.9.3026-3032.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland I. B., Blight M. A., Kenny B. The mechanism of secretion of hemolysin and other polypeptides from gram-negative bacteria. J Bioenerg Biomembr. 1990 Jun;22(3):473–491. doi: 10.1007/BF00763178. [DOI] [PubMed] [Google Scholar]
- Inzana T. J., Todd J., Ma J. N., Veit H. Characterization of a non-hemolytic mutant of Actinobacillus pleuropneumoniae serotype 5: role of the 110 kilodalton hemolysin in virulence and immunoprotection. Microb Pathog. 1991 Apr;10(4):281–296. doi: 10.1016/0882-4010(91)90012-y. [DOI] [PubMed] [Google Scholar]
- Inzana T. J. Virulence properties of Actinobacillus pleuropneumoniae. Microb Pathog. 1991 Nov;11(5):305–316. doi: 10.1016/0882-4010(91)90016-4. [DOI] [PubMed] [Google Scholar]
- Jansen R., Briaire J., Kamp E. M., Gielkens A. L., Smits M. A. Cloning and characterization of the Actinobacillus pleuropneumoniae-RTX-toxin III (ApxIII) gene. Infect Immun. 1993 Mar;61(3):947–954. doi: 10.1128/iai.61.3.947-954.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Briaire J., Kamp E. M., Smits M. A. Comparison of the cytolysin II genetic determinants of Actinobacillus pleuropneumoniae serotypes. Infect Immun. 1992 Feb;60(2):630–636. doi: 10.1128/iai.60.2.630-636.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Ledley F. D. Production of discrete high specific activity DNA probes using the polymerase chain reaction. Gene Anal Tech. 1989 Jul-Aug;6(4):79–83. doi: 10.1016/0735-0651(89)90020-4. [DOI] [PubMed] [Google Scholar]
- Kamp E. M., Popma J. K., Anakotta J., Smits M. A. Identification of hemolytic and cytotoxic proteins of Actinobacillus pleuropneumoniae by use of monoclonal antibodies. Infect Immun. 1991 Sep;59(9):3079–3085. doi: 10.1128/iai.59.9.3079-3085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny B., Taylor S., Holland I. B. Identification of individual amino acids required for secretion within the haemolysin (HlyA) C-terminal targeting region. Mol Microbiol. 1992 Jun;6(11):1477–1489. doi: 10.1111/j.1365-2958.1992.tb00868.x. [DOI] [PubMed] [Google Scholar]
- Konings R. N., Verhoeven E. J., Peeters B. P. pKUN, vectors for the separate production of both DNA strands of recombinant plasmids. Methods Enzymol. 1987;153:12–34. doi: 10.1016/0076-6879(87)53045-2. [DOI] [PubMed] [Google Scholar]
- Kraig E., Dailey T., Kolodrubetz D. Nucleotide sequence of the leukotoxin gene from Actinobacillus actinomycetemcomitans: homology to the alpha-hemolysin/leukotoxin gene family. Infect Immun. 1990 Apr;58(4):920–929. doi: 10.1128/iai.58.4.920-929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo R. Y., Strathdee C. A., Shewen P. E. Nucleotide sequence of the leukotoxin genes of Pasteurella haemolytica A1. Infect Immun. 1987 Sep;55(9):1987–1996. doi: 10.1128/iai.55.9.1987-1996.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendal S., Devenish J., MacInnes J. I., Lumsden J. H., Watson S., Xun H. Evaluation of heat-sensitive, neutrophil-toxic, and hemolytic activity of Haemophilus (Actinobacillus) pleuropneumoniae. Am J Vet Res. 1988 Jul;49(7):1053–1058. [PubMed] [Google Scholar]
- Rycroft A. N., Williams D., Cullen J. M., Macdonald J. The cytotoxin of Actinobacillus pleuropneumoniae (pleurotoxin) is distinct from the haemolysin and is associated with a 120 kDa polypeptide. J Gen Microbiol. 1991 Mar;137(3):561–568. doi: 10.1099/00221287-137-3-561. [DOI] [PubMed] [Google Scholar]
- Sebunya T. N., Saunders J. R. Haemophilus pleuropneumoniae infection in swine: a review. J Am Vet Med Assoc. 1983 Jun 15;182(12):1331–1337. [PubMed] [Google Scholar]
- Smits M. A., Briaire J., Jansen R., Smith H. E., Kamp E. M., Gielkens A. L. Cytolysins of Actinobacillus pleuropneumoniae serotype 9. Infect Immun. 1991 Dec;59(12):4497–4504. doi: 10.1128/iai.59.12.4497-4504.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P., Koronakis V., Hughes C. Mutational analysis supports a role for multiple structural features in the C-terminal secretion signal of Escherichia coli haemolysin. Mol Microbiol. 1991 Oct;5(10):2391–2403. doi: 10.1111/j.1365-2958.1991.tb02085.x. [DOI] [PubMed] [Google Scholar]